Sir,
A recent study by Bannwarth et al. (2014) implicated CHCHD10 as a novel gene for amyotrophic lateral sclerosis/frontotemporal lobar degeneration (ALS/FTLD), reporting a p.S59L substitution (c.176C > T; NM_213720.2) in a large French kindred. Affected family members were presented with a complex phenotype that included symptoms of amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD), cerebellar ataxia, Parkinson’s disease and a mitochondrial myopathy associated with multiple mitochondrial DNA deletions. So far, seven missense CHCHD10 mutations have been reported in patients with a broad phenotypic range, including ALS/FTLD (p.S59L and p.P34S) (Bannwarth et al., 2014; Chaussenot et al., 2014), ALS (p.R15L and p.G66V) (Johnson et al., 2014; Muller et al., 2014), myopathy (p.R15S and p.G58R) (Ajroud-Driss et al., 2015) and late-onset spinal motor neuronopathy (p.G66V) (Penttila et al., 2015). All of them affect exon 2 (a mutational hotspot of CHCHD10).
Notably, mitochondrial dysfunction has been implicated in several neurodegenerative diseases (Lin and Beal, 2006; Cozzolino et al., 2013); however, there are no studies evaluating the contribution of CHCHD10 to pure FTLD, Parkinson’s disease or Alzheimer’s disease. Hence, we sequenced CHCHD10 exon 2 in 204 ALS, 153 Parkinson’s disease and 141 Alzheimer’s disease patients from Canada and 158 FTLD patients from Italy in addition to 497 control subjects from USA/UK, Canada and Italy. The cases of ALS and FTLD were free from mutations in SOD1, GRN, FUS, TARDBP and MATR3 or a repeat expansion in C9orf72.
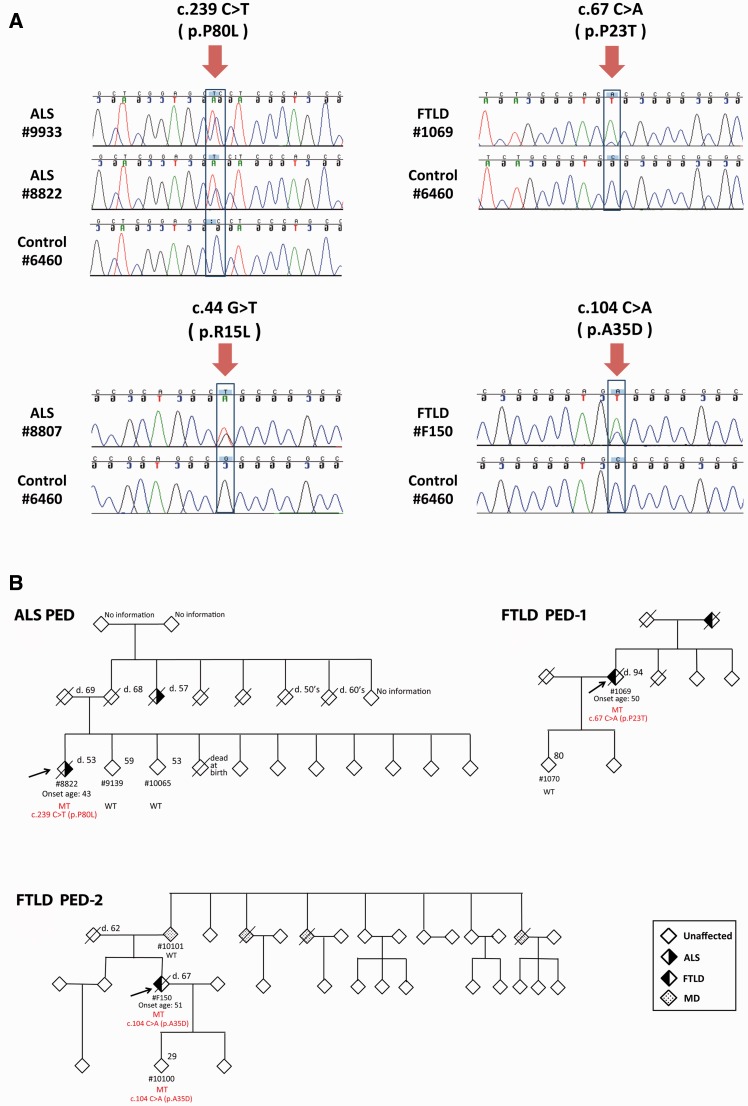
We identified a known CHCHD10 pathogenic p.R15L mutation (Johnson et al., 2014; Muller et al., 2014) in a patient with sporadic ALS (Patient 8807) (Fig. 1A), who developed symptoms involving his upper limb at 54 years of age and remains alive 12 years later. The p.R15L mutation was absent in our control samples and public databases, further indicating its pathogenic role in ALS. We also detected a heterozygous substitution (c.239C > T, p.P80L) in a patient with familial ALS (Patient 8822) of German/Dutch origin (Fig. 1A), who initially developed symptoms in the upper limbs at the age of 43 years and had ALS for 9 years. The p.P80L mutation was not present in unaffected siblings Patients 9139 and 10065, aged 59 and 53 years, respectively (Fig. 1B). Moreover, the same p.P80L mutation was identified in a patient with sporadic ALS (Patient 9933, Fig. 1A), who developed bulbar symptoms at the age of 58 years. Genotypes