Abstract
The 16.5 kb human mitochondrial genome encodes for 13 polypeptides, 22 tRNAs and 2 rRNAs involved in oxidative phosphorylation. Mitochondrial DNA (mtDNA), unlike its nuclear counterpart, is not packaged into nucleosomes and is more prone to the adverse effects of reactive oxygen species (ROS) generated during oxidative phosphorylation. The past few decades have witnessed an increase in the number of proteins observed to translocate to the mitochondria for the purposes of mitochondrial genome maintenance. The mtDNA damage produced by ROS, if not properly repaired, leads to instability and can ultimately manifest in mitochondrial dysfunction and disease. The base excision repair (BER) pathway is employed for the removal and consequently the repair of deaminated, oxidized, and alkylated DNA bases. Specialized enzymes called DNA glycosylases, which locate and cleave the damaged base, catalyze the first step of this highly coordinated repair pathway. This review focuses on members of the four human BER DNA glycosylase superfamilies and their subcellular localization in the mitochondria and/or the nucleus, as well as summarizes their structural features, biochemical properties, and functional role in the excision of damaged bases.
Keywords: Reactive oxygen species, Base excision repair, DNA glycosylases, Mitochondrial DNA damage and response, Oxidative phosphorylation, Mitochondrial dysfunction
Introduction
Damage to DNA occurs frequently within a cell and can be caused by both spontaneous reactions that originate within the cell or by exogenous agents from the environment (reviewed in [De Bont and van Larebeke, 2004; Duclos et al., 2012]). Endogenous damaging agents include mismatches generated during DNA replication, deamination of bases, depurination or depyrimidination, and oxidative damage that occurs from the generation of ROS within the cell through normal metabolism. Exogenous factors such as ionizing radiation cause toxic double-strand DNA breaks (DSBs), ultraviolet (UV) radiation results in the formation of cyclobutane pyrimidine dimers, and alkylating agents such as cisplatin lead to unwanted alkylation and DNA crosslinks [Friedberg et al., 2004].
Like the extensively studied nuclear DNA, mtDNA is also subject to the harmful effects of ROS. MtDNA is condensed into spheroid bodies called nucleoids and is proximal to sites of ROS production at the inner membrane of the mitochondria [Bogenhagen, 2012]. MtDNA is therefore 10 – 20X more susceptible to DNA damage than its nuclear counterpart [Cadenas and Davies, 2000; Richter et al., 1988]. The human mitochondrial genome comprises 16,569 bp of circular double-stranded DNA, which encodes 2 rRNAs, 22 tRNAs, and 13 polypeptides involved in oxidative phosphorylation via the electron transport chain [Anderson et al., 1981]. Base substitutions, deletions, and missense mutations that alter the protein coding of mitochondrial genes are leading causes of the diseases associated with mtDNA [Druzhyna et al., 2008; Wallace, 2012]. Disease states in the mitochondria still remain an enigma not only because of the nature of inheritance of mtDNA, but also due to the fact that mitochondrial diseases can arise from mutations in nuclear genes [Shaughnessy et al., 2014; Wallace, 2012].
In the nucleus, several repair mechanisms function either to restore or bypass disruptive DNA damage and some of these pathways have also been described in the mitochondria. The highly conserved BER pathway is involved in the repair of non-bulky lesions produced by oxidation, alkylation, deamination, and single-strand DNA breaks (SSBs) (reviewed in [Fromme and Verdine, 2004; Krokan and Bjoras, 2013; Liu and Demple, 2010]). This pathway is well documented in the nucleus and was the first repair pathway to be described in the mitochondria. The nucleotide excision repair (NER), mismatch repair (MMR), and the double-strand break (DSB) repair pathways including Homologous Recombination (HR) and Non-Homologous End Joining (NHEJ) are all present in the nucleus. While components of MMR and the DSB repair pathways have been described in the mitochondria and likely aid in mtDNA repair, to date the NER pathway has not been shown to take place in this organelle [Kazak et al., 2012].
In the BER pathway, DNA glycosylases catalyze the first step in the process by removal of the damaged base. These enzymes are highly conserved among species and a significant number of these have been shown to translocate to the mitochondria. The mitochondrial localization of proteins and enzymes can be predicted using algorithms such as the TargetP 1.1 server [Emanuelsson et al., 2000], MitoProt II [Claros and Vincens, 1996], and PSORTII [Nakai and Horton, 1999]. Below we discuss components of the BER pathway describing both nuclear and mitochondrial proteins that are involved in the repair process.
BER in the Nucleus and Mitochondria
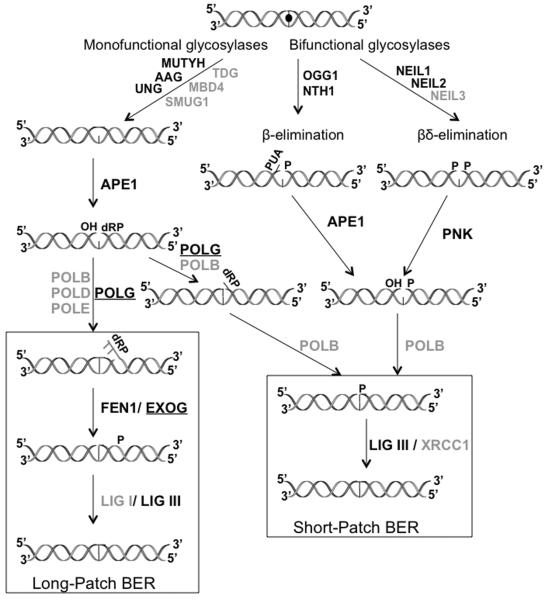
BER can proceed as either short-patch (1-nt) or long-patch (2 or more nt) and is carried out in the five basic steps summarized in Fig. 1: (a) recognition and excision of the damaged DNA base, (b) removal of the resulting abasic (AP) site, (c) end processing, (d) gap filling, and (e) ligation. The initiation step of BER is carried out by DNA glycosylases, which catalyze the cleavage of the N-glycosidic bond between the damaged base and its deoxyribose resulting in an abasic (AP) site [Fromme and Verdine, 2004; Krokan and Bjoras, 2013; Liu and Demple, 2010]. These enzymes can be categorized by one of two mechanistic types, monofunctional or bifunctional, depending on whether they possess an associated intrinsic lyase activity. The DNA glycosylases are encoded by nuclear genes with some of these containing a mitochondrial targeting signal (MTS) that allows for translocation to the mitochondria [Larsen et al., 2005; Takao et al., 1998].
