Abstract
Mosquito-based malaria transmission-blocking vaccines (mTBVs) target midgut-surface antigens of the Plasmodium parasite's obligate vector, the Anopheles mosquito. The alanyl aminopeptidase N (AnAPN1) is the leading mTBV immunogen; however AnAPN1's role in Plasmodium infection of the mosquito and how anti-AnAPN1 antibodies functionally block parasite transmission remains elusive. Here we present the 2.65 Å crystal structure of AnAPN1 and the immunoreactivity and transmission-blocking profile of three AnAPN1 monoclonal antibodies (mAb), including mAb 4H5B7, which effectively block transmission of natural strains of Plasmodium falciparum. Utilizing the AnAPN1 structure we map the conformation-dependent 4H5B7 neo-epitope to a previously uncharacterized region on domain 1, and further demonstrate that non-human primate neo-epitope-specific IgG also block parasite transmission. We discuss the prospect of a novel biological function of AnAPN1 as a receptor for Plasmodium in the mosquito midgut and the implications for redesigning the AnAPN1 mTBV.
Malaria exacts a deadly toll on human populations worldwide. A new era in the fight against the disease has seen multiple malaria vaccines entering or completing advanced clinical study1, including malaria transmission-blocking vaccines (mTBVs)2. Malaria transmission requires the establishment of Plasmodium in the Anopheles mosquito, which is contingent on the parasite's ookinete stage successfully traversing the midgut epithelium to initiate sporogonic development3,4. TBVs disrupt this obligatory step in the parasite life cycle5, limiting the number of infectious mosquito vectors and introducing local herd immunity6. The concept of mTBVs is simple: antibodies against specific, mosquito midgut antigens circulating in the peripheral blood are ingested by the mosquito while feeding on immunized hosts. These antibodies, as well as complement, can survive in the mosquito midgut for up to 24 hours post-blood feeding and prevent parasite access to host ligands that mediate midgut cell adhesion and invasion. Unable to establish infection in the vector, progression of parasite development and transmission to new human hosts is arrested or reduced.
We have shown that AnAPN1, an alanyl aminopeptidase N present on the Anopheles gambiae midgut apical surface is a potent mTBV candidate7-9. Importantly, parallel studies in the field, using P. falciparum collected from parasitized individuals corroborated laboratory-derived data9, thus demonstrating the strain- and species-transcending potency of anti-AnAPN1 antibodies. However, the role of AnAPN1 in Plasmodium infection of the mosquito gut, and how anti-AnAPN1 antibodies functionally block parasite transmission remains elusive. To identify cryptic AnAPN1 conformational epitopes and gain insight into functional versus decoy vaccine domains we solved the crystal structure of AnAPN1. Here, we describe the immunoreactivity and transmission-blocking profile of AnAPN1 monoclonal antibodies (mAbs), and together with the AnAPN1 structure, map a novel transmission-blocking epitope. These findings deepen our understanding of vector-Plasmodium interactions and ultimately fuel the continued development and optimization of the AnAPN1 mTBV.
Results
Structure determination
AnAPN1 is comprised of an N-terminal signal peptide (residues 1-19) and C-terminal ectodomain (residues 22-993) that contains a putative mucin O-glycosylated region (residues 952-993). A glycosylphosphatidylinositol (GPI)-anchor (residues 997-1020) resides at the C-terminus. We determined the crystal structure of near full-length AnAPN1 (residues 22-942) to 2.65 Å with a crystallographic R-factor of 20.3% (Rfree 25.4%). The crystallographic refinement statistics are reported in Table 1. There were two AnAPN1 molecules within the asymmetric unit (molecules A and B) and superposition of the Cα backbone atoms gave an overall root mean square deviation (RMSD) of 0.26 Å. The first 35 residues and residues 531-541 could not be modeled in either molecule due to poor electron density. An additional disordered region included residues 826-830 in molecule A. Although no attempt was made to obtain ligand-bound AnAPN1, a peptide was coordinated within the active site of both molecules and modeled according to the electron density as AAAKL and AAK for molecules A and B, respectively (Supplementary Fig. 1a,b). Anomalous diffraction difference maps14 confirmed the presence of a zinc metal ion in the active site of both molecules (Supplementary Table 1, Supplementary Fig. 1c).
Table 1. Data collection and refinement statistics.
| AnAPN1 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 113.48, 132.9, 146.01 |
| α β γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 98.0-2.65 (2.72-2.65)a |
| Rmerge | 0.111 (0.842) |
| I / σI | 13.5 (2.3) |
| Completeness (%) | 99.5 (99.2) |
| Redundancy | 8.8 (8.0) |
| Refinement | |
| Resolution (Å) | 64.0 -2.65 |
| No. reflections | 61048 |
| Rwork / Rfree | 20.3/25.4 |
| No. atoms | |
| Protein | 14067 |
| Ligand/ion | 96 |
| Water | 146 |
| B factors | |
| Protein | 59.5 |
| Ligand/ion | 69.5 |
| Water | 49.7 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.0120 |
| Bond angles (°) | 1.52 |
Values in parentheses are for highest-resolution shell.
Data was collected from a single crystal.
Overall structure
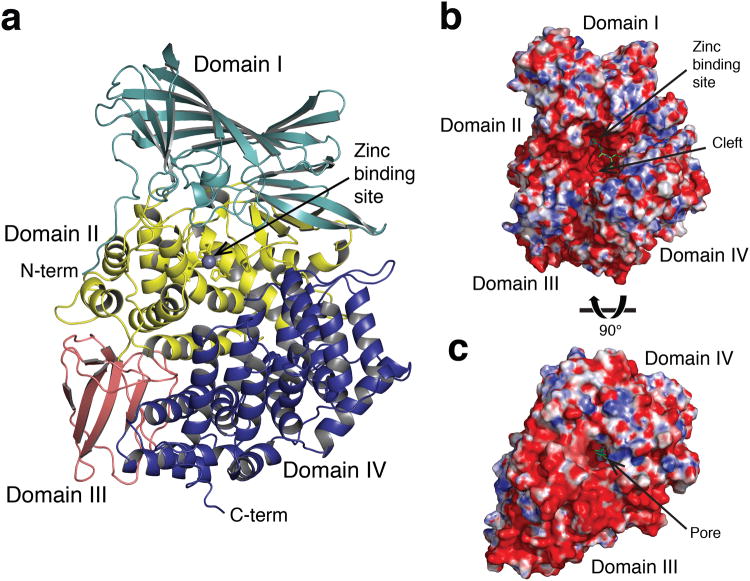
The tertiary structure of the AnAPN1 ectodomain (residues 57-942) exhibited the classical four-domain assembly of M1-family metallopeptidases, designated domains I-IV (Fig. 1a). The N-terminal domain I, visible from residue 57 (residues 57-270), comprised a 15-stranded β-barrel fold, nine strands of which define the subunit vaccine immunogen (NT135aaAnAPN1; residues 60-194). Domain II, or the catalytic domain (residues 271-523), is involved in substrate recognition and contains the G330AMEN motif common to many exopeptidases, in addition to the zinc-binding motifs HEXXH (H366EYAH370) and NEXFA (NE389GFA). This domain had a thermolysin-like fold and comprised a four-stranded β-sheet with connecting α-helix (helix α4) and an eight α-helical superhelix subdomain. Domain III (residues 524-613) adopted a β-sandwich architecture in which two separate anti-parallel β-sheets packed together. The C-terminal domain IV (residues 614-942) adopted the most variable conformation compared to other M1-family metallopeptidases and comprised a 17 α-helical super-helix. According to a DALI structural alignment, AnAPN1 was most similar to porcine APN (PDB ID 4FKE10; RMSD 2.5 Å) with 32% sequence identity and 48% sequence similarity (Supplementary Fig. 2).
Figure 1. Overview of AnAPN1 structure.
(a) Cartoon representation of the AnAPN1 structure. Domains I-IV are shown in light blue, yellow, pink and dark blue respectively. The zinc ion is depicted as a gray sphere. (b) Surface representation of AnAPN1 from a side and (c) underneath view, highlighting openings to the active site. Electrostatic potential is coloured from the most negative (red) to positive (blue) and ranges from (± 10 kT/e). Peptide bound in the active site is shown in green ball-and-stick.
