Abstract
Background & Aims
Many companies provide genetic tests for obesity-related polymorphisms (nutrigenetics) and make dietary recommendations for weight loss based on the results. We performed a randomized controlled trial to determine whether more participants who followed a nutrigenetic-guided diet lost ≥5% of their body weight than participants on a standard balanced diet, for 8 and 24 weeks.
Methods
We performed a prospective study of 51 obese or overweight US veterans on an established weight management program at the Veterans Administration San Diego Healthcare System (the MOVE! Program). Participants were randomly assigned to groups placed on a nutrigenetic-guided diet (balanced, low-carbohydrate, low-fat, or Mediterranean; n=30) or a standard balanced diet (n=21). Nutrigenetic diets were selected based on results from the Pathway FIT test (Pathway Genomics; San Diego, CA).
Results
There was no significant difference in the percentage of participants on the balanced diet vs the nutrigenetic-guided diet who lost 5% of their body weight at 8 weeks (35.0%±20.9% vs 26.9%±17.1%, respectively; P=.28) or at 24 weeks. Both groups had difficulty adhering to the diets. However, adherence to the nutrigenetic-guided diet correlated with weight loss (r=0.74; P= 4.0 × 10−5), but not adherence to standard therapy (r=0.34; P=.23). Participants who had low-risk polymorphisms for obesity lost more weight than all other participants at 8 weeks (5.0% vs 2.9%, respectively; P=.02), and had significantly greater reductions in body mass index (6.4% vs 3.6% respectively; P=.03) and waist circumference (6.5% vs 2.6% respectively; P=.02) at 24 weeks.
Conclusions
In a prospective study, a nutrigenetic-based diet did not increase weight loss compared with a standard balanced diet. However, genetic features can identify individuals most likely to benefit from a balanced diet weight loss strategy; these findings require further investigation. ClincialTrials.gov number: NCT01859403
Keywords: BMI, nutrigenomics, diet, personalized medicine
INTRODUCTION
More than one third (34.9%) of the US adult population is obese,1 and it is estimated to cost $147 billion dollars to the healthcare system annually.2 However, there is a lack of effective, sustainable, non-surgical treatments of obesity.3 This difficulty is in part due to the multi-genetic nature of obesity, where heritable factors can provide up to 70% of the estimated risk in some individuals.4 While genome-wide association studies have led to the identification of at least 32 gene loci associated with obesity,5-9 whether an individual's genetic profile can play a role in personalized obesity therapy is still unknown.
Nevertheless, many US and European companies provide targeted genetic testing for obesity-related polymorphisms and make dietary and other intervention recommendations based on their results. These tests are marketed often directly to patients and can range in cost from approximately $100 to $1000.10, 11 Published data on the use and market of nutrigenetic testing is sparse, however direct-to-consumer genetic testing is a growing industry, projected to reach $233 million by 2018.12 Although there are questions about the usefulness of these tests in patient care,11 there is also potential in improving and individualizing therapy in obesity and, as a result, decreasing overall healthcare costs.13
Several observational studies have shown that those with high risk polymorphisms of a few specific genes have improved weight loss or metabolic profiles by changing to a particular diet (e.g. low fat diet, Mediterranean diet, etc.).14-21 Specifically, the negative consequences associated with the high risk polymorphisms in these seven genes can be mitigated by a change in diet: apolipoprotein A-II gene (APOA2),18, 22 adiponectin gene (ADIPOQ),19, 23, 24 fat mass and obesity-associated protein gene (FTO),17, 25, 26 potassium channel tetramerization domain containing 10 gene (KCTD10),21 hepatic triglyceride lipase gene (LIPC),16, 27 methylmalonic aciduria (cobalamin deficiency) cblB type gene (MMAB),21 and peroxizome proliferative activated receptor gamma gene (PPARG)20, 28 (Table S1). Still, evidence to support a strategy of nutrigenetic-guided weight loss intervention is limited.
In this prospective randomized control, clinical trial, participants’ genetic profile was used to provide a personalized diet recommendation to see if education and support for the genetic-based diet would improve weight loss and metabolic measurements compared to standard therapy in an established weight management program. This is a feasibility study to determine whether it would be fruitful to implement this strategy, and its potential efficacy. The main objective was to determine whether more participants in the genetics-guided therapy (GT) group lost ≥5% of their weight after 8 weeks compared to those in the standard therapy (ST) group. The secondary objectives were to evaluate whether more GT participants lost ≥5% of their weight after 24 weeks.
SUBJECTS AND METHODS
The Supplementary Materials and Methods section describes the full details of the clinical trial including methodology, patient eligibility, measures taken, and statistical analysis.
In brief, the study was a prospective, randomized controlled, feasibility trial of an 8-week diet counseling intervention for veterans enrolled in the MOVE! program with continued assessment to week 24, between November 2012 to March 2014. The MOVE! program is an 8-week, evidence-based weight management program for overweight and obese veterans that is established in all Veterans Administration hospitals.29 This study received IRB approval (protocol # H130174). It was also registered with ClinicalTrials.gov (NCT01859403). All authors had access to the study data and had reviewed and approved the final manuscript.
