Abstract
The identification of specific genetic alterations that drive the initiation and progression of cancer and the development of targeted drugs that act against these driver alterations has revolutionized the treatment of many human cancers. While substantial progress has been achieved with the use of such targeted cancer therapies, resistance remains a major challenge that limits the overall clinical impact. Hence, despite progress, new strategies are needed to enhance response and eliminate resistance to targeted cancer therapies in order to achieve durable or curative responses in patients. To date, efforts to characterize mechanisms of resistance have primarily focused on molecular events that mediate primary or secondary resistance in patients. Less is known about the initial molecular response and adaptation that may occur in tumor cells early upon exposure to a targeted agent. Although understudied, emerging evidence indicates that the early adaptive changes by which tumor cells respond to the stress of a targeted therapy may be crucial for tumor cell survival during treatment and the development of resistance. Here, we review recent data illuminating the molecular architecture underlying adaptive stress signaling in tumor cells. We highlight how leveraging this knowledge could catalyze novel strategies to minimize or eliminate targeted therapy resistance, thereby unleashing the full potential of targeted therapies to transform many cancers from lethal to chronic or curable conditions.
Introduction
The identification of specific somatic oncogenic alterations that drive tumor growth1-4 and the development of targeted therapies that act against these oncogenic drivers have transformed the treatment of many cancer patients. Common oncogenic signaling components and pathways are depicted in Figure 1. Targeted therapies often elicit substantial initial tumor responses in patients with advanced-stage cancers in which conventional cytotoxic chemotherapy is largely inactive (Table 1). Paradigm-defining examples of this approach include the use of BRAF inhibitors in BRAF V600E mutant melanoma patients5 and the use of EGFR or ALK tyrosine kinase inhibitors (TKIs) in EGFR mutant and ALK fusion positive non-small cell lung cancer (NSCLC) patients6-9, respectively. However, these targeted agents do not induce durable or curative responses in patients, with a few notable exceptions, because of therapy resistance that emerges after an initial response (secondary or acquired resistance10-12). Furthermore, many patients whose tumors harbor a genetic driver of tumor growth fail to respond initially to the relevant targeted agent and exhibit primary (or innate) resistance11,13,14.
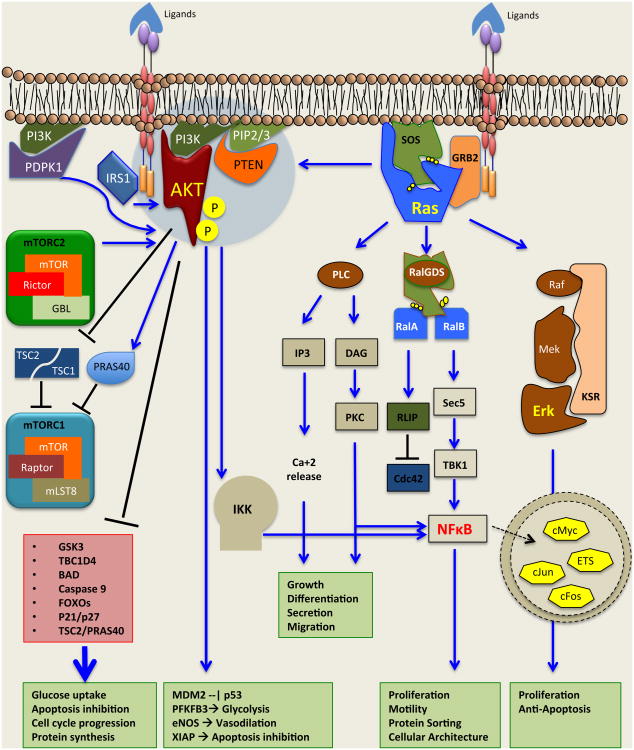
Figure 1. Oncogenic signaling in tumor cells.
Shown are major signaling pathways involved in the initiation and progression of many tumors. Pathway crosstalk can occur at multiple levels from signaling emanating from the plasma membrane to the mitochondrial and nuclear events. Thus, there is significant potential for stress response signaling such that inhibition of one pathway results in the engagement of a distinct pathway that maintains tumor cell homeostasis and promotes escape from therapy. This profound robustness is a critical feature of the evolution of tumors both in the absence and the presence of therapy, consistently enhancing tumor survival in response to various stresses. Abbreviations: Raf, Raf proto-oncogene serine/threonine kinase; Mek, mitogen activated protein kinase kinase; Erk, extracellular signal related kinase; PI3K, phosphoinositide 3-kinase; AKT, v-akt murine thymoma viral oncogene homolog; IRS1, insulin receptor substrate; mTORC1/2, mammalian target of rapamycin; GRB2, growth factor receptor-bound protein; SOS, son of sevenless homolog; PTEN, phosphatase and tensin homolog; PDPK1, 3-phosphoinositide dependent protein kinase 1, TSC1/2, tuberus sclerosis; PLC, phospholipase C; RalGDS, ral guanine nucleotide dissociation stimulator; IKKα/β, inhibitor of nuclear factor kappa-B kinase subunit alpha/beta. NFκB, nuclear factor kappa-light-chain-enhancer of activated B cells
Table 1. Targeted genetic events, targeted agents in clinical use, and adaptive response mechanisms identified in tumor cells.