of eight polymorphic microsatellite markers (D22S420, D22S539, D22S1174, D22S1174, D22S315, D22S1154, D22S1163 and D22S280) revealed that Patients 8822 and 9933 share a haplotype stretching from 6.4 Mb upstream to 9 Mb downstream of CHCHD10, suggesting a common ancestor. The p.P80L mutation changes a conserved amino acid (proline) and is predicted to be disease-causing (MutationTaster). It was absent in our control subjects, Single Nucleotide Polymorphism Database (build 141), 1000 Genome project (n = 5006 chromosomes) and NHLBI Exome Sequencing project (n = 13 006 chromosomes). It was found only in the ExAC database (n = 121 412 chromosomes), but at a much lower frequency (minor allele frequency = 0.0003) compared to our ALS cohort (minor allele frequency = 0.005). Furthermore, our segregation analysis revealed the p.P80L mutation only in an affected family member (Fig. 1B). Importantly, in the process of preparing this report, the p.P80L mutation was independently identified in two Italian patients with sporadic ALS and muscle mitochondrial pathology (Ronchi et al., 2015). These data collectively support the pathogenic role of p.P80L substitution.
Figure 1.
Sequencing results and pedigrees of CHCHD10 mutation carriers. (A) Sequencing results detecting four CHCHD10 mutations. A c.239C > T (p.P80L) mutation was found in a patient with sporadic ALS (Patient 9933) and a patient with familial ALS (Patient 8822). A c.44G > T (p.R15L) mutation was found in a patient with sporadic ALS Patient 8807. Novel mutations were found in a patient with sporadic FTLD (c.67C > A, p.P23T, Patient F150), and a patient with familial FTLD (c.104C > A, p.A35D, Patient 1069). (B) Three pedigrees of CHCHD10 mutation carriers. The arrow indicates index patient. Detailed information of CHCHD10 mutation is shown in red font beneath the diamond symbol of the corresponding carrier. MT = mutant allele; WT = wild-type allele; patterned diamond symbols = individuals with ALS, FTLD or major depression (MD).
Among the cohort with FTLD, we detected two novel heterozygous CHCHD10 substitutions in two FTLD cases of Italian origin, including a p.P23T (c.67C > A) mutation in a patient with familial FTLD (Patient 1069, Fig. 1) and a p.A35D (c.104C > A) mutation in a patient with sporadic FTLD (Patient F105, Fig. 1). The p.A35D variant alters a conserved amino acid and is predicted to be damaging (MutationTaster), whereas the p.P23T variant changes a less conserved amino acid. Both mutations were absent in our control samples and public databases.