Veterans with a physician's referral to weight management clinic and a BMI ≥ 30.0 were recruited from those enrolled in the Veterans Administration San Diego Healthcare System's MOVE! program. Participants entered the study on their normal diet. The baseline visit was 3-4 weeks prior to start of MOVE! program initiation. After baseline measurements were taken, participants provided saliva for genetic analysis (Pathway Genomics, Inc., San Diego, CA; Figure S1). Participants were then randomly assigned to either the genetics-guided therapy (GT) group or standard therapy (ST) groups. Randomization, which was performed prior to receipt of nutrigenetic report, was non-stratified, two-group, concealed allocation, using the Research Randomizer website.30
In the GT group, participants and researchers were unblinded to the diet match and participants were informed of their nutrigenetic report. GT participants were matched to one of four possible diet types: balanced, low-carbohydrate, low-fat, or Mediterranean based on their report. They received a meal plan, lists of foods to incorporate in the plan, and samples of menus (similar to the MOVE! packet of literature given to ST group) to assist adherence to their diet and to obtain their caloric goal (Online Material: Meal Plan). The macronutrient guidelines of the different diets for the GT participants are shown in Table S2. The macronutrient composition of each diet plan was based on a compilation of research studies that showed the benefit of that particular diet plan on patients with a high risk polymorphism. For example, the macronutrient composition of the Mediterranean diet plan was based on references 17-19 and 20-22 (see Table S1).
In the ST group, participants and researchers were blinded to the nutrigenetic report. These participants were given the balanced diet plan. The ST group were provided similar education and resources as the GT group for the balanced diet plan and provided the same amount of educational time as those in the GT group. To aid in simplicity and adherence, all diet plans (for both ST and GT participants) incorporated Healthy Choice (ConAgra Foods®, Inc.) entrees at lunch and dinner (Online Material: Sample Menu) for the first 8 weeks of the study, for which participants were fully reimbursed upon delivery of receipts. At the conclusion of the study, ST participants were provided their nutrigenetic reports.
Salivary samples from participants were sent to Pathway Genomics and the Pathway Fit Test® (a genomic array) was performed. Based on the SNP alleles for seven genes, and using a proprietary algorithm, the Pathway Fit Test® made a recommendation to a specific diet (Figure S1). The genes (and reference SNP [rs] number) used to make these dietary recommendations were APOA2 (rs5082), ADIPOQ (rs17300539), FTO (rs9939609), KCTD10 (rs10850219), LIPC (rs1800588), MMAB (rs2241201), and PPARG (rs1801282); (Table S1).
RESULTS
Primary Outcomes
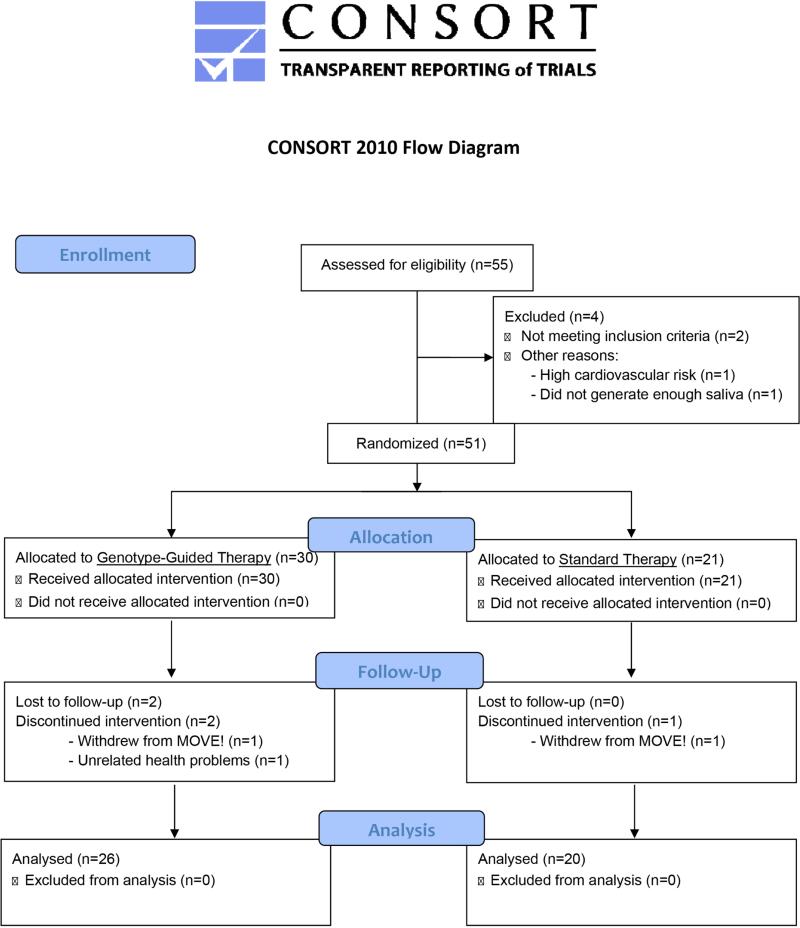
A total of 51 participants were randomized. At the end of 8 weeks, 46 participants remained enrolled in the study. At the end of 24 weeks, 32 participants completed the study; 14 were from the ST group and 18 were from the GT group. (Figure 1). Baseline characteristics were similar between the GT and the ST groups (Table 1). The trial ended once the target number of participants were recruited.
Figure 1.
CONSORT Diagram.
Table 1.