Shown are major genetic alterations that drive the growth of many human cancers as well as the cognate targeted therapies approved or in clinical development and, where known, specific stress signaling adaptations that occur in response to targeted therapy. Abbreviations: NSCLC, non-small cell lung cancer; CLL, chronic lymphoblastic leukemia; AML, acute lymphoblastic leukemia; GBM, glioblastoma, RCC, renal cell carcinoma; HCC, hepatocellular carcinoma; GIST, gastrointestinal stromal tumor
| Oncogen ic Driver |
Alteration Type |
Targeted Therapy | Cancer Type | Adaptive response |
|---|---|---|---|---|
| EGFR | Mutation or Amplification | Erlotinib, Afatinib, Gefitinib, Lapatinib, Neratinib, Rociletinib, Dacometinib, Sapitinib, Cetuximab, Panitumumab, | NSCLC, Colorectal, Glioblastoma | PDGFR, STAT3, NFκB |
| ALK | Translocation | Crizotinib, Ceritinib | NSCLC | Non-receptor kinase, STAT3 |
| ERBB2 | Mutation or Amplification | Lapatinib, Sapitinib, Dacometinib, Neratinib, Afatinib, Trastuzumab, Pertuzumab | Breast, Gastric | ERBB3, NFκB, mTORC1/2,AKT |
| ERBB3 | Mutation | Anti-ERBB, MM-121, AV-203 | Colorectal, Gastric | Unknown |
| ERBB4 | Mutation or Translocation | Dacomitinib | Melanoma | Unknown |
| ABL | Translocation | Imatinib, Ponatinib, Nilotinib, Dasatinib, Bosutinib | CLL | Unknown |
| KIT | Mutation | Sunitinib, Imatinib, Dasatinib, Nilotinib | Melanoma, AML, GIST | Unknown |
| MET | Mutation or Amplification | Crizotinib, Tivantinib, Foretinib, Volitinib, MK-8083, INC280, Cabozantinib, MGCD-265 | NSCLC, Gastric | Unknown |
| FGFR1 | Amplification | BGJ398, AZD4547, Dovitinib, E-3810, PD173074, Lucitanib, Ponatinib, BAY1163877 | NSCLC (sq) Breast | Unknown |
| FGFR2 | Mutation or Amplification | BGJ398, AZD4547, Dovitinib, Ponatinib, BAY1163877 | Gastric, Breast, Endometrial | Unknown |
| FGFR3 | Mutation or Translocation | BGJ398, AZD4547, Dovitinib, Ponatinib, BAY1163877 | NSCLC, Bladder | Unknown |
| FGFR4 | Mutation or Amplification | Ponatinib | Rhabdomyosarco ma | Unknown |
| NTRK1 | Translocation | LOXO-101, ARRY-470, MGCD-516, RXDX-101, Crizotinib, TSR-011 | NSCLC, GBM Colorectal, Thyroid | Unknown |
| BRAF | Mutation | Vemurafenib, Dabrafenib, Encorafenib, Sorafenib | NSCLC, Thyroid Melanoma | EGFR, IGF1R, AXL,PDGFRB, DDR1/2, KDR, metabolic, phenotype switcing |
| JAK2 | Mutation | Ruxolitinib, Fedratinib, AZD1480, Pacritinib, Momelotinib, Baricitinib, | Myelofibrosis | Unknown |
| FLT3 | Mutation | Lestaurtinib, Quizartinib, Sunitinib, Sorafenib | AML | Unknown |
| PIK3CA | Mutation or Amplification | GDC-941, GDC-0890, BKM-120, BYL719, XL147, XL765, Dactolisib | Colorectal, GBM NSCLC, Breast | IRS1,mTORC2, EGFR, IGF1R, AXL, ERBB2, NFκB, STAT3/6 |
| AKT1/2/3 | Mutation, Amplification, Translocation | MK-2206, AZD5363, GDC-0068, Perifosine, GSK690693 | Pancreatic, Gastric, Breast, Ovarian | ERBB3, IGF1R, IR, ERK, mTORC |
| VEGFR2 | Amplification or Mutation | Bevacuzimab, Sorafenib, Sunitinib, Cabozantinib, Pazopanib, Cetiranib, Dovitinib, Linifanib, MGCD-265, Ramucirumab | RCC, Colorectal, HCC, Gastric, Thyroid | Unknown |
| ROS1/RET | Mutation or Translocation | Cabozantinib, Crizotinib, Sorafenib, Regorafenib, | NSCLC, Thyroid | Unknown |
| AR | Amplification | Enzalutamide, Bicalutamide | Prostate | PI3K, GR, β-catenin, |
To date, efforts to understand the basis of therapy resistance have largely focused on uncovering mechanisms of this secondary or primary resistance many of which consist of genetic or epigenetic events that pre-exist before treatment (recently extensively reviewed elsewhere15,16). These mechanisms are broadly categorized into 3 main groups: (1) on-target mutations that compromise binding or inhibition of the drug to the target (for example, the EGFR T790M resistance mutation in EGFR mutant lung cancer17-19); (2) bypass signaling, in which the target remains inhibited by the targeted drug but compensatory engagement (or disengagement) of other critical signaling components rescues the tumor cells from death and enables proliferation and survival (for example, upregulation of MET or AXL receptor kinase signaling in resistance to EGFR targeted therapy20-23); (3) phenotypic transformation from one histology or morphology to another (for example, lung adenocarcinoma-to-small cell lineage transformation, or prostate adenocarcinoma to neuroendocrine small-cell morphology24-26). In contrast, the signaling events that occur dynamically and immediately in tumor cells in response to initial therapy that may adaptively enable tumor-cell survival and drive resistance are less well understood. In order to unleash the full potential of targeted cancer therapy to transform cancers from lethal to chronic or curable conditions, it is essential to understand the biological mechanisms by which tumor cells adapt and survive the stress of initial therapy that enable initial or eventual escape from death. Recent studies have begun to fill this critical knowledge gap, and herein we categorize these adaptive signaling events as “adaptive stress signaling”. We review emerging findings that unveil how tumor cells respond dynamically and adapt to the stress of targeted therapy, emphasizing the promise this new knowledge holds for enabling truly transformative advances in cancer patient survival. Critically, we focus here not on mechanisms of signaling crosstalk or primary or secondary resistance to targeted therapy but rather more specifically on summarizing the biological events that have been shown to constitute an adaptive stress response in tumor cells treated with targeted therapy.
