Abstract
A balance of van der Waals, electrostatic, and hydrophobic forces drive the folding and packing of protein side chains. Although such interactions between residues are often approximated as being pairwise additive, in reality, higher-order many-body contributions that depend on environment drive hydrophobic collapse and cooperative electrostatics. Beginning from dead-end elimination, we derive the first algorithm, to our knowledge, capable of deterministic global repacking of side chains compatible with many-body energy functions. The approach is applied to seven PCNA x-ray crystallographic data sets with resolutions 2.5–3.8 Å (mean 3.0 Å) using an open-source software. While PDB_REDO models average an Rfree value of 29.5% and MOLPROBITY score of 2.71 Å (77th percentile), dead-end elimination with the polarizable AMOEBA force field lowered Rfree by 2.8–26.7% and improved mean MOLPROBITY score to atomic resolution at 1.25 Å (100th percentile). For structural biology applications that depend on side-chain repacking, including x-ray refinement, homology modeling, and protein design, the accuracy limitations of pairwise additivity can now be eliminated via polarizable or quantum mechanical potentials.
Introduction
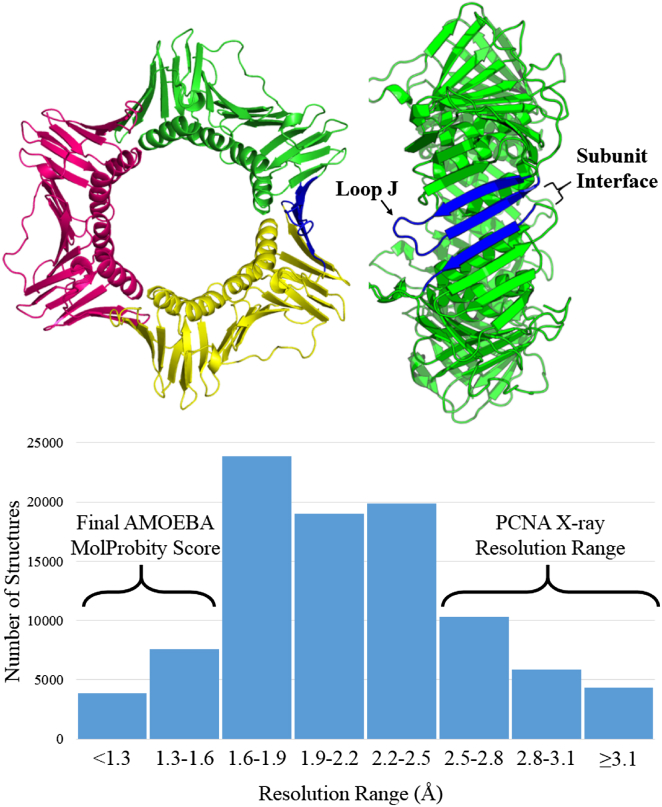
The Protein Data Bank (PDB; http://www.rcsb.org/pdb/home/home.do) (1) now contains biomolecular structural models derived from >90,000 x-ray diffraction experiments conducted over the last half century. More than 80,000 of these structures have been deposited with their original diffraction data, which permits the experiments to be more fully interpreted as biomolecular refinement programs improve (2,3). Only a small fraction of PDB structures result from diffraction data to atomic resolution (i.e., ∼1 Å). Mid- to low-resolution data sets, such as those for proliferating-cell nuclear antigen (PCNA) studied here, are much more common (Fig. 1). For these data, attainment of high-quality models relies heavily on the use of both systematic validation tools such as MOLPROBITY (4) and the prior chemical knowledge contained in molecular mechanics force fields (5). It is also possible to leverage previously solved structures to parameterize restraints based on elastic networks (6,7), although this level of coarse-graining is incapable of repacking side chains as networks deform or come together to form the interface of a complex.
Figure 1.
The biological unit of PCNA (above) is a trimeric ring with a central hole for binding double-strand DNA. Viewing PCNA from the side shows the subunit interface between two PCNA monomers (blue). Histogram (below) showing the number of x-ray diffraction data sets within resolution bins for structures deposited in the PDB as of January 20, 2015. The experimental resolution and deposited MOLPROBITY scores for the PCNA data sets used in this study averaged 2.96 and 2.86 Å, respectively. Structures determined by the algorithm described here yielded a mean MOLPROBITY score of 1.25 Å.
