Abstract
Viral nanoparticles (VNPs) are a novel class of bionanomaterials that harness the natural biocompatibility of viruses for the development of therapeutics, vaccines, and imaging tools. The plant virus, cowpea mosaic virus (CPMV), has been successfully engineered to create novel cancer-targeted imaging agents by incorporating fluorescent dyes, polyethylene glycol (PEG) polymers, and targeting moieties. Using straightforward conjugation strategies, VNPs with high selectivity for cancer-specific molecular targets can be synthesized for in vivo imaging of tumors. Here we describe the synthesis and purification of CPMV-based VNPs, the functionalization of these VNPs using click chemistry, and their use for imaging xenograft tumors in animal models. VNPs decorated with fluorescent dyes, PEG, and targeting ligands can be synthesized in one day, and imaging studies can be performed over hours, days, or weeks, depending on the application.
Keywords: Cowpea mosaic virus (CPMV), Bionanomaterials, CPMV-based viral nanoparticles, Molecular imaging agents, Chemical conjugation, Click chemistry, Tumor-homing nanoparticles, Peptide-based affinity probes
1 Introduction
Advancements in nanotechnology have been fueled in part by the development of affinity probes for ligand-mediated targeting. The generation of these “smart” targeted nanoparticles has advanced the field of molecular imaging [1–5] and drug delivery (as reviewed in [6]). Nanoparticles of different sizes and formulations can be visualized and quantified as they flow in the bloodstream, extravasate from tumor-associated vessels, and accumulate at the tumor site using advanced techniques such as high-resolution intravital imaging [1, 7, 8]. In this chapter, we describe the functionalization of cowpea mosaic virus (CPMV) with near-infrared fluorescent dyes, targeting ligands, and/or polyethylene glycol (PEG) polymers [1, 7, 9, 10] and their use as molecular imaging agents.
CPMV comprises an icosahedral protein cage that is approximately 31 nm in diameter [7]. The exterior of the viral capsid surface displays 300 accessible lysine residues (Fig. 1b) that can be exploited for chemical conjugation with functional moieties. Moieties such as fluorescent dyes [1, 8, 9], metals [9, 11], and quantum dots [12] can be added to the viral nanoparticle (VNP) surface for imaging. Functional groups such as antibodies [13] or peptides [8, 10] can be conjugated to the VNP for molecular targeting, while pharmacokinetics, clearance, and immunogenicity can be influenced by affixing PEG [3, 10, 14, 15]. Standard coupling procedures using N-hydroxysuccinimide (NHS) ester- activated reagents to target Lys side chains have slow reaction kinetics, and large excesses of reagents have to be used to facilitate efficient labeling. NHS coupling therefore is only efficient for conjugation of small molecules such as dyes or ligation handles. For conjugation of peptides or PEG, a bio-orthogonal reaction approach such as click chemistry is preferred. Click reactions have been widely used for bioconjugation and are a popular strategy because of their specificity, high yield, and wide range of solvents and pH stabilities [16]. Click reactions require low concentration and excess of the reagent or ligand of interest. This is important when reagents are scarce, if solubility in aqueous conditions is a problem, or during conjugation of high-molecular-weight ligands.
Fig. 1.
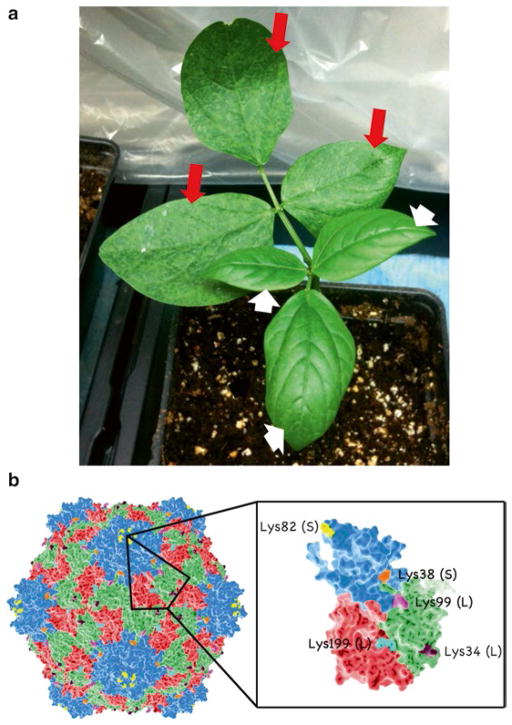
Cowpea mosaic virus (CPMV) nanoparticle. (a) Black-eyed pea plant that contains leaves that have been infected with CPMV (indicated by long red arrows). Infection symptoms are prominent on secondary leaves 7–10 days after inoculation with mosaic virus. Uninfected primary leaves are indicated by white short arrows. (b) Molecular model of CPMV nanoparticles with Lys side chains available as ligation handles. Space filling models are created using PyMol (atomic coordinates are available in the protein databank (PDB ID 1NY7)
For the purpose of generating tumor-homing nanoparticles, we have focused our efforts on the use of peptide-based affinity probes as they are small, easy to synthesize, and can be designed for straightforward conjugation to VNPs. Cancer-homing peptides have been used extensively for tumor targeting [17], and novel affinity peptides have been successfully discovered through rationale design approaches [18, 19] or through screening approaches such as phage display [20] or one-bead-one-compound library screening approaches [4, 21]. We have shown previously that VNPs decorated with affinity peptide probes against the gastrin-releasing peptide receptor (GRPR) [8] or vascular endothelial growth factor receptor 1 (VEGFR1) [10] are useful for molecular imaging of tumors in vivo. Methods described here are suitable for a wide range of peptide-based affinity ligands. Additionally, protocols are described to incorporate shielding from unwanted uptake and immune responses. We have found that it is necessary to shield VNPs using at least 60 copies of PEG500 or 30 copies of PEG2000, PEG3400, or PEG5000 [1, 8, 14, 15].
Effective imaging agents such as VNPs must be optimized to maximize tumor uptake and minimize background. Current technologies for tumor imaging, such as ultrasound, MRI, PET, and CT, do not provide high enough resolution to adequately assess nanoparticle uptake in tumors. To address this, we describe a high-resolution intravital imaging approach to evaluate nanoparticle uptake in human tumor xenografts in a modified, shell-less chicken embryo model. The chicken embryo chorioallantoic membrane (CAM) model is particularly appropriate for in vivo intravital and tumor imaging studies using targeted nanoparticles as it is inexpensive and supports the growth of human tumors [22]. Tumor cells implanted in the CAM are able to form fully vascularized xenografts within 7 days [23]. VNPs can be injected directly into the bloodstream and imaged for over several hours or days.