Active site access
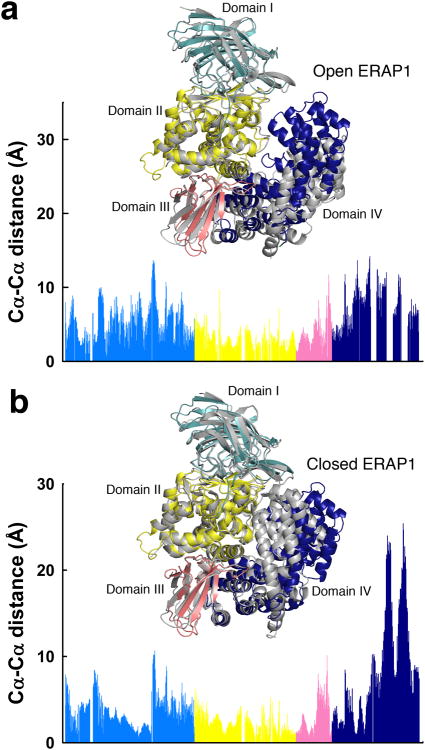
Each AnAPN1 monomer revealed two openings to the active site (Fig. 1b,c). The first was a large electronegative cleft, 35 Å wide, 20 Å deep and 40 Å long, that was predominantly flanked by domains II and IV and allowed access to the active site. The second, located on domain IV, was a small 11 Å diameter pore that led to a 38 Å-long channel. An analogous pore, or C-terminal channel, was reported for PfA-M1 (ref. 11) and postulated to permit substrate entry, whereas that of ERAP1 was considered a putative C-terminal substrate-binding site12. Given the exposed, solvent accessible active site, AnAPN1 was described as adopting an open conformation as per ERAP1 (Fig. 2a versus b; Supplementary Fig. 3). However, the orientation of domain IV relative to the other domains, which determines the extent of the aperture, lay between the closed (PDB ID 2YD0; ref. 12) and open state of ERAP1 (PDB ID 3MDJ13) (Fig. 2). This was reflected by a DALI structural alignment in which AnAPN1 had a RMSD of 4.2 Å for the open conformation (PDB ID 3MDJ13) and similarly, 3.9 Å for the closed conformation (PDB ID 2YD0; ref. 12). The most pronounced differences between the open conformations of AnAPN1 and ERAP1 were in domain IV, specifically, four helix-loop-helix motifs (α22-α23, α24-α25, α27-α28 and α29-α30) differed in orientation (Fig. 2). The hinge between the conformations originated between helices α20 and α21 with two disulfide bridges connecting helix α19 to α20 (Cys736-Cys743) and α21 to α23 (Cys772-Cys808). The bridge between α19 and α20 restricted the angle of rotation between these two helices and limited the movement of domain IV. The surface representation of AnAPN1, with a large rift between domains II and IV, was most similar to porcine APN and the open conformation of ERAP1 (Supplementary Fig. 3a versus b,c), but was distinct from human APN and closed ERAP1 (Supplementary Fig. 3a versus d,e).
Figure 2. Structural comparison of AnAPN1 with the open and closed forms of ERAP1.
(a,b) Cα-Cα distance plot of equivalent residues of AnAPN1 with the open (a) and closed (b) forms of ERAP1, PDB IDs 3MDJ13 and 2YD0 (ref. 12) respectively. Inset, cartoon representation of the structural alignment of AnAPN1 (domains I, II, III and IV coloured pale blue, yellow, pink and dark blue respectively) with ERAP1, either open or closed, coloured in gray.
The active site of AnAPN1
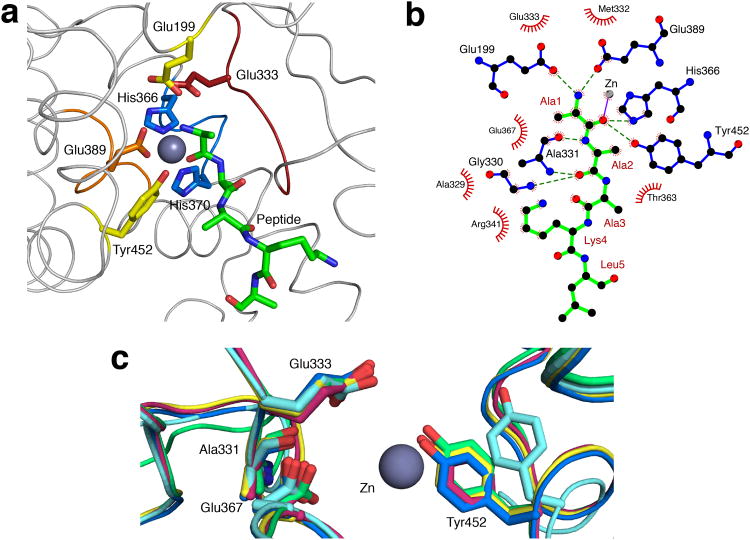
The active site residues of AnAPN1 were within domain II (Fig. 3a). The zinc ion was coordinated by Nε2 atoms of His366 and His370 from the H366EXXH370 motif, the carboxyl Oε of Glu389 of the NE389XFA motif and the N-terminus of the modeled peptide (Fig. 3a,b, Supplementary Fig. 1a,b). The N-terminal residue of this peptide was anchored to the active site, coordinated to the highly conserved Glu333 of the GAMEN motif in addition to Glu199 and Glu389. This residue also formed hydrogen bonds with His366 and the highly conserved Tyr452 (Fig. 3a,b), which is proposed to stabilize the oxyanion generated in the transition state15. The alanine at the second position of the peptide also formed contacts with residues that lined the electronegative cleft and formed hydrogen bonds with Gly330 and Ala331 from the GAMEN motif (Fig. 3b). The conformation of Tyr452 (equivalent to Tyr438 in ERAP1) is often used as a partial indicator of whether M1-family metalloenzymes are in an active or inactive state12. Tyr452 pointed towards the active site in AnAPN1 (Fig. 3c) and suggested an active enzyme conformation.
Figure 3. Catalytic site.
(a) Active site of AnAPN1 showing positions of key catalytic motifs including G330AMEN (red), H366EYAH370 (blue) and NE389GFA (orange). Tyr452 (yellow) and Glu199 (yellow), are also shown, as well as the peptide bound in the active site (green). (b) LigPlot+ representation of interactions between peptide (green) and residues within the AnAPN1 active site. Hydrogen bonded residues are shown in blue (hydrogen bonds represented by a dashed green line) and hydrophobic interaction residues are shown as red half circles. Peptide residues are numbered in dark red. (c) Structural alignment of the active site of AnAPN1 (pink), ERAP1 in a catalytically inactive conformation (pale blue, PDB ID 3MDJ13) and active conformation [bright blue, PDB ID 2YD0 (ref. 12)], as well as human APN (yellow, PDB ID 4FYQ17) and porcine APN (green, PDB ID 4FKE10). The zinc ion is depicted as a gray sphere.
Potential for AnAPN1 dimerization
Given that solved structures of M1-family metalloenzymes are reported as either monomeric or dimeric, and two AnAPN1 molecules were in the AnAPN1 crystal asymmetric unit, we determined the quaternary structure of AnAPN1 in aqueous solution. Sedimentation velocity experiments showed a single sedimenting boundary (Supplementary Fig. 4a) and subsequent c(s) analysis (Supplementary Fig. 4b) showed that AnAPN1 existed primarily as a single species with a standardized sedimentation coefficient of 9.2 S. c(M) analysis (Supplementary Fig. 4c) revealed this species had a molecular weight of approximately 230 kDa, consistent with an AnAPN1 dimer (Mr = 214 kDa). Intact human aminopeptidase A (APA) and APN dimers have been observed in vivo16,17.