Baseline Characteristics of the Participants in the Study
| Genotype-based Therapy (GT) | Standard Therapy (ST) | P-value | |
|---|---|---|---|
| Participants (n) 26 | 20 | ||
| Demographics | |||
| Female [n (%)] | 10 (38%) | 3 (15%) | 0.08 |
| Age (years) | 48.4 (±2.6) | 54.6 (±2.7) | 0.11 |
| Latino [n (%)] | 7 (27%) | 4 (20%) | 0.73 |
| African-American [n (%)] | 5 (19%) | 3 (15%) | 0.99 |
| Asian-American [n (%)] | 2 (8%) | 0 (0%) | 0.50 |
| Caucasian [n (%)] | 12 (46%) | 13 (65%) | 0.24 |
| Genotype Diet Recommendation | |||
| Balanced Diet [n (%)] | 6 (23%) | 8 (40%) | 0.33 |
| Low Fat Diet [n (%)] | 15 (58%) | 10 (50%) | 0.77 |
| Low Carbohydrate Diet [n (%)] | 2 (8%) | 1 (5%) | 1.00 |
| Mediterranean Diet [n (%)] | 3 (12%) | 1 (5%) | 0.62 |
| Weight | |||
| Weight (Kg) | 112.6 (±4.9) | 114.3 (±4.6) | 0.80 |
| BMI (kg/m2) | 39.3 (±1.3) | 37.3 (±1.4) | 0.31 |
| Abdominal Circumference (cm) | 120.5 (±3.8) | 120.0 (±2.8) | 0.92 |
| Lipid Profile | |||
| LDL (mg/dL) | 96.3 (±4.9) | 105.9 (±7.1) | 0.28 |
| HDL (mg/dL) | 46.1 (±2.7) | 44.2 (±1.7) | 0.55 |
| TG (mg/dL) | 125.1 (±11.9) | 166.0 (±17.5) | 0.06 |
| Glucose Homeostasis | |||
| Fasting Blood Glucose (mg/dL) | 101.8 (±3.9) | 100.4 (±3.7) | 0.79 |
| Hemoglobin A1c (%) | 5.72 (±0.18) | 5.78 (±0.18) | 0.80 |
| Fasting Serum Insulin (uIU/mL) | 15.9 (±7.1) | 25.0 (±2.3) | 0.23 |
| Blood Pressure | |||
| Systolic Blood Pressure (mm Hg) | 127.8 (±2.6) | 130.0 (±3.0) | 0.59 |
| Diastolic Blood Pressure (mm Hg) | 81.1 (±1.9) | 83.0 (±2.5) | 0.56 |
| Mean Arterial Pressure (mm Hg) | 96.7 (±1.8) | 98.6 (±2.3) | 0.51 |
| Bioelectrical impedance | |||
| Body Fat (%) | 41.4 (±1.3) | 37.7 (±2.0) | 0.14 |
| Lean Mass (kg) | 63.7 (±3.2) | 70.5 (±3.3) | 0.15 |
| Med Gem Analysis | |||
| Resting Metabolic Rate (Cal) | 1927.7 (±118.3) | 1975.0 (±65.5) | 0.73 |
In the primary comparison, there was no significant difference between the GT and ST groups in percentage achieving the 5% weight loss at difference at 8 weeks (26.9% ± 17.1 vs. 35.0% ± 20.9, p = 0.28 for GT and ST respectively; difference in proportion 8.1% with CI of - 17.5% to 33.5%) (Figure 2A, Table 2). There was also no significant difference in proportion achieving 5% weight loss at 24 weeks (38.9% ± 22.5 vs. 35.7% ± 25.1, p = 0.77; difference in proportion 3.2% with CI of -32.1% to 36.3%) (Figure 2A, Table 2). In addition, there was no significant difference in the relative amount of percent weight lost by participants in the study between the GT and ST groups (3.2% ± 0.6 vs. 4.0% ± 0.7, p = 0.36, at 8 weeks and 4.3% ± 1.1 vs. 4.4% ± 1.3, p = 0.93, at 24 weeks, respectively) (Figure 2B, Table 2). Notably, observed results of both groups were better than previously published results of the MOVE! program (~15-20%) 31.
Figure 2.
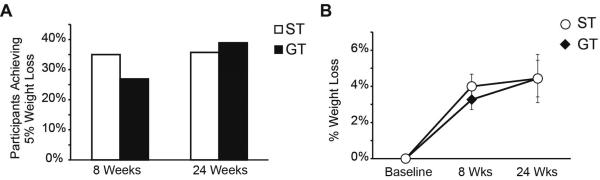
Weight loss in the genotype guided therapy (GT) and the standard therapy (ST) groups. (A) Percentage of participants who achieved at least 5% weight loss. (B) Percentage weight lost by participants at 8 and 24 weeks. Adherence is a predictor of weight loss by both the genotype guided therapy (GT) and the standard therapy (ST) groups.
Table 2.
