Abstract
The fluid mosaic model of Singer and Nicolson correctly predicted that the plasma membrane (PM) forms a lipid bi-layer containing many integral trans-membrane proteins. This model also suggested that most of these proteins were randomly dispersed and freely diffusing moieties. Initially, this view of a dynamic and rather unorganized membrane was supported by early observations of the cell surfaces using the light microscope. However, recent studies on the PM below the diffraction limit of visible light (~ 250 nm) revealed that, at nanoscale dimensions, membranes are highly organized and compartmentalized structures. Lymphocytes are particularly useful to study this nanoscale membrane organization because they grow as single cells and are not permanently engaged in cell:cell contacts within a tissue that can influence membrane organization. In this review, we describe the methods that can be used to better study the protein:protein interaction and nanoscale organization of lymphocyte membrane proteins, with a focus on the B cell antigen receptor (BCR). Furthermore, we discuss the factors that may generate and maintain these membrane structures.
Abbreviations: BN-PAGE, blue native polyacrylamide gel electrophoresis; PM, plasma membrane; TM, transmembrane; dSTORM, direct stochastic optical reconstruction microscopy; PALM, photoactivated localization microscopy; TEM, transmission electron microscopy; PLA, proximity ligation assay; BCR, B cell antigen receptor; Syk, spleen tyrosine kinase
Keywords: B cell antigen receptor, Nanocluster, Protein islands
Highlights
-
•
Fab-PLA, a new method to study principles of nanoscale organization of the PM
-
•
Nanoscale protein island organization of resting B cells alters upon activation
-
•
BCR dissociation and signal amplification involves Syk-mediated inside-out signaling
1. Introduction
The plasma membrane (PM) of eukaryotes is composed of several hundred lipid species and thousand of different proteins. Approximately 30% of all cellular proteins are membrane proteins [1–3]. The composition and complexity of biological membranes varies, not only between different cell types, but also between the PM and the membranes of different intracellular organelles [4]. The Singer and Nicolson fluid mosaic model predicts that all proteins and lipids are equally distributed over a biological membrane [5]. This view was challenged by the discovery of a compartmentalization of the PM into different lipid domains [6,7]. At micrometer (μm) distances, specialized structures of the PM such as membrane ruffles [8], primary cilia and synapses have already been discovered with the light microscope and have been attributed to different functions [9,10]. However, at nanometer (nm) distances, a much higher organization of membranes has been observed, suggesting that most membrane proteins are localized in specific compartments that are variably be called as nanoclusters, nanoislands or protein islands [11–13].
The B cell antigen receptor (BCR) plays a central role in the antigen-specific activation of B-lymphocytes leading to the production of antibodies. We have studied the nanoscale organization of the BCR and its coreceptors on the surface of B-lymphocytes at 10 to 20 nm distances [11]. These studies show that the two different antigen receptor classes on mature B cells, namely the IgM-BCR and the IgD-BCR, reside in different protein islands with a distinct protein and lipid composition. B-lymphocytes can be easily isolated and activated by well established activation protocols [14]. This makes these cells an ideal object for the study of the nanoscale organization of membrane proteins and their alteration during cell activation. Here we describe the methods used to study the organization of biological membranes in Section 2 and summarize the current knowledge of PM nanoscale organization in Section 3. This is then followed, in Section 4, by a discussion of the molecular arrangement of the B cell PM and its alteration during the activation.
2. Methods to study nanoscale membrane organizations
The proteins inside membrane nanoclusters are restrained only by weak intermolecular protein:protein and protein:lipid interactions [15,16]. Often these interactions exist in dynamic equilibrium of associated and dissociated states [17,18]. The mechanism that establishes this dynamic equilibrium depends on the type of membrane protein and the constituents of lipid bi-layer. For example, the syntaxin-1 containing nanodomains on neuroendocrine cells are maintained by a critical balance between self-association and steric repulsion [17]. Furthermore, rhodopsins on artificial membrane assemble by transient dimerization [19]. The isolation of these protein nanoclusters and their biochemical characterization is challenging. Depending on the detergents used for permeabilization and solubilization of the membrane and subsequent purification protocol, one can get very different information about the protein:protein interactions within a particular membrane. In addition, the purification of native membrane proteins for biochemical studies requires large amounts of starting materials and highly specific antibodies raised against the membrane proteins involved in nanocluster formation [20]. Therefore, in recent years, live cell imaging techniques employing the confocal microscope and fluorescent protein-tagged (FP-tagged) proteins were frequently used to study the composition and dynamics of proteins on the membrane [21,22]. However, due to the diffraction barrier of visible light at 250 nm [23], most of these confocal microscopy studies failed to reveal the nanoscale organization of proteins in biological membranes (Fig. 1A, upper panel). Thus a better understanding and study of the nano-world of membrane organization requires the development and combination of novel techniques. In the following sections we describe how improved biochemical methods, together with super-resolution approaches and proximity ligation assays (PLA) can be employed for a better study of membrane nano-world.
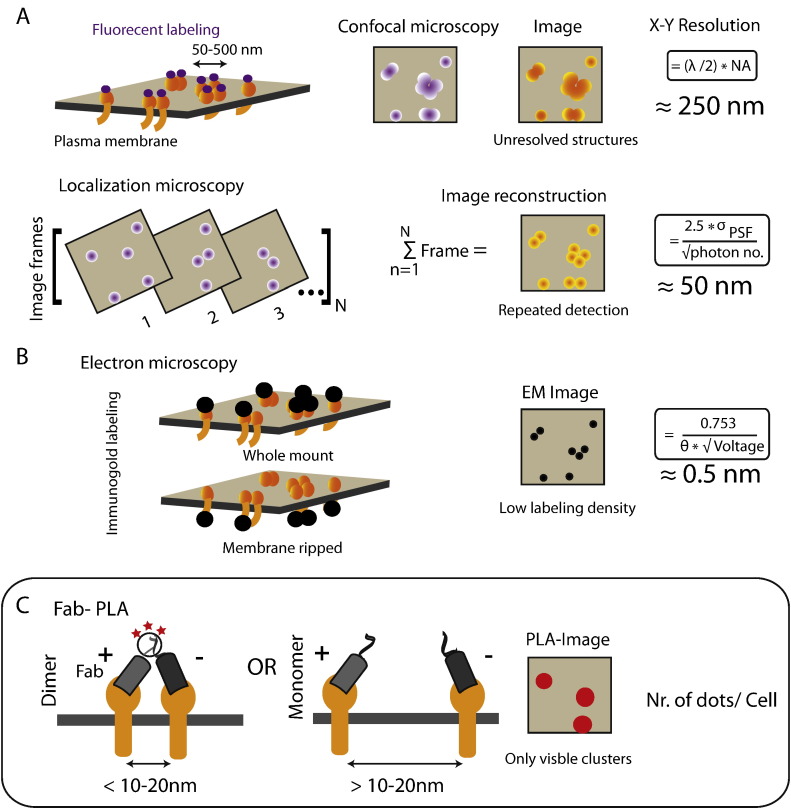
Fig. 1.
Nanoscale detection of membrane protein complexes using different microscopy techniques. A. Fluorescence-based microscopy. Upper, confocal microscopy imaging of fluorescently labeled molecules and its spatial (X–Y) resolution (given by Abbe's equation). λ, wavelength of light. NA, Numerical aperture of the objective. Lower, localization microscopy resolves the overlapping point spread functions (PSFs) by reducing the number of detections per frame in an image-series and finally reconstructs the image by overlaying. The resolution of the image is proportional to standard deviation of PSF (σPSF) and inversely proportional to the square-root of the number of photons detected.
B. Electron microscopy Membrane protein immunogold labeling by whole-mount (above) or membrane ripping protocol (below), followed by imaging. The spatial resolution is determined by a modified Abbe's equation. θ, half-aperture angle. Voltage, accelerating velocity of the electrons.
C. Fab-PLA method to visualize nanoscale protein:protein interactions in protein islands on the plasma membrane. The PLA method detects nanoscale protein islands by the close proximity of two target proteins using antibodies coupled to DNA-oligonucleotides that can direct the ligation of two oligonucleotides into a circle. The signal is amplified by rolling cycle amplification PCR and detected by fluorophore-coupled complementary oligonucleotides. The method identifies pairs of molecules closely spaced within 10–20 nm (left) compared to undetectable disperse monomers (middle) in a fluorescence microscopy image (right). PLA data is quantified by counting the number of PLA signal per cells by using BlobFinder software.
2.1. Biochemical studies of membrane protein complexes
Isolation and enrichment of membrane protein complex in its native state is an important step for further biochemical studies. To maintain the native lipid environment, it is preferable to extract the protein complex without the use of detergent or only with mild detergent. Using a nitrogen cavitation system Harder's group, demonstrated the detergent-free isolation of native TCR complex from homogenized T cells [24,25]. However this method has weak signal to noise ratio due to high background binding of other cellular materials, incomplete homogenization and low avidity binding of antibody coated beads [25]. Apart from isolating lipid–protein complexes from the plasma membrane, the nitrogen cavitation method is also useful for isolating intracellular vesicles [26]. Most standard methods of biochemical analysis of membrane proteins are dependent on a detergent lysis of the membrane that can be quite variable. For example, the affinity purification of the BCR from 1% Triton-X100 B cell lysates allows the isolation of only the membrane bound-IgM (mIgM) molecule but not the mIgM-associated signaling subunit Igα and Igß [27]. In contrast the complete BCR complex can be co-purified from 1% digitonin B cell lysates. Another commonly used mild detergent is Brij98, a member of the polyoxyethylene family [28,29]. This detergent was used to isolate low-density detergent resistant membranes from the buoyant fractions of a sucrose density gradient [28,30]. Using this method, key players of the T cell antigen receptor (TCR) signaling machinery were studied [28]. It was found that on resting T cells, the TCR/CD3 complex, the co-receptor CD4 in association with the Src-family kinase p56 Lck, and the linker for activation of T-cell (LAT), are each organized in different membrane compartments. Upon TCR ligation, the different compartments apparently combine to organize a signaling platform where the phosphorylation of the CD3 and TCR-zeta chains by the respective kinases Lck and ZAP70 results in the recruitment of ZAP70 to the TCR and induces phosphorylation of LAT and other adapters such as SLP-76 [28]. These biochemical data were confirmed by transmission electron microscopy (TEM) and super-resolution microscopy studies showing that, upon T cell activation, the different nanoscale membrane patches or protein islands move closer together [31].