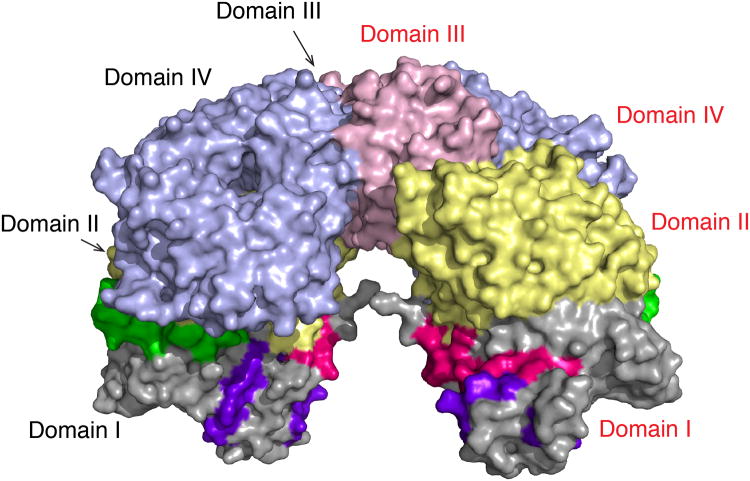
We predicted two molecules from adjacent asymmetric units formed a putative AnAPN1 dimer, with contacts via domains III and IV (Fig. 4, Supplementary Table 2). Several salt bridges and hydrogen bonds were formed between Asp653 (domain IV) and Arg531 (domain III). Lys551 (domain III) from each monomer formed hydrogen bonds with each other, as well as an extensive network of hydrophobic contacts (Supplementary Table 2). As opposed to the dimer arrangement in the asymmetric unit, this dimer allowed free movement of domain IV into an open and closed conformation and an unobstructed cleft for peptide entry and exit. In addition the dimer was symmetrical, buried 864 Å2 and had ΔiG = -7.9 kcal/mol (P-value = 0.21). Moreover peptides 1, 5, 7 and 9 discussed below were equally solvent accessible in the monomeric versus putative dimeric form of AnAPN1 (Fig. 4).
Figure 4. Proposed AnAPN1 dimer.
The putative AnAPN1 dimer is formed via interactions between domains III and IV. Domains I-IV are shown in gray, light yellow, light pink and light blue respectively. Molecule A is labeled in black and molecule B in red. Peptides 1, 7 and 9, referred to later in the text, are shown in pink, purple and green, respectively.
Biochemical characterization of AnAPN1 activity
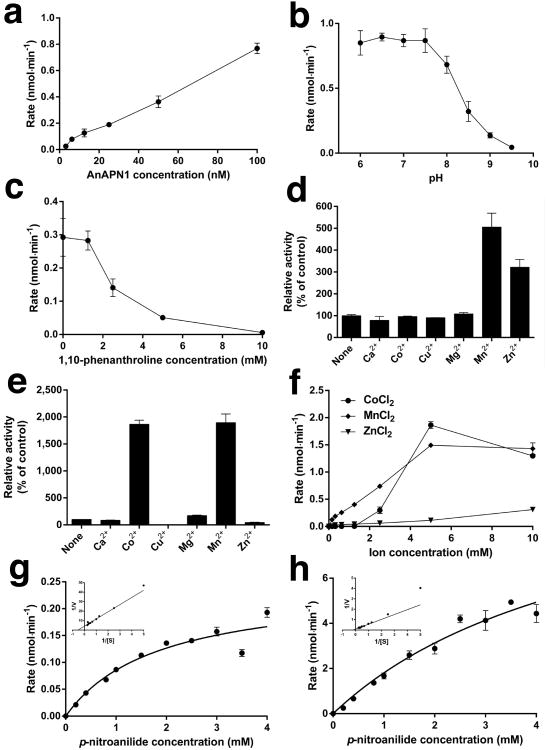
The enzymatic activity of recombinant near full-length S2-expressed AnAPN1 (residues 22-942) was verified with in vitro spectrophotometric-based assays by measuring the continuous increase in absorbance at 405 nm due to cleavage of the substrate L-leucine-p-nitroanilide and release of 4-nitroaniline (Fig. 5a). We observed a pH optimum of 6.0-7.5 (Fig. 5b), the latter of which is consistent with that of the blood fed mosquito midgut18. AnAPN1 was inactive in the presence of the metalloprotease inhibitor 1,10-phenanthroline (Fig. 5c), but was substantially activated by Co2+, Mn2+, and Zn2+ cations (Fig. 5d-f). The Km for L-leucine-p-nitroanilide at pH 7.5 was estimated to be 1.84 mM in the presence of 1 mM Mn2+ (Fig. 5g), and 5.46 mM at 10 mM Mn2+ (Fig. 5h).
Figure 5. Enzymatic characterization of AnAPN1.
(a) Rate (nmol/min) of L-leucine-p-nitroanilide hydrolysis by S2-expressed recombinant near full-length AnAPN1. AnAPN1 concentration (nM) is indicated on the x-axis. (b) Rate (nmol/min) of L-leucine-p-nitroanilide hydrolysis by AnAPN1 between pH 6.0 and 9.5, as indicated on the x-axis. (c) Rate (nmol/min) of L-leucine-p-nitroanilide hydrolysis by AnAPN1 in the presence of 1,10-phenanthroline. (d, e) Relative activity of AnAPN1 in the presence of (d) 1 mM or (e) 10 mM divalent metal cations, as indicated on the x-axis. Data are expressed as the percentage activity compared with a control of AnAPN1 in the presence of 1,10-phenanthroline and no ions. (f) Rate (nmol/min) of L-leucine-p-nitroanilide hydrolysis by AnAPN1 in the presence of cobalt, manganese, and zinc (II) ions. (g,h) Michaelis-Menten plots of the hydrolysis of L-leucine-p-nitroanilide [S] by AnAPN1 in the presence of (g) 1 mM and (h) 10 mM MnCl2. L-leucine-p-nitroanilide concentration is plotted against velocity (nmol/min). Insets show Lineweaver-Burk plots used to determine Km. Data points for all experiments (a-h) are the mean ± s.d. of three technical replicates.
Peptide-epitope mapping ELISA
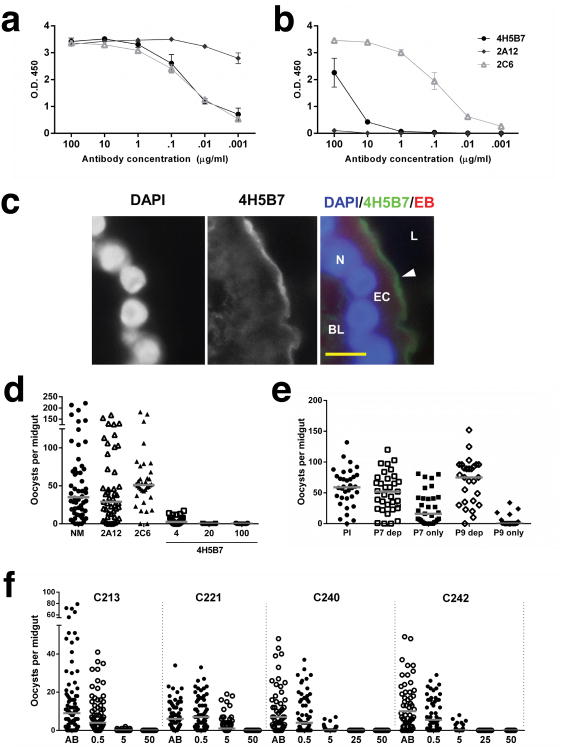
The CD spectra of the original AnAPN1 subunit vaccine immunogen (NT135aaAnAPN1) confirmed the predicted secondary structure as primarily β-sheet with a strong negative signal at 218 nm (data not shown). The NT135aaAnAPN1 immunogen contains nine predicted B and T cell epitopes that were synthesized and probed with individual mouse, rabbit, and non-human primate (NHP) AnAPN1 antisera by ELISA8. In this study, we generated three new mAbs against this immunogen: 2A12, 2C6 and 4H5B7. As expected, all mAbs recognized NT135aaAnAPN1 (Fig. 6a). However, of the three, only 4H5B7 and 2C6 reacted with An. gambiae midgut lysate (Fig. 6b), and importantly, only 4H5B7 recognized native AnAPN1 in midgut cryosections (Fig. 6c). We observed that 2A12 recognized peptide 1 (Supplementary Table 3, Supplementary Fig. 5a), while 4H5B7 showed weak reactivity with peptides 4 and 5 and negligible reactivity to peptide 7 (Supplementary Table 3, Supplementary Fig. 5b). These data suggest poor presentation of the epitope-context to 4H5B7 in ELISAs. Of the three mAbs, only 4H5B7 blocked cultured P. falciparum development in An. gambiae, and at concentrations of 4-100 μg/mL (Fig. 6d, Supplementary Table 4), while 2A12 and 2C6 had no blocking activity (Fig. 6d, Supplementary Fig. 5c, Supplementary Table 4). By immunoblot, 4H5B7 recognized both NT135aaAnAPN1 and near full-length AnAPN1; although for the latter, more protein was required (Supplementary Fig. 5d). However, in a more complex sample, such as whole mosquito midgut lysate, 4H5B7 can barely stain AnAPN1 (data not shown); suggesting that in immunoblots, there is either (a) potential masking of the linear portion of the complete conformational epitope for 4H5B7 by several midgut proteins of the same relative molecular mass or (b) 4H5B7 recognizes a conformational epitope (Fig. 6c).