Figure 1.
Overview of the BER pathway. Nuclear and mitochondrial enzymes are indicated at various stages of the repair process. The lesion (indicated by a filled circle) is excised by both monofunctional and bifunctional DNA glycosylases. The resulting AP site gets processed by either APE1 or PNK leaving suitable ends for the gap-filling polymerase (either Polβ in the nucleus or Polγ in the mitochondria). DNA ligase (either I or III) seals the gap and completes the repair process. Additional steps involving the FEN1 endonuclease are required for the long-patch repair process. Gray color indicates enzymes in the nucleus alone, black includes both nuclear and mitochondrial enzymes, and underlined black text represents enzymes present in the mitochondria alone. (This diagram was adapted from [Duclos et al., 2012]).
Monofunctional glycosylases target non-oxidative damage such as alkylated and deaminated DNA bases. These enzymes excise the damaged base but lack lyase activity and must rely on AP endonuclease (APE1) to hydrolyze the phosphate backbone. These processed DNA ends are then suitable substrates for the dRP-lyase and gap-filling step of the process performed by a DNA repair polymerase [Demple and Sung, 2005]. Glycosylases involved in the removal of oxidized DNA bases are bifunctional and possess an associated lyase activity whereby the DNA backbone is nicked 3' to the lesion after removal of the damaged base. Polynucleotide kinase phosphate (PNKP) processes the DNA ends prior to nucleotide insertion by a polymerase [Das et al., 2006; Wiederhold et al., 2004]. In the nucleus, Polymerase β (POLB) is involved in incorporating the correct nucleotide into the DNA whereas polymerase γ (POLG) performs this function as the sole polymerase transported to the mitochondrion [Wilson et al., 2000; Yakubovskaya et al., 2006]. Ligation is carried out primarily by DNA ligase I in the nucleus and by ligase III in the mitochondria [Gao et al., 2011; Simsek et al., 2011]. Even though the enzymes discussed above are sufficient for the in vitro reconstitution of BER, interplay between BER enzymes and proteins involved in other facets of DNA metabolism is necessary for the coordinated repair of DNA lesions [Hegde et al., 2010].
Many crystal structures of DNA glycosylases both liganded and in a complex with DNA containing their respective lesions have been analyzed and provide insights into lesion-recognition by glycosylases (reviewed in [Brooks et al., 2013; Prakash et al., 2012]). In cases where human enzymes have resisted crystallization attempts, orthologous enzymes from bacteria, viruses, or plants have served as useful models. Single-molecule studies and the ability to trap intermediates via disulfide-crosslinking have significantly advanced our understanding of DNA glycosylases [Prakash et al., 2012]. Current structural information for the mammalian DNA glycosylases has led to a proposed common mechanism of damaged base extrusion into the active site of the enzyme. However, each glycosylase family uses structurally distinct motifs for base recognition, “flipping”, and stabilization of the DNA. In the following section, we briefly summarize information about the mammalian DNA glycosylases in the context of their subcellular localization, targeted substrates, and the structural motifs used in DNA binding.
DNA Glycosylase Families in the Mitochondria: Biochemical Function and Structural Properties
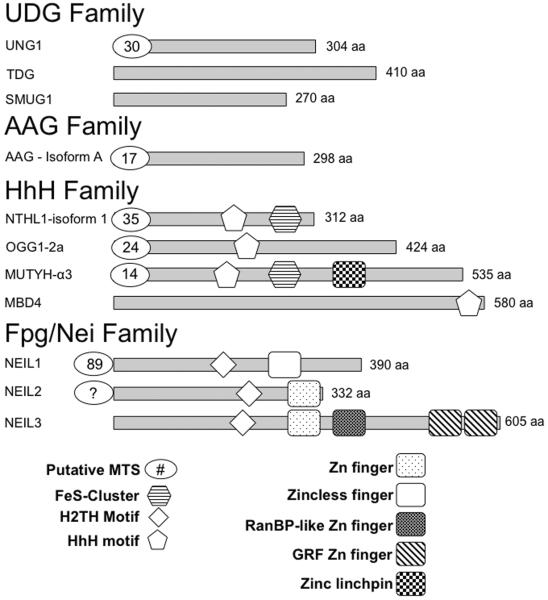
DNA glycosylases are evolutionarily conserved through all domains of life and numerous tools have been utilized to probe their function in both the nucleus and mitochondria [Jacobs and Schar, 2012]. Studies employing in vitro overexpression, purification and enzymatic assays, co-immunoprecipitation, fluorescent labeling, subcellular and co-localizations, knockout mouse models, in vitro single-molecule experiments, and structure-based functional analysis have provided a wealth of information regarding these enzymes. These tools have identified and characterized 11 mammalian DNA glycosylases and differentiated them into 4 superfamilies based on conserved structural motifs and the substrates they recognize (see Table 1 and Fig. 2) [Jacobs and Schar, 2012]. These are the Uracil DNA Glycosylase (UDG) family, the Alkyladenine DNA Glycosylase (AAG) family, the Helix-Hairpin-Helix family (HhH), and the Formamidopyrimidine DNA Glycosylase (Fpg)/ Endonuclease VIII (Nei) or Helix-Two-Turns-Helix (H2TH) family. Thus far, 7 of the 11 mammalian glycosylases have been observed in the mitochondria (Table 1) with at least one representative from each of the four superfamilies being identified in this organelle.
Table 1.