Most Western blot studies of proteins are done with anionic sodium dodecyl sulfate containing polyacrylamide gel electrophoresis (SDS-PAGE), a procedure that denatures proteins and thus cannot be used to directly analyze non-covalently associated protein complexes. Thus, for the analysis of membrane protein complexes, native gel methods such as the blue native polyacrylamide gel electrophoresis (BN-PAGE) are more suited [32]. Schamel et al. used this method to study monomeric and oligomeric complexes of BCR and TCR [29,33]. For this, they used different detergents such as Digitonin, Thesit, Brij96 and Triton-X100 and found that, cell lysis with a low percentage of Thesit or Brij96 is more suitable to detect and isolate oligomeric receptor complexes.
The results obtained in biochemical studies of membrane protein organization depend on the type of detergent used. This makes the characterization of membrane complexes difficult. One way around this problem is to use chemical or photo-active crosslinkers prior to a detergent lysis [20]. For example, the Strep-tagged membrane protein interaction experiment (SPINE) uses formaldehyde to crosslink co-localized membrane proteins prior to their affinity purification with Streptavidin coupled beads [34,35]. Another useful strategy for stabilizing the transient protein:protein interactions is by incorporating photo-activatable amino acids into cellular proteins, followed by a short UV exposure to activate the crosslinker [36]. Using this method, Suchanek et al. identified the interaction of progesterone-binding membrane protein PGRMC1 and Insulin-induced gene-1, Insig-1 in a membrane-residing complex of the endoplasmic reticulum.
2.2. Super-resolution and localization microscopy
Currently, the most popular methods for exploring the distribution of membrane proteins at nanometer resolution utilize localization microscopy. They include photoactivated localization microscopy (PALM) [37] and stochastic optical reconstruction microscopy (STORM) [38]. Apart from localization microscopy, stimulated emission depletion (STED) microscopy [39], a method based on RESOLFT technique [40,41], is another commonly used method for exploring the distribution of membrane proteins. These new imaging techniques are commonly known as super-resolution microscopy. In contrast to classical light microscopy, which constantly monitors the light emitted from all fluorophores in a picture, the localization microscopy techniques reduce the number of fluorophores and increase their spatial separation by imaging the samples in multiple frames, where each frame has only a subset of a few active fluorophores (Fig. 1A, lower panel). The light signals from more than 5000 frames are then combined to reconstruct the complete picture. Both PALM and STORM methods use the same underlying principle to reduce the number of active fluorophores. In PALM, the protein of interest is tagged with a photo-activatable protein such as paGFP [42] or photo-switchable proteins such as mEOS [43,44]. The photo-activatable proteins are converted to active fluorophores by exposure to low intensity short wavelength light and bleached with a high intensity excitation laser used during imaging. The photo-switchable proteins undergo a green to red photo-shift of emission wavelength by exposure to low intensity, short wavelength light. In the PALM method, the use of tagged fluorescent proteins instead of a labeled antibody enables time-lapse imaging of living cells down to 50 nm (or < 50 nm) resolution [45].
The STORM method uses either a tandem pair of organic fluorophores or a single organic fluorophore that is maintained in a meta-stable dark state by a reducing chemical environment. The latter approach is frequently called direct stochastic optical reconstruction microscopy (dSTORM) [46,47]. A spontaneous switching or low intensity, short wavelength light induced switching of individual fluorophores between visible and dark states is needed in dSTORM imaging [48]. The low intensity, short wavelength light converts a few fluorophores to their active state and the emitted photons are collected in multiple frames. After each round of imaging, the activated fluorophores are bleached by the excitation laser used during imaging and a new subset of fluorophores appears on the subsequent frames. Thus, the localization of all molecules in the field of view requires the sequentially acquisition of many frames (Fig. 1A, lower panel). This process can take up to a few minutes, and therefore super-resolution microscopy is an inherently slow method and is mainly applied on fixed samples. However, the technical advances in optical microscopy, the increasing speed of the used acquisition camera and continuous optimization of image analysis algorithms improved the speed and photon detection efficiency of super-resolution microscopy. This allows one to study the nanoscale movements of molecules in living cells, albeit at a lower resolution (40-50 nm) [49]. Another limitation of localization microscopy is the lack of three-dimensional (3D) information. Recently, various approaches have been introduced to obtain z-position information for each localized fluorophore [50]. These include the splitting of different z-planes onto different regions of the detectors and use of interferometry. Such a three-dimensional localization approach was used to characterize the dimensions of clathrin-dependent endocytic vesicles [51].
In contrast to PALM and dSTORM, STED microscopy uses a specialized depletion laser, in combination with the excitation light. The shape of the depletion laser is engineered such that it only partially overlaps with the diffraction-limited spot created by excitation laser (donut-shaped depletion profile). In this way, the emission from the non-overlapping portion becomes visible [52]. As the intensity of the depletion laser is increased, the signal obtained from each fluorophore can be localized below the diffraction limit [39,40]. Using this method, small, 50–60 nm nanocusters of the SNARE protein syntaxin-1 were found on the PM of the neuroendocrine cell line PC12 [17]. Interestingly, syntaxin-1 molecules are only transiently associated with their nanoclusters and rapidly exchange with freely diffusing syntaxin-1 molecules. Live cell STED imaging also revealed the transient, nanoscale organization of sphingolipids and glycosylphosphatidylinositol (GPI)-anchored proteins within a 20 nm cholesterol rich membrane region [53]. These studies support the notion that the PM organization is controlled by dynamic molecular interactions of associated and dissociated states.
Apart from live cell super-resolution microscopy methods, described above, other microscopy based techniques such as fluorescence correlation spectroscopy (FCS) [54], total internal reflection microscopy (TIRFM) [55] and single particle tracking (SPT) [56,57] are also used for exploring the distribution of membrane proteins in living cells. The FCS method determines the molecular dynamics (diffusion rate) of fluorescently labeled membrane proteins [18,58]. Several variations of classical FCS method such as scanning-FCS and spot-variation FCS were introduced by different groups to study the nanoscale organization of membrane proteins within the complexity of PM (for review read [59]). TIRFM allows detection of molecules to a thin slice of illuminated field, about 100 nm, adjacent to the glass-sample interface. Thereby, the membrane localized and membrane proximal cytoplasmic features can be specifically studied in this system. However, the depth of the TIRFM acquisition is still larger than the thickness of the PM lipid bi-layer (~ 5 nm). It is thus not always clear with this technique whether the observed molecular process occurs at or below the membrane. For example, the movement of PM-proximal membrane vesicles carrying a GFP-tagged protein could be misinterpreted as the movement of the GFP-tagged proteins inside the PM [60]. The SPT method records the trajectory of fluorescently labeled molecules within a given time interval. Combining a high-speed camera to a TIRFM, the SPT method provides insight into the dynamic behavior of membrane proteins on the cell surface [56,57]. Unlike super-resolution microscopy, both FCS and SPT method cannot resolve the nanoscale structures of the PM but quantify the molecular dynamics and zone of confinement.
2.3. Electron microscopy
One well established technique that can be used to explore nanoscale organization of membrane proteins below the diffraction barrier of visible light (~ 250 nm) is TEM. Depending on the voltage of the accelerating electrons, the resolution of a TEM can be in the range of 0.5 to 0.2 nm, thus reaching well below the dimensions of most membrane proteins (Fig. 1B). However, TEM requires fixation, dehydration and embedding of the biological object being studied and these procedures can potentially distort the organization of proteins on the membrane. Furthermore, proteins cannot be directly visualized on fixed samples but require antibodies coupled to different-sized gold particles (immunogold) for their detection. Nevertheless, the high-resolution of TEM images with gold particles on cellular or membrane preparations allows the study of membrane protein compartmentalization. Two major sample preparation methods are currently used to study the spatial organization of membrane proteins by immunogold-TEM. The whole-mount method that detects membrane protein on the surface of fixed intact cells, uses primary antibodies followed by secondary immunogold antibodies [61,62]. This technique revealed the nanocluster organization of several membrane proteins, including the potassium channel Kv1.3 [62], leucocyte-specific integrin LFA-1 [63], and the HIV-1 receptor of antigen presenting cells DC-SIGN [61]. Furthermore, an oligomeric organization of the BCR was detected on the B cell surface by a TEM study combined with mathematical modeling [64]. A complementary method is the immunogold labeling of isolated membrane sheets [65]. For the generation of these membranes, the cells are first sandwiched between a poly-l-lysine (PLL) coated electron microscopy (EM) grid and a glass coverslip. The rapid removal of the coverslip generates shearing forces that rupture the cells, leaving behind the membrane, bound to the EM grid with an inside-out orientation [65]. After fixing the membrane sheets with paraformaldehyde or paraformaldehyde/glutaraldehyde, the membrane proteins are detected with antibodies specific for their cytosolic regions or attached peptide tags, followed by gold-conjugated secondary antibodies. The use of two alternatively tagged proteins in combination with corresponding anti-tag antibodies, followed by secondary antibodies labeled with different size gold particles (10 nm and 5 nm gold) allows the determination of the relative distribution of two different membrane proteins [12]. In addition, standard EM staining reagents such as osmium tetroxide, tannic acid and uranyl acetate are used to add contrast and to stain membrane-associated structures such as the cortical cytoskeleton or clathrin coated-pits. In this way, the spatial distribution of receptors such as the FcεRI on mast cells [66] and the TCR and LAT on T cells [12] has been studied. Furthermore, this technique revealed the nanoscale clustering of small G-proteins such as Ras and their association with lipid-ordered domains [13,67]. These studies support the model of PM compartmentalization and the lateral segregation of most membrane proteins.