Figure 6. Anti-AnAPN1 mAb 4H5B7 recognizes AnAPN1 and blocks development of P. falciparum via a novel epitope.
MAb recognition of (a) NT135aaAnAPN1 and (b) An. gambiae midgut lysates by ELISA. Error bars indicate s.e.m from triplicate assays. (c) Immunofluorescence micrograph of a transverse cryosection of a female, sugar-fed An. gambiae midgut demonstrating 4H5B7 staining of the luminal (L), apical midgut epithelium. In the merged image, the nucleus (N) stained with DAPI appears blue, 4H5B7 is green and Evans Blue (EB), a general protein counterstain, appears red. The basal lamina (BL) is indicated for orientation. (d) P. falciparum (NF54) oocysts per An. gambiae midgut following blood feeding in the presence of normal mouse IgG (NM), 2A12, 2C6, and 4H5B7 anti-AnAPN1 mAbs in SMFA. (e) P. falciparum (NF54) oocysts per An. gambiae midgut following blood feeding in the presence of non-human primate (NHP) pre-immune IgG, peptide 7-depleted (P7 dep), peptide 7-specific (P7 only), peptide 9-depleted (P9 dep), or peptide 9-specific (P9 only) anti-AnAPN1 IgG in SMFA. (f) P. falciparum oocysts per An. gambiae (Ngousso) midgut following membrane feeding with gametocytemic blood from four children (C213, C221, C240, and C242). Control human AB serum alone (AB) and 4H5B7 mAb concentrations between 0.5-50 μg/mL, as indicated on the x-axis, were tested. (d-f) Statistical analyses used zero-inflated GLMM (see Methods) and horizontal bars indicate median oocyst intensity. Raw data and statistical results of representative studies of three or four biological replicate experiments are shown in Supplementary Table 4.
Polyclonal antiserum from NHPs showed strong recognition of peptide 5 and 7 epitopes8, and 4H5B7 shared similar recognition of peptide 5, which is part of the larger peptide 7 fragment (Supplementary Table 3, Supplementary Fig. 5b). Although the reactivity in the peptide ELISA was low, given the high signal-to-noise, the data suggest that the 4H5B7 epitope is potentially located in peptide 7. We depleted peptide 5-specific antibodies from pooled NHP antiserum (post-NT135aaAnAPN1 immunization) and observed that anti-peptide 5 IgG does not confer transmission-blocking activity (Supplementary Fig. 5e), and that weak reactivity of 4H5B7 to peptide 5 in the peptide ELISA was not correlated with 4H5B7 transmission-blocking activity. However, depletion of peptide 7-specific IgG resulted in the loss of transmission-blocking activity (Fig. 6e, Supplementary Table 4). Using only 10 μg/mL of the peptide 7-specific IgG from that depletion experiment restored transmission-blocking activity (Fig. 6e). This was surprising in that we anticipated a reduction in oocyst number since we had not depleted peptide 9-specific IgG from the pooled serum (Supplementary Table 3), and we have shown that antibodies directed only to the peptide 9 epitope demonstrate potent transmission-blocking efficacy8. The 57% reduction in mean oocyst intensity by peptide 7-specific IgG (P-value = 0.31) was low compared to 95% reduction by peptide 9-specific IgG (P-value = 3.00 × 10-8); however in a follow-up study, reduction in mean oocyst intensity by peptide 7-specific IgG increased to 93% (Supplementary Fig. 5f, Supplementary Table 4). It is possible that anti-peptide 7 IgG may not all recognize the same critical sub-epitope or that IgG concentration could not be adequately controlled. In an independent study, total IgG from BALB/c mice immunized with peptide 7 significantly reduced P. falciparum oocyst intensity by 97.5% (Supplementary Fig. 5f, Supplementary Table 4). These results further suggest cooperative transmission-blocking activity between critical anti-peptide 7 and anti-peptide 9 IgG, when concentrations of either IgG in total IgG antisera are too low. Furthermore, the data suggest that peptide 4, which is part of the peptide 7 epitope (Supplementary Table 3), is likely the sub-epitope on peptide 7 that is recognized by 4H5B7 on native AnAPN1.
Peptide 7 appears to be highly conserved across Old and New World anophelines8. However, among lab populations of An. stephensi, differences in transmission-blocking activity of mouse anti-NT135aaAnAPN1 IgG suggest nuanced polymorphisms that may exist between laboratory populations (data not shown). We therefore tested 4H5B7 activity against P. falciparum NF54 in two An. stephensi strains: NIH and Nijmegen (Supplementary Fig. 5g,h, respectively), and found equivalent blocking activity between the strains (Supplementary Table 4). Moreover, given known genetic and phenotypic differences between laboratory and field parasites, we conducted a parallel study in Cameroon to confirm laboratory observations and observed complete transmission-blocking activity against naturally circulating strains of P. falciparum in the An. gambiae Ngousso strain (Fig. 6f, Supplementary Table 4). Clearly, if only a single specific antibody at concentrations greater than 10 μg/mL against either peptide 7 or 9 are used in these assays (Fig. 6d, 6f, Supplementary Fig. 5f, Supplementary Table 4), we can expect complete reduction in P. falciparum oocyst intensity. Unfortunately, humans do not elicit a single ‘monoclonal’ antibody response following vaccination and these results have clear implications in vaccine re-design.
Constructing an epitope map of domain I of AnAPN1
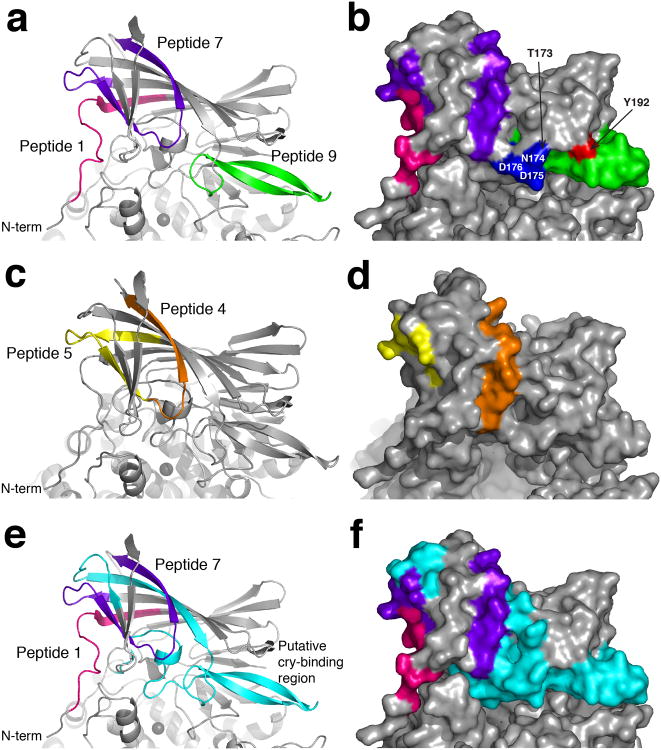
The antibodies described in this study recognized peptides 1, 4, 5, 7 and 9 (Supplementary Table 3) and these peptides were thus mapped to the AnAPN1 monomer (Fig. 7). The 2A12 mAb appears to bind peptide 1 and, as expected, lacks any transmission-blocking activity against P. falciparum. In the AnAPN1 structure, the majority of this peptide was unstructured (residues 60-68), but residues 69-74 were within the first β-strand of domain I and situated in a cleft formed within the β-barrel (Fig. 7a, b). Whilst the unstructured portion of peptide 1 is surface exposed, notably ectodomain residues 20-56, almost immediately preceding peptide 1, were absent in the structure and may alter solvent accessibility. Accordingly, 2A12 could bind native NT135aaAnAPN1 by ELISA (Fig. 6a, c) and denatured, near full-length AnAPN1 by western blot (data not shown). However, 2A12 did not bind native AnAPN1 in midgut lysates by ELISA (Fig. 6b).