Nuclear and Mitochondrial Human DNA Glycosylases
| FAMILY Fold | ISOFORMS | |||
|---|---|---|---|---|
| GLYCOSYLASE | Nuclear | Mitochondrial | SUBSTRATES | |
|
| ||||
| UDG | UNG | UNG2 | UNG1 | U, 5-FU & ssDNA, dsDNA |
| TDG | TDG | NF | T:G, U:G, εC:G, 5-FU, Tg:G & dsDNA>>ssDNA | |
| SMUG1 | SMUG1 | NF | U, 5-hmU, 5-hmC, 5-FU & ssDNA, dsDNA | |
|
| ||||
| AAG | AAG/MPG | A,B,C | A, B | 3-meA, 7-meG, 1-meG, Hx, U, εG, εA & ssDNA, dsDNA |
|
| ||||
| Helix-hairpin-helix | NTHL1 | NTHL1 | NTHL1 | Tg, 5-hC, 5-hU, Fapy lesions & ssDNA, dsDNA |
| OGG1 | 1a | 1b, 1c; 2a – 2e | 8-oxoG, Fapy lesions & dsDNA | |
| MUTYH | β, γ | α | A:8-oxoG, A:G, and A:C, 2-OHA:G & dsDNA | |
| MBD4 | MBD4 | NF | T:G, U:G, 5-MeC, halogenated pyrimidines, 5-FU, Tg:G & dsDNA | |
|
| ||||
| Fpg/Nei Helix-two turns-helix | NEIL1 | NEIL1 | NEIL1 | Sp, Gh, Tg, DHU, 5-OHU, 5-OHC, DHT, FapyG, FapyA & dsDNA>bubble, bulge, fork>ssDNA |
| NEIL2 | NEIL2 | NEIL2 | Sp, Gh, DHT, DHU, 5-OHU, 5-OHC & ssDNA>bubble, bulge, fork>dsDNA | |
| NEIL3 | NEIL3 | NF | Sp, Gh, FapyG, FapyA, MeFapyG, DHU, DHT, 5-OHU, 5-OHC, Tg & ssDNA>dsDNA | |
Isoforms specific for the mitochondria or the nucleus have been described so far for AAG, OGG1 and MUTYH. The substrate preferences for each glycosylase listed in Table 1 have been reviewed extensively [Brooks et al., 2013; Jacobs and Schar, 2012; Liu et al., 2013; Prakash et al., 2012]. NF, not found in the mitochondria.
Figure 2.
Domain map of the 11 human DNA glycosylases. The mature form of the most common isoform of each of these enzymes is shown as a grey rectangle. The number of amino acids displayed is based on these deposited sequences (Uniprot IDs: UNG1: P13051-2, TDG: Q13569, SMUG1: Q53HV7, AAG: P29372, MBD4: O95243, MUTYH: Q9UIF7, NTH1: P78549, OGG1-1a: O15527, NEIL1: Q96FI4, NEIL2: Q969S2, NEIL3: Q8TAT5). The number of putative N-terminal amino acids in the leader sequence that get cleaved upon mitochondrial localization were determined by MitoProt II [Claros and Vincens, 1996] and are indicated in an oval in this diagram. For NEIL2, there is no predicted N-terminal MTS thus far reported in the literature and thus it is indicated by “?”.
The UDG family
Overview
Udg from Escherichia coli was the first DNA glycosylase identified by Thomas Lindahl in 1974 [Lindahl, 1974]. Since then, the UDG superfamily has come to comprise 6 subfamilies: family I, uracil N-glycosylase (UNG); family II, thymine DNA glycosylase (TDG) or mismatch uracil DNA glycosylase (MUG) family; family III, single-strand-specific monofunctional uracil DNA glycosylase (SMUG); and families IV – VI glycosylases found in thermophilic and hyperthermophillic eubacteria and archaea. Of these, subfamilies I, II, and III are found in higher eukaryotes and only UNG has been found in the human mitochondria to date [Schormann et al., 2014].
The best-documented substrates for the family I Ung enzymes are uracil and 5-fluoro-uracil (5-FU), which is cleaved at a reduced rate. Human UNG1 and UNG2 are the mitochondrial and nuclear isoforms of this enzyme, respectively, and are generated via both alternative splicing and transcription from different start sites (Table 1). UNG enzymes are monofunctional and cleave substrates from both single-stranded (ss) DNA and double-stranded (ds) DNA with a slight preference for ss over ds substrates. The mitochondrial UNG1 has a MTS comprising a 30-amino acid leader sequence at the N-terminal end of the enzyme (according to MitoProt II, Fig. 2). This sequence gets cleaved upon entry into the inner membrane of the mitochondria yielding a mature enzyme [Neupert, 1997].
TDG is monofunctional and belongs to the family II MUG enzymes. TDG cleaves a broad range of substrates including thymine from G:T mismatches, bulky etheno (ε) adducts of cytosine and adenine, 5-FU, and thymine glycol (Tg) opposite G. Lesions in dsDNA appear to be the best substrates for this enzyme [Sjolund et al., 2013]. The SMUG family, like its name suggests, was originally thought to function only on ssDNA. However, reports of its ability to cleave lesions such as uracil, 5-FU, 5-hydroxymethyluracil (5-hmU), and 5-hydroxymethylcytosine (5-hmC) from dsDNA have been published [Schormann et al., 2014]. In the nucleus, it thought to serve as a backup enzyme for UNG2 due to its substrate overlap with the latter. Both TDG and SMUG are currently not reported to be present in the mitochondria (Table 1) [Schormann et al., 2014].