The molecular organization of membrane proteins studied by using immunogold-TEM and super-resolution imaging can be subjected to various quantitative analysis that can reveal the dimension of individual nanoclusters and their separation. One of the classical methods is Ripley's K function analysis, which estimates spatial clustering or co-clustering of each signal point as a function of length [67,68]. This method has been widely used to quantify immunogold-TEM and super-resolution images. An alternate method is pair correlation analysis [69,70], which is widely used for super-resolution image analysis. The pair-correlation analysis also measures spatial clustering among the groups compared to Ripley's cumulative method. Thus, unlike Ripley's K function, the pair-correlation analysis is not obscured by the presence of molecules at the center. This method also takes into account the reoccurrences of the same fluorophores in consecutive frames as a result of blinking in localization microscopy. Therefore, it is more suitable to super-resolution images. In addition, pair correlation analysis can more accurately estimate cluster size and molecular density within the cluster. The distribution and interaction of two different molecular species can be evaluated by bivariate Ripley's K function analysis and pair cross-correlation analysis [70,71].
2.4. Proximity ligation assay
Despite their popularity and usefulness, immunogold-TEM and super-resolution microscopy have several problems, as these approaches require the expression of tagged and modified proteins in cell lines or a specific labeling strategy for the protein of interest. This raises issues about the expression of modified proteins in an altered cellular and membrane environment. Alternatively, endogenous proteins can be detected in TEM by a specific antibody [72]. However the reactivity of the antibody against an endogenous protein must endure the rigorous membrane preparation and fixation steps necessary for TEM methods. In comparison, the proximity ligation assay (PLA) [73] has the advantage that it can be performed with any cell type, including isolated primary cells or even cells inside a tissue, and thus can be used for clinical specimens [74,75]. Furthermore most antibodies that work in immunoflourescence assays can also be used in the PLA method. The in situ PLA enables studies of endogenous proteins in their natural environment and can also be used to analyze post-translational modification of the proteins [76]. Prior to PLA analysis, cells are placed on non-stimulatory PLL-coated glass slides and fixed by formaldehyde. The PLA method detects the close proximity of two target proteins using antibodies coupled to DNA-oligos that can direct the ligation of two oligonucleotides into a circle (Fig. 1C). One of these antibody-coupled oligos is then amplified 100,000-fold by a rolling circle amplification mechanism and detected by fluorophore-coupled complementary oligonucleotides [73,74]. For this reaction to occur, the oligo-coupled antibodies must be close (< 10 nm) to each other and this makes the assay depend on the nanoscale proximity of the two components under study [74,75]. The amplified PLA signal can be easily visualized and documented by simple fluorescence microscopy. Quantification of the PLA data involves counting the number of red dots per cell, which can be performed by using simple image analysis algorithms such as the BlobFinder [77]. The PLA method can also be automated by using standardized reagents and a microfluidic chip [78]. The limitation of this assay is that it requires antibodies against the targeted protein pair and the cells need to be fixed. Thus, like most other super-resolution studies, PLA cannot be performed on living cells.
A positive PLA signal does not require a direct protein:protein interaction but only close proximity. Therefore, PLA enables the detection of protein assemblies or protein clusters that do not withstand detergent extraction and immunoprecipitation procedures. This makes PLA particularly useful to study the nanoscale protein organization on cell surfaces. Furthermore, by fixing but not permeabilizing the cells, PLA can be used to study the protein organization, specifically on the cell surface. However, using a cell-permeabilizing-reagent such as Saponin or Triton-X100, one can also test the proximity of intracellular proteins or their interaction with membrane-bound receptors [79]. The classical PLA method (2-PLA) involves DNA-oligo coupled secondary antibodies and detects the proximity of two molecules in the 10–80 nm range. By coupling the DNA-oligos directly to primary antibodies (1-PLA), or to Fab fragments (Fab-PLA), the detection range can be reduced to 10–40 nm and 10–20 nm, respectively [11]. We have found that the Fab-PLA method with its 10–20 nm detection range is particularly useful to explore the nanoscale membrane organization on the surface of resting or activated B cells (Fig. 1C). Only with Fab-PLA, but not the other PLA methods, we were able to demonstrate the opening of the BCR and the reorganization of BCR with its coreceptors following B cell activation [11]. The use of single-chain Fv fragments, or single VH domains in the form of nanobodies, can further improve the resolution of the PLA method. However, one drawback of the monovalent binding PLA approach is that it is less sensitive than 2-PLA and 1-PLA. Importantly, in this context, the PLA technique can be used to study not only protein:protein but also protein:lipid interactions. This can be done by coupling one of the PLA oligos directly to a lipid-binding domain, for example to the choleratoxin B domain, and the second oligo to an anti-receptor antibody. In this way, we found that the IgD-BCR is localized in close proximity to raft-like lipids in the PM of resting B cells [11].
3. The nanoscale compartmentalization of the PM
Progress in biological science is connected with the discovery of order at all levels of biological phenomena. In opposition to the second law of thermodynamics, living organisms accumulate order in to the face of the chaotic universe around it. Most biological processes that were first thought to occur randomly, such as the immunoglobulin V gene assembly, were later found to be highly regulated. For a long time, it was thought that biological membranes were an exception to this rule. The fluid mosaic model of Singer and Nicolson predicted a free unrestrained movement of all membrane proteins in the lipid bi-layer of the PM [5]. Higher organization of membrane proteins seemed to occur only after the activation of cells or within specialized areas of cell:cell contact, such as the neuronal or immunological synapses [80,81]. However, research of the last 20 years has helped to establish the view that the PM of all cells is ordered and highly compartmentalized [82,83]. Three cellular components contribute to this membrane order namely, the lipids, the membrane proteins, and the cytoskeleton.
3.1. The lipids
Experiments with biological and artificial membranes have shown that the different lipids making up the PM self-assemble and separate into disordered and ordered phases [84]. The later phase, when enriched with sphingolipids and cholesterol, is also known as a “lipid raft” [85,86]. GPI-anchored proteins are preferentially associated with such lipid raft domains and this can help to better define these domains [87,88]. One problem with the raft hypothesis, is that the methods to isolate and study these ordered lipid domains are quite variable and are not always appropriate. For example, biochemical studies of these domains uses detergent extraction and sucrose gradients to isolate the low-density, detergent-insoluble fraction enriched in cholesterol and glycosphingolipids [89]. However, these detergent-resistant membrane fractions can be quite heterogeneous and only poorly represent the organization of these domains on the membrane of living cells [90,91]. Another problem was that lipid rafts were thought to be tightly associated with active signaling processes. Newer data show, however, that ordered lipid domains are already abundant on the surface of resting non-signaling cells and that they are more diverse than was previously thought.
The historical merit of the lipid raft model is that this hypothesis postulated, from early on, that the PM is compartmentalized. Originally, the lipid raft hypothesis was postulated to explain the compartmentalized delivery of glycosphingolipid and cholesterol to PM from the trans-Golgi network [92]. Later, Simons and Ikonen redefined the lipid raft as a PM compartmentalizing unit and a platform for the assembly of proteins involved in signal transduction [86]. Newer studies using high-resolution microscopy techniques such as single particle tracking (SPT) and fluorescence resonance energy transfer (FRET), confirmed the existence of nanoscale lipid raft domains, albeit at much smaller dimensions than previous models [53,56,93]. The nanoscale organization of the membrane is, however, not restricted to ordered lipid domains but applies also to other lipid domains [12].
3.2. The transmembrane proteins
Membrane proteins can either be loosely associated with the PM or can cross the lipid bi-layer with one or several transmembrane (TM) domains, thus being integral membrane proteins. In most cases, the TM domains form α-helixes of 18–25 amino acids (aa). Functioning as a membrane anchor, the aa sequence of a TM domain can be quite diverse, apart from containing predominantly hydrophobic aa to be compatible with the hydrophobic core of the lipid bi-layer. This led to the view that TM domains are evolutionarily less conserved than the rest of the proteins. In contrast to this assumption, the TM domains are often the most conserved part of a TM protein. This finding suggests that the TM domains have additional functions and are not just serving as a membrane anchor. For example, the TM domains can be directly involved in the formation of dimers and oligomers of TM proteins. The TM domains of the IgD, part of the IgD-BCR on the B cell surface, are evolutionarily highly conserved and it has been shown that they are involved in the oligomerization of this receptor [94,95]. The clustering of TM proteins may occur as a response to a hydrophobic mismatch, as it reduces the number of lipids bound to the non-matching membrane inclusion [82,96]. Thus, the interactions of hydrophilic residues inside the TM sequence may drive the formation of higher ordered protein oligomers. In addition, the TM domains of the TCR and BCR contain conserved charged and polar aa residues, respectively that seem to play important roles in the assembly and function of these receptor complexes [97–100].