Figure 7. Location of predicted B and T cell epitopes on domain I of AnAPN1.
Peptides 1, 7 and 9 are shown in pink, purple and green respectively in (a) cartoon and (b) surface representation. The surface exposed residues of the first 5 residues (blue) and the last 3 residues (red) of peptide 9 are labeled. Peptides 4 and 5, which together form peptide 7, are shown in orange and yellow respectively in (c) cartoon and (d) surface representation. Equivalent residues of the Cry1Aa toxin binding site on Bombyx mori (Supplementary Fig. 6) are mapped onto AnAPN1 in both (e) cartoon and (f) surface representation and shown in cyan. The Cry1Aa toxin binding site overlaps peptide 9 and is situated alongside peptide 7. Peptide 1 is shown in pink for reference.
The epitope of the transmission-blocking 4H5B7 mAb mapped to peptide 7 (residues 98-123), which incorporated peptides 5 (residues 95-109) and 4 (residues 107-121) (Fig. 7c, d). Whilst the N- (residues 95-109) and C-terminal (residues 116 onwards) regions of peptide 7 (mapping to peptide 5 and 4, respectively) were predominantly surface exposed, the central portion sat beneath and was occluded by the β6 and β7-strands of AnAPN1 domain I (Supplementary Fig.2, Fig. 7c, d). Peptide 7 doubled back on itself and had proximal N- and C-termini (closest distance 7.2 Å), which partly explains the peptide ELISA results (Fig. 7c, d).
Peptide 9 (residues 173-194) is a transmission-blocking epitope and anti-peptide 9 antibodies recognized the first five (residues 173-177) and last three residues (192-194) of peptide 9 as well as native and denatured protein8. Peptide 9 predominantly formed a β-hairpin, with immunoreactive residues 173-176 and 192 being surface exposed and proximal to one another (Fig. 7b). Peptide 9 flanked an opening of the active site and formed a small number of contacts with both catalytic domain II (specifically, hydrogen bonds to Asn285, Asp327 and Asp326) and domain IV, but as previously determined, anti-peptide 9 AnAPN1 antibodies do not interfere with AnAPN1 aminopeptidase activity8.
AnAPN1 structure defines a novel transmission-blocking epitope
APN molecules have been identified as Cry-binding proteins in lepidopteran (e.g. B. mori, P. xylostella) and dipteran (e.g. An. gambiae, Ae. aegypti) species19,20, suggesting a conserved mode of action for Cry toxins. Binding of Cry1Aa toxin to B. mori APN has been localized to residues 135-198 (B. mori numbering; NP_001037013.1)21, a region proposed as a common Cry-binding region in other APN isoforms22,23. We mapped the Cry-binding region of B. mori onto the AnAPN1 structure. Intriguingly, the homologous Cry-binding region entirely encompassed peptide 9 and directly abutted the two surface exposed regions of peptide 7, particularly the peptide 4 sub-epitope (Fig. 7e,f, Supplementary Fig. 6). This structure-guided observation provides a link between the two somewhat distal transmission-blocking epitopes within domain I (peptides 7 and 9) and suggests transmission-blocking anti-APN1 antibodies function by sterically hindering the binding of a Plasmodium pore-forming protein.
Discussion
Malaria remains an unabated global health burden and eradication requires the development of enabling tools and interventions such as TBVs. To maximize investment in TBVs, fundamental knowledge of immunogen structure is paramount, including identification of neutralizing, polymorphic and decoy epitopes that in turn can inform rational, efficacious vaccine design24-26.
To our knowledge, we are the first to report on the three-dimensional structure and enzymatic characterization of APN1 from adult An. gambiae. The overall structure resembled that of APNs from other species and exhibited classic four-domain architecture. The active site, accessible to solvent and harboring a peptide was described as open, with residue Tyr452 assuming a catalytically active position. X-ray fluorescence scans of the crystal revealed the presence of a zinc metal ion in the active site, and divalent cation-mediated activation was confirmed in vitro. As AnAPN1 was crystallized at pH 9.7, and despite the inward-facing orientation of Tyr452, we proposed that the AnAPN1 structure resembled a catalytically inactive state. This is supported by biochemical experiments with recombinant AnAPN1, which we found to be optimally active at pH 6.0–7.5, and evidence that the adult midgut at steady state has a pH range of 8-9.5 but seldom >10 (ref. 27). AnAPN1 is structurally most similar to porcine APN, including the orientation and placement of the highly variable domain IV, which influences the degree to which the active site cleft opens. Despite similarities in the solvent exposure of the porcine and AnAPN1 active sites, porcine APN1 was reported to be in the closed form and active based on mutagenesis, the conformation of the Tyr452 equivalent residue within the active site and the pH of crystallization16. Overall these two studies highlight the need to exercise caution in assigning APN enzymes, which are capable of adopting transient and intermediate states, to an open or closed state in which the enzyme is inactive or active, respectively. In further support of this notion an inward-facing active site tyrosine is not necessarily synonymous with catalytic activity.
Sedimentation velocity experiments confirmed that recombinant AnAPN1 (spanning residues 22-945) is a dimer in aqueous solution. Our predicted dimer has characteristics typical of biological assemblies such as symmetry, a large buried surface area and a statistically significant ΔiG value28,29. Importantly, in the dimer configuration, all peptides relevant to this study, including 1, 4, 5, 7 and 9, were exposed. None of the potential dimer interfaces observed in the AnAPN1 crystal structure corresponded to previously published aminopeptidase A (APA) or APN dimers; however, each of those published are also distinct16,17,30. The absence of the N-terminal 35 residues of the AnAPN1 ectodomain makes it difficult to predict the orientation of a putative AnAPN1 dimer on the membrane. Nevertheless, the structure defines the atomic coordinates of transmission-blocking AnAPN1 epitopes identified in this study, a pivotal step towards optimizing the AnAPN1 mTBV.
The 4H5B7 mAb blocked parasite development in mosquitoes at low concentrations and focused reactivity to peptides 5 and 7 by ELISA similar to NHP antiserum. The peptide ELISA is very sensitive but it does not quantify the concentration of specific IgG to a given linearized peptide in total IgG, it cannot capture antibodies against conformation-dependent epitopes, nor can it predict functional activity. As a result, we observed that a strong NHP IgG response to peptide 5 or the mAb 2A12 response to peptide 1 in this assay did not correlate with blocking activity. This is further borne out in our use of 2A12 as an isotype matched control IgG in the standard membrane feeding assays (SMFA). Moreover, we would have predicted that a strong peptide 7 and 9 response in the assay would not have resulted in a reduction in blocking activity when only peptide 7-specific IgG was depleted. These results suggest that peptide 1 and 5 are highly immunogenic decoy epitopes. Fortunately, the NHP recognition profile and transmission-blocking activity as a whole also suggest that peptide 7 alone can be targeted in different ways by antibodies and that IgG may be potentially directed to different sub-epitopes of peptide 7, beyond the arbitrary selection of peptides 4 and 5. Considering that only some peptide 7 residues are exposed, the structure suggests new sub-epitopes; straddling peptides 4 and 5 may be recognized. The data also supports the notion that the mechanism underlying the potent transmission-blocking activity of 4H5B7 is the recognition of a conformation-dependent epitope that is predominantly found on peptide 4. This is important to know since peptide 4 was predicted to be an immunogenic hotspot for a human humoral response8. By utilizing the structure of AnAPN1 we reveal peptides 7 and 9 map to the same surface of AnAPN1, but make no direct contacts and that cooperative binding of antibodies to these epitopes are critical for maximal transmission-blocking efficacy.