Absolute values and percent reduction in patient weight, serum biomarkers, anthropomorphic measures, and resting metabolic rate. (n/a - not applicable)
| Absolute Values | Percent Reduction | |||||||
|---|---|---|---|---|---|---|---|---|
| GT (8 wks) | ST (8 wks) | GT (24 wks) | ST (24 wks) | GT (8 wks) | ST (8 wks) | GT (24 wks) | ST (24 wks) | |
| Weight | Weight | |||||||
| Achieving 5% weight loss [n, (%)] | 7 /26 (27%) | 7/20 (35%) | 7/18 (39%) | 5 /14 (36%) | n/a | n/a | ||
| Weight Loss (kg) | 3.7 (±0.8) | 4.6 (±0.8) | 5.0 (±01.3) | 5.2 (±1.5)) | 3.2 (±0.6) | 4.0 (±0.7) | 4.3 (±1.1) | 4.4 (±1.3) |
| BMI (kg/m2) | 38.2 (±1.2) | 35.8 (±1.4) | 38.0 (±1.7) | 34.7 (±1.9) | 2.6 (±0.8) | 4.0 (±0.7) | 4.4 (±1.0) | 4.4 (±1.3) |
| Waist Circumference (cm) | 118.2 (±3.6) | 116.7 (±3.0) | 118.7 (±4.9) | 114.6 (±2.9) | 1.8 (±0.7) | 2.8 (±1.0) | 4.3 (±1.0) | 3.2 (±1.6) |
| Lipid Profile | Lipid Profile | |||||||
| LDL (mg/dL) | 93.0 (±5.9) | 88.7 (±6.8) | 100.8 (±8.2) | 113.6 (±7.0) | 4.4 (±4.4) | 16.3 (±3.1) | −5.3 (±5.2) | −5.6 (±4.1) |
| HDL (mg/dL) | 43.1 (±2.6) | 44.6 (2.9) | 46.4 (±3.5) | 49.1 (±3.8) | 5.2 (±2.9) | 4.1 (±2.7) | −2.7 (±4.1) | −3.1 (±2.4) |
| TG (mg/dL) | 132.7 (±11.7) | 147.1 (±14.4) | 141.8 (±13.8) | 144.6 (±13.6) | −14.4 (±8.7) | 6.6 (±6.3) | −26.1 (±13.6) | 4.4 (±8.8) |
| Glucose Homeostasis | Glucose Homeostasis | |||||||
| Fasting Serum Glucose (mg/dL) | 96.0 (±2.3) | 96.0 (±3.2) | 96.8 (±3.8) | 93.0 (±3.7) | 3.8 (±2.4) | 3.8 (±2.3) | 4.7 (±3.0) | 2.9 (±4.0) |
| Fasting Serum Insulin (uIU/mL) | 21.3 (±4.4) | 15.9 (±2.3) | 18.7 (±3.7) | 13.4 (±1.7) | −12.7 (±10.0) | 1.8 (±11.0) | 12.0 (±10.2) | 7.5 (±15.2) |
| Hemoglobin A1c (%) | 5.37 (±0.10) | 5.58 (±0.13) | 5.54 (±0.15) | 5.54 (±0.11) | 4.0 (±1.7) | 3.1 (±1.3) | 0.6 (±1.7) | 0.7 (±1.3) |
| Blood Pressure | Blood Pressure | |||||||
| Systolic Blood Pressure (mm Hg) | 124.1 (±2.0) | 124.4 (±2.3) | 127.4 (±3.1) | 124.6 (±2.7) | 1.9 (±1.7) | 4.0 (±2.2) | 1.4 (±2.5) | 3.9 (±2.7) |
| Diastolic Blood Pressure (mm Hg) | 80.1 (±1.8) | 79.4 (±1.7) | 81.0 (±2.6) | 81.2 (±21.6) | 1.0 (±2.0) | 3.6 (±2.2) | 0.5 (±2.6) | 4.7 (±2.0) |
| Mean Arterial Pressure (mm Hg) | 94.8 (±1.7) | 94.4 (±1.7) | 96.5 (±2.4) | 95.7 (±1.7) | 1.3 (±1.7) | 4.0 (±2.0) | 0.7 (±2.3) | 2.2 (±2.0) |
| Bioelectrical impedance | Bioelectrical impedance | |||||||
| Body Fat (%) | 41.9 (±2.2) | 35.8 (±1.2) | 39.7 (±1.9) | 36.6 (±2.6) | 2.0 (±1.1) | 4.3 (±1.9) | 4.1 (±1.6) | 7.2 (±2.3) |
| Lean Mass (kg) | 61.3 (±3.0) | 69.4 (±3.2) | 61.4 (±3.5) | 66.4 (±2.6) | 2.0 (±0.8) | 1.3 (±1.3) | 1.9 (±1.5) | −0.5 (±1.8) |
| MedGem Analysis | MedGem Analysis | |||||||
| RMR (Cal) | 1905.8 (±113.2) | 1865.3 (±77.3) | 2020.9 (±123.5) | 1942.9 (±60.5) | 0.1 (±3.1) | 4.2 (±3.0) | −5.7 (±4.0) | −2.8 (±2.8) |
Post Hoc Analyses
Since there were no differences in the primary objective of the study, we investigated whether the GT group had improvements in biomarkers of metabolic disease associated with obesity. However, no differences were found in the lipid profile or glucose homeostasis in the GT vs. ST groups at 24 weeks (Figure S2, Figure S3, Table 2). Furthermore, no differences were found in measured parameter (Table 2).