Adaptive stress response to targeted inhibition of oncogenic receptors
In this section, we discuss important findings that demonstrate how tumor cells with a particular oncogenic receptor can rewire intracellular signaling pathways as a stress response to survive initial targeted therapy. Oncogenic alterations in EGFR drive the growth of several tumor types, most notably NSCLC27-29 and glioblastoma multiforme30 (GBM). Inhibition of oncogenic EGFR can elicit compensatory signaling events that contribute to tumor cell survival. Inhibition of oncogenic EGFR (EGFRvIII) in GBM cells resulted in de-repression of the platelet-derived growth factor receptor, beta polypeptide (PDGFRB). This EGFR inhibitor induced upregulation of PDGFRB occurred via relief of mammalian target of rapamycin complex 1 (mTORC1) and ERK mediated suppression of PDGFRB expression. PDGFRB, when upregulated, provided survival signaling that limited the anti-tumor effects of EGFR oncogene inhibition in GBM cells31. The data raise the possibility that co-inhibition of EGFR and PDGFR using potent and selective targeted agents that cross the blood-brain barrier could enhance response in GBM patients.
More recently, inhibition of oncogenic EGFR was shown to increase STAT3 signaling and thereby rescue tumor cells from death upon EGFR targeted therapy in NSCLC cells32. These effects occurred via inhibition of MEK downstream of EGFR that led to activation of STAT3 and downstream IL6 signaling to promote cell survival and eventual (acquired) resistance32,33. This signaling axis may be a more general stress response, as some tumor cells with oncogenic forms of the anaplastic lymphoma kinase (ALK) or MET exhibited similar upregulation of STAT3 upon oncogene inhibition32,34.
Additional work by our group has revealed another layer of complexity in the adaptive response to EGFR inhibitor treatment. We found that EGFR targeted therapy elicits immediate activation of NFκB survival signaling via an NFκB activating biochemical complex in NSCLC cells that increased IL6-STAT signaling (Blakely, Pazarentzos & Bivona, manuscript under revision). This NFκB driven stress response enforced a cell survival circuit that was required for the development of acquired EGFR inhibitor resistance. NFκB activation occurred as a consequence of oncogene-inhibitor induced ubiquitination of TRAF2, which in turn activates RIP1. Subsequently, RIP1 activates NEMO which provides the scaffolding for the activation of IKKβ and ultimately the phosphorylation and degradation of IκBα. Within minutes of EGFR inactivation the RelA subunit of NFκB translocates to the nucleus and initiates an extensive transcriptional survival program. In summary, the oncogene-driven cell rewires the signaling to compensate immediately for inhibition of the oncogene and assembles a TRAF2-RIP1-IKK-EGFR complex to activate anti-apoptotic and pro-survival NFκB targets. These studies provide new insight into the resiliency in tumor cells with oncogenic EGFR and reveal an interesting role for dynamic modulation of ubiquitination as a molecular switch in this context. Together, these findings indicate that targeting NFκB or IL6-STAT signaling in combination with oncogenic EGFR initially may deprive tumor cells the opportunity to adapt and survive primary therapy and thereby eliminate the eventual emergence of drug-resistance.
The lessons learned through the study of oncogenic EGFR also extend to other oncogenic ERBB family members. Overexpression of ERBB2 by genomic amplification occurs in approximately 15% of breast cancers and ERBB2 targeted therapies such as lapatinib are approved for use in patients35. However, patients almost inevitably develop resistance during therapy. Recently mechanisms of adaptive stress response have been identified that contribute to the development of acquired resistance. For example, inhibition of amplified ERBB2 can lead to transcription upregulation of ERBB336. ERBB3 upregulation is caused by ERBB2 inhibitor treatment, which leads to de-repression of ERBB3 expression that occurs via PI3K-AKT signaling operating downstream of ERBB2 in breast cancer cells. ERBB3 upregulation is dependent on FOXO3A that is activated upon PI3K-AKT signaling inhibition and leads to compensatory activation of ERBB3 signaling and tumor cell survival37,38. Thus, dual inhibition of ERBB2 and ERBB3 may subvert this adaptive survival circuit and enhance response in patients. Another example of such adaptive stress response mechanisms is hyper-activation of NFκB39,40. Treatment of ERBB2 positive tumors led to activation of NFκB through NIBP upregulation. The findings were also validated in ERBB2 expressing esophageal cancers40. Interestingly, lapatinib-induced NFκB activation created a dependency on NFκB signaling that was exploited therapeutically using proteasome inhibitors, which were effective against these tumor cells41.
Beyond oncogenic receptor kinases, inhibition of hormone receptors that drive the growth of endocrine cancers can lead to adaptive signaling events that promote tumor cell survival and thereby limit targeted therapy efficacy. The proliferation and progression of prostate cells to a malignant state is mainly driven by the stimulatory effects of the androgen receptor (AR). Recent evidence revealed that inhibition of the AR with small molecule AR-targeted agents led to rapid activation of PI3K-AKT signaling in prostate adenocarcinomas42. This effect of AR inhibition occurred via downregulation of the AKT phosphatase PHLPP whose expression is controlled, in part, by AR. This dynamic activation of PI3K-AKT limited response to AR blockade and provided rationale for co-targeting AR and PI3K-AKT signaling in prostate adenocarcinoma patients. Recently, another mechanism that seems to appear acutely following AR inhibition is the immediate upregulation of the glucocorticoid receptor (GR), which in turn leads to a transcriptional program that promotes resistance to anti-androgen therapy43. Activation of WNT-β catenin signaling is another mechanism of adaptive response to androgen deprivation therapy that promotes reactivation of AR output44. This study revealed that prostate-specific antigen (PSA) is re-expressed via transcriptional upregulation by β-catenin which binds to PSA promoter.