To address the protein side-chain repacking problem, a brute force search over discrete conformations is computationally intractable for even small proteins due to a combinatorial explosion of conformational possibilities. However, by considering the relative energetics of discrete side-chain conformations (rotamers) for a single residue in the context of its interactions with the rest of the protein structure, unfavorable rotamers can be eliminated by proving that they cannot be part of the global minimum energy conformation (GMEC). The eliminated conformations are dead-ends in the search; therefore, the algorithm used to eliminate rotamers, rotamer pairs, and so on, is known as dead-end elimination (DEE).
The combination of low-energy side-chain rotamer libraries (8–10) with DEE (11,12) global optimization has been widely used for protein electrostatic network optimization and sequence design (13–17). However, rotamer elimination criteria have only been defined for pairwise-additive energy functions such as the OPLS-AA (18), AMBER (19), and CHARMM (20) families of fixed partial-charge force fields and pairwise decomposable continuum solvents (21–23). Explicit inclusion of many-body effects has been neglected such that the strength of the interaction between two residues must be independent of their mutual environment. Therefore, important molecular driving forces such as the hydrophobic effect (24) and electronic polarization (25), which are fundamentally many-body in nature, have been implicitly approximated or neglected entirely (Fig. 2). Here we overcome the restriction to pairwise energy functions by showing that both the DEE criteria (11) and more-stringent Goldstein criteria (12) can be derived in the context of many-body energy functions such as polarizable force fields (25,26) as well as quantum mechanical potentials and continuum solvents (27–30).
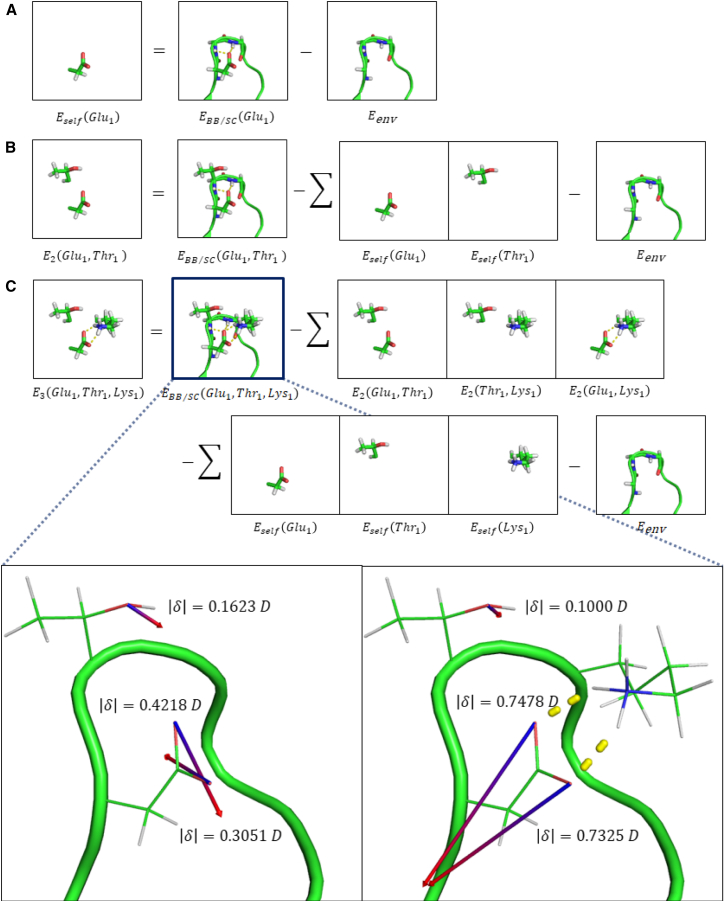
Figure 2.
This diagram explains energy terms used in Eq. 1 including the (A) self-energy, (B) two-body energy, and (C) three-body energy. The induced dipole vectors for three side-chain oxygen atoms in wild-type PCNA are shown in the absence of Lys107 (left) and in its presence (right). A vector length of 7 Å corresponds to 1 Debye. Changes in induced dipole direction and magnitude reflect the AMOEBA electronic polarization response to the total electrostatic field, which results in polarization energy that is not pairwise. For this example, the three-body energy is 1.55 kcal/mol.
Rotamer and rotamer pair elimination criteria compatible with many-body energy functions are given below and their derivations supplied in the Supporting Material. For a pairwise decomposable energy function, the new expressions simplify to the established elimination criteria. The approach is used to refine a series of PCNA structures in the context of a many-body x-ray crystallographic target function Etot = Echem + wAex-ray. Here Echem is a parallelized implementation of the polarizable AMOEBA force field that supports space group symmetry (31), Ex-ray is a real-space electron density function (32,33), and wA is used to weight the importance of the force field and x-ray terms (33). The resulting AMOEBA structures are compared to PDB_REDO (34) and pairwise DEE refinements based on the OPLS-AA/L(18) fixed charge force field.