Patient 1069 may have inherited the p.P23T substitution from their mother affected by FTLD. At age 50 years, the proband had mild memory lapses and disorientation in time. At 82 years of age, the patient manifested impaired planning, absence of critical thinking and spatial disorientation. At age 84 years, the patient became aggressive, delirious, had visual hallucinations, dressing apraxia and incontinence; and a year later, signs of bradykinesia appeared. Aged 88 years, the patient had bulimia, reduced verbal fluency, hypersomnia, disinhibition, confabulation, mutism, inertia and hyperreflexia. Rigidity and focal signs were not observed. Mini-Mental State Examination score was 15/30. The patient died at 94 years of age. The proband’s son (Patient 1070) did not inherit the p.P23T mutation and does not have FTLD at age 80 years (Fig. 1B).
The p.A35D carrier (Patient F150) had initial symptoms at 51 years of age with progressive withdrawal from social activities (without effect from antidepressants for 2 years). The patient subsequently developed subtle memory complaints and behavioural disturbances; and 5 years later was diagnosed with behavioural variant FTLD. Mini-Mental State Examination indicated moderate dementia (15/30). MRI revealed asymmetrical frontotemporal atrophy, and cerebral 99 mTc SPECT showed reduced tracer uptake in the frontal and temporal region. At the last visit, mild axial extrapyramidal signs were observed. The patient died at age 67 years secondary to pneumonia (FTLD disease duration = 16 years). The patient did not have familial history of FTLD or other neurodegenerative diseases (Fig. 1). The mother did not carry the p.A35D mutation, suggesting that this variant was likely inherited from the father, who died of lung cancer at 62 years of age.
Finally, we detected a heterozygous p.P34S substitution (c.100 C > T) in a Canadian patient with Parkinson’s disease (Patient 8625) of Norwegian origin with no symptoms of muscle weakness, dementia or ALS (minor allele frequency = 0.0028). We also identified the p.P34S variant in two cases of Alzheimer’s disease (Patients 2205 and 1249) (minor allele frequency = 0.0071), which did not segregate with Alzheimer’s disease phenotype in the available family. The p.P34S variant was previously reported in two unrelated French patients with FTLD/ALS (Chaussenot et al., 2014) and one Italian patient with sporadic ALS (Ronchi et al., 2015). However, the p.P34S substitution was found at comparable frequencies in our control samples (minor allele frequency = 0.0074), the 1000 Genome project (minor allele frequency = 0.001) and the ExAC database (minor allele frequency = 0.003), suggesting that this variant is not pathogenic.
CHCHD10 encodes a protein enriched at cristae junctions of mitochondria (Bannwarth et al., 2014). Mutations in CHCHD10 may disrupt the morphology of mitochondrial cristae, affect the stability of mitochondrial DNA, induce mitochondrial fragmentation, and reduce mitochondrial complex-IV activities (Bannwarth et al., 2014; Ajroud-Driss et al., 2015). Two novel variants identified in the current study, p.A35D and p.P23T, affect amino acids in the non-structured N-terminal region of CHCHD10 protein. Therefore, they may affect mitochondria by disrupting protein stability or localization (Chaussenot et al., 2014). Further functional studies are needed to confirm their pathogenic roles.
Among the identified mutation carriers, three died and all presented with slow disease progression regardless of disease phenotype (disease duration: ALS = 9 years; FTLD = 16 and 44 years). Moreover, the two living mutation carriers have ongoing disease durations of >3 and >12 years. These findings agree with the previous observation that CHCHD10 mutations are associated with slow progression and long disease duration (6–17 years) (Muller et al., 2014). In the current study, carriers developed ALS in their 40s or 50s (43, 54 and 58 years of age), similar to the reported range of 50–60 years in other CHCHD10 mutation carriers (Bannwarth et al., 2014; Chaussenot et al., 2014). The site of ALS onset varied among patients carrying the same p.P80L mutation, with upper limb-onset and predominant bulbar-onset observed in the current study, and flail arm-onset and bulbar onset observed in a recent study (Ronchi et al., 2015). These findings argue against a previous suggestion that CHCHD10 mutation carriers may have a restricted site of onset (Bannwarth et al., 2014; Chaussenot et al., 2014; Muller et al., 2014).