In this chapter, we describe the generation and characterization of molecular targeted VNPs and the optimization of these nanoparticles using intravital tumor imaging in vivo using the chicken embryo CAM model. Optimized VNP formulations can then be validated in the appropriate human tumor models.
2 Materials
2.1 CPMV Propagation, Purification, and Conjugation
Reach-in indoor plant chamber (Percival Scientific, Perry, Iowa).
Vigna unguiculata seeds (California black-eye no. 5; Burpee).
Pro-mix BX potting soil (Premier Horticulture).
Jack’s Professional 20-10-20 Peat-Lite Fertilizer (JR Peters, Allentown, Pennsylvania).
Carborundum (Fisher Scientific, Pittsburgh, Pennsylvania).
Sorvall RC-6 Plus centrifuge (Thermo Scientific, Asheville, North Carolina) with SLA-3000 and SS-34 rotor.
Optima L-90 K ultracentrifuge (Beckman, Brea, California) with 50.2 Ti rotor and SW 32 Ti rotor.
Sucrose (Fisher Scientific, Pittsburgh, Pennsylvania).
Chloroform (Fisher Scientific, Pittsburgh, Pennsylvania).
1-Butanol (Fisher Scientific, Pittsburgh, Pennsylvania).
NaCl (Fisher Scientific, Pittsburgh, Pennsylvania).
PEG (MW 8,000) (Fisher Scientific, Pittsburgh, Pennsylvania).
Potassium phosphate dibasic (Fisher Scientific, Pittsburgh, Pennsylvania).
Potassium phosphate monobasic (Fisher Scientific, Pittsburgh, Pennsylvania).
NanoDrop (NanoDrop 2000c) (Thermo Scientific, Asheville, North Carolina).
AKTA Explorer 100 Chromatograph with Superose6 column (GE Healthcare, Pittsburgh, Pennsylvania).
2.2 Peptide Synthesis
Manual reaction vessel (Peptides International, Louisville, Kentucky).
Hand-powered blower (Fisher Scientific, Ottawa, Ontario).
15 and 50 mL falcon tubes (VWR Inc., Radnor, Pennsylvania).
Shaker (IKA, Wilmington, North Carolina).
Centrifuge (Beckman Coulter, Brea, California).
Lyophilizer (Labconco, Kansas City, Missouri).
High-performance liquid chromatography (HPLC) (Waters, Milford, Massachusetts).
9-Fluorenylmethoxycarbonyl (Fmoc)-protected rink amide 4-methylbenzhydrylamine (MBHA) resin (loading ~0.5 meq/g) (Aapptec, Louisville, Kentucky).
N,N-dimethylformamide (DMF) (Fisher Scientific, Ottawa, Ontario).
Methylene chloride (DCM) (Fisher Scientific, Ottawa, Ontario).
Piperidine (Sigma Aldrich, St. Louis, Missouri).
N-Fmoc amino acids (Aapptec, Louisville, Kentucky).
2-(1H-benzotriazole-1-yl)-1, 1, 3, 3-tetramethyluronium hexafluorophosphate (HBTU) (Aapptec, Louisville, Kentucky).
N,N-diisopropylethylamine (DIPEA) (Sigma Aldrich, St. Louis, Missouri).
550 MW N3-(PEG)7-COOH (Novabiochem, Gibbstown, New Jersey).
Trifluoroacetic acid (TFA) (Sigma Aldrich, St. Louis, Missouri).
Phenol (Sigma Aldrich, St. Louis, Missouri).
Triisopropylsilane (Sigma Aldrich, St. Louis, Missouri).
Tert-butyl methyl ether (Sigma Aldrich, St. Louis, Missouri).
0.8 mL micro bio-spin columns (Bio-Rad, Hercules, California).
2.3 CPMV Conjugation and Characterization
Alexa Fluor 647 carboxylic acid, succinimidyl ester (Life Technologies, Grand Island, New York).
NHS-alkyne (N-(4-pentynoyloxy) succinimide) (Life Technologies, Grand Island, New York).
Dimethyl sulfoxide, DMSO (Fisher Scientific, Pittsburgh, Pennsylvania).
L-ascorbic acid sodium salt (Acros Organics, New Jersey).
Aminoguanidine hydrochloride (Acros Organics, New Jersey).
Copper sulfate pentahydrate (Acros Organics, New Jersey).
Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA), gift from MG Finn, The Scripps Research Institute (La Jolla, California).
4–12 % Bis-Tris NuPAGE SDS gel (Life Technologies, Grand Island, New York).
MPOS buffer (Life Technologies, Grand Island, New York).
Coomassie Brilliant Blue R-250 (Fisher Scientific, Pittsburgh, Pennsylvania).
4× LDS (Life Technologies, Grand Island, New York).
Methanol (Fisher Scientific, Pittsburgh, Pennsylvania).
Acetic Acid (Fisher Scientific, Pittsburgh, Pennsylvania).
Agarose (Fisher Scientific, Pittsburgh, Pennsylvania).
6× loading dye for native agarose gels (Fisher Scientific, Pittsburgh, Pennsylvania).
1× Tris–boric acid–EDTA (TBE buffer; pH 8.0; Tris: 90 mM; boric acid: 90 mM; EDTA: 2.5 mM).
TEM grids (Ted Pella, Redding, California).
Uranyl Acetate (Fisher Scientific, Pittsburgh, Pennsylvania).
Transmission electron microscope (Zeiss, Libra 200FE, North Chesterfield, Virginia).
Amicon Ultra-0.5 mL Centrifugal filters (10 kDa-cut-off) (Millipore, Billerica, Massachusetts).
Ultracentrifuge and rotors as described above.
2.4 Preparation of Eggs and Ex Ovo Culture of Chicken Embryos
Eggs (Rochester Hatchery, Alberta) (incubated at 38 °C at 60 % humidity).
Digital hatcher (GQF Manufacturing Co, Savannah, Georgia).
Humidified incubator (Caron, Ohio).
Plastic container (with holes drilled on the sides) (Rubbermaid).
Dremel drill with circular wheel (36 cutoff).
Distilled water.
70 % ethanol.
Sterile polystyrene weigh boats (VWR).
Sterile square plastic cover (Simport, Quebec).
2.5 Preparation of Cancer Cell Lines for Inoculation into Chicken Embryo
Cancer cell line of interest (see Note 1).
Growth medium (containing 10 % fetal bovine serum (FBS) and 1 % Pen/Strep) (specific for each cancer cell line).
FBS (heat-inactivated) (Invitrogen, Burlington, Ontario).