Finally, functional insights into changes in PCNA stability due to single amino-acid mutations are discussed. PCNA plays an essential role in the maintenance of genome stability. It is a replication accessory factor that interacts with and regulates the activities of proteins involved in DNA replication, DNA repair, DNA recombination, chromatin modifications, sister chromatid cohesion, and cell-cycle control (35). Each PCNA subunit consists of two domains, which interact in a head-to-tail arrangement to form a ring-shaped homo-trimer possessing pseudo-sixfold symmetry (Fig. 1) (36). The PCNA trimer binds double-stranded DNA through the central pore of the ring. PCNA function is regulated in part by posttranslational modifications. For example, ubiquitylation of PCNA on Lys164 promotes translesion synthesis (TLS), which is the replicative and generally mutagenic bypass of damaged DNA (37). Several separation-of-function mutations in PCNA have been identified that inhibit various cellular processes including DNA mismatch repair as well as TLS (38,39). X-ray structures of wild-type PCNA, ubiquitin-modified PCNA, SUMO-modified PCNA, two separation-of-function mutant PCNA proteins that block mismatch repair, and two separation-of-function mutant proteins that block TLS have been determined (40–42).
Materials and Methods
Theory
Side-chain repacking with a pairwise potential
For a potential energy function that approximates nonbonded interactions as being a pairwise sum over residues, the total energy of a protein E(r) is given by
| (1) |
where Eenv is the energy of the environment (i.e., the protein backbone and residues that are not being optimized), Eself(ri) is the self-energy of residue including its intramolecular bonded energy terms and nonbonded interactions with the backbone, and E2(ri, rj) is the two-body nonbonded interaction energy between residues i and j with other residues turned off. The self-energy and two-body terms, diagrammed in Fig. 2, are calculated as
| (2) |
| (3) |
where EBB/SC(ri) is the energy of the protein backbone with only the side chain of residue i attached. Likewise, EBB/SC(ri,rj) is the energy of the backbone and only side chains i,j. Eenv is subtracted from each self and two-body term to avoid double counting. The original elimination criteria for rotamers and rotamer pairs (11), respectively, under the approximation of a pairwise decomposable force field, are
| (4) |
| (5) |
| (6) |
where riα and riβ are different rotamers of the same residue i. The prime notation indicates that the summation occurs over all residues i ≠ j; similarly, k′ implies k ≠ i, k ≠ j.
Side-chain repacking with a many-body potential
Under a many-body potential, the total energy of a protein E(r) can be defined to arbitrary precision using the expansion
| (7) |
where the three- and four-body contributions, respectively, are given by
| (8) |
| (9) |
The DEE rotamer and rotamer pair elimination equations, respectively, can be extended to arbitrary order as follows:
| (10) |
| (11) |
where the ellipses signify the presence of further higher-order terms up to n-body (see the Supporting Material for the derivation).
Although terms based on interactions between three or more residues are zero for pairwise decomposable energy functions such as OPLS-AA/L, for the polarizable AMOEBA force field the three-body term E3(ri, rj, rk) captures energetic changes due to mutual polarization between the three residues and their environment (Fig. 2). In the context of continuum solvation, higher-order terms additionally capture energetic changes that result from modifications to the protein-solvent dielectric boundary.
Goldstein elimination
More stringent elimination criteria were introduced by Goldstein (12), which for a pairwise energy function are given by
| (12) |
and
| (13) |
for rotamer and rotamer pair elimination, respectively. The Goldstein elimination criteria, extended to include higher-order energy components for rotamers and rotamer pairs, respectively, are given by
| (14) |
and
| (15) |
Many-body x-ray refinement function
Pairwise molecular mechanics force fields have been used in tandem with experimental x-ray diffraction data to refine protein structural models for more than two decades (43,44). To quantify agreement between the experimental and model electron densities, and avoid overfitting, both R and Rfree values are monitored (45). To measure agreement between the structural model and prior chemical knowledge, the MOLPROBITY structure validation tool (4) compares van der Waals contacts, hydrogen-bond distances, side-chain rotamers, and peptide backbone conformation with tabulated values from high-resolution protein structures. The overall MOLPROBITY score was calibrated against the PDB to reflect the x-ray diffraction resolution that, on average, is needed to produce a structure of a given quality. For example, the average MOLPROBITY score for the original seven PCNA models indicates structure quality consistent with 2.86 Å diffraction data, which is near the actual 2.96 Å experimental resolution of the data. MOLPROBITY clash scores were corrected based on experimental evidence (46) and quantum mechanical calculations for the optimal CH…O hydrogen-bond distance (47). Although the ideal distance is reported to be 2.3 Å, MOLPROBITY incorrectly reports this separation as a clash (31). The corrected scores are denoted uwith a footnote as MOLPROBITYa.