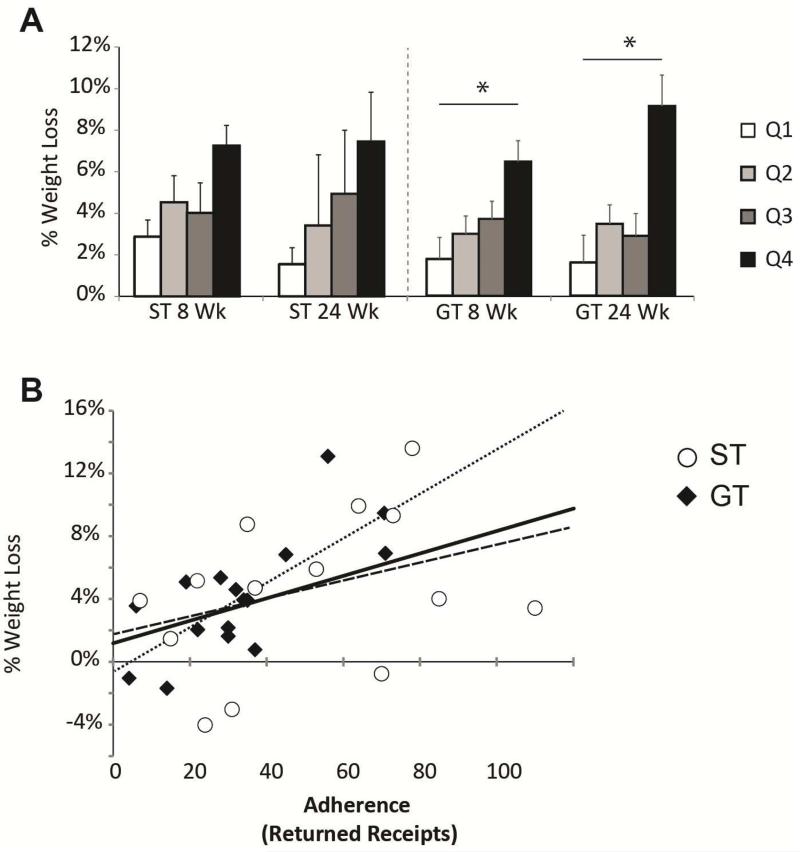
Since the diets recommended by the genetic testing may be more difficult to maintain, the role of adherence was investigated in participants’ weight loss. Adherence was measured through receipts returned for Healthy Choice meals in the first 8 weeks. The average returned receipt was 39 ± 4.1 (range = 0-110; note that maximum possible is 112). Participants were split into quartiles to analyze the results of effects of adherence. Across both groups there was a significant relationship between adherence and weight loss (p = 0.001). However, when the ST group was analyzed alone, no significant difference was found between the quartiles of the ST group (p = 0.45; Figure 3A). In the GT group, there was a significant relationship between adherence and weight loss (p = 0.002) and those in the top quartile of the GT groups had lost a significant amount of weight compared to those in the lower quartile (p = 0.03). There were no differences in weight loss within subgroups defined by adherence to the intervention between the ST and GT groups (e.g. top quartile of adherence in GT versus top quartile of adherence in ST). The correlation between returned receipts and weight loss was quite strong across both groups (r = 0.44, respectively, p = 0.001; Figure 3B). However, when each group was analyzed separately, only the GT had a significant correlation between adherence and weight loss at 24 weeks (r = 0.74, p = 4.0 × 10−5 for GT; r = 0.34, p = 0.23 for ST; Figure 3B dotted line).
Figure 3.
Adherence and weight loss. (A) Adherence was measured by the amount of receipts of Healthy Choice meals returned for reimbursement during the first 8 weeks. Those in the top quartile of the GT participants lost significantly more weight than those in the bottom two quartiles, whereas this wasn't true for the ST group. (* p <0.05). (B) Correlation plot showing the relationship between returned receipts (a measure of adherence) and percent weight loss. Solid line shows trend line for all participants, (R = 0.47; p = 0.001). Dotted line shows trend line for GT participants ((R = 0.74, p = 4.0 × 10−5) and dashed line shows trend line for ST participants (R = 0.34, p = 0.23).
About a third of the participants who were in the ST group would have been matched to the balanced diet based on their genetic profile (Table 1). Hence, all the data was reanalyzed, grouping participants into those who were matched to their genetic-guided diet and those who were not matched. Again no significant differences were found between these two groups in weight loss, serum biomarkers, or anthropometric measures. In addition, there was no significant difference in adherence or weight loss by diet received (Figure S4).