Sterile 1× PBS (Invitrogen, Burlington, Ontario).
2.5 % Trypsin–EDTA (Invitrogen, Burlington, Ontario).
Centrifuge (Eppendorf, Mississauga, Ontario).
Serological pipettes and dispenser (Mandel Scientific Company, Guelph, Ontario).
Micropipettes (Mandel Scientific Company, Guelph, Ontario).
15-mL falcon tube (Cedarlane, Burlington, Ontario).
Hemocytometer (for cell counting) (Bright-line, Bridgeville, Pennsylvania).
Tissue culture flow hood.
Humidified microbiological CO2 incubator (37 °C and 5 % CO2) (Nuaire, Plymouth, Manhattan).
2.6 Establishment of Solid Tumors in the Chicken Embryo CAM
Vertical pipette puller (model 720) (KOPF Instruments, Tujunga, California).
Pulled glass needle (made using pipette puller) (Sutter Instrument Co, Novato, California).
Humidified incubator (Caron, Marietta, Ohio).
Fine-point forceps (VWR, Edmonton, Alberta).
Circuit Inspection Zoom Power Stereo Microscope (AmScope, Irvine, California).
18-gauge needle (BD, Mississauga, Ontario).
1 mL syringe (BD, Mississauga, Ontario).
Tygon R-3603 tubing (1/32 in. inner diameter, 3/32 in. outer diameter, 1/32 in. wall thickness) (VWR, Edmonton, Alberta).
Kimwipe (VWR, Edmonton, Alberta).
Chicken embryo at day 9 of development.
Cancer cell line of interest (resuspended in 1× PBS).
2.7 Intravenous Injection of Fluorescently Labeled CPMV Nanoparticles
Assembled microinjector.
Day-16 chicken embryo.
Embryo imaging unit (Innovascreen Inc., Halifax, Nova Scotia).
Fluorescent CPMV nanoparticles (800 mg/mL) (synthesized as described above).
AxioExaminer Z1 upright microscope (Zeiss, Toronto, Ontario).
Spinning disk head (Yokogawa, Tokyo, Japan).
9100–12 ImageEM CCD camera (Hamamatsu, Bridgewater, New Jersey).
3 Methods
3.1 Propagation and Purification of CPMV
All reactions were carried out at room temperature unless specifically indicated.
CPMV propagation: Set the indoor plant chamber controls to 15 day-h (100 % light, 25 °C, 65 % humidity) and 9 night-h (0 % light, 22 °C, 60 % humidity).
Plant three cowpea seeds/pot on day 0; infect leaves with CPMV (5 μg/5 μl/leaf) on day 10 by mechanical inoculation by light dusting of carborundum (Fig. 1a); on day 20, harvest leaves and store at −80 °C.
Homogenize 100 g of frozen leaves in a standard blender using two volumes of cold 0.1 M potassium phosphate buffer (pH 7.0). Filter through 2–3 layers of cheesecloth.
Centrifuge crude plant material at 5,500 × g for 20 min. Collect supernatant.
Extract plant material by adding 0.7 volumes of 1:1 (v/v) chloroform:1-butanol. Stir mixture for 30–60 min.
Centrifuge solution at 5,500 × g for 20 min. Collect the upper aqueous phase.
Add NaCl to 0.2 M and 8 % (w/v) PEG (MW 8,000). Stir for at least an hour, and then let sit for at least an hour.
Centrifuge solution at 15,000 × g for 15 min. Resuspend pellet in 10 mL of 0.1 M potassium phosphate buffer (pH 7.0).
Centrifuge at 8,000 × g for 30 min, and collect supernatant.
Ultracentrifuge supernatant at 160,000 × g for 3 h. Resuspend pellet in 5 mL 0.1 M potassium phosphate buffer (pH 7.0) and stir overnight. Prepare a 10–40 % sucrose gradient using an equal volume of 10, 20, 30, and 40 % sucrose in buffer (heaviest first). Allow the gradient to equilibrate overnight at room temperature.
Overlay resuspended pellet over sucrose gradient and ultracentrifuge at 100,000 × g for 2 h.
Collect light scattering band and dialyze against 500 mL 0.1 M potassium phosphate buffer (pH 7.0).
Perform UV/visible spectroscopy to determine the concentration of VNPs. Measure the absorbance of 2 μL of sample using the NanoDrop (see Subheading 3.4).
Perform size-exclusion fast protein liquid chromatography (FPLC) using a Superose 6 size-exclusion column and the ÄKTA Explorer (see Subheading 3.4).
Store the purified CPMV at 4 °C. For long-term storage, store at −80 °C. The molecular model of CPMV nanoparticle is shown in Fig. 1b.
3.2 Synthesis of Peptides for Conjugation
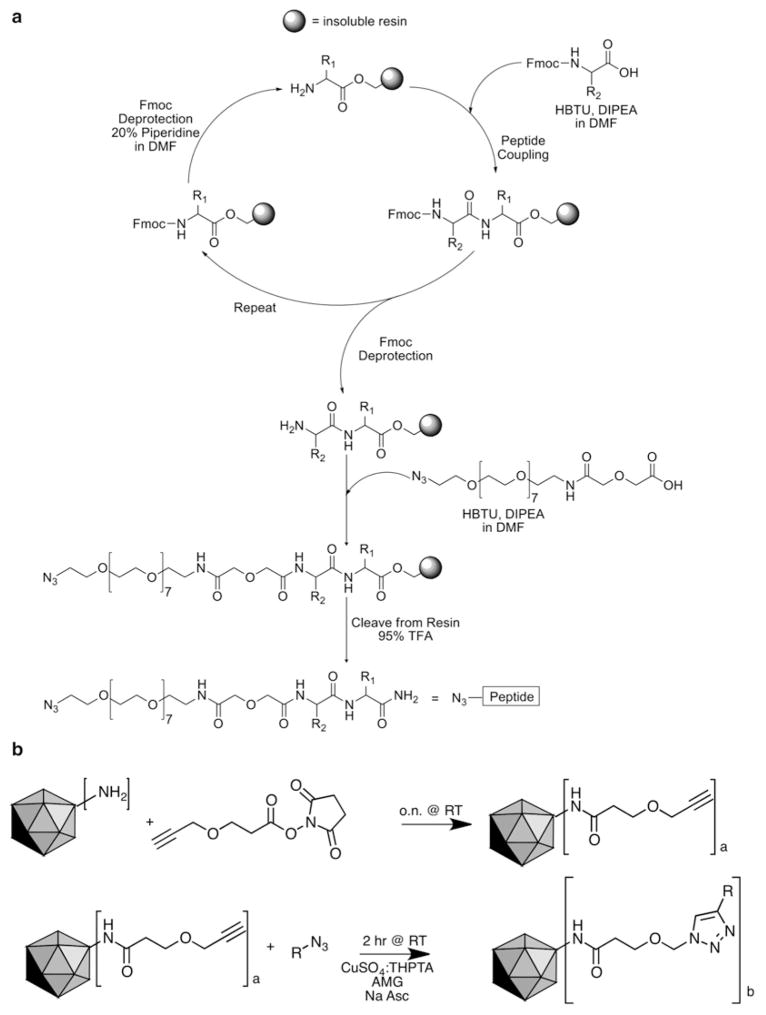
Fully protected resin-bound peptides are synthesized via standard Fmoc solid-phase peptide chemistry from C-terminus to N-terminus [24] on a 0.1 mmol scale (Fig. 2a). All peptide couplings are performed at room temperature in a manual reaction vessel. Alternatively, an automated peptide synthesizer may be used.