Newer studies suggest that the TM domains are not only involved in protein:protein but also in protein:lipid interactions and the sum of these interactions maybe involved in the sorting of TM proteins into specific nanoclusters or protein islands with a defined lipid composition [101]. Wedlich–Söldner's group tested a large set of yeast TM domains in swapping experiments and found that these sequences could influence the nanoscale location of the corresponding membrane proteins on the PM of yeast cells [102]. A functional segregation is also found in the PM of mammalian cells, even for TM proteins of the same family such as the Lipid Phosphate Phosphatases LPP1 and LPP2 [103] or for the SNARE protein syntaxin-1 [17]. In TEM studies of the inner leaflet of the PM of mast cells, Wilson et al. found that many membrane proteins are not uniformly distributed but are clustered in nanoscale membrane areas [65,66]. Specifically, they showed that the high-affinity Fc-receptor for IgE is localized in different protein clusters than the TM adaptor LAT. Lillemeier et al. extended these studies by demonstrating that most proteins on the PM of T lymphocytes are confined in so-called protein islands with a size of 80–150 nm [31]. These pre-organized domains coalesce after T cell activation. The molecular mechanism for the specific sorting of TM proteins into these protein islands is unknown at present. Each TM domain may bind to a specific set of lipids, thus generating a lipid shell of distinct composition around them. Those TM proteins with the same or a similar lipid shell will then be sorted or form a specific protein island. The sorting event and the place where the sorting process take place are discussed in Section 3.4.
3.3. The cytoskeleton
The formation and stability of protein islands are not only dependent on the lipids and TM proteins but also on the cytoskeleton. This notion is based on the observation that the treatment of cells with Latrunculin-A (Lat-A) not only inhibits F-actin formation but also the clustering of raft-associated TM proteins [104–106]. The nanoscale confinement of GPI-anchored proteins and several other membrane proteins including the glycine receptor and the BCR is dependent on the proper formation of cortical actin, and to lesser extent on lipids such as cholesterol [107–110]. High-resolution imaging and SPT studies showed that TM proteins move freely only within confined zones between 30 and 700 nm in diameter [57,111]. At the borders of these confinement zones, diffusion is reduced. For long-range movements on the cell surface, the proteins have to jump between adjacent confinement zones [111,112]. Theses studies resulted in the “picket-fence” model, suggesting that the membrane-proximal cortical cytoskeleton imposes effective barriers for the diffusion of TM proteins [111]. Facundo Batista and colleagues also found that the diffusion of the BCR on the B cell surface is restricted by a membrane proximal actin cytoskeleton meshwork. The BCR complexes located in actin-rich regions exhibited slower diffusion compared to those located in actin-poor regions [109]. These SPT studies suggest that, at least in part, the diffusion barrier for the BCR is derived from the ezrin–radixin–moesin (ERM) family of cytoskeleton proteins [113]. Furthermore, they showed that treatment of B cells with the F-actin inhibitor Lat-A, activates the B cells [114]. The confinement zones of protein diffusion may be identical or similar to the protein islands detected by other methods. Consistent with this view, Mark Davis and colleagues found that the actin cytoskeleton is involved in the formation and maintenance of cholesterol-enriched, protein islands on the T cell surface [12].
3.4. The origin of protein islands
Studies from several groups using diverse techniques show that TM proteins and lipids are highly organized inside nanoscale protein islands [31,65,85,87]. How are these structures formed and what forces maintain them? The finding that the same proteins are often clustered together whereas others are excluded from specific protein islands is a clear indication for a sorting process [17,102]. There are two scenarios where this sorting could take place: either the sorting happens on the PM or, more likely, sorting already occurs during the intracellular transport of the TM proteins and lipids to the PM. All membrane proteins that leave the ER first pass through the Golgi apparatus before they are placed on the cell surface. The Golgi has well-known cellular protein sorting capabilities. In polarized cells, it assembles different cargo vesicles that are then transported either to the apical or the basolateral cell surface [115,116]. These vesicles differ not only in their protein content, but also in their lipid composition [117,118]. The protein and lipid sorting process in the Golgi maybe more diverse and specific than currently appreciated and could potentially generate hundreds of different cargo vesicles. It is thus feasible that all protein islands detected on the cell surface have their origin in precisely sorted transport vesicles that are assembled in the Golgi apparatus. Currently, it is thought that once a cargo vesicle fuses with the PM, the transported TM proteins are equally distributed over the whole cell surface. An alternative scenario is that after the vesicle-PM fusion, the pre-sorted proteins and lipids stay together to form a nanoscale protein islands. In line with this view, Soares et al. have shown that the constituents of the TCR signaling machinery, viz. TCRζ, Lck and LAT traffic through distinct vesicular compartments [119]. Fusion of these TCRζ and LAT containing vesicles to the immune synapse controls the nanoscale organization of phosphorylated TCRζ and LAT clusters. This observation suggests a directed transport of vesicles; however the model requires further validation.
4. Nanoscale organization and reorganization of receptors on the B cell surface
Surface expression and assembly of BCR requires a membrane-bound immunoglobulin (mIg) molecule and the Igα/Igβ heterodimer, acting as antigen-binding and signaling subunits, respectively [33]. The proper assembly of all BCR components in the ER is required for the expression of this receptor on the cell surface. Each mature B cell carries roughly 120,000 BCR complexes on its cell surface [64]. How these many receptors are organized and kept silent on resting B cells is still not resolved. During B cell activation, the tails of Igα and, to lesser extent, that of Igβ become phosphorylated on two tyrosines that are part of the immunoreceptor tyrosine-based activation motif (ITAM) [120]. It is dominantly the spleen tyrosine kinase (Syk) that phosphorylates the two ITAM tyrosines [121,122]. Phosphorylated ITAMs are binding-sites for the tandem SH2 domain of Syk [123]. This process couples the BCR to an active tyrosine kinase and amplifies BCR signaling as discussed in Section 4.4.
4.1. Biochemical studies of the BCR
A first hint that the BCR forms oligomers on the surface of resting B cells came from biochemical studies employing blue native polyacrylamide gel electrophoresis (BN-PAGE) and differently tagged Igα proteins [33]. In these studies, the BCR was solubilized with low concentrations of non-ionic detergents to extract the receptor complex from the cell membrane. It was shown that both the IgM-BCR and the IgD-BCR form oligomers that differ in size [33]. Mutation of several conserved aa residues of the TM sequences of the mIgD molecule reduced the size of the IgD-BCR oligomer. This finding suggested that the TM regions of the mIg molecules are involved in the formation of the BCR oligomer. Interestingly, one side of the TM regions of the mIgM and mIgD molecules are quite different from each other. As the IgM-BCR and the IgD-BCR do not form mixed oligomers, it is likely that this side of the TM region is required for the class-specific formation of BCR oligomers [33]. This notion was further supported by TM region mutation analysis. The discovery of BCR oligomers on resting B cells lead to a model postulating that it is the opening of BCR oligomers rather than cross-linking the BCR that results in B cell activation [94]. However, the model of an oligomeric organization of the resting BCR was not well accepted at the time, and primarily regarded as artifact of detergent lysis [124].
4.2. Fluorescence complementation studies of the BCR
The bimolecular fluorescence complementation assay (BiFC) is a method that allows the detection of protein dimer formation in living cells [125]. In this method, the fluorescent protein (FP) is split in two parts (N-FP and C-FP) and used to generate two fusion proteins, each carrying one FP part. If the two proteins under study dimerize, a functional FP domain is reconstructed and is detected by the emitted fluorescence. In this way, one can test for hetero and homo-dimerization in living cells expressing the two fusion proteins. Using the S2 Schneider cells, we followed the assembly of N-YFP- or C-CFP-tagged Igα proteins together with the other BCR components in a pulse-chase like fashion [126]. This, together with a stringent quantification of reconstructed FP, showed that BCR dimerization is an efficient and spontaneously occurring process, and that BCR dimers are stably expressed on the cell surface [95]. With this assay, we were also able to identify an IgD-BCR mutant with a defect in the oligomerization process. This monomeric IgD-BCR carries mutations on the class-specific side of the TM region and lacks the disulfide bridge between Igα and Igβ [127]. This finding showed that the conserved amino acids in the TM region of mIgD are required for BCR dimerization and thus confirmed the conclusions of our previous BN-PAGE studies [33]. Interestingly, the monomeric BCR mutant signals more actively and is less stably expressed on the B cell surface, thereby suggesting that BCR oligomers are autoinhibited, inactive forms of the receptor [95]. In line with this, we found that the BiFC-stabilized BCR-dimers are less active in signaling and less well internalized. These studies led to the formulation of the dissociation activation model (DAM), suggesting that it is the dissociation of BCR oligomers, rather than the cross-linking of BCR monomers, that initiates B cell activation [128].
4.3. Fab-PLA studies of BCR activation
The dissociation and opening of a BCR oligomer involves movements of receptor monomers relative to each other in the 10 nm to 20 nm range (Fig. 2). These receptor alterations cannot be monitored by classical light microscopy with a diffraction limit of 250 nm. Even with TEM, it is challenging to reliably detect such tiny receptor alterations. Fab-PLA is here the method of choice. In an analysis of monomeric and pentameric soluble IgM bound to antigen-coupled beads, we showed that only Fab-PLA but not 1-PLA can distinguish pentameric and monomeric IgM from each other [11]. Apparently, Fab-PLA requires the close spacing of molecules inside the IgM pentamer for the amplification of the PLA reaction. Therefore, Fab-PLA is a specific assay for the detection of IgM multimers, either in the form of pentameric IgM, or as a receptor oligomer. Similarly, the monomeric IgD-BCR mutant, when expressed in S2 Schneider cells, does not give a signal in Fab-PLA. To study nanoscale alterations of the IgM-BCR and IgD-BCR upon B cell activation, we performed IgM:IgM Fab-PLA as well as IgD:IgD Fab-PLA. The result was the same for both BCR classes. The assay detects a strong Fab-PLA signal on resting B cells that was lost upon B activation. Therefore, BCR activation is accompanied by the opening of BCR oligomers so that they are more distantly spaced Ig monomers and can no longer be detected by Fab-PLA. This finding supports the DAM hypothesis and suggests that upon antigen binding, the BCR oligomers dissociate and form clusters of active BCR monomers. Importantly, the same observations were made in studies of B cell lines or primary B cells from the mouse spleen or from human peripheral B cells, showing that BCR opening is a conserved process of B cell activation. Apart from antigen, the dissociation of BCR oligomers was also induced by the treatment of B cells with activating drugs such as pervanadate (Perv) and Lat-A, which alter the redox equilibrium and inhibit actin polymerization, respectively. The latter finding suggests that the cytoskeleton stabilizes BCR oligomers on resting B cells.