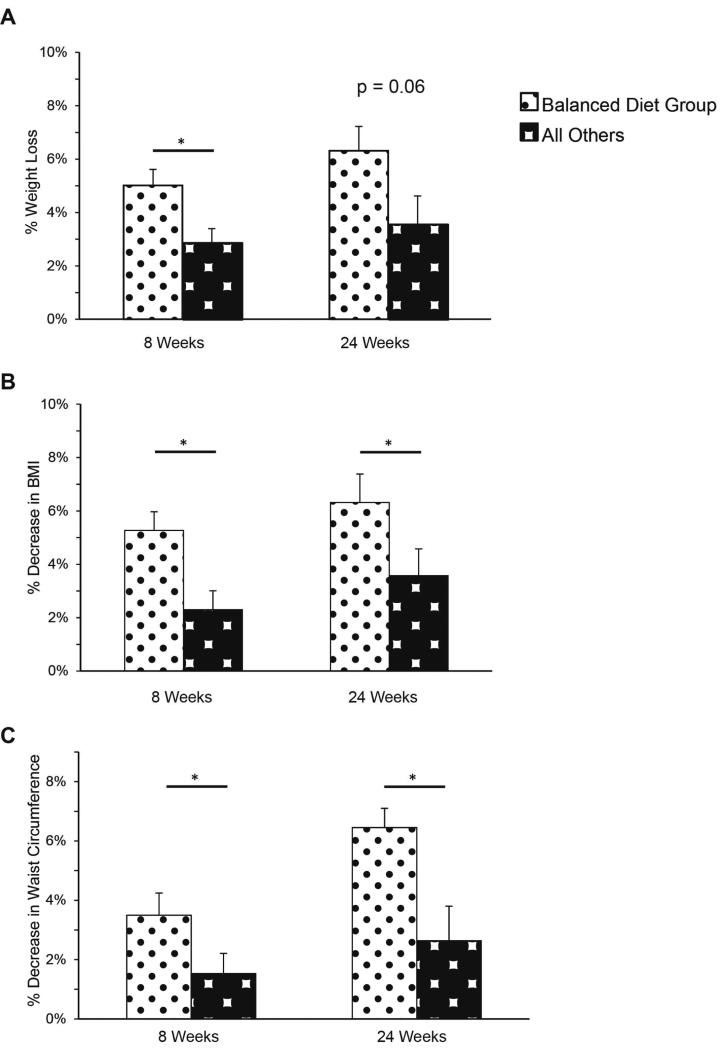
In order to find predictors of successful weight loss, a post-hoc analysis was done to evaluate specific genetic profiles. Participants who were matched to the balanced diet based on their nutrigenetic profile (balanced diet genotype, BDG; n = 14) regardless of which group they were randomized to (GT or ST) lost a significant amount of weight at 8 weeks when compared to other participants (5.0% ± 0.6, vs 2.9% ± 0.5, p = 0.02; Figure 4A) and trended toward significance at 24 weeks (6.3% ± 0.9, vs 3.5 ± 1.0, p = 0.06 at 24 weeks; Figure 4A). Furthermore, the BDG participants had a significant decrease in their BMI (6.4% ± 1.2 vs. 3.6 ± 1.1 % reduction, p = 0.02) (Figure 4B) and waist circumference (6.5% ± 0.6 vs. 2.6% ± 0.1 reduction, p = 0.03) at 24 weeks (Figure 4C). Non-BDG participants who were consuming a balanced diet did not have as significant improvements as the BDG group did.
Figure 4.
Balanced Diet Genotype (BDG) performed better with lifestyle modification. (A) Percentage weight lost by BDG participants (versus all others) at 8 and 24 weeks (p = 0.02 and p =0.06, respectively). (B) Percentage change in BMI by BDG participants (versus all others) at 8 and 24 weeks (p = 0.02 and p = 0.03, respectively). (C) Percentage decrease in waist circumference BDG participants (versus all others) at 8 and 24 weeks. (p = 0.01 and p = 0.02).
Since those with the BDG did particularly well in the MOVE! program, we measured whether nutrigenetic testing could play a role in prognosticating success (Table S3). The sensitivity and specificity of nutrigenetic testing in detecting which patients would lose 3% of baseline weight, at 24 weeks, where the test performed best, was 47% and 100% respectively. The positive predictive value was 100%, and negative predictive value was 50%.
DISCUSSION
The aim of this randomized control trial was to determine if there was a difference in weight loss in a group that had the advantage of education on their genetically-guided recommended diets compared to a group that were guided to follow a general balanced diet. GT and ST participants were not significantly different in any outcome measures, and, as expected, that diet adherence was a much more important factor in weight loss. Since this was a feasibility study, the sample size was small (n=18 for the GT, and n=14 for the ST group at 24 weeks). Analysis of the confidence intervals around the estimate of absolute benefit of GT at 24 weeks (which was 3.2% with CI of −32.1% to 36.3%) is consistent with there being no difference at all, or a difference of up to 30% in either direction. This suggests that the sample size of this feasibility trial is too small to exclude all clinically significant differences. However, based on the observed 3.2% difference in this study, planning a sufficiently powered clinical trial would require 336 participants for each group (80% statistical power; α-level of 0.05). This would involve either a considerable commitment of resources in a future study or methods that could enhance the efficacy of the current treatment. Furthermore, future research investigating nutrigenetic treatment effects on metabolic parameters might also consider using dietary intervention groups with larger macronutrient differences relative to the current study. In absence of new data, use of nutrigenetic-based diet management in usual practice is unlikely to be highly clinically effective.
There is some suggestion, based on the correlative data, that nutrigenetic guided dietary recommendation may offer a benefit to those who are most adherent to their recommended diet plan. The relationship between diet adherence and weight loss was very strong in participants in the GT group. Adherence was also important for the ST group, but our study may have had a sample size was too small to have sufficient power to detect a correlation.
Nevertheless, the problem with nutrigenetic-based personalized diet therapy is that recommendations to alter dietary intake remains a poor treatment for obesity because of non-adherence. Even when given their nutrigenetic information with guided education regarding their nutrigenetic-based diet, GT participants are no more adherent to their diet as those in the ST group. In the post-hoc analysis, there is some suggestion that nutrigenetics might be used as a potential predictor of individuals who would benefit from lifestyle modification and dietary intervention. In the BDG group, 100% of participants were able to lose at least 3% of their body weight, whereas only 50% of participants who were genotyped to other diets lost that amount of weight. These absolute differences in weight are likely clinically significant since even minimal to moderate weight loss has been found to confer health advantages.32 Hence, nutrigenetics role in personalized therapy against obesity may be to give clinicians an idea of whether the participant will be successful with lifestyle modification therapy or if a more aggressive therapy is needed at an early stage.