Nearly 75% of breast cancers are positive for the estrogen receptor (ER) and tamoxifen has revolutionized the treatment of ER-positive tumors by antagonizing ligand binding. In ER-positive breast cancer, activation of the PI3K–AKT-mTOR pathway appears to be an important mechanism of resistance that occurs early after tamoxifen therapy45. Additionally, IGF1R, which activates PI3K-AKT-mTOR signaling, has been shown to provide an immediate escape mechanism from tamoxifen therapy via the upregulation of an IGF1R transcription program and the ultimate development of resistance in ER-positive breast cancers46. Altogether, these emerging findings provide further impetus to explore the dynamic response to the stress induced by both receptor and non-receptor targeted therapies across a broad range of tumor types.
Adaptive stress response to targeted inhibition of cancer-driving non-receptor kinases
Here, we review important findings that reveal how tumor cells with a particular cancer-promoting non-receptor kinase can rewire intracellular signaling pathways as a stress response to survive initial targeted therapy. The RAS-RAF-MEK-ERK signaling pathway is critical for the initiation and progression of a wide spectrum of human cancers2,3. This pathway is activated in tumor cells either through somatic alterations in pathway components, most commonly RAS and RAF, or via oncogenic activation of an upstream receptor kinase such as EGFR. Although direct inhibitors of RAS remain under investigation, to date no direct inhibitor of RAS has been clinically effective47-49. However, RAF and MEK inhibitors50 are approved for use in patients with advanced stage BRAFV600E mutant melanoma although therapy resistance remains a major challenge that limits the overall clinical impact of these agents.
Rapid activation of upstream receptor kinases has recently been shown to limit response to targeted inhibition of BRAF or MEK in different tumor types. Primary resistance to BRAF inhibition in BRAFV600E colon cancer cells has been attributed to compensatory activation of EGFR51. EGFR was activated as a consequence of RAF inhibitor induced suppression of ERK and CDC25C, a phosphatase that negatively regulates EGFR52. Importantly, EGFR inhibition together with BRAF inhibitor treatment counteracted this stress response and enhanced efficacy, providing rationale for combination therapy trials in colon cancer patients. Recent studies have extended this paradigm, indicating that BRAF or MEK inhibition leads to activation of multiple receptor kinases including EGFR in melanoma53. In this context, increased receptor kinase expression and signaling was a consequence of suppression of the transcription factor sex determining region Y-box10 (SOX10) following BRAF or MEK targeted therapy53. Interestingly, the supra-physiologic activation of MEK signaling that occurs as a consequence of this receptor kinase activation was detrimental to tumor cell growth. However, this signaling adaptation impaired tumor cell proliferation but not survival. Therefore, this study revealed that this adaptive compensatory activation may provide a context-specific survival advantage in melanoma, in that a subpopulation of tumor cells may survive RAF-MEK targeted therapy at a fitness cost that nevertheless enables the emergence of a drug-resistant tumor over time.
Parallel findings have been observed in various cancer cell lines treated with a MEK inhibitor. Indeed, inhibition of MEK can induce rapid dephosphorylation of EGFR and ERBB2 on inhibitor sites that phosphorylated by ERK54,55. Increased EGFR and ERBB2 activation, in turn, promotes ERBB3 upregulation and signaling that buffers the cells against the effects of MEK inhibition. Similarly, MEK inhibition has been shown to induce activation of multiple receptor kinases beyond those in the ERBB family by a variety of adaptive, rapid transcriptional and post-translational mechanisms in breast cancer and other tumor types. These receptor kinases induced upon MEK inhibition include IGF1R56, AXL57, DDR1/257, PDGFRB53,57, and KDR. Interestingly, the molecular basis of this stress response included MYC-driven upregulation of these receptor kinases in triple negative breast cancer, colorectal, and NSCLC cells58. Combined MEK plus receptor kinase inhibition could abolish compensatory survival signaling that occurred through both ERK and AKT57-59, providing rationale for potential combination therapies in the relevant patient subsets.