Structure and biochemistry of UDG enzymes
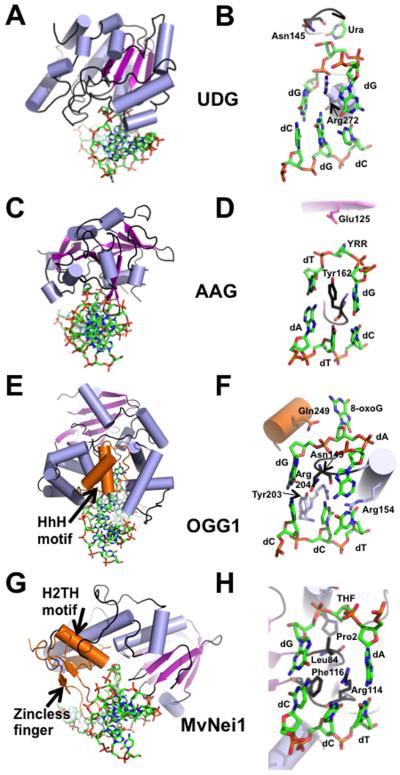
Structural and mechanistic insights into the UDG family of DNA glycosylases have been obtained by several groups and reviewed in [Zharkov et al., 2010]. A single domain constructed from a β-sheet comprising 4 parallel β-strands sandwiched between two sets of α-helices is characteristic of the UNG DNA glycosylases and the DNA binding groove is narrow and shallow (example PDB ID 4SKN, Fig. 3A [Slupphaug et al., 1996]). In this structure and others, the mechanism of cleavage by an UNG enzyme is described as the concerted action of four loops (a water-activating loop, pro-rich loop, gly-ser loop, and a leu-intercalation loop) involved in base flipping into the active site, kinking of the DNA, nucleophilic attack and cleavage of the uracil base [Slupphaug et al., 1996]. The active site residue, Asp145, activates a water molecule making it the nucleophile that initiates the catalytic cleavage of the N-glycosidic bond. Another residue in UNG important for DNA binding is Leu272, which is inserted into the minor groove of the DNA helix and causes local disruption of the DNA aiding in the eversion of uracil into the active site binding pocket [Slupphaug et al., 1996]. Curiously, a double mutation of the active site Asp145 to asparagine and Leu272 to arginine does not completely inactivate the enzyme. The crystal structure of UNG containing this double mutation bound to uracil-containing DNA indicates that the uracil gets cleaved but remains bound to the enzyme (Fig. 3B).
Figure 3.
Representative crystal structures from each of the 4 families of DNA glycosylases. (A) Overall DNA-bound structure of UDG bound to DNA (PDB ID: 4SKN [Slupphaug et al., 1996]). (B) Close-up view of the active site of UDG where the active site nucleophile Asp145 is mutated to Asn, Leu272 is mutated to Arg, and the cleaved uracil (Ura) remains bound in the active site pocket. (C) Human AAG (PDB ID: 1BNK [Lau et al., 1998]) bound to DNA. (D) Active site view of the AAG-DNA complex indicating an abasic pyrrolidine nucleotide (YRR) that is extruded into the active site, active site nucleophile Glu125, and Tyr162 that causes a severe kink in the DNA. (E) Overall structure of human OGG1 bound to 8-oxoG containing DNA (PDB ID: 1EBM [Bruner et al., 2000]), an example of the HhH glycosylase family. (F) Close-up view of the active site residues of OGG1 emphasizes the extrahelical 8-oxoG lesion, the active site Lys249 mutated to Gln, and the four residues that contact the estranged dC. (G) Overall structure of the viral ortholog of human NEIL1, MvNei1, bound to an abasic site analog (THF), representing the Fpg/Nei family (PDB ID: 3A46 [Imamura et al., 2009]). (H) Zoomed-in view of the MvNei1-THF complex depicting the three void-filling residues and the active site nucleophile, Pro2. For all the structures, the DNA is shown in green as a stick model, and colored by element; the α-helices are colored in light blue; β-strands are purple; loops are shown in black. The HhH motif (OGG1), the H2TH motif (MvNei1) and the zincless finger motif (MvNei1) are highlighted in orange.
The AAG/MPG family
Overview
AAG, also referred to as MPG or MDG, is monofunctional and recognizes alkylated and deaminated DNA bases and translocates to the mitochondria via an N-terminal MTS ([van Loon and Samson, 2013] Fig. 2). Post-transcriptional processing is thought to result in three isoforms AAG-A, -B and -C. Of the three, isoforms A and B contain a putative MTS and translocate to the mitochondria using a 17 and 12-aa MTS, respectively [van Loon and Samson, 2013]. The best substrates include 3-methyladenine (3-meA), 7-methylguanine (7-meG), 1-methylguanine (1-meG), hypoxanthine (Hx), 1,N2-ethenoguanine (εG), and ethenoadenine (εA) in both ss and dsDNA (Table 1) [Jacobs and Schar, 2012].
Structure and biochemistry of AAG
The structure of human AAG, like UDG, reveals a single domain but with mixed α/β topology comprising a positive DNA-binding groove (Fig. 3C, PDB ID: 1BNK [Lau et al., 1998]), shown in Fig. 3C. Human AAG recognizes similar substrates as E. coli AlkA, but differs from the latter in that it lacks the HhH motif involved in DNA binding. From the crystal structure of AAG, it is evident that Glu125 is poised to mediate nucleophilic attack on the N-glycosidic bond via a water-mediated interaction (Fig. 3D). An aromatic residue, Tyr162, interrogates the minor groove of the DNA and causes a kink in the DNA (Fig. 3D) [Hollis et al., 2000].
The HhH family
Overview
HhH family members are a diverse group comprising six subfamilies of DNA glycosylases comprising endonuclease III (Nth), 8-oxo-7,8-dihydroguanine (8-oxoG) DNA glycosylase 1 (Ogg1), A/G mismatch-specific adenine glycosylase (MutY/Mig), alkyladenine DNA glycosylase (AlkA), 8-oxoG DNA glycosylase 2 (Ogg2), and N-methylpurine-DNA glycosylase II (MpgII). Members of the AlkA subfamily exist in many bacterial and eukaryotic genomes but are lost in the mammalian genomes. Mammals instead possess AAG/MPG enzymes, which perform similar functions to that of AlkA, but belong to the structurally distinct AAG/MPG family described above. Ogg2 enzymes are found in archaeal genomes and MpgII enzymes are found in both bacteria and archaea. There are four HhH DNA glycosylases in the human genome, namely, NTHL1, OGG1, MUTYH, and methyl-binding domain protein 4, MBD4.