Bacillus thuringiensis
Cry toxins form pores in mosquito larval midguts, a process likely involving receptors such as cadherins, alkaline phosphatases, glucosidases and aminopeptidases20,31-36. A predicted common Cry-binding region in APN isoforms was determined in B. mori and mapped on the AnAPN1 structure. The Cry-binding region completely encompassed peptide 9 and sat adjacent to peptide 4, potentially linking these transmission-blocking epitopes. Although a portion of the Cry-binding region sits adjacent to peptide 5, antibodies to the peptide 5 sub-epitope do not have any blocking activity, presumably because they are distant from the main site. As Cry proteins are pore-forming toxins, we predict antibodies to peptides 4 and 9 block an ookinete micronemal pore-forming protein, which is a member of the Plasmodium Perforin-like Protein (PPLP) family37-40, from binding AnAPN1; critically influencing transmission by essentially preventing pore-formation in the midgut during cell invasion. Consistent with this hypothesis, antibodies to peptide 1, again remote from the above-described binding site, do not block P. falciparum, nor does the peptide 1-binding mAb 2A12 described herein8.
We know that outbred mice generate a vigorous humoral response to NT135aaAnAPN1 that is devoid of anti-peptide 7 or peptide 9 antibodies, and as would be expected, these antisera fail to block P. falciparum transmission8. Considering both peptides 1 and 5 are immune-decoy epitopes in outbred mice and NHPs, we are now compelled to re-examine the current immunogen design. Preclinical studies predominantly depend on mouse immunization studies to generate evidence to inform “go or no-go” decisions. We anticipate that in future studies of our candidate mTBV antigen, ELISAs that demonstrate a robust humoral response following immunization of outbred animals (as humans are outbred by definition) will only confirm what we already know about AnAPN1's potent immunogenicity, but such data has poor predictive value for functional activity. The structure-guided mapping of transmission-blocking epitopes on AnAPN1 strongly suggest that the next generation AnAPN1 immunogen should preclude peptides 1 and 5, and shift the B cell epitope window to immunofocus the humoral immune response to what we predict is a larger neo-epitope comprised of the sub-epitope peptides 7 and 9.
Online Methods
Cloning, expression and purification of near full-length AnAPN1
The near full-length coding sequence of An. gambiae AnAPN1 was amplified from midgut cDNA (pMTAPN1F 5′-TACCTACCATGGCCGCCATACAAGAGTAGTGGA-3′ and pMTAPN1R 5′-GATATGGCGGCCGCCTCGGCTAGGAAGTTGGACAG-3′) and cloned into the pMT-Bip-V5-His C Drosophila expression vector using NcoI and NotI restriction enzymes. Drosophila S2 cells (Life Technologies) were stably transfected with the expression vector using Effectene Transfection Reagent (Qiagen) and grown in the presence of hygromycin B (300 μg/mL). Expression was induced upon addition of copper sulfate (600 μM), and near full-length recombinant AnAPN1 (rFL-APN1) was recovered from the supernatant 24 hours later by concentration with PEG-8000 in Spectra/Por 2 dialysis sacks (12-14 kDa molecular weight cut off) (Spectrum Labs). The protein was subsequently purified using Ni-NTA agarose (Qiagen). For further purification, 50 μl of recombinant protein was directly injected onto a Bio SEC-3 (Agilent, 150 Å, 7.8×300 mm) HPLC size-exclusion column equilibrated with 150 mM sodium phosphate, pH 7.0 (Supplementary Fig. 7), followed by buffer exchange into 10 mM Tris, 150 mM NaCl, pH 7.5, using a 3 kDa Amicon filter (EMD Millipore, Billerica, MA). Purity was assessed via SDS-PAGE and purified AnAPN1 was subsequently used in functional analyses and crystallization trials.
Enzymatic characterization of the near full-length, S2-expressed AnAPN1
Aminopeptidase assays were performed as described previously41,42, using a final concentration of 50 nM recombinant near full-length AnAPN1 and 1 mM of L-leucine-p-nitroanilide (Sigma) in a total volume of 200 μl of APN buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl) at 25°C, unless described otherwise. Activity was measured spectrophotometrically by following the continuous increase in absorbance at 405 nm due to the release of 4-nitroaniline every 30 seconds over the course of 30 minutes. For determination of pH optimum, activity was measured in APN buffer adjusted to a pH between 6.0 and 9.5. The ability of various metal cations to re-activate AnAPN1 was determined following one hour incubation at room temperature with 10 mM 1,10-phenanthroline at final ion concentrations of 1 mM and 10 mM. The effect of cobalt, magnesium, and zinc (II) ions to reactivate AnAPN1 activity following incubation with 10 mM 1,10-phenanthroline was measured over a range of ions from 0–10 mM. Kinetics of AnAPN1 activity was determined in the presence of either 1 mM or 10 mM MnCl2 for a range of L-leucine-p-nitroanilide concentrations (0-4 mM).
Crystallization
Crystals of near full-length AnAPN1 (residues 22-942) were grown at 294K using the hanging drop vapour diffusion method. Crystals grew at 9.5 mg/mL in 0.1 M CHES pH 9.7, 22% (w/v) PEG 8000. Crystals, pre-equilibrated in crystallization solution containing 15% (v/v) glycerol as the cryoprotectant, were flash-cooled in a liquid nitrogen stream at 110K.
Structure determination and refinement
Data were collected (λ = 0.954, 110K) from a single crystal using an ADSC Q315r CCD detector on the MX2 micro-crystallography beamline at the Australian Synchrotron. The data were integrated and scaled with MOSFLM43 and SCALA, respectively, both within the CCP4 program suite44. The crystals belong to space group P212121 with unit cell dimensions a=113.48 Å, b=132.90 Å, c=146.01 Å, α=β=γ=90°, consistent with two AnAPN1 molecules per asymmetric unit. The structure of AnAPN1 was determined by molecular replacement using PHASER45 with uncomplexed porcine aminopeptidase-N (PDB ID 4FKE10) as the search model. Refinement of the AnAPN1 model to 2.65 Å was performed in REFMAC546 with iterative cycles of manual model building in COOT47. The quality of the model was monitored by the Rfree value, which represented 5% of the data. The structure was validated using the MolProbity server48 and figures created using PyMOL (http://www.pymol.org). Ramachandran statistics showed 94% of residues in the most favoured region, 4% in the additionally allowed regions and 1% in the disallowed region. Details of data processing statistic and final refinement values are summarized in Table 1.
Identification of active site metal ion
An X-ray fluorescence scan (5.5-15.5 keV) was performed to identify metal ions within the crystal. Both zinc and copper were identified, so complete anomalous diffraction data were collected at either end of the K-absorption edge for both Zn (λ = 1.281, 1.286) and Cu (λ = 1.378, 1.384) (Supplementary Table 1). Element specific anomalous difference DANO maps were calculated using both datasets and the CCP4 supported program FFT14.
Analytical ultracentrifugation
Sedimentation velocity experiments were performed in a Beckman Coulter model XL-I analytical ultracentrifuge. Double sector quartz cells were loaded with 400 μL of buffer (10 mM Tris pH 7.5, 150 mM NaCl) and 380 μL AnAPN1 at an initial concentration of 0.15 mg/mL. The cells were loaded into an An50-Ti rotor and left to equilibrate at 20°C. Initial scans were performed at 3,000 rpm to determine the optimal wavelength and radial positions for the experiment. Absorbance readings were subsequently collected every two mins at 230 nm and 40,000 rpm at 20°C. Solvent density, solvent viscosity and estimates of the partial specific volume of AnAPN1 (0.7325 mL/g) at 20°C were calculated using SEDNTERP49. Data were analysed using c(s) and c(M) using SEDFIT50,51.
Monoclonal and polyclonal antibody production
A panel of monoclonal antibodies were generated following immunization of female BALB/c mice (N=10) with E. coli expressed 135 amino acid N-terminal AnAPN1 (NT135aaAnAPN1) at GenScript (Piscataway, New Jersey) following their proprietary protocol. Three hybridomas were produced and the secreted antibodies were analyzed as described below. Five female Swiss Webster mice (6-8 weeks of age) were immunized with incomplete Freund's adjuvant (IFA) as a control or NT135aaAnAPN1 (N=10) were primed via subcutaneous injection and boosted three times at 2-week intervals via intraperitoneal injection with 5 μg/mL of NT135aaAnAPN1 in 15% sucrose, 10 mM Tris pH 7.85, 0.2% Tween 80 (ST/T80 buffer) emulsified (1:1) in IFA, as described previously8. All mice studies were approved by the Johns Hopkins Animal Care and Use Committee (MO12H232).