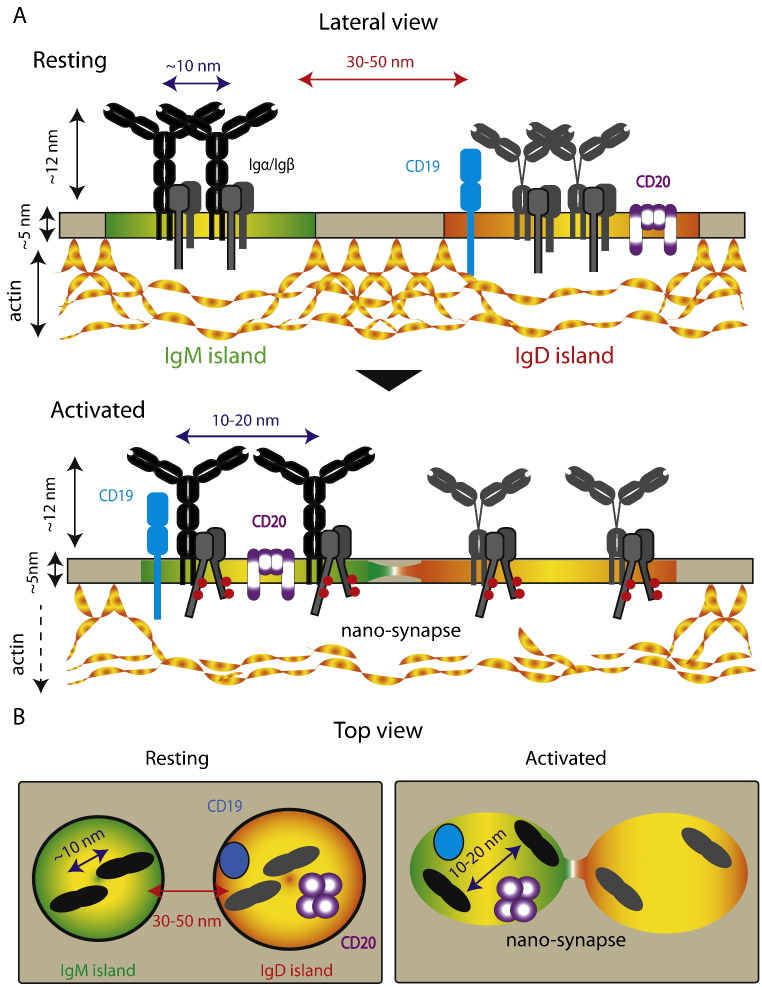
Fig. 2.
Reorganization of BCR nano-islands during B cell activation. A. Lateral view, a unified model depicts class-specific nanocluster organization of IgM-BCR (dark) and IgD-BCR (grey) in separated nano-islands in resting B cells (top). IgD-BCR reside in same nano-island (yellow-red region) with co-receptors CD19 (blue) and CD20 (purple). The close proximity (~ 10 nm) of IgM:IgM and IgD:IgD within their specific nano-islands in resting B cells keeps the Igα/Igß cytoplasmic tails inaccessible and maintains the silent state. Upon B cell activation, BCRs dissociate (10–20 nm), sub-membrane actin-cytoskeleton dissociates and the nano-island reorganizes (bottom). CD19 and CD20 are exchanged from IgD-BCR nano-islands to IgM-BCR nano-islands (yellow-green region) via a nano-synapse.
B. Top view, reorganization of IgM and IgD-BCR nano-islands upon B cell activation. The two large shaded circles surrounding the visible Fab-arms of IgM-BCR (dark) and IgD-BCR (grey) show the boundary of the nano-islands. Left, co-receptor CD19 (blue) and CD20 (purple) reside in IgD-BCR nano-island in resting B cells. Right, BCRs dissociate (10–20 nm), the sub-membrane actin-cytoskeleton dissociates, and IgM- and IgD-BCR nano-islands meet each other to form a nano-synapse.
4.4. Discovery of inside-out Syk/BCR signaling by Fab-PLA
So far, all read-outs for BCR activation measured downstream events, such as tyrosine phosphorylation of kinase substrate proteins or the release of calcium. With the Ig:Ig Fab-PLA, we have, for the first time, an assay that can directly monitor nanoscale alterations in the BCR itself after B cell activation. This allows us to study the molecular requirements of BCR opening. Interestingly, we found that the inhibition of Syk activity with a pharmacological inhibitor prevents BCR opening on B cells, exposed either to antigen or Lat-A. In contrast, inhibition of the Src-family kinase Lyn only delays but does not prevent BCR opening upon stimulation. The same results were obtained from Lyn- or Syk-deficient splenic B cells, showing that this is not an off-target effect of the inhibitors used. Therefore, the BCR-interacting-kinase Syk is not only required for downstream signaling but also for the opening of the BCR, suggesting that a Syk-BCR inside-out signaling mechanism amplifies the BCR signal in activated B cells [11]. With other words, this process involves the binding of Syk to the phosphorylated ITAMs of Igα and Igß at the inner side of the PM, thus changing the BCR so that it resumes an open conformation on the cell surface. The extent of BCR signal amplification by Syk was studied by an assay involving the pre-incubation of B cells with the Syk inhibitor R406 for different times, followed by stimulation with increasing doses of antigen. B cells exposed for 60 min to the Syk inhibitor loose the IgM:IgM Fab-PLA signal at 250 ng/ml or higher doses of antigen, whereas untreated cells require only 10 ng/ml of antigen to loose the IgM:IgM Fab-PLA signal. The result of this experiment supports the notion that the Syk/BCR inside-out signaling increases the sensitivity of B cells to low doses of antigen.
To study the molecular details of the Syk-BCR inside-out signaling mechanism, we rebuilt the BCR, together with wild type or mutant forms of Syk in S2 Schneider cells. This experiment tested whether it is the phosphorylation of the BCR tails by Syk or the binding of Syk to the phosphorylated receptor that opens the BCR oligomer. The result of these experiments showed that the latter is the case and suggests a two step mechanism of B cell activation (Fig. 3). In the first step, limited amounts of antigen bind and open only few BCR dimers by outside-in signaling and active BCR/Syk seed complexes are formed. In the second step, these BCR/Syk seed complexes diffuse in the membrane and phosphorylate neighboring BCR oligomers. This results in more Syk recruitment and BCR dissociation and the amplification of the BCR signal via an inside-out signaling mechanism.
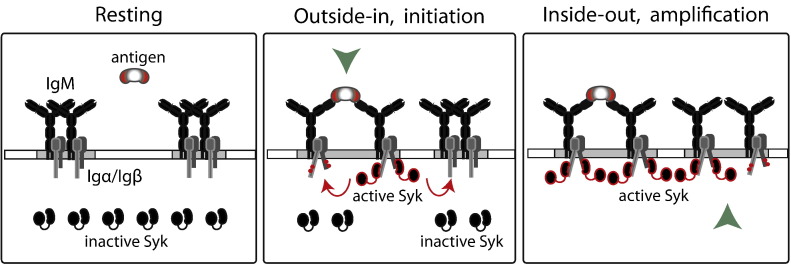
Fig. 3.
Signal amplification and dissociation of BCR by Syk-mediated inside-out signaling. The model depicts a two-step mechanism of Syk dependent amplification of BCR signaling and dissociation of the BCR. Left to right, in the resting state, Syk remains closed inactive and resides in the cytoplasm. During signal initiation and/or in presence of low concentrations of antigen, small numbers of BCR become active, allowing a low level of active Syk binding to the phosphorylated ITAMs of Igα/Igß cytoplasmic tails to form a seed complex. The process corresponds to the transmission of a signal across the plasma membrane by an outside-in mechanism. Activated BCR dissociate from each other and this increased mobility leads to phosphorylation of neighboring BCRs. This amplifies the signal by allowing more active Syk to bind to the phosphorylated tails. Binding of Syk to the phosphorylated ITAM opens the receptor from the inside and results in further signal spreading by an inside-out mechanism.
4.5. Fab-PLA studies of the B cell surface
The Fab-PLA can be used not only to detect the opening of the BCR oligomer but also to monitor its association with coreceptor molecules such as CD19 and CD20. For this, we conducted IgD:CD19 and IgM:CD19 Fab-PLA and found that on resting B cells, CD19 is in close proximity to the IgD-BCR whereas on activated B cells it is associated with the IgM-BCR and no longer close to the IgD-BCR (Fig. 2). The same class specific associations were also found for the tetraspanin protein CD20. These data support the notion that most proteins on the B cell surface reside in nanoscale protein islands. In line with this, it was recently found that CD19 forms nanoclusters on the surface of B cells [114]. The formation of these CD19 clusters requires the expression of the tetraspanin molecule CD81, which is known to form a complex with CD19 and CD21 in mature B cells [114,129]. However, the co-localization of CD19 with other receptors such as the IgD-BCR was not addressed by this one-color super-resolution microscopy study. Interestingly, with Fab-PLA, we found that the IgD-BCR islands on resting B cells contain not only CD19, CD81 and CD20, but also ordered lipid domains, as indicated by the close proximity of GM1 gangliosides and the GPI-linked protein CD52 to the IgD-BCR [11]. We currently are using the Fab-PLA technique to determine the location of other B cell surface proteins in relation to the IgD and IgM islands. For example, in preliminary studies, we have found that the integrin component CD18 is in close association with both BCR classes on activated but not on resting B cells (unpublished data).