Recent work has highlighted a role for energy metabolism in the adaptive response to RAF-MEK targeted therapy in some tumors. The primary molecular circuit to produce ATP in normal cells resides at mitochondria where pyruvate enters and is converted to ATP. The process is called oxidative phosphorylation (OXPHOS) and utilizes glucose and oxygen60. Otto Warburg in 1956 described an alternative process by which oxygen is not used even if present and glucose is instead converted to lactate, generating ATP albeit inefficiently 61. The exact reason that cancer cell switch to this inefficient process of ATP generation is currently not completely understood and is not likely to be due to loss of OXPHOS62. However recent reports demonstrate that Warburg glycolysis is required to overcome oncogene-induced senescence and this has been clearly shown in cells with mutant KRAS or mutant BRAF63. Recently RAF inhibition was shown to promote a rapid switch from Warburg glycolysis to OXPHOS in melanoma64. This adaptive response involved metabolic reprogramming that enabled mutant BRAF melanoma cells to survive anti-RAF therapy. Patients treated with vemurafenib showed increase ATP production and expression of an OXPHOS genetic program. This metabolic reprogramming was mediated through EKR1/2 inhibition, which promoted upregulation of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α). Specifically in melanoma cells, PGC1α upregulation coincided with stabilization and increase in the expression of the microphthalmia-associated transcription factor (MITF). The upregulation of PGC1α and MITF resulted in decrease in the glycolytic flux with co-current enhancement of OXPHOS. The data suggest that ERK1/2 inhibition either by RAF or MEK inhibitors forced the cells to induce an adaptive mechanism that at first glance seems unfavorable for cancer cells. However, melanoma cells shift their dependency from glycolysis to OXPHOS to dynamically enable tumor initiation by an oncogene or adapt to inhibition of that oncogene. Another adaptive response mechanism involving a metabolic switch to OXPHOS was utilized by the BRAF mutant melanoma cells, namely the upregulation of lysine-specific demethylase 5B (JARID1B)65. JARID1B upregulation occurred within hours of RAF inhibition or chemotherapy, demonstrating that cells can quickly engage adaptive mechanisms to resist the lethal consequences of oncogene inhibition or DNA damage. PGC1α-mediated MITF upregulation and overexpression of JARID1B seem to have the same consequences to the cells, indicating that metabolic adaptation is a mechanism of adaptive response that is possibly utilized by other tumor types where MITF is not involved. Interestingly in EGFR mutant non-small cell lung cancer cells, increased histone demethylase function, specifically that of JARID1A, has been observed in a subpopulation of cells that are treated with gefitinib65. Although in that study the effects of JARID1B were attributed to IGF1R and not OXPHOS, it is possible that that energy regulation is a more general means by which tumor cells of distinct lineages adapt to therapy and survive. Inhibitors of OXPHOS in combination with RAF inhibitors, or perhaps other targeted therapies, may be a useful and successful strategy to decrease tumor burden and suppress drug resistance.
Beyond the RAS-RAF-MEK-ERK pathway, resiliency in the face of targeted inhibition of PI3K-AKT-mTOR signaling has also been observed in some tumor cells. Indeed, pharmacologic suppression of mTORC1 with rapamycin relieves inhibition of the upstream adaptor protein IRS-166, resulting in mTORC2-mediated AKT activation in breast cancer and multiple myeloma cells67,68. This rapid activation of the pathway occurs because mTORC1 inhibition decreases phosphorylation of the downstream target S6K which, when active, phosphorylates and inhibits IRS-1. Dual inhibition of mTORC1 and mTORC2 can overcome this adaptive signaling circuit. However, this dual suppression of mTORC1-2 then led to upstream activation of receptor kinases via their transcriptional upregulation69-74. Thus, combined inhibition of PI3K-AKT-mTOR and receptor kinase signaling may be effective clinically if a sufficient therapeutic window can be achieved in patients.
Additional adaptive responses to inhibition of PI3K were recently described and, interestingly, involved regulation of structural components of tumors, including the extracellular matrix and basement membrane72. In studies in ovarian and breast cancer cells, adaptive protection from the suppressive effects of PI3K-AKT-mTOR inhibition involved upregulation of several receptor kinases including EGFR, ERBB2, IGF1R, AXL and alternative signaling pathways such as JAK-STAT3/6 signaling72. Interestingly Bcl-2 was also found to be upregulated in this context. As Bcl-2 is an established NFκB target gene, these findings suggest another potential role for NFκB in the adaptive remodeling of the extracellular matrix during adaptation to therapy. The combination of PI3K-AKT-mTOR targeted drugs together with BH3 mimetic agents targeting anti-apoptotic proteins such as Bcl-2 may overcome this adaptive response and enhance anti-tumor efficacy.
Hence, intra and inter-pathway crosstalk in the RAS-RAF-MEK-ERK and PI3K-AKT-mTOR signaling pathways enables dynamic rewiring and adaptation to targeted therapy in cancer cells. Leveraging this knowledge clinically to enhance therapy efficacy will require not only knowledge of the tumor-type selective and context- specific signaling network features that enable tumor cell escape from treatment but also the appropriate use of combinatorial drug strategies that are safe and well-tolerated in patients.
Adaptive stress response to targeted therapy by phenotype switching
Here, we review intriguing recent evidence linking drug response in cancer to cellular phenotype switching. Phenotype switching can be observed after inhibition of receptor or non-receptor kinases as well as in response to chemotherapy and represents an additional manifestation of adaptive stress signaling, and one that may be functionally related to the adaptive signaling events discussed above in oncogene-driven tumor cells. Lineage-specific factors that regulate cellular phenotype and oncogenesis have been identified in certain cancers, including melanoma. The microthalmia-associated transcription factor is one such factor that has been shown to contribute to the pathogenesis of melanoma75,76. Interestingly, suppression of MITF has been shown to not only decrease cell cycle progression but also simultaneously promote a stem-cell like invasive phenotype characterized by loss of differentiation and tyrosinase expression77. These findings suggested that agents that decrease MITF levels may have dual, opposing effects on tumor growth and that activation of MITF may be beneficial in some contexts. In a recent study, treatment with the chemotherapy agent methotrexate adaptively and rapidly induce MITF expression to suppress invasiveness and promote differentiation of melanoma cells regardless of the genetic status of BRAF or other common somatic genetic alterations present in melanoma. These effects were accompanied by increased tyrosinase expression and consequent tumor cell specific hypersensitivity to a tyrosinase-processed antifolate drug77. Together, the data indicate context-specific modulation of therapy-induced, adaptive signaling changes that mediate cellular phenotype switching can unveil therapy strategies to enhance responses and limit systemic toxicity in patients.