The optimization procedure used here operates on a hybrid target function based on maximum-likelihood principles (48). The target function (ETot) is composed of a weighted sum of force-field (25,49,50) (Echem) and x-ray (Ex-ray) energy terms, where the latter is a measure of the agreement between a real-space map and the electron density of the model:
| (16) |
Calculation of real-space density maps followed the formalism of Read (51) and implementation of Cowtan (52) to compute σA and figure-of-merit coefficients for structure factors. Real-space density values at specific coordinates were computed using a Catmull-Rom spline (τ = 0.25). OPLS-AA/L and AMOEBA electrostatics were evaluated using particle-mesh Ewald summation as described previously in Schnieders et al. (31).
Methods
The rotamer elimination criteria were implemented in the FORCE FIELD X (FFX) molecular biophysics software package (http://ffx.biochem.uiowa.edu) (31,33) and applied in an iterative fashion, such that rounds of rotamer and rotamer-pair elimination were performed until no new eliminations were produced. The target function for all remaining permutations was then evaluated to determine the GMEC. For all AMOEBA stages of this work, the electron density and potential energy terms were weighted equally (33). The electron density weight was doubled for OPLS-AA/L refinements (wA = 2) as this was observed to yield output structures with a better balance between R and other quality metrics.
Seven structures of PCNA were optimized according to the following protocol: input structures were first minimized (in coordinates and temperature factors) to remove clashes, and then the coordinates of each side chain were recorded. Each unit cell was divided into subvolumes with axis lengths of 4 Å that were placed with 3 Å overlap between neighboring subvolumes. Residues were placed into any box containing their Cα atom. Side-chain optimization via DEE was performed on each box using the Richardson rotamer library augmented by the initial coordinates of each residue as an additional rotameric choice (Ponder and Richards (8)). Pairwise DEE was applied for OPLS-AA/L, while many-body DEE truncated after trimer interactions was used for AMOEBA. After another round of minimization in both coordinates and temperature factors, residues that remained in poor rotameric positions were optimized individually using the same criteria, but without using the initial coordinates as a rotameric choice. This final step was performed iteratively until no further improvement in structure quality metrics was achieved, which accounted for <5% of the final side-chain positions.
A conservative approximation was employed to significantly reduce the computational expense of applying the elimination criteria. All rotamers whose self-energy was 30 kcal/mol larger than that residue’s self-energy minimum were pruned before continuing on to two- and three-body calculations. This approach is based on the observation that rotamers with self-energy disparities of this magnitude, which often arise from side-chain van der Waals clashes with the protein backbone, are inconsistent with a well-packed GMEC (11). Such prunable rotamers are also eliminated during application of the rotamer elimination criterion; however, removing them immediately after self-energy calculation drastically reduces the required number of two- and three-body energies. This pruning strategy produced even more benefit under a hybrid target function because in many cases the density map is well fit by only a handful of rotamers.
Results and Discussion
Atomic-resolution quality from mid- to low-resolution diffraction
PCNA data sets (Table 1) are ideal for demonstrating the ability of the refinement approaches described above to achieve atomic-resolution structural quality from mid- to low-resolution diffraction data. Overall, many-body DEE with the AMOEBA polarizable force field yielded higher quality PCNA models than PDB_REDO, local minimization, or traditional two-body DEE using OPLS-AA/L (Table 2). Although each strategy was able to improve the original PDB models, many-body DEE displayed the most significant gains across all major quality metrics. Mean improvement in Rfree was 3.0 for AMOEBA DEE versus 2.5 for pairwise OPLS-AA/L DEE and almost no reduction for PDB_REDO. The locally minimized structures were used as a baseline for comparing force-field energy, against which both OPLS-AA/L and AMOEBA DEE models were favored by an average of >200 kcal/mol per structure. These large increases in stability may favorably affect downstream computational methods such as molecular-dynamics simulations, which generally begin from a crystal structure after local optimization using a chosen force field, but without side-chain repacking. A more targeted analysis on the effects of the three-body term is available in Table S3 in the Supporting Material, which compares structure quality for the two- versus three-body approximation under AMOEBA. Three-body optimization under AMOEBA is shown to yield additional improvements not obtained by any other combination.