The activation of B cells results in the reorganization of several surface proteins, such that CD19 and CD20, as well as raft-like lipids, all of which find their way from the IgD-BCR to the IgM-BCR. This exchange of coreceptors between the two BCR classes may be facilitated by the concatenation of IgD and IgM islands. Such a concatenation of protein islands was found in a TEM study of TCR and LAT islands on activated T cells [31]. Indeed, the IgD:IgM Fab-PLA test detects a positive PLA-signal only on activated but not on resting B cells, supporting the notion that the IgD and IgM islands move closer together only upon B cell activation (unpublished data). In their concatenated arrangement, the two different protein islands may form specific nano-synapses for the exchange proteins and lipids with each other (Fig. 2). How the IgD and IgM islands are kept apart on resting cells and are joined together on activated B cell is not known at present and shows that much remains to be learned about the nanoscale organization of the lymphocyte surface and its reorganization upon the activation of these cells. As described above, the Fab-PLA technique is an important new tool to explore this nano-world.
5. Conclusions and further perspectives
The cellular membrane is not only a border separating the inside of the cell from the outside. It is also an important organelle for the exchange of molecules and information between the cell and its environment. For this, the cell carries many different transporter and receptor proteins on its cell surface. Light microscopy studies monitoring the movements of the many TM proteins on the cell surface draw a view of a rather dynamic and unorganized PM [57,111]. However, this view is now corrected by super-resolution studies that reveal nanoscale confinements of the TM proteins into patchy domains or protein islands [17,31,114]. From these new studies, a picture is emerging that biological membranes at sizes well below the 250 nm diffraction barrier of visible light are compartmentalized and highly organized structures. This view invites a rethinking of many membrane processes that are likely to be exercised or controlled not by single TM proteins but rather protein–lipid clusters of defined composition and function. However, as discussed here, the study of these nanoscale protein–lipid organization structures is still challenging and requires the development of many new methods. Fab-PLA, which can detect molecular proximity of TM proteins at 10 nm to 20 nm distances, is one such method that can be used to better explore the nanoworld of the PM. With this method, we have discovered that the two antigen receptor classes on mature B cells, namely the IgM- and IgD-BCR reside in different protein islands that show a class-specific association with the coreceptor molecules CD19 and CD20. Furthermore, with Fab-PLA, we found that the different IgD and IgM islands are reorganized upon B cell activation. It will be important to elucidate whether the other BCR classes (IgG, IgA and IgE) and the many other B cell coreceptors are also organized in protein islands and how many different islands exist on the B cell surface. In preliminary Fab-PLA studies of human B cell tumor cells, we have already found several alterations in their nanoscale membrane organization compared to normal human B cells. Such nanoscale studies may lead to new diagnostic tools for these diseases.
Acknowledgements
We thank Dr. Peter Nielsen and Dr. Wolfgang Schamel for the critical reading of this manuscript. This study was supported by the Excellence Initiative of the German Federal and State Governments (EXC 294), by ERC-grant 322972 and by the Deutsche Forschungsgemeinschaft through SFB746 and TRR130.
Author contribution
P.M. reviewed literature, prepared manuscript, revised manuscript and conducted PLA experiments. J.Y. conducted BiFC experiments. K.K. conducted PLA experiments. M.R. reviewed literature, prepared and revised manuscript.
Footnotes
This article is part of a Special Issue entitled: Nanoscale membrane organisation and signalling.
Contributor Information
Palash Chandra Maity, Email: maity@immunbio.mpg.de.
Michael Reth, Email: michael.reth@bioss.uni-freiburg.de.
References
- 1.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens T.J., Arkin I.T. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39:417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 3.Dupuy A.D., Engelman D.M. Protein area occupancy at the center of the red blood cell membrane. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2848–2852. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Meer G. Cellular lipidomics. EMBO J. 2005;24:3159–3165. doi: 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer S.J., Nicolson G.L. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 6.Jacobson K., Sheets E.D., Simson R. Revisiting the fluid mosaic model of membranes. Science. 1995;268:1441–1442. doi: 10.1126/science.7770769. [DOI] [PubMed] [Google Scholar]
- 7.Maxfield F.R. Plasma membrane microdomains. Curr. Opin. Cell Biol. 2002;14:483–487. doi: 10.1016/s0955-0674(02)00351-4. [DOI] [PubMed] [Google Scholar]
- 8.Razani B., Woodman S.E., Lisanti M.P. Caveolae: from cell biology to animal physiology. Pharmacol. Rev. 2002;54:431–467. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 9.Leventis P.A., Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- 10.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 11.Klasener K., Maity P.C., Hobeika E., Yang J., Reth M. B cell activation involves nanoscale receptor reorganizations and inside-out signaling by Syk. Elife. 2014;3:e02069. doi: 10.7554/eLife.02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lillemeier B.F., Pfeiffer J.R., Surviladze Z., Wilson B.S., Davis M.M. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plowman S.J., Muncke C., Parton R.G., Hancock J.F. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15500–15505. doi: 10.1073/pnas.0504114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brummer T., Elis W., Reth M., Huber M. B-cell signal transduction: tyrosine phosphorylation, kinase activity, and calcium mobilization. Methods Mol. Biol. 2004;271:189–212. doi: 10.1385/1-59259-796-3:189. [DOI] [PubMed] [Google Scholar]
- 15.Kusumi A., Suzuki K. Toward understanding the dynamics of membrane-raft-based molecular interactions. Biochim. Biophys. Acta. 2005;1746:234–251. doi: 10.1016/j.bbamcr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 16.Kusumi A., Koyama-Honda I., Suzuki K. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 2004;5:213–230. doi: 10.1111/j.1600-0854.2004.0178.x. [DOI] [PubMed] [Google Scholar]
- 17.Sieber J.J., Willig K.I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G., Rammner B., Eggeling C., Hell S.W., Grubmüller H., Lang T. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 18.Lenne P.F., Wawrezinieck L., Conchonaud F., Wurtz O., Boned A., Guo X.J., Rigneault H., He H.T., Marguet D. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J. 2006;25:3245–3256. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusumi A., Hyde J.S. Spin-label saturation-transfer electron spin resonance detection of transient association of rhodopsin in reconstituted membranes. Biochemistry. 1982;21:5978–5983. doi: 10.1021/bi00266a039. [DOI] [PubMed] [Google Scholar]
- 20.Hunke Sabine, Müller Volker S. Approaches to analyze protein–protein interactions of membrane proteins. In: Cai Jianfeng., editor. Protein Interactions. InTech; 2012. (ISBN: 978-953-51-0244-1) [Google Scholar]
- 21.Harms G.S., Cognet L., Lommerse P.H., Blab G.A., Kahr H., Gamsjager R., Spaink H.P., Soldatov N.M., Romanin C., Schmidt T. Single-molecule imaging of l-type Ca(2 +) channels in live cells. Biophys. J. 2001;81:2639–2646. doi: 10.1016/S0006-3495(01)75907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iino R., Koyama I., Kusumi A. Single molecule imaging of green fluorescent proteins in living cells: E-cadherin forms oligomers on the free cell surface. Biophys. J. 2001;80:2667–2677. doi: 10.1016/S0006-3495(01)76236-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang B., Babcock H., Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Wet B., Zech T., Salek M., Acuto O., Harder T. Proteomic characterization of plasma membrane-proximal T cell activation responses. J. Biol. Chem. 2011;286:4072–4080. doi: 10.1074/jbc.M110.165415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harder T., Kuhn M. Immunoisolation of TCR signaling complexes from Jurkat T leukemic cells. Sci. STKE. 2001;2001:l1. doi: 10.1126/stke.2001.71.pl1. [DOI] [PubMed] [Google Scholar]
- 26.Engelke M., Pirkuliyeva S., Kuhn J., Wong L., Boyken J., Herrmann N., Becker S., Griesinger C., Wienands J. Macromolecular assembly of the adaptor SLP-65 at intracellular vesicles in resting B cells. Sci. Signal. 2014;7:ra79. doi: 10.1126/scitranslmed.2005104. [DOI] [PubMed] [Google Scholar]
- 27.Schamel W.W., Reth M. Stability of the B cell antigen receptor complex. Mol. Immunol. 2000;37:253–259. doi: 10.1016/s0161-5890(00)00025-0. [DOI] [PubMed] [Google Scholar]
- 28.Drevot P., Langlet C., Guo X.J., Bernard A.M., Colard O., Chauvin J.P., Lasserre R., He H.T. TCR signal initiation machinery is pre-assembled and activated in a subset of membrane rafts. EMBO J. 2002;21:1899–1908. doi: 10.1093/emboj/21.8.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schamel W.W., Arechaga I., Risueno R.M., van Santen H.M., Cabezas P., Risco C., Valpuesta J.M., Alarcon B. Coexistence of multivalent and monovalent TCRs explains high sensitivity and wide range of response. J. Exp. Med. 2005;202:493–503. doi: 10.1084/jem.20042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hueber A.O., Bernard A.M., Herincs Z., Couzinet A., He H.T. An essential role for membrane rafts in the initiation of Fas/CD95-triggered cell death in mouse thymocytes. EMBO Rep. 2002;3:190–196. doi: 10.1093/embo-reports/kvf022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lillemeier B.F., Mörtelmaier M.A., Forstner M.B., Huppa J.B., Groves J.T., Davis M.M. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 2010;11:90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schagger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 33.Schamel W.W., Reth M. Monomeric and oligomeric complexes of the B cell antigen receptor. Immunity. 2000;13:5–14. doi: 10.1016/s1074-7613(00)00003-0. [DOI] [PubMed] [Google Scholar]