The Nth subfamily is named after its bacterial prototype and comprises homologs observed in several species. The human NTHL1 enzyme is a bifunctional glycosylase involved in the excision of oxidized DNA bases such as Tg, 5-hydroxycytosine (5-hC), 5-hydroxyuracil (5-hU), and the ring-opened 2,6-diamino-4-hydroxy-5-formamidopyrimidine (Fapy) lesions (Table I). Whereas human NTHL1 is found in both nucleus and mitochondria, mouse Nth1 translocates primarily to the mitochondria [Sampath, 2014].
The Ogg subfamily of enzymes predominantly excises 8-oxoG, one of the most potent oxidative lesions generated in the cell [Faucher et al., 2012]. Human OGG1 is a bifunctional DNA glycosylase and several isoforms of OGG1 have been documented in recent years [Boiteux and Radicella, 2000]. There appears to be 8 isoforms of OGG1 generated from alternative splicing: OGG1-1a-c and -2a-e [Nishioka et al., 1999]. The OGG1 glycosylases have a common N-terminal MTS but varying C-terminal domains. OGG1-1a also called OGG1-α is the most abundant isoform and possesses a nuclear localization signal (NLS) and an MTS but is predominantly thought to function in the nucleus. However, OGG1-1b, c and OGG1-2a-e have an N-terminal MTS and translocate to the mitochondria [Boiteux and Radicella, 2000]. The precise role for each isoform still requires further scrutiny.
Bacterial MutY was first discovered in 1988 as an enzyme that cleaves adenine from A:G mispairs (reviewed in [Markkanen et al., 2013]). This subfamily of enzymes is unique in that they cleave an undamaged base from DNA instead of a damaged base. The human homolog, MUTYH, is monofunctional and excises adenine opposite 8-oxoG, guanine, and cytosine representing an additional mode of eliminating mutagenic oxidized guanine from cells. There are 3 primary transcripts of MUTYH generated from alternative splicing, (α, β, and γ) which give rise to an estimated >15 transcripts [Oka and Nakabeppu, 2011]. MUTYH-α3 is the primary mitochondrial transcript that contains an N-terminal 14-aa MTS (Fig. 2). The primary nuclear isoform of MUTHYH seems to be encoded by the β3, β5, or γ3 transcripts. A complete understanding of the role of each isoform of MUTYH is necessary to fully comprehend the function of this enzyme in nuclear and mitochondrial BER. There exists yet another layer of defense against 8-oxoG. The human homolog of bacterial MutT, MTH1, is an oxidized purine nucleoside triphosphatase that cleaves oxidized purine nucleotides before a DNA polymerase inserts them into DNA. MTH1 is also present in the mitochondria where 8-oxoG levels are predicted to be high [Nakabeppu et al., 2006].
MBD4 is unique among other HhH family members in that it has two functional domains, an N-terminal methyl-binding domain (MBD) and a C-terminal glycosylase domain [Sjolund et al., 2013]. Therefore this enzyme belongs not only to the HhH family of DNA glycosylases but also is classified under the MBD family of proteins. Some of the preferred lesions of this monofunctional glycosylase include T and U opposite G within CpG cites. Halogenated pyrimidines, 5-hmU, and 5-FU are also good substrates for this enzyme (Table I) [Sjolund et al., 2013]. Human MBD4 is the only member of the HhH family not observed in the mitochondria thus far.
Structure and biochemistry of HhH family members
Even though the HhH family comprises several subfamilies each with distinct substrate specificities they are typified by a common HhH motif. Overall, these glycosylases harbor two domains with a α-helical character. The interface between these two domains creates a binding groove for the DNA. Residues within the HhH motif make extensive H-bond contacts with the DNA. As OGG1 has been extensively studied, it is used here as an example to describe members of the HhH family (Fig. 3E, PDB ID: 1EBM [Bruner et al., 2000]). The structure of OGG1 bound to DNA describes a role for the HhH motif in making H-bond contacts with the DNA 3' to the lesion where the DNA is predominantly B-form [Bruner et al., 2000]. A similar arrangement for the HhH motif is observed in the structure of AlkA bound to DNA [Hollis et al., 2000]. The structure of human OGG1 also reveals an antiparallel β-sheet domain in addition to the two α-helical domains (Fig. 3E). The major contribution of this domain to DNA binding is interaction between the carbonyl oxygen of Gly42, which makes a H-bond contact to the hydrogen at the N7 position of 8-oxoG that distinguishes it from guanine (Fig. 3F) [Bruner et al., 2000]. The active site nucleophile is a lysine at position 249. Mutation of this residue to Gln249 renders a catalytically inactive glycosylase that still binds tightly to DNA (Fig. 3F). The structure of OGG1 bound to 8-oxoG-containing DNA reveals four residues, Asn149, Tyr203, Arg154, and Arg204 that are involved in binding to the DNA in the vicinity of the DNA lesion (Fig. 3F). Asn149 fills the void created upon 8-oxoG extrusion into the active site and makes H-bonds contacts with the estrange dC opposite the lesion. A “wedge” residue, Tyr203, invades the DNA helix from the minor groove resulting in buckling of the target base pair and bending of the DNA (Fig. 3F). The two arginine residues, Arg154 and Arg204, make stabilizing H-bond contacts with the orphaned dC base [Bruner et al., 2000].