Beyond melanoma, phenotype switching has been associated with drug resistance in several epithelial cancers. Indeed, the epithelial-mesenchymal transition (EMT) and transformation to small cell histology has been associated with resistance to targeted therapies against EGFR in NSCLC, AR in prostate adenocarcinoma, and targeted agents in different tumor types. In the case of EMT, the drug resistant cells are often hyper-invasive and acquire other features associated with increased metastasis such as stem-like molecular profiles. The extend to which this phenotype switching is triggered early and dynamically in response to targeted therapy and the underlying molecular basis remains to be deeply explored. Interestingly, several studies have implicated increased expression of the receptor kinase AXL in phenotype switching in NSCLC, breast cancer, and melanoma. This finding raises the possibility the stress response to targeted therapy may involve a conserved molecular pathway involving AXL and the acquisition of stem-like molecular properties that culminates in a cellular phenotype switch coupling metastasis and drug resistance. Future studies are needed to shed light in this important area, as the knowledge gained could provide tumor-type selective and context-specific strategies to subvert both drug resistance and metastatic tumor progression simultaneously.
Conclusions and Future Perspectives
As targeted cancer therapy gains a broader foothold in the clinic, it will become increasingly important to define the immediate molecular and cellular responses by which tumor cells adapt and buffer against these potentially-lethal insults. The genetic and epigenetic evolution of tumor cells endows them with substantial resiliency, confounding even our most potent targeted therapy attacks. In order to eliminate the presence of innate and the emergence of acquired resistance to treatment, therapies that pre-empt the stress response enabling tumor cell survival early during therapy are needed (Figure 2). A systematic approach, perhaps through the use of coupled genetic and proteomic profiling together with functional genomics screens, to define these molecular escape routes is critical for progress. Additionally, access to tumor specimens from patients not only before treatment and after the development of resistance but also early during therapy is necessary to define and validate the most clinically relevant molecular and cellular adaptions for subsequent therapeutic targeting.
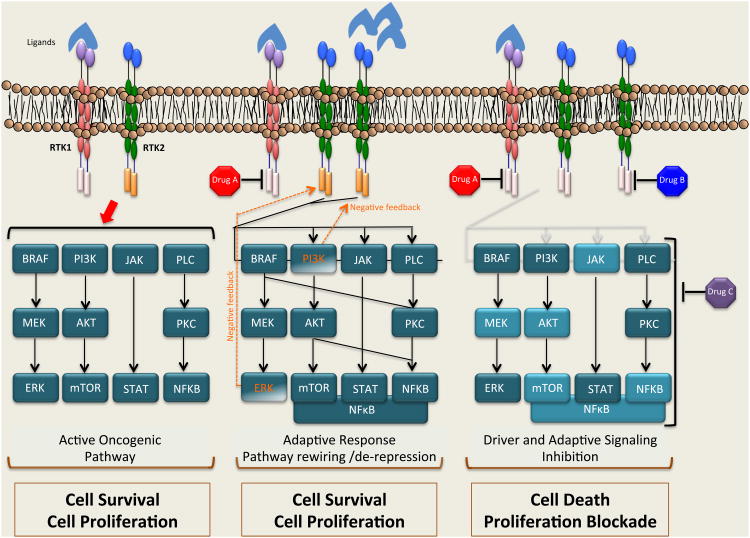
Figure 2. Model for blocking primary and adaptive molecular events underpinning tumor cell survival in order to induce profound and durable responses in patients.
Ligand stimulation, mutation, amplification or crosstalk between receptor tyrosine kinases (RTK1, RTK2) leads to their constitutive activation, which in turn activates downstream effector pathways. Inhibition of the driver oncogene leads to an immediate, adaptive stress response. Subsequently, signaling pathways are rewired in order to adapt to the new condition in which signaling from the oncogene is inhibited and sustain cell proliferation and survival. A new paradigm for cancer treatment would be the use of upfront combination therapies along with the oncogenic driver inhibition. While Drug A is normally used for inhibition of the driver oncogene, Drug B or Drug C can be used in combination upfront to prevent cancer cell adaptation. The outcome of this combination therapy approach would be enhanced killing of tumor cells and delay or prevention of therapy resistance. Abbreviations: Raf, Raf proto-oncogene serine/threonine kinase; Mek, mitogen activated protein kinase kinase; Erk, extracellular signal related kinase; PI3K, phosphoinositide 3-kinase; AKT, v-akt murine thymoma viral oncogene homolog; mTORC1/2, mammalian target of rapamycin; PLC, phospholipase C; NFκB, nuclear factor kappa-light-chain-enhancer of activated B cells
While this review focuses on the adaptive stress responses induced by targeted therapeutics, it is worth noting that another interesting area of adaptive stress signaling that has been relatively unexplored is the response to radiotherapy. Interestingly, activation of a HER2-NFκB signaling axis that induced additional HER2 in a feedback loop manner was observed in cancer stem cells in response to radiotherapy in breast cancer cells 78. This work is particularly interesting because many breast cancers respond initially to radiation therapy but relapse occurs and may be linked to residual or emergent stem cells resident within the tumor. In this recent work, radiotherapy rapidly induced NFκB activation, which in turn promoted the expression of HER2.79 Another interesting example of radiation-induced adaptive-stress response is the activation and cytosolic sequestering of cyclin D1 as well as activation of NFκB. In keratinocytes treated with low dose radiation therapy NFκB was rapidly activated and induced radioresistance80. In a similar model of keratinocytes, radiotherapy induced cytosolic levels of cyclin D1 that interacted with the pro-apoptotic Bax and prevented apoptosis. 81 Induction of cyclin D1 levels was rapid and did not coincide with translocation of cyclin D1 into the nucleus. Instead, a complex between cyclin D1 and BAX occurred that prevented each protein from translocating to the appropriate cellular locale, namely the mitochondria for Bax and nucleus for cyclin D1.81 More work is required to understand the adaptive stress response to radiotherapy and the role of stem cells in adaptive stress responses more generally.