- 34.Herzberg C., Weidinger L.A., Dorrbecker B., Hubner S., Stulke J., Commichau F.M. SPINE: a method for the rapid detection and analysis of protein–protein interactions in vivo. Proteomics. 2007;7:4032–4035. doi: 10.1002/pmic.200700491. [DOI] [PubMed] [Google Scholar]
- 35.Muller V.S., Jungblut P.R., Meyer T.F., Hunke S. Membrane-SPINE: an improved method to identify protein–protein interaction partners of membrane proteins in vivo. Proteomics. 2011;11:2124–2128. doi: 10.1002/pmic.201000558. [DOI] [PubMed] [Google Scholar]
- 36.Suchanek M., Radzikowska A., Thiele C. Photo-leucine and photo-methionine allow identification of protein–protein interactions in living cells. Nat. Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- 37.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 38.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hell S.W., Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission–depletion fluorescence microscopy. Opt. Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 40.Klar T.A., Hell S.W. Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett. 1999;24:954–956. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- 41.Heintzmann R., Jovin T.M., Cremer C. Saturated patterned excitation microscopy—a concept for optical resolution improvement. J. Opt. Soc. Am. A Opt. Image Sci. Vis. 2002;19:1599–1609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- 42.Patterson G.H., Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 43.McKinney S.A., Murphy C.S., Hazelwood K.L., Davidson M.W., Looger L.L. A bright and photostable photoconvertible fluorescent protein. Nat. Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiedenmann J., Ivanchenko S., Oswald F., Schmitt F., Rocker C., Salih A., Spindler K.D., Nienhaus G.U. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandez-Suarez M., Ting A.Y. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- 46.Heilemann M., van de Linde S., Schuttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M. Subdiffraction–resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 47.Heilemann M., van de Linde S., Mukherjee A., Sauer M. Super-resolution imaging with small organic fluorophores. Angew. Chem. Int. Ed. Engl. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- 48.van de Linde S., Loschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., Sauer M. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 2011;6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 49.Shroff H., Galbraith C.G., Galbraith J.A., Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quirin S., Pavani S.R., Piestun R. Optimal 3D single-molecule localization for superresolution microscopy with aberrations and engineered point spread functions. Proc. Natl. Acad. Sci. U. S. A. 2012;109:675–679. doi: 10.1073/pnas.1109011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang B., Wang W., Bates M., Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klar T.A., Jakobs S., Dyba M., Egner A., Hell S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eggeling C., Ringemann C., Medda R., Schwarzmann G., Sandhoff K., Polyakova S., Belov V.N., Hein B., von Middendorff C., Schonle A., Hell S.W. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–1162. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 54.Magde D., Elson E.L., Webb W.W. Fluorescence correlation spectroscopy. II. An experimental realization. Biopolymers. 1974;13:29–61. doi: 10.1002/bip.1974.360130103. [DOI] [PubMed] [Google Scholar]
- 55.Axelrod D. Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 1981;89:141–145. doi: 10.1083/jcb.89.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dietrich C., Yang B., Fujiwara T., Kusumi A., Jacobson K. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 2002;82:274–284. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Simson R., Sheets E.D., Jacobson K. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys. J. 1995;69:989–993. doi: 10.1016/S0006-3495(95)79972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wawrezinieck L., Rigneault H., Marguet D., Lenne P.F. Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys. J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He H.T., Marguet D. Detecting nanodomains in living cell membrane by fluorescence correlation spectroscopy. Annu. Rev. Phys. Chem. 2011;62:417–436. doi: 10.1146/annurev-physchem-032210-103402. [DOI] [PubMed] [Google Scholar]
- 60.Ross J.A., Digman M.A., Wang L., Gratton E., Albanesi J.P., Jameson D.M. Oligomerization state of dynamin 2 in cell membranes using TIRF and number and brightness analysis. Biophys. J. 2011;100:L15–L17. doi: 10.1016/j.bpj.2010.12.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cambi A., de Lange F., van Maarseveen N.M., Nijhuis M., Joosten B., van Dijk E.M., de Bakker B.I., Fransen J.A., Bovee-Geurts P.H., van Leeuwen F.N., Van Hulst N.F., Figdor C.G. Microdomains of the C-type lectin DC-SIGN are portals for virus entry into dendritic cells. J. Cell Biol. 2004;164:145–155. doi: 10.1083/jcb.200306112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Panyi G., Bagdany M., Bodnar A., Vamosi G., Szentesi G., Jenei A., Matyus L., Varga S., Waldmann T.A., Gaspar R., Damjanovich S. Colocalization and nonrandom distribution of Kv1.3 potassium channels and CD3 molecules in the plasma membrane of human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 2003;100:2592–2597. doi: 10.1073/pnas.0438057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cambi A., Joosten B., Koopman M., de Lange F., Beeren I., Torensma R., Fransen J.A., Garcia-Parajo M., van Leeuwen F.N., Figdor C.G. Organization of the integrin LFA-1 in nanoclusters regulates its activity. Mol. Biol. Cell. 2006;17:4270–4281. doi: 10.1091/mbc.E05-12-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fiala G.J., Kaschek D., Blumenthal B., Reth M., Timmer J., Schamel W.W. Pre-clustering of the B cell antigen receptor demonstrated by mathematically extended electron microscopy. Front. Immunol. 2013;4:427. doi: 10.3389/fimmu.2013.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wilson B.S., Pfeiffer J.R., Oliver J.M. Observing FcepsilonRI signaling from the inside of the mast cell membrane. J. Cell Biol. 2000;149:1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wilson B.S., Pfeiffer J.R., Oliver J.M. FcepsilonRI signaling observed from the inside of the mast cell membrane. Mol. Immunol. 2002;38:1259–1268. doi: 10.1016/s0161-5890(02)00073-1. [DOI] [PubMed] [Google Scholar]
- 67.Parton R.G., Hancock J.F. Lipid rafts and plasma membrane microorganization: insights from Ras. Trends Cell Biol. 2004;14:141–147. doi: 10.1016/j.tcb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 68.Kiskowski M.A., Hancock J.F., Kenworthy A.K. On the use of Ripley's K-function and its derivatives to analyze domain size. Biophys. J. 2009;97:1095–1103. doi: 10.1016/j.bpj.2009.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sengupta P., Jovanovic-Talisman T., Skoko D., Renz M., Veatch S.L., Lippincott-Schwartz J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veatch S.L., Machta B.B., Shelby S.A., Chiang E.N., Holowka D.A., Baird B.A. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE. 2012;7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prior I.A., Muncke C., Parton R.G., Hancock J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Melo R.C., Morgan E., Monahan-Earley R., Dvorak A.M., Weller P.F. Pre-embedding immunogold labeling to optimize protein localization at subcellular compartments and membrane microdomains of leukocytes. Nat. Protoc. 2014;9:2382–2394. doi: 10.1038/nprot.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.-J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.-G., Landegren U. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- 74.Söderberg O., Leuchowius K.-J., Gullberg M., Jarvius M., Weibrecht I., Larsson L.-G., Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 75.Bellucci A., Fiorentini C., Zaltieri M., Missale C., Spano P. The “in situ” proximity ligation assay to probe protein–protein interactions in intact tissues. Methods Mol. Biol. 2014;1174:397–405. doi: 10.1007/978-1-4939-0944-5_27. [DOI] [PubMed] [Google Scholar]
- 76.Brychtova V., Vojtesek B. In situ proximity ligation assay for detection of proteins, their interactions and modifications. Klin. Onkol. 2014;27(Suppl.):87–91. doi: 10.14735/amko20141s87. [DOI] [PubMed] [Google Scholar]
- 77.Allalou A., Wahlby C. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 2009;94:58–65. doi: 10.1016/j.cmpb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 78.Blazek M., Betz C., Hall M.N., Reth M., Zengerle R., Meier M. Proximity ligation assay for high-content profiling of cell signaling pathways on a microfluidic chip. Mol. Cell. Proteomics. 2013;12:3898–3907. doi: 10.1074/mcp.M113.032821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Infantino S., Benz B., Waldmann T., Jung M., Schneider R., Reth M. Arginine methylation of the B cell antigen receptor promotes differentiation. J. Exp. Med. 2010;207:711–719. doi: 10.1084/jem.20091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Freiberg B.A., Kupfer H., Maslanik W., Delli J., Kappler J., Zaller D.M., Kupfer A. Staging and resetting T cell activation in SMACs. Nat. Immunol. 2002;3:911–917. doi: 10.1038/ni836. [DOI] [PubMed] [Google Scholar]