The value of the use of nutrigenetic testing to predict who will be a poor responders to lifestyle modification could be clinically significant in the treatment of obesity. Clinicians may be able to shift the focus of intervention as the preferred treatment modality to earlier medication use or bariatric surgery in those predicted to be poor responders. This could potentially provide results more quickly, shortening the period of time that the patient will have obesity or its associated metabolic disorders, and with less distress to those who would first need to fail dietary intervention prior to advancing to other treatment options. However, not only does nutrigenetics have to be proven to be a good predictor of who will fail lifestyle modification, but also, there needs to be an alternative, more aggressive therapy from which those who are predicted to be poor responders will gain a therapeutic benefit. Whether the most effective aggressive therapy is meal replacement, pharmacotherapy, or bariatric surgery, will need to be stipulated in future studies.
The use of the VA population participating in the standardized weight loss program MOVE! provided a great enrollment pool, but this group is very different from the general population.33 In a study of the MOVE! program's effectiveness for providing weight loss, less than 1 in 5 veterans lose 5% or more of their body weight with only an average of 3.6 lbs weight loss at 6 months.31 It is notable that participants lost more weight at 6 months and a greater percentage achieved 5% weight loss in both groups compared to the standard MOVE! results, likely from more aggressive follow up and meal replacement in the initial 8 weeks.
With the rising interest in personalized medicine from both providers and participants, and physicians increasing inclination to use genetic-guided therapies,34 nutrigenetics will remain a growing factor in those treating obesity and its related disease. However, many problems still remain. There is yet no consistency in nutrigenetic reports from various companies and costs remain high.11 This study shows that personalized nutrigenetic recommendations for diet is still premature and cost-ineffective. Though the expectation remains that the costs of nutrigenetic testing to continue to fall with advances in sequencing technology,12 the lack of effective remedies for obesity remains the main hurdle for nutrigenetic guided personalized therapy.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Charles Ha, B.S. from VA San Diego Healthcare Systems, who helped collect data during participant visits. We would also like to thank Robert Henry, M.D. from VA San Diego Healthcare Systems, and Samuel Ho, M.D., from VA San Diego Healthcare Systems, for allowing generous use of space and resources to conduct this study.
Grant Support: This was an investigator-initiated study funded by an industry sponsor, Pathway Genomics Corporation. Additional funding was provided by NIH grants P50 GM085764, KL2 TR00099, and R24 DK080506. JE was supported by NIH R25 MH71544 and the UCSD Sam and Rose Stein Institute for Research on Aging. AZ is supported by AASLD Liver Scholar Award.
Abbreviations
- BDG
balanced diet genotype
- BIA
Bioelectrical impedance analysis
- GT
genotype-guided therapy
- RMR
resting metabolic rate
- ST
standard therapy
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
This was an investigator-initiated study funded by an industry sponsor, Pathway Genomics Corporation. MLK has worked as a contract dietician for Pathway Genomics, the study sponsor, for work distinct from this study. KLH is non-compensated member of the scientific advisory board for Pathway Genomics. The remaining authors declare no conflict of interest. The funders had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Contributions: KLH provided the study concept, designed the study, and obtained funding; MLK, KAF, PSL, KLH, and AZ conducted research and acquired data; MLK is the study dietician who created individual meal plans; JE, AZ, and TRR analyzed and interpreted the data; KAF, AZ, JE, and MLK drafted the manuscript; SG provided critical revision of the manuscript for important intellectual content; AZ had primary responsibility for final content and supervised the study. All authors read and approved the final manuscript.
REFERENCES
- 1.Ogden CL, Carroll MD, Kit BK, et al. Prevalence of childhood and adult obesity in the United States, 2011-2012. Jama. 2014;311:806–14. doi: 10.1001/jama.2014.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkelstein EA, Trogdon JG, Cohen JW, et al. Annual medical spending attributable to obesity: payer-and service-specific estimates. Health Aff (Millwood) 2009;28:w822–31. doi: 10.1377/hlthaff.28.5.w822. [DOI] [PubMed] [Google Scholar]
- 3.Picot J, Jones J, Colquitt JL, et al. The clinical effectiveness and cost-effectiveness of bariatric (weight loss) surgery for obesity: a systematic review and economic evaluation. Health Technol Assess. 2009;13:1–190, 215-357, iii-iv. doi: 10.3310/hta13410. [DOI] [PubMed] [Google Scholar]
- 4.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–51. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 5.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willer CJ, Speliotes EK, Loos RJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos RJ, Lindgren CM, Li S, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorleifsson G, Walters GB, Gudbjartsson DF, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 10.Conti R, Veenstra DL, Armstrong K, et al. Personalized medicine and genomics: challenges and opportunities in assessing effectiveness, cost-effectiveness, and future research priorities. Med Decis Making. 2010;30:328–40. doi: 10.1177/0272989X09347014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.U.S. Government Accountability Office . Direct-To-Consumer Genetic Tests: Misleading Test Results Are Further Complicated by Deceptive Marketing and Other Questionable Practices (GAO Publication No. 10-847T) U.S. Government Printing Office; Washington, DC: 2010. [Google Scholar]
