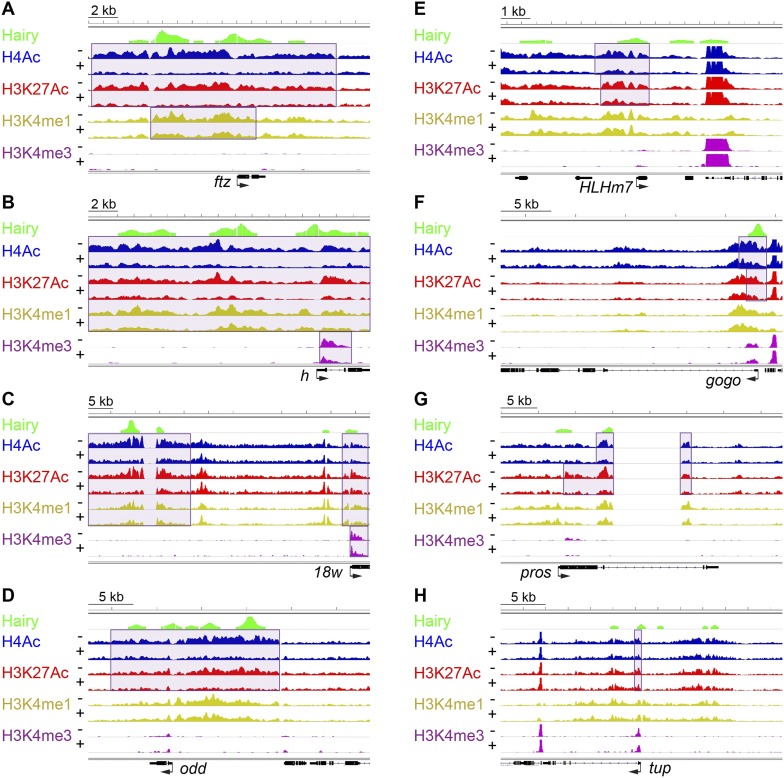
Figure 2. Examples of coupled, large-scale chromatin changes mediated by Hairy.
Chromatin immunoprecipitation-high throughput sequencing (ChIP-seq) tracks for H4Ac, H3K27Ac, H3K4me1 and H3K4me3 are shown at repressed genes before (−) and after Hairy (+) induction, with gene models below. (A–D) Coupled reduction of active histone marks was observed in a wide-spread fashion on ftz, h, 18w and odd genes (scale at top left). (E–H) Relatively smaller blocks of chromatin changes were detected on HLHm7, gogo, pros and tup genes. Significantly changed regions (shaded boxes) were identified by the diffReps program. Hairy binding (top track) from MacArthur et al. (2009).