Fig. 2.
Targeted CPMV nanoparticle synthesis strategy. (a) Peptides are synthesized using standard Fmoc solid-phase peptide chemistry using both manual synthesis methods and an APEX 396 automated peptide synthesizer. N3-(PEG)7-COOH spacer is used to install the azide functionality at the N-terminus of the peptide. DMF dimethylformamide, HBTU 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate, DIPEA N,N-diisopropylethylamine, TFA trifluoroacetic acid. (b) Targeted viral nanoparticles are synthesized via the following steps: (1) (N-(4-pentynoyloxy)succinimide) is reacted with available Lys side chains, and (2) azide-activated functional group (R) is coupled via copper(I)-catalyzed azide-alkyne cycloaddition. R represents PEG, peptide, or PEG–peptide, and Na Asc is sodium ascorbate (5 mM)
Swell resin in reaction vessel with DCM. Swirl to form a suspension and leave for 30 min. Remove DCM with hand-powered blower.
Removal of Fmoc protecting group from resin: Treat resin with a solution of 20 % v/v piperidine in DMF (see Note 2). Shake for 5 min (see Note 3). In order to remove soluble impurities, filter and rinse the resin with DCM and DMF alternating 3× each. Treat resin a second time with 20 % piperidine in DMF. Shake for 20 min. Rinse the resin again as above (see Note 4).
Amino acid coupling to resin: Pre-activate three equivalents of desired amino acid in a 50 mL falcon tube with three equivalents of coupling agent HBTU and six equivalents of DIPEA in DMF and shaking for approximately 5 min (see Note 5). Add solution to resin and shake for 30 min. Rinse resin as above. Prepare a second pre-activated solution as above and add to resin. Shake for 2 h. Rinse resin as in step 1 (see Note 4). Repeat steps 2 and 3 until desired length of peptide is reached (see Note 6).
Coupling of azide to N-terminus of peptide: Perform a final Fmoc deprotection prior to coupling of azide (step 2). Pre-activate three equivalents of N3-(PEG)7-COOH in a 50 mL falcon tube with three equivalents of HBTU and six equivalents of DIPEA in DMF (see Note 5). Shake for approximately 5 min. Add solution to resin and shake for 2 h. Rinse resin as in step 1.
Deprotection and cleavage of peptide from resin: Treat resin with a solution of TFA containing 5 % v/v water, 5 % m/v phenol, and 2 % v/v triisopropylsilane as scavengers. Shake for 4 h. Filter into a 15 mL falcon tube and rinse resin with a small amount of TFA. To precipitate the peptide from the TFA, add cold TBME. Cool the solution on ice for approximately 5 min, centrifuge for 5 min, and decant the remaining ether. Resuspend resulting solid in cold TBME, and repeat cooling and centrifugation. Redissolve solid in water and lyophilize to obtain crude peptide.
Purification of peptide is performed by preparative HPLC on a reverse-phase C-18 preparative column (for example Waters Sunfire RP-C18 19 × 150 mm, 5 μm column) (see Note 7).
3.3 Conjugation of VNPs and Characterization
-
Conjugation of fluorescent dye and alkyne functional group to CPMV surface Lys residues (Fig. 2b):
Add 2,000 M equivalents each of Alexa Fluor 647 succinimi-dyl ester and N-(4-pentynoyloxy) succinimide dissolved in DMSO to CPMV in 0.1 M potassium phosphate buffer. Adjust the buffer and DMSO volumes such that the final concentration of CPMV is 2 mg/mL and DMSO content is 10 % of the total reaction volume. Incubate the reaction mixture overnight on an overhead shaker at room temperature protected from light. Purify the samples using centrifugal 10-kDa cutoff filters and ultrapelleting (Beckman 50.2 Ti rotor, 42,000 rpm, 3 h, 4 ° C), and resuspend CPMV in 0.1 M potassium phosphate buffer pH 7.0 to obtain a concentration of 10 mg/mL.
Covalent attachment of azide-functional PEG and PEG–peptide: To a buffered solution of CPMV-A647-alkyne (0.4 μM final concentration at 2 mg/mL) add azide–PEG or azide–PEG–peptide (150 μM) in DMSO, respectively, in a molar excess of 625:1 per CPMV. Add the following reagents for the click reaction: amino guanidine (AMG, 5 mM), CuSO4:THPTA in a molar ratio of 1:5 (500 μM CuSO 4, 2.5 mM THPTA), and sodium ascorbate (Na Asc) (5 mM). Incubate the reaction mixture at room temperature for 60 min on a shaker. Purify by ultrapelleting (see Subheading 3.1, steps 11–13). Resuspend in 0.1 M potassium phosphate buffer and store at 4 °C at a desired concentration (recommended 1–5 mg/mL).
3.4 Characterization of CPMV Conjugates
3.4.1 Perform UV/visible spectroscopy to determine the concentration of VNPs and number of dyes attached to CPMV
Measure the absorbance of 2 μL of sample using the NanoDrop.
Determine the concentration of particles and dyes using the Beer–Lambert law (A =εcl, where A is the absorbance, ε is the extinction coefficient, c is the concentration, and l is the path length). The path length is 0.1 cm for the NanoDrop. The extinction coefficient for CPMV is 8.1/cm/mg mL (at 260 nm).
3.4.2 Perform Native Gel Electrophoresis to Confirm Covalent Attachment of Dye, PEG, and Peptides (Fig. 3a)
Fig. 3.