- 12.Global Industry Analysts Future of Direct-to-Consumer (DTC) Genetic Testing Market Remains Fraught with Challenges. 2014:2012. [Google Scholar]
- 13.Vakili S, Caudill MA. Personalized nutrition: nutritional genomics as a potential tool for targeted medical nutrition therapy. Nutr Rev. 2007;65:301–15. doi: 10.1111/j.1753-4887.2007.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 14.Stryjecki C, Mutch DM. Fatty acid-gene interactions, adipokines and obesity. Eur J Clin Nutr. 2011;65:285–97. doi: 10.1038/ejcn.2010.277. [DOI] [PubMed] [Google Scholar]
- 15.Lairon D, Defoort C, Martin JC, et al. Nutrigenetics: links between genetic background and response to Mediterranean-type diets. Public Health Nutr. 2009;12:1601–6. doi: 10.1017/S1368980009990437. [DOI] [PubMed] [Google Scholar]
- 16.Ordovas JM, Corella D, Demissie S, et al. Dietary fat intake determines the effect of a common polymorphism in the hepatic lipase gene promoter on high-density lipoprotein metabolism: evidence of a strong dose effect in this gene-nutrient interaction in the Framingham Study. Circulation. 2002;106:2315–21. doi: 10.1161/01.cir.0000036597.52291.c9. [DOI] [PubMed] [Google Scholar]
- 17.Sonestedt E, Roos C, Gullberg B, et al. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am J Clin Nutr. 2009;90:1418–25. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- 18.Corella D, Peloso G, Arnett DK, et al. APOA2, dietary fat, and body mass index: replication of a gene-diet interaction in 3 independent populations. Arch Intern Med. 2009;169:1897–906. doi: 10.1001/archinternmed.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warodomwichit D, Shen J, Arnett DK, et al. ADIPOQ polymorphisms, monounsaturated fatty acids, and obesity risk: the GOLDN study. Obesity (Silver Spring) 2009;17:510–7. doi: 10.1038/oby.2008.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memisoglu A, Hu FB, Hankinson SE, et al. Interaction between a peroxisome proliferator-activated receptor gamma gene polymorphism and dietary fat intake in relation to body mass. Hum Mol Genet. 2003;12:2923–9. doi: 10.1093/hmg/ddg318. [DOI] [PubMed] [Google Scholar]
- 21.Junyent M, Parnell LD, Lai CQ, et al. Novel variants at KCTD10, MVK, and MMAB genes interact with dietary carbohydrates to modulate HDL-cholesterol concentrations in the Genetics of Lipid Lowering Drugs and Diet Network Study. Am J Clin Nutr. 2009;90:686–94. doi: 10.3945/ajcn.2009.27738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith CE, Tucker KL, Arnett DK, et al. Apolipoprotein A2 polymorphism interacts with intakes of dairy foods to influence body weight in 2 U.S. populations. J Nutr. 2013;143:1865–71. doi: 10.3945/jn.113.179051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goyenechea E, Collins LJ, Parra D, et al. The - 11391 G/A polymorphism of the adiponectin gene promoter is associated with metabolic syndrome traits and the outcome of an energy-restricted diet in obese subjects. Horm Metab Res. 2009;41:55–61. doi: 10.1055/s-0028-1087204. [DOI] [PubMed] [Google Scholar]
- 24.AlSaleh A, O'Dell SD, Frost GS, et al. Single nucleotide polymorphisms at the ADIPOQ gene locus interact with age and dietary intake of fat to determine serum adiponectin in subjects at risk of the metabolic syndrome. Am J Clin Nutr. 2011;94:262–9. doi: 10.3945/ajcn.111.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ortega-Azorin C, Sorli JV, Asensio EM, et al. Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc Diabetol. 2012;11:137. doi: 10.1186/1475-2840-11-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang T, Qi Q, Li Y, et al. FTO genotype, dietary protein, and change in appetite: the Preventing Overweight Using Novel Dietary Strategies trial. Am J Clin Nutr. 2014;99:1126–30. doi: 10.3945/ajcn.113.082164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nettleton JA, Steffen LM, Ballantyne CM, et al. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and White adults. Atherosclerosis. 2007;194:e131–40. doi: 10.1016/j.atherosclerosis.2006.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouchard-Mercier A, Godin G, Lamarche B, et al. Effects of peroxisome proliferator-activated receptors, dietary fat intakes and gene-diet interactions on peak particle diameters of low-density lipoproteins. J Nutrigenet Nutrigenomics. 2011;4:36–48. doi: 10.1159/000324531. [DOI] [PubMed] [Google Scholar]
- 29.Kinsinger LS, Jones KR, Kahwati L, et al. Design and dissemination of the MOVE! Weight-Management Program for Veterans. Prev Chronic Dis. 2009;6:A98. [PMC free article] [PubMed] [Google Scholar]
- 30.Urbaniak GC, Plous S. Research Randomizer (Version 4.0) [Computer software] 2013:2013. [Google Scholar]
- 31.Kahwati LC, Lance TX, Jones KR, et al. RE-AIM evaluation of the Veterans Health Administration's MOVE! Weight Management Program. Transl Behav Med. 2011;1:551–60. doi: 10.1007/s13142-011-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de las Fuentes L, Waggoner AD, Mohammed BS, et al. Effect of moderate diet-induced weight loss and weight regain on cardiovascular structure and function. J Am Coll Cardiol. 2009;54:2376–81. doi: 10.1016/j.jacc.2009.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Del Re AC, Maciejewski ML, Harris AH. MOVE: weight management program across the Veterans Health Administration: patient- and facility-level predictors of utilization. BMC Health Serv Res. 2013;13:511. doi: 10.1186/1472-6963-13-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanek EJ, Sanders CL, Taber KA, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91:450–8. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.