It is tempting to speculate that immediate adaptations to therapy occur not only in tumor cells but also in tumor micro-environmental cells that impact tumor growth, drug response, and metastatic progression. Hence, it is critical to extend studies of the dynamic response to therapy to stromal and immune cells that reside within the broader tumor ecosystem. Indeed, recent data indicate that inhibition of MEK may lead to activation of cytotoxic T cells in some melanomas, providing rationale to further explore this largely uncharted role of the adaptive response to therapy in cancer. Such investigations could yield novel combinatorial treatment strategies that suppress adaptive survival signaling in tumor cells while simultaneously engaging the host anti-tumor response to enhance the magnitude and duration of response in patients.
In summary, a deeper understanding of the survival mechanisms engaged rapidly in response to the stress of targeted therapy promises to offer not only increased fundamental knowledge but also improved treatment strategies capable of unleashing the full potential of targeted cancer therapy to transform cancer patient survival.
Acknowledgments
The authors thank members of the Bivona lab for critical review and thoughtful comments on the manuscript. We are grateful to the following funding sources: NIH Director's New Innovator Award, NIH R01 CA169338, Howard Hughes Medical Institute, Doris Duke Charitable Foundation, Searle Scholars Program, California Institute for Quantitative Biosciences, QB3@UCSF (to T.G.B).
References
- 1.Weinstein IB Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 3.Dhillon AS, Hagan S, Rath O, Kolch W. MAP kinase signalling pathways in cancer. Oncogene. 2007;26:3279–3290. doi: 10.1038/sj.onc.1210421. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 5.Chapman PB, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maemondo M, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 7.Rosell R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The Lancet Oncology. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 8.Sequist LV, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 9.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. The New England journal of medicine. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 10.Diaz LA, Jr, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misale S, Di Nicolantonio F, Sartore-Bianchi A, Siena S, Bardelli A. Resistance to Anti-EGFR Therapy in Colorectal Cancer: From Heterogeneity to Convergent Evolution. Cancer discovery. 2014;4:1269–1280. doi: 10.1158/2159-8290.CD-14-0462. [DOI] [PubMed] [Google Scholar]
- 12.Misale S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lovly CM, Shaw AT. Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:2249–2256. doi: 10.1158/1078-0432.CCR-13-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goltsov A, Langdon SP, Goltsov G, Harrison DJ, Bown J. Customizing the therapeutic response of signaling networks to promote antitumor responses by drug combinations. Frontiers in oncology. 2014;4:13. doi: 10.3389/fonc.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garraway LA, Janne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer discovery. 2012;2:214–226. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 16.Glickman MS, Sawyers CL. Converting cancer therapies into cures: lessons from infectious diseases. Cell. 2012;148:1089–1098. doi: 10.1016/j.cell.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi S, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 18.Pao W, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godin-Heymann N, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Molecular cancer therapeutics. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 20.Bardelli A, et al. Amplification of the MET receptor drives resistance to anti-EGFR therapies in colorectal cancer. Cancer discovery. 2013;3:658–673. doi: 10.1158/2159-8290.CD-12-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bean J, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman JA, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nature genetics. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sequist LV, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Science translational medicine. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe S, et al. Transformation to small-cell lung cancer following treatment with EGFR tyrosine kinase inhibitors in a patient with lung adenocarcinoma. Lung cancer. 2013;82:370–372. doi: 10.1016/j.lungcan.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Aparicio A, Logothetis CJ, Maity SN. Understanding the lethal variant of prostate cancer: power of examining extremes. Cancer discovery. 2011;1:466–468. doi: 10.1158/2159-8290.CD-11-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 28.Paez JG, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 29.Pao W, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang PH, Xu AM, White FM. Oncogenic EGFR signaling networks in glioma. Science signaling. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 31.Akhavan D, et al. De-repression of PDGFRbeta transcription promotes acquired resistance to EGFR tyrosine kinase inhibitors in glioblastoma patients. Cancer discovery. 2013;3:534–547. doi: 10.1158/2159-8290.CD-12-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HJ, et al. Drug resistance via feedback activation of Stat3 in oncogene-addicted cancer cells. Cancer cell. 2014;26:207–221. doi: 10.1016/j.ccr.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 33.Ishiguro Y, Ishiguro H, Miyamoto H. Epidermal growth factor receptor tyrosine kinase inhibition up-regulates interleukin-6 in cancer cells and induces subsequent development of interstitial pneumonia. Oncotarget. 2013;4:550–559. doi: 10.18632/oncotarget.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell. 2011;145:19–24. doi: 10.1016/j.cell.2011.03.026. [DOI] [PubMed] [Google Scholar]
- 35.Scaltriti M, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 36.Garrett JT, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirouac DC, et al. Computational modeling of ERBB2-amplified breast cancer identifies combined ErbB2/3 blockade as superior to the combination of MEK and AKT inhibitors. Science signaling. 2013;6:ra68. doi: 10.1126/scisignal.2004008. [DOI] [PubMed] [Google Scholar]
- 38.McDonagh CF, et al. Antitumor activity of a novel bispecific antibody that targets the ErbB2/ErbB3 oncogenic unit and inhibits heregulin-induced activation of ErbB3. Molecular cancer therapeutics. 2012;11:582–593. doi: 10.1158/1535-7163.MCT-11-0820. [DOI] [PubMed] [Google Scholar]