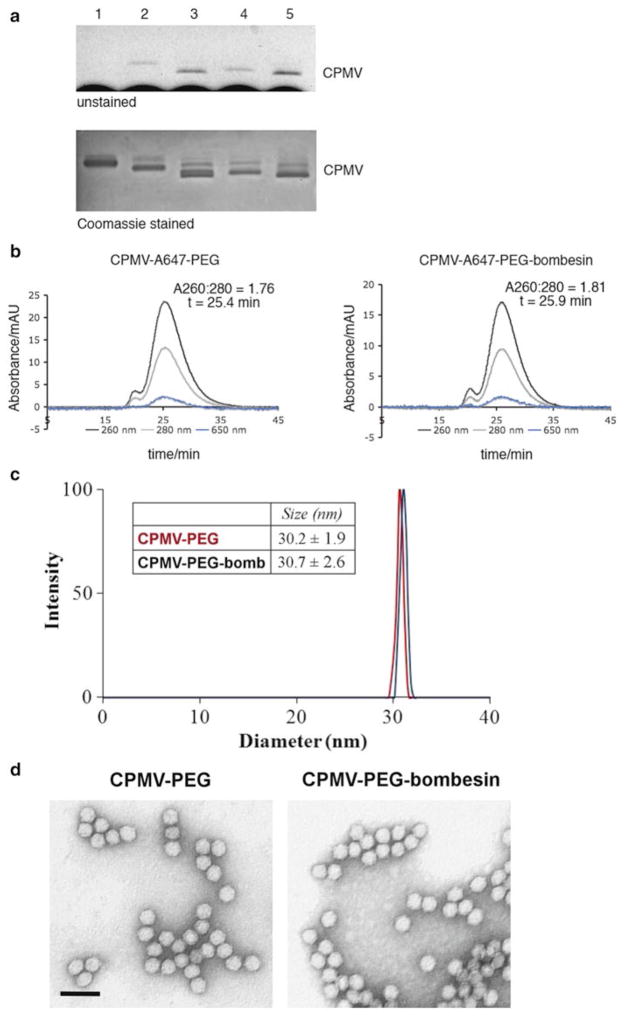
Characterization of CPMV nanoparticles. Native gel electrophoresis of intact CPMV particles (1.2 % w/v agarose gel). Top panel: Dye-labeled particles are visualized as blue bands under white light. Bottom panel: Coomassie staining reveals native and modified CPMV. Lane 1=CPMV, Lane 2=CPMV-A647, Lane 3=CPMV-A647-alkyne, Lane 4=CPMV-A647-PEG-peptide, Lane 5=CPMV-A647-PEG. (b) Size-exclusion chromatography of CPMV-A647-PEG-peptide and CPMV-A647-PEG using Sepharose6 column. Black line indicates absorbance at 260 nm (encapsidated RNA), grey line indicates absorbance at 280 nm (protein coat), while blue line indicates Alexa Fluor 647. (d) TEM images of negative-stained CPMV-PEG and CPMV-PEG-peptide particles (scale bar = 60 nm) (color figure online)
Add 2 μL of 6× sample buffer to 10 μg of the CPMV particles in 10 μL of potassium phosphate buffer.
Load samples to 1.2 % agarose gel, and run samples at 100 V for 30 min in 1× TAE buffer.
Document the gel under UV light if samples are fluorescent.
Stain with Coomassie blue (0.25 % (w/v) Coomassie Brilliant Blue R-250, 30 % (v/v) methanol, 10 % (v/v) acetic acid) for 1 h.
Destain with 30 % methanol and 10 % acetic acid overnight. Change the solution if required.
Document the gel under white light.
3.4.3 Perform Denaturing SDS Gel Electrophoresis to Analyze Conjugation of Dyes, PEG, and Peptides to Individual Coat Proteins (Fig. 3a)
Add 3 μL of 4× LDS sample buffer to 10 μg of the particles in 9 μL of potassium phosphate buffer.
Incubate in heat block for 5 min at 100 °C.
Load samples onto an SDS gel.
Run samples at 200 V for 1 h in 1× MOPS running buffer.
Document the gel under UV light if samples are fluorescent.
Stain with Coomassie blue, destain, and document as described above. Use band analysis tool to determine the ratio of labeled versus native non-modified proteins.
3.4.4 Analyze Particles by Size-Exclusion Fast Protein Liquid Chromatography (Fig. 3b)
Using a Superose 6 size-exclusion column and the ÄKTA Explorer, load 50–100 μg of VNPs in 200 μL of 0.1 M potassium phosphate buffer (pH 7.0).
Set detectors to 260 nm (nucleic acid), 280 nm (protein), and the excitation wavelength of any dyes attached.
Run at a flow rate of 0.5 mL/min for 72 min.
The elution profile and A260:A280 nm indicate whether the VNP preparation is pure and whether particles are intact and assembled. The A260:280 ratio that equals to 1.8 ± 0.1 indicates a pure CPMV preparation.
3.4.5 Analyze Integrity of Particles by Transmission Electron Microscopy (Fig. 3c)
Dilute samples to 0.1–1 mg/mL in 20 μL of DI water, place 10 μL drops of a transmission electron microscopy (TEM) grid, and let sit for 2 min. Remove excess solution on the grid with filter paper.
Wash grid with a drop of DI water.
Stain grid by placing on a 10 μL drop of 2 % (w/v) uranyl acetate for 2 min. Remove excess stain with filter paper.
Wash grid once more in water.
Observe grid under a transmission electron microscope.
3.5 Intravital Imaging in the Xenograft Ex Ovo Chicken Embryo Tumor Model
3.5.1 Preparation of Eggs and Ex Ovo Culture of Chicken Embryos [3]
Incubate fertilized eggs (day 0) at 38 °C at 60 % humidity in a digital hatcher with rotation (set to turn every 30 min) for 4 days (day 4). Before then, fertilized eggs should be stored in the refrigerator at 4 °C for no longer than 10 days before incubation in the hatcher so that the development of the embryo is arrested.
Assemble Dremel drilling tool with circular wheel (36 cutoff). Use a clamp on a retort stand to hold the Dremel tool in place. Sterilize the Dremel tool with 70 % ethanol to prevent contamination while working with the shell-less embryos.
On day 4, remove the eggs from the rotating hatcher and leave the eggs on a benchtop undisturbed for approximately 10 min. Without rotating the egg, hold the egg up with both hands towards the Dremel drilling tool. De-shell the embryo by creating shallow cuts at equidistance along the circumference of the shell while holding the egg firmly with both hands. Apply pressure on the shell so as to split it into two halves, releasing the contents of the egg into a sterilized weighing dish (see Note 8). A small heartbeat should be visible within the yolk to indicate a healthy embryo. Discard the egg if this heartbeat is not present.