In addition to the HhH motif, some members of this family like human NTHL1 have an iron-sulfur (4Fe-4S type) cluster formed by the N- and C- terminal ends of the enzyme (reviewed in [Brooks et al., 2013; Lukianova and David, 2005]). As there is no current available structural information for the human NTHL1 enzyme, a potential role for the residues in the Fe-S cluster in DNA binding was proposed given the polar, and positively charged nature of the residues in the vicinity of the DNA in the crystal structure of Nth from Bacillus stearothermophilus [Fromme and Verdine, 2003]. MUTYH, another HhH family member, also possesses an Fe-S cluster within its catalytic domain [Lukianova and David, 2005]. Studies with both MutY and Nth indicate that the redox potential of the Fe-S cluster is not necessary for glycosylase activity, but upon DNA binding, a shift in redox potential occurs that maybe utilized by these enzymes to detect DNA lesions [Grodick et al., 2014].
The Fpg/Nei family
Overview
The Fpg/Nei family members were named after their bacterial prototypes Fpg and Nei. E. coli Nei was discovered in the Wallace Laboratory in 1994 and exhibited sequence similarity to the Fpg enzymes, prompting the classification of these enzymes together in the Fpg/Nei family [Melamede et al., 1994]. While there are no Fpg homologs in humans, there are three mammalian Nei-like (Neil) DNA glycosylases belonging to the Fpg/Nei family (reviewed in [Prakash et al., 2012]). The human NEIL1, NEIL2, and NEIL3 enzymes are all found in the nucleus, whereas evidence exists for the presence of only NEIL1 and NEIL2 in the mitochondria [Mandal et al., 2012; Vartanian et al., 2006]. NEIL1 excises lesions in dsDNA, bubble, bulge, and fork structures, and to a lesser extent in ssDNA, whereas NEIL2 prefers lesions in ssDNA, bubble, fork, and bulge substrates compared to duplex DNA [Prakash et al., 2012]. NEIL3 also exhibits a preference for ssDNA over duplex substrates (reviewed in [Liu et al., 2013]). There is significant overlap in the substrates recognized by the NEIL enzymes. The best substrates for NEIL1 primarily include oxidized pyrimidines such as Tg, 5-hyroxyuracil (5-OHU), dihydrouracil (DHU), 5-hydroxycytosine (5-OHC), 5,6-dihydrothymine (DHT), as well as the ring opened Fapy lesions and the further oxidation products of 8-oxoG namely spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh). However, 8-oxoG itself is not a preferred substrate for the NEIL enzymes. NEIL2 and NEIL3 also have a broad substrate recognition spectrum where Sp and Gh lesions are the best substrates for these enzymes (summarized in Table 1) [Liu et al., 2013]. Based on the available sequence for transcript variant 1 of NEIL1 (NCBI reference # NM_001256552.1), the MTS appears to be 89-aa at the N-terminal end (as determined by MitoProt II [Claros and Vincens, 1996]). This leader sequence as well as the N-terminal Met residue must be cleaved for this enzyme to be functional [Zharkov et al., 1997]. For NEIL2, the precise location and sequence of the MTS remains unknown from available sequence data (Fig. 2).
Structure and biochemistry of Fpg/Nei enzymes
Structural information has been obtained for several Fpg/Nei family members (reviewed in [Prakash et al., 2012]). Overall, the structures indicate a classic 2-domain architecture where the N- and C-terminal domains are connected by a flexible interdomain linker with the DNA binding groove lying orthogonal to the long axis of the protein. The N-terminal domain harbors a 2-layered β-sandwich capped on either end by an α-helix. The C-terminal domain comprises two highly conserved structural motifs, namely the H2TH motif and the zinc (or zinc-less) finger motifs, which are characteristic of this family and are involved in binding to the DNA. The residues within the H2TH motif are critical for binding to the phosphates in the DNA backbone. The zinc-finger motif comprises two anti-parallel β-strands and four residues (typically cysteines, or cysteines and a histidine) that coordinate a zinc ion. While there is currently no structural information for human NEIL2, the unliganded structure of human NEIL1 is available [Doublié et al., 2004]. Crystal structures of the viral ortholog of human NEIL1, Mimivirus Nei1 bound to DNA lesions (MvNei1, PDB ID 3A46, Fig. 3G [Imamura et al., 2009]) have served as a models to describe how the human enzymes might bind to DNA (reviewed in [Prakash et al., 2012]). Both NEIL1 and MvNei1 lack the residues that coordinate a zinc atom (termed a zincless finger motif) whereas NEIL2 harbors a zinc finger comprising three cysteine residues and one histidine (C-H-C-C-type) residue that contact the zinc atom. This zinc (zincless) finger motif contains an absolutely conserved Arg residue involved in making critical H-bond contacts with the DNA backbone. Mutating this conserved Arg results in a glycosylase with reduced glycosylase activity [Doublié et al., 2004]. Of the three NEIL enzymes, NEIL3 is the longest and comprises three additional zinc finger motifs including a RanBP-like zinc finger and two GRF zinc finger motifs of unknown function but predicted to be involved in nuclear localization (Fig. 2) [Liu et al., 2013].
The active site nucleophile is highly conserved among Fpg/Nei family members and is typically an N-terminal proline (Pro2 in NEIL1 and NEIL2) or a valine (Val2 in NEIL3). Mutating the N-terminal Pro2 or the neighboring Glu3 of NEIL1 to glycine and glutamine, respectively, yields an inactive glycosylase. These family members also have a conserved lysine residue that is also required for glycosylase activity. In the case of NEIL1 this corresponds to residue Lys54 [Vik et al., 2012]. Fpg/Nei enzymes possess highly conserved residues that fill the void upon lesion extrusion into the active site thereby stabilizing the DNA and the orphaned base. In MvNei1, Phe116 serves as the wedge residue while Leu84 takes the place of the damaged base and Arg114 stabilizes the orphaned base opposite the lesion (Fig. 3H). The corresponding void-filling residues in NEIL1 are Phe120, Met81, and Arg118 [Prakash et al., 2012].