- 39.Wetterskog D, et al. Identification of novel determinants of resistance to lapatinib in ERBB2-amplified cancers. Oncogene. 2014;33:966–976. doi: 10.1038/onc.2013.41. [DOI] [PubMed] [Google Scholar]
- 40.Bailey ST, et al. NF-kappaB activation-induced anti-apoptosis renders HER2-positive cells drug resistant and accelerates tumor growth. Molecular cancer research : MCR. 2014;12:408–420. doi: 10.1158/1541-7786.MCR-13-0206-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen YJ, et al. Lapatinib-induced NF-kappaB activation sensitizes triple-negative breast cancer cells to proteasome inhibitors. Breast cancer research : BCR. 2013;15:R108. doi: 10.1186/bcr3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carver BS, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora VK, et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell. 2013;155:1309–1322. doi: 10.1016/j.cell.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schweizer L, et al. The androgen receptor can signal through Wnt/beta-Catenin in prostate cancer cells as an adaptation mechanism to castration levels of androgens. BMC cell biology. 2008;9:4. doi: 10.1186/1471-2121-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fox EM, et al. A kinome-wide screen identifies the insulin/IGF-I receptor pathway as a mechanism of escape from hormone dependence in breast cancer. Cancer research. 2011;71:6773–6784. doi: 10.1158/0008-5472.CAN-11-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baker NM, Der CJ. Cancer: Drug for an ‘undruggable’ protein. Nature. 2013;497:577–578. doi: 10.1038/nature12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zimmermann G, et al. Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, Adjei AA. The clinical development of MEK inhibitors. Nature reviews Clinical oncology. 2014;11:385–400. doi: 10.1038/nrclinonc.2014.83. [DOI] [PubMed] [Google Scholar]
- 51.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 52.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer discovery. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun C, et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature. 2014;508:118–122. doi: 10.1038/nature13121. [DOI] [PubMed] [Google Scholar]
- 54.Li X, Huang Y, Jiang J, Frank SJ. ERK-dependent threonine phosphorylation of EGF receptor modulates receptor downregulation and signaling. Cellular signalling. 2008;20:2145–2155. doi: 10.1016/j.cellsig.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turke AB, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer research. 2012;72:3228–3237. doi: 10.1158/0008-5472.CAN-11-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flanigan SA, et al. Overcoming IGF1R/IR resistance through inhibition of MEK signaling in colorectal cancer models. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:6219–6229. doi: 10.1158/1078-0432.CCR-13-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun C, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 59.Lito P, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papa S, et al. The oxidative phosphorylation system in mammalian mitochondria. Advances in experimental medicine and biology. 2012;942:3–37. doi: 10.1007/978-94-007-2869-1_1. [DOI] [PubMed] [Google Scholar]
- 61.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 62.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. The FEBS journal. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 63.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 64.Haq R, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1alpha and MITF. Cancer cell. 2013;23:302–315. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haruta T, et al. A rapamycin-sensitive pathway down-regulates insulin signaling via phosphorylation and proteasomal degradation of insulin receptor substrate-1. Molecular endocrinology. 2000;14:783–794. doi: 10.1210/mend.14.6.0446. [DOI] [PubMed] [Google Scholar]
- 67.O'Reilly KE, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer research. 2006;66:1500–1508. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A. Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Molecular cancer therapeutics. 2005;4:1533–1540. doi: 10.1158/1535-7163.MCT-05-0068. [DOI] [PubMed] [Google Scholar]
- 69.Chandarlapaty S. Negative feedback and adaptive resistance to the targeted therapy of cancer. Cancer discovery. 2012;2:311–319. doi: 10.1158/2159-8290.CD-12-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chandarlapaty S, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ducker GS, et al. Incomplete inhibition of phosphorylation of 4E-BP1 as a mechanism of primary resistance to ATP-competitive mTOR inhibitors. Oncogene. 2014;33:1590–1600. doi: 10.1038/onc.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Muranen T, et al. Inhibition of PI3K/mTOR leads to adaptive resistance in matrix-attached cancer cells. Cancer cell. 2012;21:227–239. doi: 10.1016/j.ccr.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodrik-Outmezguine VS, et al. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Serra V, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30:2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carreira S, et al. Mitf regulation of Dia1 controls melanoma proliferation and invasiveness. Genes & development. 2006;20:3426–3439. doi: 10.1101/gad.406406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheli Y, et al. Mitf is the key molecular switch between mouse or human melanoma initiating cells and their differentiated progeny. Oncogene. 2011;30:2307–2318. doi: 10.1038/onc.2010.598. [DOI] [PubMed] [Google Scholar]
- 77.Saez-Ayala M, et al. Directed phenotype switching as an effective antimelanoma strategy. Cancer cell. 2013;24:105–119. doi: 10.1016/j.ccr.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 78.Duru N, Candas D, Jiang G, Li JJ. Breast cancer adaptive resistance: HER2 and cancer stem cell repopulation in a heterogeneous tumor society. Journal of cancer research and clinical oncology. 2014;140:1–14. doi: 10.1007/s00432-013-1494-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Duru N, et al. HER2-associated radioresistance of breast cancer stem cells isolated from HER2-negative breast cancer cells. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:6634–6647. doi: 10.1158/1078-0432.CCR-12-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X, et al. Activation of nuclear factor kappaB in radioresistance of TP53-inactive human keratinocytes. Cancer research. 2002;62:1213–1221. [PubMed] [Google Scholar]
- 81.Ahmed KM, Fan M, Nantajit D, Cao N, Li JJ. Cyclin D1 in low-dose radiation-induced adaptive resistance. Oncogene. 2008;27:6738–6748. doi: 10.1038/onc.2008.265. [DOI] [PMC free article] [PubMed] [Google Scholar]