Place a sterile square plastic cover over the weighing dishes containing the embryo. Add approximately 1 cm of distilled water into the container, and place square plastic covers in the humidified rubber maid container with holes drilled in the sides. This is done to elevate the weighing dishes containing the embryo to avoid submerging the dishes in water. Transfer the embryos onto these square plastic covers into the humidified container, and incubate the containers in a clean humidified incubator at 38 °C at 60 % humidity.
3.5.2 Preparation of Cancer Cell Lines for Inoculation into Chicken Embryo (Fig. 4a)
Fig. 4.
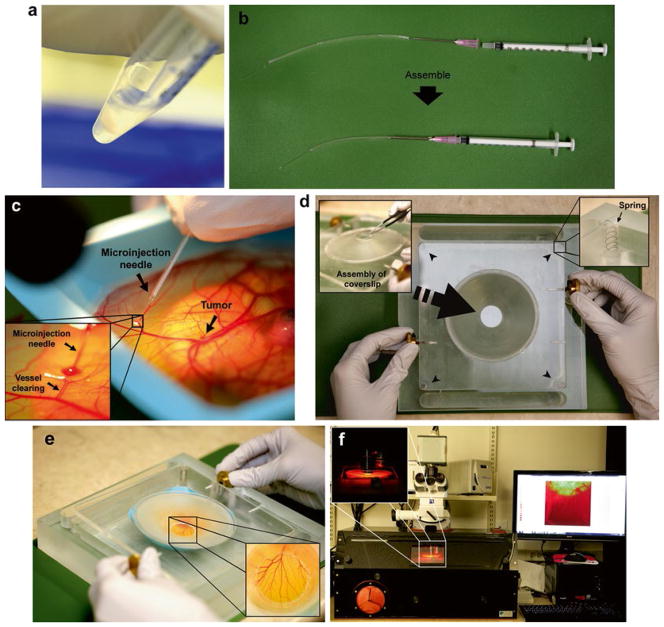
The ex ovo chicken embryo model as a VNP optimization platform. (a) Preparation of cancer cell line. Cancer cells are detached from flask using 2.5 % trypsin–EDTA, neutralized with serum-containing media, washed with 1× PBS, and resuspended in 1× PBS at a final concentration of 1 × 107 cells/mL. (b) Assembly of microinjection syringe by connecting an 18-G needle attached to a piece of Tygon tubing onto a 1 mL syringe. A microinjection glass needle is inserted at the end of the tubing. (c) Intravenous injection where the microinjection needle is inserted into the lumen of a vessel, and nanoparticles are injected and visualized while entering the blood flow by clearing of the vessel. (d) Chicken embryo incubation chamber for intravital imaging. The cover slip is fitted onto the port of the imaging unit. Springs are indicated with short black arrows. (e) Chicken embryo mounted within the imaging unit. The cover slip is placed directly on the CAM on the area of interest for imaging. (f) Confocal microscopy of the chicken embryo held within the imaging unit is performed in a temperature-regulated enclosure set to 37 °C
Culture cancer cell line of interest in serum-containing growth media to 70–80 % confluency. Cells were typically grown in growth medium supplemented with 10 % FBS and 1 % Pen/Strep in a cell culture incubator at 37 °C with 5 % CO2.
Wash cells once with 10 mL of 1× PBS (pH 7.4). In order to detach adherent cells from the flask, aspirate the PBS, and then add 2.5 % trypsin–EDTA (1 mL in a T75 flask and 1.5 mL into a T175 flask) onto the cells. Incubate the flask at 37 °C for 2–5 min until most cells have detached.
Add media (supplemented with serum) to neutralize the trypsin, and transfer cell suspension to 15 mL falcon tube. Centrifuge at 325 × g for 4 min. Aspirate the supernatant, and resuspend the cell pellet with 10 mL of PBS. Centrifuge again at 1,300 rpm for 4 min.
Aspirate the supernatant, and resuspend the cell pellet with 0.5–1 mL of 1× PBS.
Count the cells with a hemocytometer, and adjust the volume of the cells to reach a final concentration of 1 × 107 cells/mL. Only use 1× PBS to dilute/resuspend cell concentrates. Store the cell suspension on ice, and proceed immediately to implantation.
3.5.3 Establishment of Solid Tumor into Chicken Embryo CAM
On day 9 or 10 of chicken embryonic development, assemble a micro-injector by connecting an 18-G needle onto a 1 mL syringe. Cut a 2–3 in. piece of Tygon tubing, and gently slide the bevel of needle into the tubing. Once the needle is completely inserted, approximately 1–2 in. of tubing should extend from the tip of the needle (Fig. 4b).
Fill the syringe (through the extended tubing) with cell suspension. Remove all air bubbles by gently tapping the syringe. Then, insert a microinjection glass needle at the end of the tubing (see Note 9) (Fig. 4b). Carefully remove any air bubbles.
Inject day-9 embryos with 50,000–100,000 cancer cells under a dissection scope with an illuminator as a bolus within the CAM. Inject carefully, ensuring that the tip of the microinjection needle has pierced through the upper membrane of the CAM and is placed properly within the CAM without penetrating the bottom membrane. Inject the cells to form a visible bolus within the CAM. Cells that drip onto the surface of the CAM can be cleaned using a Kimwipe or a cotton applicator.
Return embryos to the humidified incubator at 38 °C with 60 % humidity, and allow tumors to grow and vascularize (up to 7 days).
3.5.4 Intravenous (IV) Injection of Fluorescently Labeled CPMV Nanoparticles
On day 16 of chicken embryo growth, assemble the micro-injector. Draw up approximately 200 μL of CPMV nanoparticle into the syringe. Carefully remove any air bubbles. Insert micro-injection glass needle at the end of the tubing (see Note 10).
Intravenously inject 50–100 μL of 800 μg/ml of fluorescent CPMV conjugates into the blood vessel of the chicken embryo containing tumors (Fig. 4c). Select a vessel that is located distal from the desired site to be visualized (see Note 11).
Image tumors immediately under an intravital spinning-disk confocal microscope that comprises an upright Zeiss AxioExaminer Z1, LUDL filter wheels, and large format motorized stage that are fitted with a Yokogawa spinning disk head and a Hamamatsu ImagEM 9100-12 EM-CCD camera (Fig. 5).