Non-human primate immunizations
Four, 10 year old female Macaca mulatta (India strain) NHPs were primed and boosted at 28 and 70 days post-priming with recombinant NT135aaAnAPN1 (0.1 mg/mL) in ST/T80 buffer, formulated with Alhydrogel (0.8 mg/mL) via intramuscular injection, as previously described8. NHP experiments were approved by the Johns Hopkins Animal Care and Use Committee (PR11H18). Anti-AnAPN1 serum titers were determined for each individual animal at various time points during the immunization regimen by ELISA.
BALB/c mouse immunizations
Five female BALB/c mice (7 weeks of age) were primed subcutaneously and subsequently boosted via intraperitoneal injection three times at two-week intervals with 5 μg/mL peptide 7 conjugated (1:1) in PBS with keyhole limpet hemocyanin (KLH) in a 1:1 emulsion with IFA. Five additional control mice were primed and boosted with 5 μg KLH in PBS emulsified 1:1 with IFA. Serum collected two weeks after the final boosting immunization was pooled for purification of total IgG.
ELISA and western blot assays
For ELISAs, 96-well Maxisorp microtiter plates (Nunc, Fisher Scientific) were coated with the NT135aaAnAPN1 or An. gambiae midgut lysate as previously described8. The optical density (O.D. 450 nm) and reciprocal serum dilutions were measured on a Molecular Devices absorbance microplate reader. End point titers were defined as the highest reciprocal serum dilution giving an O.D. reading greater than that of pre-immune serum plus 3 standard deviations. For immunoblots, NT135aaAnAPN1 or the near full-length S2-expressed AnAPN1 were resolved on a 4-20% Tris-Glycine SDS-PAGE gel and transferred to nitrocellulose membrane. The blot was incubated with 4H5B7 mAb (30 μg/mL) overnight at 4°C and then detected with goat anti-mouse IgG 680LT Ab (1:50,000) for 1 hour at room temp using the Li-Cor Odyssey Near-infrared imager (Li-Cor). Peptide epitope-mapping ELISAs were performed as described8.
Peptide-specific antibody purification
Protein G purified from pooled NHP IgG was fractionated for peptide 5, 7, and 9-specific IgG by affinity chromatography using Iodoacetyl Coupling Resin (Pierce) coupled to 1 mg of peptide. IgG was quantified by BCA protein assay (Pierce).
Immunofluorescence Assays
Transverse cryosections of 5-6 day old, sugar-fed, female Anopheles gambiae midguts on a slide were blocked with 2% BSA for 30 minutes and then probed with 4H5B7 mAb (10 μg/mL) and detected by Alexa Fluor 488-conjugated anti-mouse secondary antibodies (Life Technologies, Frederick, MD), as previously described7. Images were acquired using a Zeiss Axioskop 2 fluorescence compound microscope fitted with a ProgRes MF camera and images were acquired and analyzed by CapturePro Software, v. 2.9 (Jenoptik, Jupiter, FL).
Standard and Direct Membrane Feeding Assays
Standard membrane feeding assays (SMFA) and Direct Membrane Feeding Assays (DMFA) were performed as described previously8 and fed to An. gambiae (Keele or Ngousso) or An. stephensi (NIH or Nijmegen) mosquitoes. Infective blood was mixed with control (normal mouse or pre-immune IgG) or human AB serum only for DMFAs, anti-AnAPN1 IgG (final concentration of 4–100 μg/ml), mouse total IgG (500 μg/ml), or peptide-specific IgG (10 μg/ml) prior to delivery into water-jacketed membrane feeders maintained at 37°C. In general, SMFA and DMFA data were analyzed by a zero-inflated Generalized Linear Mixed-Model statistical model (GLMM)52, and when inappropriate for a comparison, by the non-parametric Mann Whitney U test (Supplementary Table 4).
P. falciparum gametocyte carriers (children ages 5 to 11) were from the Mfou district in Cameroon and were enrolled in the study upon receiving informed consent from their legal guardians. The experimental procedures were approved by the National Ethics Committee of Cameroon (46/CNE/SE/2012). Infective, venous blood was collected and prepared as described previously8. DMFAs were performed using mAb 4H5B7 and diluted in non-immune human AB serum. AB serum alone was used as a control. 4H5B7 or AB serum was added directly to the infective blood meal prior to feeding to mosquitoes through a membrane feeder. No statistical difference was observed in oocyst development following feeding of infected blood with either pre-immune or AB serum in pilot experiments. In each experiment, mAb concentrations between 0.5-50 μg/mL were tested against P. falciparum isolated from a single carrier. Colony mosquitoes, established from a field-caught founding population of An. gambiae (Ngousso) mosquitoes, were used in Cameroon.
Supplementary Material
Acknowledgments
We thank the staff at the MX2 beamline of the Australian Synchrotron for assistance with X-ray data collection and the Monash Macromolecular Crystallization Facility for initial crystallization experiments. We also thank Y. Mok at the Macromolecular Interactions Facility at the University of Melbourne for assistance with sedimentation velocity experiments and J. Plieskatt at George Washington University for valuable comments and suggestions. We thank P. Eggleston and H. Hurd for the An. gambiae KEELE strain. Last but not least, we thank the children, parents and the community of Mfou for their eager participation in this study. The work was funded in part by the PATH-Malaria Vaccine Initiative and Bloomberg Family Foundation through the Johns Hopkins Malaria Research Institute (to R.R.D.). J.S.A. was supported as a Johns Hopkins Malaria Research Institute predoctoral fellow and D.K.M. was supported as a Calvin and Helen Lang Postdoctoral Fellow in the Biological Sciences. M.M.S. is supported by an Institut de Recherche pour le Développement Fellowship. B.B.T is funded by the Ifakara Health Institute. S.C.A is supported by an Australian National Health and Medical Research Council Early Career Fellowship (1072267). N.A.B is supported by an Australian Research Council Future Fellowship (110100223). This publication was also made possible by the NIH National Center for Research Resources (UL1 RR 025005).
Footnotes
Accession Codes: AnAPN1 atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID 4WZ9).
Author Contributions: S.C.A, J.S.A, D.K.M, M.M.S, D.T, N.B.D, and I.M designed and performed experiments. S.C.A, J.S.A, R.R.D and N.A.B analyzed structural and biochemical data. B.B.T, I.M, and R.R.D analyzed all functional data. S.C.A, J.SA, R.R.D and N.A.B led the manuscript preparation with critical contributions from all authors.