In conclusion, our data revealed two novel mutations (p.P23T and p.A35D) in two Italian patients with FTLD and two known mutations (p.P80L and p.R15L) in Canadian ALS patients, but no pathogenic mutations in patients with Alzheimer’s disease or Parkinson’s disease. Notably, our study does not support the pathogenic nature of the p.P34S variant, which is important in the utility of genetic screening in patient care. The estimated mutation frequencies in our data sets are 2.6% for familial ALS, 1.2% for sporadic ALS, 1.6% for familial FTLD and 1% for sporadic FTLD. All mutation carriers are characterized by atypically long duration and slow disease progression. Our study supports the causal role of CHCHD10 mutations in both ALS and FTLD, suggesting a common mechanism of mitochondrial dysfunction. It is conceivable that other proteins structurally similar to CHCHD10 could also contribute to neurodegenerative disorders. In agreement with such a possibility, mutations in CHCHD2 were recently reported to cause Parkinson's disease in Japanese patients (Funayama et al., 2015).
Funding
This work was supported by the W. Garfield Weston Foundation (E.R.), and Cassa di Risparmio di Pistoia e Pescia (CRPT 2013/0347) (B.N.), Cassa di Risparmio di Firenze (CRF 2013/0199), the Ministry of Health RF-2010-2319722 (S.S.), the Canadian Institutes of Health Research, Wellcome Trust, Medical Research Council, National Institute of Health Research, Ontario Research Fund and Alzheimer Society of Ontario (P.S.G-H).
References
- Ajroud-Driss S, Fecto F, Ajroud K, Lalani I, Calvo SE, Mootha VK, et al. Mutation in the novel nuclear-encoded mitochondrial protein CHCHD10 in a family with autosomal dominant mitochondrial myopathy. Neurogenetics. 2015;16:1–9. doi: 10.1007/s10048-014-0421-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannwarth S, Ait-El-Mkadem S, Chaussenot A, Genin EC, Lacas-Gervais S, Fragaki K, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137(Pt 8):2329–45. doi: 10.1093/brain/awu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaussenot A, Le Ber I, Ait-El-Mkadem S, Camuzat A, de Septenville A, Bannwarth S, et al. Screening of CHCHD10 in a French cohort confirms the involvement of this gene in frontotemporal dementia with amyotrophic lateral sclerosis patients. Neurobiol Aging. 2014;35:2884.e1–4. doi: 10.1016/j.neurobiolaging.2014.07.022. [DOI] [PubMed] [Google Scholar]
- Cozzolino M, Ferri A, Valle C, Carri MT. Mitochondria and ALS: implications from novel genes and pathways. Mol Cell Neurosci. 2013;55:44–9. doi: 10.1016/j.mcn.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Funayama M, Ohe K, Amo T, Furuya N, Yamaguchi J, Saiki S, et al. CHCHD2 mutations in autosomal dominant late-onset Parkinson's disease: a genome-wide linkage and sequencing study. Lancet Neurol. 2015;14:274–82. doi: 10.1016/S1474-4422(14)70266-2. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Glynn SM, Gibbs JR, Nalls MA, Sabatelli M, Restagno G, et al. Mutations in the CHCHD10 gene are a common cause of familial amyotrophic lateral sclerosis. Brain. 2014;137(Pt 12):e311. doi: 10.1093/brain/awu265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–95. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Muller K, Andersen PM, Hubers A, Marroquin N, Volk AE, Danzer KM, et al. Two novel mutations in conserved codons indicate that CHCHD10 is a gene associated with motor neuron disease. Brain. 2014;137(Pt 12):e309. doi: 10.1093/brain/awu227. [DOI] [PubMed] [Google Scholar]
- Penttila S, Jokela M, Bouquin H, Saukkonen AM, Toivanen J, Udd B. Late onset spinal motor neuronopathy is caused by mutation in CHCHD10. Ann Neurol. 2015;77:163–72. doi: 10.1002/ana.24319. [DOI] [PubMed] [Google Scholar]
- Ronchi D, Riboldi G, Del Bo R, Ticozzi N, Scarlato M, Galimberti D, et al. CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain. 2015 doi: 10.1093/brain/awu384. Advance Access published on January 9, 2015, doi: 10.1093/brain/awu384. [DOI] [PubMed] [Google Scholar]