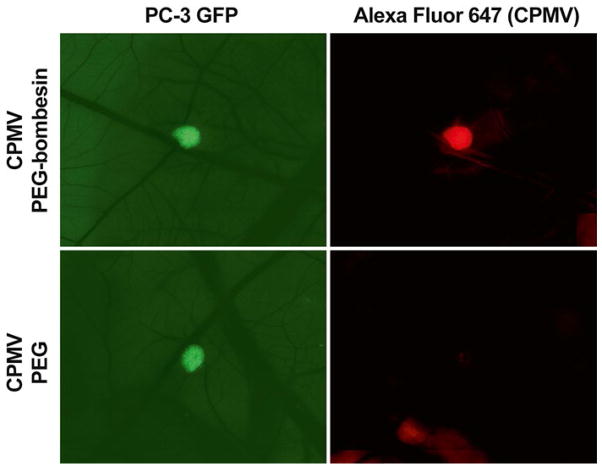
Fig. 5.
Intravital imaging of VNP uptake in tumors. Human prostate tumors expressing GFP are visualized in the CAM of the ex ovo chicken embryo using intravital fluorescence microscopy (left panels). The uptake CPMV- based VNPs labeled with Alexa Fluor 647 are visualized using near-infrared imaging (right panels) with bombe-sin peptide-mediated targeting (upper panels) or PEG alone (lower panels)
3.5.5 Real-Time Intravital and Tumor Imaging
Set the temperature within the regulated microscope chamber to 37 °C.
Assemble the ex ovo chicken embryo imaging unit [3, 25]. Apply a thin layer of vacuum grease using a cotton swab around the circumference of the lid of the imaging port where the cover slip will fit, and then place an 18 or 22 mm glass cover slip (depending on the embryo imaging unit used) onto the port. Ensure that the cover slip has tightly sealed the port (Fig. 4d).
Place the embryo within the imaging unit (see Note 12). For long-term imaging, add approximately 1 mL of distilled water outside the dish containing the embryo within the imaging unit to prevent the embryo from dehydration. Position the embryo within the imaging unit such that the cover slip can be placed onto the desired area for imaging. Slowly lower the lid until the cover slip gently makes contact with the CAM of the embryo. Tighten the screws on the unit lid to hold the lid in place (Fig. 4e).
Add distilled water outside of the embryo-containing compartment, and then place the unit onto the microscope stage within the environmental chamber equilibrated to 37 °C. For confocal microscopy, we use the Zeiss AxioExaminer Z1 upright microscope with LUDL filter wheels and large format motorized stage, fitted with a Yokogawa spinning disk head and a Hamamatsu ImagEM 9100-12 EM-CCD camera.
Position the imaging unit containing the embryo so that the cover slip is directly under the objective of the confocal fluorescence microscope. Center the tumor within the field of view, and acquire high-resolution three-dimensional Z-stacks of the tumor and surrounding vasculature to visualize detailed structural analyses at specific time points. Flatten the Z-stack of images of the tumor to create a single image using Volocity (Perkin Elmer). Acquire three-dimensional stacks at regular time-points to map detailed structural changes in the tumor vasculature over time (Fig. 4f).
Quantify the uptake of CPMV nanoparticles by calculating the mean fluorescence signal within the tumor or comparing it with the mean fluorescence signal in the stroma (non-tumor area) using image quantitation software such as Volocity (Perkin Elmer) or ImageJ (National Institutes of Health). Calculate the tumor:stroma ratio by dividing the mean fluorescence signal in the tumor by the mean fluorescence signal in the stroma. A tumor:stroma ratio higher than 1 indicates that the nanoparticle is being taken up by the tumor [5].
Acknowledgments
This work was supported by Prostate Cancer Canada Grant #2011- 742 to JDL, Natural Sciences and Engineering Research Council of Canada (NSERC) grant #326972 to LGL, and NIH/NIBIB grant R00 EB009105 and Mt. Sinai Foundation to NFS. All experiments were performed in accordance with the regulations and guidelines of the Institutional Animal Care and Use Committee at Case Western Reserve University and at the University of Alberta. We thank Desmond Pink for his photography.
Footnotes
In our hands, the following cell lines form tumors in the chicken embryo model: HT1080 (fibrosarcoma), HEp3 (squamous carcinoma), PC3 (prostate cancer), HT29 (colon adenocarcinoma), and MDA231 (breast carcinoma).
Once prepared, 20 % piperidine in DMF can be stored at room temperature for extended periods of time.
Peptide should only be deprotected if amino acid coupling is immediately following. Resin should not be stored after Fmoc deprotection, but only after coupling of an amino acid. If storing peptide, two extra rinses with DCM should be performed after normal rinses, and the peptide should be dried very well with the hand-powered blower. The peptide should then be stored in the −20 °C freezer.
In order to test for the presence of free amine (absence of Fmoc), a Kaiser test can be performed [26]. Place a few resin beads in a small test tube, and add a couple of drops of each of the three solutions: 400 mg phenol in 100 mL ethanol, 5 g ninhydrin in 100 mL ethanol, and 2 mL 0.001 M KCN solution diluted to 100 mL with pyridine. Heat to 120 °C for approximately 5 min. If the resin contains a free amine (Fmoc has been removed) resin beads should turn blue. This can also be done after coupling of an amino acid. In this case, the resin should not change color as the Fmoc protecting group should still be attached; thus, there is no free amine.
Prepare pre-activated amino acid immediately before use by dissolving amino acid and HBTU in DMF prior to adding DIPEA.
At any point in the synthesis a microcleave can be performed in order to ensure that the synthesis is going as planned. Remove a small amount of resin and place in a 0.8 mL micro bio-spin column. If an Fmoc is present at the end of the peptide, removal following steps 4–7 should be performed first. Deprotection and cleavage can then be performed following steps 17–22. Analysis of the peptide can then be performed by analytical HPLC on a reverse-phase C-18 analytical column (for example Waters Sunfire RP-C18 4.6 × 250 mm, 5 μm column) (see Note 6).
Use a gradient system consisting of CH3CN + 0.1 % TFA (solvent A) and H2O + 0.1 % TFA (solvent B).
The weighing dishes need to be sterilized by spraying with 70 % ethanol and allowed to dry in a tissue-culture fume hood prior to embryo preparation.
Glass needles are produced from borosilicate glass capillary tubes using a pipette puller (720 KOPF model; settings: 16.3 (heater) and 2.3 (solenoid)). Carefully break the tip of the glass micro-injector with a pair of sterilized forceps. This break should be between 20 and 50 μm in diameter in order to accommodate the size of cells and prevent clogging of the needle while allowing effective piercing of the embryo CAM and blood vessels without causing much injury to the surrounding tissues.
For intravenous injection, ensure that the needle is as long and tapered as possible so that it can easily pierce through ectoderm and penetrate the vessel.
Successful cannulation of CAM vein is evident by the clearing of blood in the path of the injection flow [3].