Concluding Remarks
Mitochondria are more than just the “energy powerhouse of the cell”. In addition to their role in energy production, these organelles are involved in several facets of cellular metabolism and function including and not limited to apoptosis, cell-cycle regulation, and immune responses [Shaughnessy et al., 2014]. The importance of mitochondrial genome maintenance is rapidly gaining more recognition with the discovery of many nuclear proteins and enzymes being translocated to this organelle. Mitochondrial dysfunction and associated diseases can result from mutations and damage directly related with mitochondrial genes as well as damaged nuclear proteins that translocate to the mitochondria. Several examples of mutations (missense mutations, rearrangements, and single-nucleotide polymorphisms) in mitochondrial genes have been linked with conditions such as type II diabetes, Leigh syndrome, ataxia, renal dysfunction, and cardiovascular disease (reviewed in [Wallace, 2005]). Accumulation of somatic mutations in mtDNA leading to the presence of both wild-type and mutant mtDNA (heteroplasmy) within a mitochondrion may also result in mitochondrial diseases such as cancer. An example of this is seen in the case of prostate cancer where the frequency of somatic mutations in mtDNA occurs at elevated levels for patients presenting with the disease [Wallace, 2005]. Given that each cell has many mitochondria and each mitochondrion has multiple mitochondrial genomes, it is not surprising that each cell has a few to several thousand copies of mtDNA depending on the cell type. These observations present a conundrum in determining the threshold between mutations that are tolerated and those that transition to a disease state.
Although much headway has been made in identifying protein factors involved in repair of lesions in the mitochondria, several questions still remain to be answered about repair pathways in this organelle. For instance, while it is known that XRCC1 serves as a scaffold for the BER repair pathway in the nucleus [Hanssen-Bauer et al., 2011], reports of a similar scaffold in the mitochondria are absent. However, BER in the mitochondria is thought to take place at the inner membrane where the DNA is condensed into nucleoid bodies. NER is the only repair pathway not described in the mitochondria (reviewed in [Cline, 2012]). This pathway is primarily involved in the repair of bulky DNA adducts, 6,4-photoproducts, UV-induced cyclobutane pyrimidine dimers, and cisplatin induced intrastrand crosslinks. These lesions interfere with POLG activity thereby resulting in a buildup of mutations within the mtDNA [Cline, 2012]. Whether mitochondria are able to cope with such damage or if they possess an “NER-like” mechanism to resolve the damage remains unclear.
In the past few years, reports in the literature of co-localization of DNA glycosylases with mitochondrial proteins such as the mitochondrial single-strand DNA binding protein (mtSSB) and POLG, have hinted at interactions between these enzymes as part of the repair process. Unlike bacterial and viral DNA glycosylases, some of the human enzymes (like NTHL1, and the NEIL enzymes) possess disordered regions that are predicted to be involved in interactions with other proteins for coordinated repair to occur. For example the C-terminal end of NEIL1 (residues 312 – 389) is involved in binding to several proteins such as replication protein A and XRCC1 [Hegde et al., 2010]. Other protein-protein interactions involving the long, flexible extensions in the DNA glycosylases may be taking place in the mitochondria as well.
In the nucleus, while substrate redundancy among DNA glycosylases exists, it is becoming increasingly apparent that some glycosylases may be involved with specialized functions such as replication or transcription. Furthermore, expression of some glycosylases appears to be tissue-specific and cell cycle regulated. For instance, human NEIL3 expression in highest in the thymus and testes [Liu et al., 2013] and NEIL1 expression is elevated in S-phase and as such appears to be involved in DNA repair during replication [Dou et al., 2008]. Moreover, specialized functions for glycosylases such as TDG in epigenetic regulation have also recently been documented (reviewed in [Sjolund et al., 2013]). The effects of post-translation modifications of DNA glycosylases such as acetylation and phosphorylation are being scrutinized in the nucleus and whether these modifications have a role in the maintenance of the mitochondrial genome is not known. In summary, although much is known about the function of the 7 mammalian DNA glycosylases, the cross talk between the nucleus and mitochondria in mediating repair in the mitochondria still remains to be elucidated.
Acknowledgements
We apologize in advance for citing review articles instead of original research articles due to the limitation on the number of citations allowed. We would like to thank Dr. Brian E. Eckenroth for critically reading this manuscript. This work is supported by National Institutes of Health Grant P01 CA098993 awarded to SD by the National Cancer Institute. AP is supported by National Institutes of Health grant 1K99ES024417 awarded by the National Institute of Environmental Health Sciences.
Footnotes
Conflict of Interest None
References
- Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. Sequence and Organization of the Human Mitochondrial Genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bogenhagen DF. Mitochondrial DNA nucleoid structure. Biochim Biophys Acta. 2012;1819:914–20. doi: 10.1016/j.bbagrm.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch Biochem Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim Biophys Acta. 2013;1834:247–71. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000;403:859–66. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- Cadenas E, Davies KJ. Mitochondrial free radical generation, oxidative stress, and aging. Free Radic Biol Med. 2000;29:222–30. doi: 10.1016/s0891-5849(00)00317-8. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim Biophys Acta. 2012;1819:979–91. doi: 10.1016/j.bbagrm.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, Weinfeld M, Tomkinson AE, Wilson SH, Mitra S, Hazra TK. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–48. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- Demple B, Sung JS. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–9. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283:3130–40. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101:10284–9. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev. 2008;129:383–90. doi: 10.1016/j.mad.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Doublié S, Wallace SS. Consequences and Repair of Oxidative DNA Damage. In: Greim H, Albertini R, editors. The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens. Royal Society; London: 2012. pp. 115–159. [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–16. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Faucher F, Doublié S, Jia Z. 8-oxoguanine DNA glycosylases: one lesion, three subfamilies. Int J Mol Sci. 2012;13:6711–29. doi: 10.3390/ijms13066711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, McDaniel LD, Schultz RA. The role of endogenous and exogenous DNA damage and mutagenesis. Curr Opin Genet Dev. 2004;14:5–10. doi: 10.1016/j.gde.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structure of a trapped endonuclease III-DNA covalent intermediate. EMBO J. 2003;22:3461–71. doi: 10.1093/emboj/cdg311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature. 2011;471:240–4. doi: 10.1038/nature09773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodick MA, Segal HM, Zwang TJ, Barton JK. DNA-mediated signaling by proteins with 4Fe-4S clusters is necessary for genomic integrity. J Am Chem Soc. 2014;136:6470–8. doi: 10.1021/ja501973c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM, 3rd, Akbari M, Otterlei M. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen. 2011;52:623–35. doi: 10.1002/em.20663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell Mol Life Sci. 2010;67:3573–87. doi: 10.1007/s00018-010-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T, Lau A, Ellenberger T. Structural studies of human alkyladenine glycosylase and E. coli 3-methyladenine glycosylase. Mutat Res. 2000;460:201–10. doi: 10.1016/s0921-8777(00)00027-6. [DOI] [PubMed] [Google Scholar]
- Imamura K, Wallace SS, Doublié S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284:26174–83. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs AL, Schar P. DNA glycosylases: in DNA repair and beyond. Chromosoma. 2012;121:1–20. doi: 10.1007/s00412-011-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Reyes A, Holt IJ. Minimizing the damage: repair pathways keep mitochondrial DNA intact. Nat Rev Mol Cell Biol. 2012;13:659–71. doi: 10.1038/nrm3439. [DOI] [PubMed] [Google Scholar]
- Krokan HE, Bjoras M. Base excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012583. doi: 10.1101/cshperspect.a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5:89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: mechanisms for nucleotide flipping and base excision. Cell. 1998;95:249–58. doi: 10.1016/s0092-8674(00)81755-9. [DOI] [PubMed] [Google Scholar]
- Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci U S A. 1974;71:3649–53. doi: 10.1073/pnas.71.9.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Doublié S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res. 2013;743–744:4–11. doi: 10.1016/j.mrfmmm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–26. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- Lukianova OA, David SS. A role for iron-sulfur clusters in DNA repair. Curr Opin Chem Biol. 2005;9:145–51. doi: 10.1016/j.cbpa.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, Sarkar PS, Hazra TK. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287:2819–29. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markkanen E, Dorn J, Hubscher U. MUTYH DNA glycosylase: the rationale for removing undamaged bases from the DNA. Front Genet. 2013;4:18. doi: 10.3389/fgene.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33:1255–64. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Kajitani K, Sakamoto K, Yamaguchi H, Tsuchimoto D. MTH1, an oxidized purine nucleoside triphosphatase, prevents the cytotoxicity and neurotoxicity of oxidized purine nucleotides. DNA Repair (Amst) 2006;5:761–72. doi: 10.1016/j.dnarep.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–6. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Nishioka K, Ohtsubo T, Oda H, Fujiwara T, Kang D, Sugimachi K, Nakabeppu Y. Expression and differential intracellular localization of two major forms of human 8-oxoguanine DNA glycosylase encoded by alternatively spliced OGG1 mRNAs. Mol Biol Cell. 1999;10:1637–52. doi: 10.1091/mbc.10.5.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka S, Nakabeppu Y. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011;102:677–82. doi: 10.1111/j.1349-7006.2011.01869.x. [DOI] [PubMed] [Google Scholar]
- Prakash A, Doublié S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog Mol Biol Transl Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Park JW, Ames BN. Normal Oxidative Damage to Mitochondrial and Nuclear-DNA Is Extensive. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath H. Oxidative DNA damage in disease-Insights gained from base excision repair glycosylase-deficient mouse models. Environ Mol Mutagen. 2014;55:689–703. doi: 10.1002/em.21886. [DOI] [PubMed] [Google Scholar]
- Schormann N, Ricciardi R, Chattopadhyay D. Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014;23:1667–85. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaughnessy DT, McAllister K, Worth L, Haugen AC, Meyer JN, Domann FE, Van Houten B, Mostoslavsky R, Bultman SJ, Baccarelli AA, Begley TJ, Sobol RW, Hirschey MD, Ideker T, Santos JH, Copeland WC, Tice RR, Balshaw DM, Tyson FL. Mitochondria, energetics, epigenetics, and cellular responses to stress. Environ Health Perspect. 2014;122:1271–8. doi: 10.1289/ehp.1408418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature. 2011;471:245–8. doi: 10.1038/nature09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund AB, Senejani AG, Sweasy JB. MBD4 and TDG: multifaceted DNA glycosylases with ever expanding biological roles. Mutat Res. 2013;743–744:12–25. doi: 10.1016/j.mrfmmm.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996;384:87–92. doi: 10.1038/384087a0. [DOI] [PubMed] [Google Scholar]
- Takao M, Aburatani H, Kobayashi K, Yasui A. Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res. 1998;26:2917–22. doi: 10.1093/nar/26.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon B, Samson LD. Alkyladenine DNA glycosylase (AAG) localizes to mitochondria and interacts with mitochondrial single-stranded binding protein (mtSSB) DNA Repair (Amst) 2013;12:177–87. doi: 10.1016/j.dnarep.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci U S A. 2006;103:1864–9. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vik ES, Alseth I, Forsbring M, Helle IH, Morland I, Luna L, Bjoras M, Dalhus B. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair (Amst) 2012;11:766–73. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, Mitra S, Hazra TK. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15:209–20. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Wilson SH, Sobol RW, Beard WA, Horton JK, Prasad R, Vande Berg BJ. DNA polymerase beta and mammalian base excision repair. Cold Spring Harb Symp Quant Biol. 2000;65:143–55. doi: 10.1101/sqb.2000.65.143. [DOI] [PubMed] [Google Scholar]
- Yakubovskaya E, Chen Z, Carrodeguas JA, Kisker C, Bogenhagen DF. Functional human mitochondrial DNA polymerase gamma forms a heterotrimer. J Biol Chem. 2006;281:374–82. doi: 10.1074/jbc.M509730200. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Mechetin GV, Nevinsky GA. Uracil-DNA glycosylase: Structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat Res. 2010;685:11–20. doi: 10.1016/j.mrfmmm.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zharkov DO, Rieger RA, Iden CR, Grollman AP. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J Biol Chem. 1997;272:5335–41. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]