References
- 1.Vannice KS, Brown GV, Akanmori BD, Moorthy VS. MALVAC 2012 scientific forum: accelerating development of second-generation malaria vaccines. Malar J. 2012;11:372. doi: 10.1186/1475-2875-11-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.malERA Consultative Group on Vaccines, T. A Research Agenda for Malaria Eradication: Vaccines. PLoS Med. 2011;8:e1000398. doi: 10.1371/journal.pmed.1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinden RE, Carter R, Drakeley C, Leroy D. The biology of sexual development of Plasmodium: the design and implementation of transmission-blocking strategies. Malar J. 2012;11:70. doi: 10.1186/1475-2875-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinglasan RR, Jacobs-Lorena M. Flipping the paradigm on malaria transmission-blocking vaccines. Trends Parasitol. 2008;24:364–70. doi: 10.1016/j.pt.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter R. Transmission blocking malaria vaccines. Vaccine. 2001;19:2309–2314. doi: 10.1016/s0264-410x(00)00521-1. [DOI] [PubMed] [Google Scholar]
- 6.Saul A. Efficacy model for mosquito stage transmission blocking vaccines for malaria. Parasitology. 2008;135:1497–506. doi: 10.1017/S0031182008000280. [DOI] [PubMed] [Google Scholar]
- 7.Mathias DK, et al. Differential roles of an Anopheline midgut GPI-anchored protein in mediating Plasmodium falciparum and Plasmodium vivax ookinete invasion. Infect Genet Evol. 2014 doi: 10.1016/j.meegid.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armistead JS, et al. Antibodies to a single, conserved epitope in Anopheles APN1 inhibit universal transmission of Plasmodium falciparum and Plasmodium vivax malaria. Infect Immun. 2014;82:818–29. doi: 10.1128/IAI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinglasan RR, et al. Disruption of Plasmodium falciparum development by antibodies against a conserved mosquito midgut antigen. Proc Natl Acad Sci USA. 2007;104:13461–6. doi: 10.1073/pnas.0702239104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Farquhar ER, Chance MR, Palczewski K, Kiser PD. Insights into substrate specificity and metal activation of mammalian tetrahedral aspartyl aminopeptidase. J Biol Chem. 2012;287:13356–70. doi: 10.1074/jbc.M112.347518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGowan S, et al. Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase. Proc Natl Acad Sci USA. 2009;106:2537–42. doi: 10.1073/pnas.0807398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kochan G, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci USA. 2011;108:7745–50. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen TT, et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat Struct Mol Biol. 2011;18:604–13. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Than ME, et al. The endoproteinase furin contains two essential Ca2+ ions stabilizing its N-terminus and the unique S1 specificity pocket. Acta Crystallogr D. 2005;61:505–12. doi: 10.1107/S0907444905002556. [DOI] [PubMed] [Google Scholar]
- 15.Luciani N, et al. Characterization of Glu350 as a critical residue involved in the N-terminal amine binding site of aminopeptidase N (EC 3.4.11.2): insights into its mechanism of action. Biochem. 1998;37:686–92. doi: 10.1021/bi971705p. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, Lin YL, Peng G, Li F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc Natl Acad Sci USA. 2012;109:17966–71. doi: 10.1073/pnas.1210123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J Biol Chem. 2012;287:36804–13. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Billker O, Miller AJ, Sinden RE. Determination of mosquito bloodmeal pH in situ by ion-selective microelectrode measurement: implications for the regulation of malarial gametogenesis. Parasitol. 2000;120:547–51. doi: 10.1017/s0031182099005946. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, et al. Aedes aegypti cadherin serves as a putative receptor of the Cry11Aa toxin from Bacillus thuringiensis subsp. israelensis. Biochem J. 2009;424:191–200. doi: 10.1042/BJ20090730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hua G, Zhang R, Abdullah MAF, Adang MJ. Anopheles gambiae Cadherin AgCad1 Binds the Cry4Ba Toxin of Bacillus thuringiensis israelensis and a Fragment of AgCad1 Synergizes Toxicity. Biochem. 2008;47:5101–5110. doi: 10.1021/bi7023578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yaoi K, et al. Bacillus thuringiensis Cry1Aa toxin-binding region of Bombyx mori aminopeptidase N. FEBS Lett. 1999;463:221–224. doi: 10.1016/s0014-5793(99)01626-9. [DOI] [PubMed] [Google Scholar]
- 22.Zhuang M, et al. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J Biol Chem. 2002;277:13863–72. doi: 10.1074/jbc.M110057200. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi K, et al. Aminopeptidase N isoforms from the midgut of Bombyx mori and Plutella xylostella – their classification and the factors that determine their binding specificity to Bacillus thuringiensis Cry1A toxin. FEBS Lett. 2002;519:215–220. doi: 10.1016/s0014-5793(02)02708-4. [DOI] [PubMed] [Google Scholar]
- 24.Lim SS, et al. Structure and Dynamics of Apical Membrane Antigen 1 from Plasmodium falciparum FVO. Biochem. 2014 doi: 10.1021/bi5012089. [DOI] [PubMed] [Google Scholar]
- 25.Chuang GY, Liou D, Kwong PD, Georgiev IS. NEP: web server for epitope prediction based on antibody neutralization of viral strains with diverse sequences. Nucleic Acids Res. 2014;42:W64–71. doi: 10.1093/nar/gku318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delany I, Rappuoli R, De Gregorio E. Vaccines for the 21st century. EMBO Mol Med. 2014;6:708–20. doi: 10.1002/emmm.201403876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.del Pilar Corena M, et al. Carbonic anhydrase in the adult mosquito midgut. J Exp Biol. 2005;208:3263–3273. doi: 10.1242/jeb.01739. [DOI] [PubMed] [Google Scholar]
- 28.Krissinel E, Henrick K. Inference of Macromolecular Assemblies from Crystalline State. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Bahadur RP, Zacharias M. The interface of protein-protein complexes: Analysis of contacts and prediction of interactions. Cell Mol Life Sci. 2008;65:1059–1072. doi: 10.1007/s00018-007-7451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Liu C, Lin YL, Li F. Structural insights into central hypertension regulation by human aminopeptidase A. J Biol Chem. 2013;288:25638–45. doi: 10.1074/jbc.M113.494955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SB, Chen J, Aimanova KG, Gill SS. Aedes cadherin mediates the in vivo toxicity of the Cry11Aa toxin to Aedes aegypti. Peptides. 2014 doi: 10.1016/j.peptides.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim MA, Griko NB, Bulla LA., Jr Cytotoxicity of the Bacillus thuringiensis Cry4B toxin is mediated by the cadherin receptor BT-R(3) of Anopheles gambiae. Exp Biol Med (Maywood) 2013;238:755–64. doi: 10.1177/1535370213493719. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q, Hua G, Bayyareddy K, Adang MJ. Analyses of alpha-amylase and alpha-glucosidase in the malaria vector mosquito, Anopheles gambiae, as receptors of Cry11Ba toxin of Bacillus thuringiensis subsp. jegathesan. Insect Biochem Mol Biol. 2013;43:907–15. doi: 10.1016/j.ibmb.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 34.Hua G, Zhang R, Bayyareddy K, Adang MJ. Anopheles gambiae alkaline phosphatase is a functional receptor of Bacillus thuringiensis jegathesan Cry11Ba toxin. Biochem. 2009;48:9785–93. doi: 10.1021/bi9014538. [DOI] [PubMed] [Google Scholar]
- 35.Zhang R, Hua G, Andacht TM, Adang MJ. A 106-kDa aminopeptidase is a putative receptor for Bacillus thuringiensis Cry11Ba toxin in the mosquito Anopheles gambiae. Biochem. 2008;47:11263–72. doi: 10.1021/bi801181g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Likitvivatanavong S, Chen J, Bravo A, Soberón M, Gill SS. Cadherin, Alkaline Phosphatase, and Aminopeptidase N as Receptors of Cry11Ba Toxin from Bacillus thuringiensis subsp. jegathesan in Aedes aegypti. Appl Environ Microbiol. 2011;77:24–31. doi: 10.1128/AEM.01852-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lal K, et al. Characterisation of Plasmodium invasive organelles; an ookinete microneme proteome. Proteomics. 2009;9:1142–1151. doi: 10.1002/pmic.200800404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ecker A, Bushell ES, Tewari R, Sinden RE. Reverse genetics screen identifies six proteins important for malaria development in the mosquito. Mol microbiol. 2008;70:209–220. doi: 10.1111/j.1365-2958.2008.06407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ecker A, Pinto SB, Baker KW, Kafatos FC, Sinden RE. Plasmodium berghei: Plasmodium perforin-like protein 5 is required for mosquito midgut invasion in Anopheles stephensi. Exp parasitology. 2007;116:504–508. doi: 10.1016/j.exppara.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kadota K, Ishino T, Matsuyama T, Chinzei Y, Yuda M. Essential role of membrane-attack protein in malarial transmission to mosquito host. Proc Natl Acad Sci USA. 2004;101:16310–16315. doi: 10.1073/pnas.0406187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua G, Tsukamoto K, Rasilo ML, Ikezawa H. Molecular cloning of a GPI-anchored aminopeptidase N from Bombyx mori midgut: a putative receptor for Bacillus thuringiensis CryIA toxin. Gene. 1998;214:177–85. doi: 10.1016/s0378-1119(98)00199-1. [DOI] [PubMed] [Google Scholar]
- 42.Banks DJ, Hua G, Adang MJ. Cloning of a Heliothis virescens 110 kDa aminopeptidase N and expression in Drosophila S2 cells. Insect Biochem Mol Biol. 2003;33:499–508. doi: 10.1016/s0965-1748(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 43.Leslie AW, Powell H. Processing diffraction data with mosflm. In: Read R, Sussman J, editors. Evolving Methods for Macromolecular Crystallography. Vol. 245. Springer; Netherlands: 2007. pp. 41–51. [Google Scholar]
- 44.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr Section D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murshudov GN, Vagin AA, Dodson EJ. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laue T, Shah B, Ridgeway T, Pelletier S. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Soc. Chem.; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 50.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–61. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Churcher TS, et al. Measuring the blockade of malaria transmission– an analysis of the Standard Membrane Feeding Assay. Int J Parasitol. 2012;11:1037–44. doi: 10.1016/j.ijpara.2012.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.