The use of the specialized embryo-imaging unit is required (available from Innovascreen Inc., Halifax, Nova Scotia, Canada). The imaging unit will hold the CAM of interest in place during imaging and retain the field of view fixed while capturing images. This enables for three-dimensional Z-stacks and time-lapse images to be captured. We acquire and analyze three-dimensional time-lapse images using Volocity (Perkin Elmer) software package.
References
- 1.Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, Stuhlmann H. Viral nanoparticles as tools for intravital vascular imaging. Nat Med. 2006;12:354–360. doi: 10.1038/nm1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan C-X, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 3.Cho CF, Ablack A, Leong HS, Zijlstra A, Lewis J. Evaluation of nanoparticle uptake in tumors in real time using intravital imaging. J Vis Exp. 2011;52:e2808. doi: 10.3791/2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho CF, Amadei GA, Breadner D, Luyt LG, Lewis J. The discovery of novel integrin ligands from combinatorial libraries using a multiplex “beads on a bead” approach. Nano Lett. 2012;12:5957–65. doi: 10.1021/nl3034043. [DOI] [PubMed] [Google Scholar]
- 5.Steinmetz NF, Cho CF, Ablack A, Lewis JD, Manchester M. Cowpea mosaic virus nanoparticles target surface vimentin on cancer cells. Nanomedicine (Lond) 2011;6:351–364. doi: 10.2217/nnm.10.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farokhzad OC, Langer R. Impact of nanotechnology on drug delivery. ACS Nano. 2009;3:16–20. doi: 10.1021/nn900002m. [DOI] [PubMed] [Google Scholar]
- 7.Leong HS, Steinmetz NF, Ablack A, Destito G, Zijlstra A, Stuhlmann H, Manchester M, Lewis JD. Intravital imaging of embryonic and tumor neovasculature using viral nanoparticles. Nat Protoc. 2010;5:1406–1417. doi: 10.1038/nprot.2010.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinmetz NF, Ablack AL, Hickey JL, Ablack J, Manocha B, Mymryk JS, Luyt LG, Lewis JD. Intravital imaging of human prostate cancer using viral nanoparticles targeted to gastrin-releasing peptide receptors. Small. 2011;7:1664–1672. doi: 10.1002/smll.201000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, Lin T. New addresses on an addressable virus nanoblock; uniquely reactive Lys residues on cowpea mosaic virus. Chem Biol. 2004;11:855–863. doi: 10.1016/j.chembiol.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Brunel FM, Lewis JD, Destito G, Steinmetz NF, Manchester M, Stuhlmann H, Dawson PE. Hydrazone ligation strategy to assemble multifunctional viral nanoparticles for cell imaging and tumor targeting. Nano Lett. 2010;10:1093–1097. doi: 10.1021/nl1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. Bio-distribution, toxicity and pathology of cowpea mosaic virus nanoparticles in vivo. J Control Release. 2007;120:41–50. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Medintz IL, Sapsford KE, Konnert JH, Chatterji A, Lin T, Johnson JE, Mattoussi H. Decoration of discretely immobilized cowpea mosaic virus with luminescent quantum dots. Langmuir. 2005;21(12):5501–5510. doi: 10.1021/la0468287. [DOI] [PubMed] [Google Scholar]
- 13.Sapsford KE, Soto CM, Blum AS, Chatterji A, Lin T, Johnson JE, Ligler FS, Ratna BR. A cowpea mosaic virus nanoscaffold for multiplexed antibody conjugation: application as an immunoassay tracer. Biosens Bioelectron. 2006;21(8):1668–1673. doi: 10.1016/j.bios.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Destito G, Yeh R, Rae CS, Finn MG, Manchester M. Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells. Chem Biol. 2007;14:1152–1162. doi: 10.1016/j.chembiol.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinmetz NF, Manchester M. PEGylated viral nanoparticles for biomedicine: the impact of PEG chain length on VNP cell interactions in vitro and ex vivo. Biomacromolecules. 2009;10:784–792. doi: 10.1021/bm8012742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40(11):2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Li ZJ, Cho CH. Peptides as targeting probes against tumor vasculature for diagnosis and drug delivery. J Transl Med. 2012;10(Suppl 1):1. doi: 10.1186/1479-5876-10-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neveu C, Lefranc B, Tasseau O, Do-Rego JC, Bourmaud A, Chan P, Bauchat P, Le Marec O, Chuquet J, Guilhaudis L, Boutin JA, Segalas-Milazzo I, Costentin J, Vaudry H, Baudy-Floc’h M, Vaudry D, Leprince J. Rational design of a low molecular weight, stable, potent, and long-lasting GPR103 aza-beta3-pseudopeptide agonist. J Med Chem. 2012;55(17):7516–7524. doi: 10.1021/jm300507d. [DOI] [PubMed] [Google Scholar]
- 19.Auzzas L, Zanardi F, Battistini L, Burreddu P, Carta P, Rassu G, Curti C, Casiraghi G. Targeting alphavbeta3 integrin: design and applications of mono- and multifunctional RGD-based peptides and semipeptides. Curr Med Chem. 2010;17(13):1255–1299. doi: 10.2174/092986710790936301. [DOI] [PubMed] [Google Scholar]
- 20.Devlin JJ, Panganiban LC, Devlin PE. Random peptide libraries: a source of specific protein binding molecules. Science. 1990;249(4967):404–406. doi: 10.1126/science.2143033. [DOI] [PubMed] [Google Scholar]
- 21.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 22.Cretu A, Fotos JS, Little BW, Galileo DS. Human and rat glioma growth, invasion, and vascularization in a novel chick embryo brain tumor model. Clin Exp Metastasis. 2005;22(3):225–236. doi: 10.1007/s10585-005-7889-x. [DOI] [PubMed] [Google Scholar]
- 23.Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13(3):221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan WC, White PD. Fmoc solid phase peptide synthesis: a practical approach. Oxford University Press; New York: 2000. p. 288. [Google Scholar]
- 25.Leong HS, Chambers AF, Lewis JD. Assessing cancer cell migration and metastatic growth in vivo in the chick embryo using fluorescence intravital imaging. Methods Mol Biol. 2012;872:1–14. doi: 10.1007/978-1-61779-797-2_1. [DOI] [PubMed] [Google Scholar]
- 26.Kaiser E, Colescott RL, Bossinger CD, Cook PI. Color test for detection of free terminal amino groups in the solid-phase synthesis of peptides. Anal Biochem. 1970;34(2):595–598. doi: 10.1016/0003-2697(70)90146-6. [DOI] [PubMed] [Google Scholar]