- 81.Kupfer A., Kupfer H. Imaging immune cell interactions and functions: SMACs and the Immunological Synapse. Semin. Immunol. 2003;15:295–300. doi: 10.1016/j.smim.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Bagatolli L.A., Ipsen J.H., Simonsen A.C., Mouritsen O.G. An outlook on organization of lipids in membranes: searching for a realistic connection with the organization of biological membranes. Prog. Lipid Res. 2010;49:378–389. doi: 10.1016/j.plipres.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 83.van Zanten T.S., Cambi A., Garcia-Parajo M.F. A nanometer scale optical view on the compartmentalization of cell membranes. Biochim. Biophys. Acta. 2010;1798:777–787. doi: 10.1016/j.bbamem.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 84.Schafer L.V., de Jong D.H., Holt A., Rzepiela A.J., de Vries A.H., Poolman B., Killian J.A., Marrink S.J. Lipid packing drives the segregation of transmembrane helices into disordered lipid domains in model membranes. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1343–1348. doi: 10.1073/pnas.1009362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sheets E.D., Lee G.M., Simson R., Jacobson K. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 1997;36:12449–12458. doi: 10.1021/bi9710939. [DOI] [PubMed] [Google Scholar]
- 86.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 87.Varma R., Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]
- 88.Sharma P., Varma R., Sarasij R.C., Ira K. Gousset, Krishnamoorthy G., Rao M., Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 89.Brown D.A., Rose J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 90.Lichtenberg D., Goni F.M., Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 91.Schuck S., Honsho M., Ekroos K., Shevchenko A., Simons K. Resistance of cell membranes to different detergents. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Simons K., van Meer G. Lipid sorting in epithelial cells. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 93.Fujita A., Cheng J., Hirakawa M., Furukawa K., Kusunoki S., Fujimoto T. Gangliosides GM1 and GM3 in the living cell membrane form clusters susceptible to cholesterol depletion and chilling. Mol. Biol. Cell. 2007;18:2112–2122. doi: 10.1091/mbc.E07-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reth M. Oligomeric antigen receptors: a new view on signaling for the selection of lymphocytes. Trends Immunol. 2001;22:356–360. doi: 10.1016/s1471-4906(01)01964-0. [DOI] [PubMed] [Google Scholar]
- 95.Yang J., Reth M. Oligomeric organization of the B-cell antigen receptor on resting cells. Nature. 2010;467:465–469. doi: 10.1038/nature09357. [DOI] [PubMed] [Google Scholar]
- 96.Hite R.K., Li Z., Walz T. Principles of membrane protein interactions with annular lipids deduced from aquaporin-0 2D crystals. EMBO J. 2010;29:1652–1658. doi: 10.1038/emboj.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Campbell K.S., Backstrom B.T., Tiefenthaler G., Palmer E. CART: a conserved antigen receptor transmembrane motif. Semin. Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- 98.Reth M. Antigen receptors on B lymphocytes. Annu. Rev. Immunol. 1992;10:97–121. doi: 10.1146/annurev.iy.10.040192.000525. [DOI] [PubMed] [Google Scholar]
- 99.Call M.E., Pyrdol J., Wiedmann M., Wucherpfennig K.W. The organizing principle in the formation of the T cell receptor-CD3 complex. Cell. 2002;111:967–979. doi: 10.1016/s0092-8674(02)01194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Call M.E., Wucherpfennig K.W. Molecular mechanisms for the assembly of the T cell receptor-CD3 complex. Mol. Immunol. 2004;40:1295–1305. doi: 10.1016/j.molimm.2003.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharpe H.J., Stevens T.J., Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Spira F., Mueller N.S., Beck G., von Olshausen P., Beig J., Wedlich-Soldner R. Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat. Cell Biol. 2012;14:640–648. doi: 10.1038/ncb2487. [DOI] [PubMed] [Google Scholar]
- 103.Kai M., Sakane F., Jia Y.J., Imai S., Yasuda S., Kanoh H. Lipid phosphate phosphatases 1 and 3 are localized in distinct lipid rafts. J. Biochem. 2006;140:677–686. doi: 10.1093/jb/mvj195. [DOI] [PubMed] [Google Scholar]
- 104.Goswami D., Gowrishankar K., Bilgrami S., Ghosh S., Raghupathy R., Chadda R., Vishwakarma R., Rao M., Mayor S. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fujita A., Cheng J., Fujimoto T. Segregation of GM1 and GM3 clusters in the cell membrane depends on the intact actin cytoskeleton. Biochim. Biophys. Acta. 2009;1791:388–396. doi: 10.1016/j.bbalip.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 106.Gowrishankar K., Ghosh S., Saha R.C., Mayor S., Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 107.Hao S., August A. Actin depolymerization transduces the strength of B-cell receptor stimulation. Mol. Biol. Cell. 2005;16:2275–2284. doi: 10.1091/mbc.E04-10-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harwood N.E., Batista F.D. The cytoskeleton coordinates the early events of B-cell activation. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Treanor B., Depoil D., Gonzalez-Granja A., Barral P., Weber M., Dushek O., Bruckbauer A., Batista F.D. The membrane skeleton controls diffusion dynamics and signaling through the B cell receptor. Immunity. 2010;32:187–199. doi: 10.1016/j.immuni.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Charrier C., Ehrensperger M.V., Dahan M., Levi S., Triller A. Cytoskeleton regulation of glycine receptor number at synapses and diffusion in the plasma membrane. J. Neurosci. 2006;26:8502–8511. doi: 10.1523/JNEUROSCI.1758-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kusumi A., Nakada C., Ritchie K., Murase K., Suzuki K., Murakoshi H., Kasai R.S., Kondo J., Fujiwara T. Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu. Rev. Biophys. Biomol. Struct. 2005;34:351–378. doi: 10.1146/annurev.biophys.34.040204.144637. [DOI] [PubMed] [Google Scholar]
- 112.Fujiwara T., Ritchie K., Murakoshi H., Jacobson K., Kusumi A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002;157:1071–1081. doi: 10.1083/jcb.200202050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Treanor B., Depoil D., Bruckbauer A., Batista F.D. Dynamic cortical actin remodeling by ERM proteins controls BCR microcluster organization and integrity. J. Exp. Med. 2011;208:1055–1068. doi: 10.1084/jem.20101125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mattila P.K., Feest C., Depoil D., Treanor B., Montaner B., Otipoby K.L., Carter R., Justement L.B., Bruckbauer A., Batista F.D. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity. 2013;38:461–474. doi: 10.1016/j.immuni.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 115.Paladino S., Lebreton S., Tivodar S., Campana V., Tempre R., Zurzolo C. Different GPI-attachment signals affect the oligomerisation of GPI-anchored proteins and their apical sorting. J. Cell Sci. 2008;121:4001–4007. doi: 10.1242/jcs.036038. [DOI] [PubMed] [Google Scholar]
- 116.Paladino S., Lebreton S., Tivodar S., Formiggini F., Ossato G., Gratton E., Tramier M., Coppey-Moisan M., Zurzolo C. Golgi sorting regulates organization and activity of GPI proteins at apical membranes. Nat. Chem. Biol. 2014;10:350–357. doi: 10.1038/nchembio.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brugger B., Sandhoff R., Wegehingel S., Gorgas K., Malsam J., Helms J.B., Lehmann W.D., Nickel W., Wieland F.T. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J. Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Manneville J.B., Casella J.F., Ambroggio E., Gounon P., Bertherat J., Bassereau P., Cartaud J., Antonny B., Goud B. COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16946–16951. doi: 10.1073/pnas.0807102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soares H., Henriques R., Sachse M., Ventimiglia L., Alonso M.A., Zimmer C., Thoulouze M.I., Alcover A. Regulated vesicle fusion generates signaling nanoterritories that control T cell activation at the immunological synapse. J. Exp. Med. 2013;210:2415–2433. doi: 10.1084/jem.20130150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. doi: 10.1038/338383b0. [DOI] [PubMed] [Google Scholar]
- 121.Pao L.I., Famiglietti S.J., Cambier J.C. Asymmetrical phosphorylation and function of immunoreceptor tyrosine-based activation motif tyrosines in B cell antigen receptor signal transduction. J. Immunol. 1998;160:3305–3314. [PubMed] [Google Scholar]
- 122.Schmitz R., Baumann G., Gram H. Catalytic specificity of phosphotyrosine kinases Blk Lyn, c-Src and Syk as assessed by phage display. J. Mol. Biol. 1996;260:664–677. doi: 10.1006/jmbi.1996.0429. [DOI] [PubMed] [Google Scholar]
- 123.Futterer K., Wong J., Grucza R.A., Chan A.C., Waksman G. Structural basis for Syk tyrosine kinase ubiquity in signal transduction pathways revealed by the crystal structure of its regulatory SH2 domains bound to a dually phosphorylated ITAM peptide. J. Mol. Biol. 1998;281:523–537. doi: 10.1006/jmbi.1998.1964. [DOI] [PubMed] [Google Scholar]
- 124.Tolar P., Sohn H.W., Pierce S.K. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat. Immunol. 2005;6:1168–1176. doi: 10.1038/ni1262. [DOI] [PubMed] [Google Scholar]
- 125.Kerppola T.K. Visualization of molecular interactions by fluorescence complementation. Nat. Rev. Mol. Cell Biol. 2006;7:449–456. doi: 10.1038/nrm1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rolli V., Gallwitz M., Wossning T., Flemming A., Schamel W.W.A., Zürn C., Reth M. Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Mol. Cell. 2002;10:1057–1069. doi: 10.1016/s1097-2765(02)00739-6. [DOI] [PubMed] [Google Scholar]
- 127.Siegers G.M., Yang J., Duerr C.U., Nielsen P.J., Reth M., Schamel W.W. Identification of disulfide bonds in the Ig-alpha/Ig-beta component of the B cell antigen receptor using the Drosophila S2 cell reconstitution system. Int. Immunol. 2006;18:1385–1396. doi: 10.1093/intimm/dxl072. [DOI] [PubMed] [Google Scholar]
- 128.Yang J., Reth M. The dissociation activation model of B cell antigen receptor triggering. FEBS Lett. 2010;584:4872–4877. doi: 10.1016/j.febslet.2010.09.045. [DOI] [PubMed] [Google Scholar]
- 129.van Zelm M.C., Smet J., Adams B., Mascart F., Schandene L., Janssen F., Ferster A., Kuo C.C., Levy S., van Dongen J.J., van der Burg M. CD81 gene defect in humans disrupts CD19 complex formation and leads to antibody deficiency. J. Clin. Invest. 2010;120:1265–1274. doi: 10.1172/JCI39748. [DOI] [PMC free article] [PubMed] [Google Scholar]