Table 1.
The PDB ID, resolution, R/Rfree values, and MOLPROBITY analyses for the deposited PCNA models
| Data Set | Resolution (Å) | Reported |
FFX |
MOLPROBITYa |
Clasha |
Ramachandran |
Poor |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R | Rfree | R | Rfree | Score | % | Score | % | Out (%) | Favorable (%) | Rotamer (%) | ||
| 3F1W | 2.90 | 22.8 | 25.5 | 23.5 | 25.9 | 2.81 | 81 | 35.3 | 65 | 0.4 | 95.2 | 3.9 |
| 3GPM | 3.80 | 27.5 | 31.2 | 35.4 | 34.3 | 3.43 | 73 | 52.9 | 51 | 4.0 | 89.3 | 7.5 |
| 3GPN | 2.50 | 23.6 | 27.3 | 23.8 | 27.3 | 2.19 | 91 | 11.8 | 92 | 0.0 | 98.0 | 6.2 |
| 3L0W | 2.80 | 27.9 | 31.4 | 31.5 | 33.2 | 3.57 | 23 | 51.0 | 20 | 0.0 | 92.3 | 15.8 |
| 3L0X | 3.00 | 24.4 | 26.7 | 24.3 | 25.7 | 2.79 | 86 | 15.0 | 97 | 0.0 | 94.4 | 9.2 |
| 3L10 | 2.80 | 27.9 | 31.4 | 31.8 | 34.4 | 3.56 | 23 | 51.8 | 20 | 0.0 | 92.3 | 15.1 |
| WT | 2.95 | 24.6 | 27.3 | 24.9 | 27.3 | 1.65 | 100 | 5.5 | 100 | 0.4 | 94.9 | 0.9 |
| Mean | 2.96 | 25.5 | 28.7 | 27.9 | 29.7 | 2.86 | 68 | 31.9 | 64 | 0.7 | 93.8 | 8.4 |
See main text for explanation.
Table 2.
The R/Rfree values, change in force-field potential energy, and MOLPROBITY analyses for the PCNA data sets are given for PDB_REDO, OPLS-AA/L, and AMOEBA refinement methods
| Data Set | Model | R | Rfree | EFF | MOLPROBITYa |
Clasha |
Ramachandran |
Poor |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Score | % | Score | % | Out (%) | Favorable (%) | Rotamer (%) | |||||
| 3F1W | PDB_REDO | 27.01 | 29.28 | 1.98 | 99 | 3.7 | 100 | 0.0 | 96.0 | 5.2 | |
| OPLS-AA/L | 23.78 | 26.86 | 1.78 | 100 | 1.0 | 100 | 0.8 | 93.3 | 5.2 | ||
| + DEE | 23.99 | 26.21 | 133 | 1.67 | 100 | 1.2 | 100 | 0.8 | 93.7 | 3.5 | |
| AMOEBA | 21.94 | 26.11 | 1.39 | 100 | 0.5 | 100 | 0.4 | 96.4 | 4.4 | ||
| + DEE | 22.12 | 26.25 | −129 | 1.03 | 100 | 1.2 | 100 | 0.4 | 96.4 | 0.4 | |
| 3GPM | PDB_REDO | 30.14 | 32.01 | 2.68 | 96 | 8.5 | 97 | 4.8 | 86.9 | 6.1 | |
| OPLS-AA/L | 26.35 | 28.25 | 1.94 | 100 | 0.5 | 100 | 4.8 | 84.5 | 6.1 | ||
| + DEE | 23.88 | 26.32 | 40 | 1.51 | 100 | 0.5 | 100 | 3.2 | 85.3 | 1.8 | |
| AMOEBA | 29.53 | 30.43 | 2.13 | 99 | 1.3 | 100 | 5.6 | 84.9 | 6.6 | ||
| + DEE | 24.42 | 27.25 | −110 | 1.33 | 100 | 0.0 | 100 | 4.4 | 85.7 | 1.8 | |
| 3GPN | PDB_REDO | 23.60 | 27.24 | 2.02 | 95 | 2.8 | 100 | 0.4 | 96.0 | 7.9 | |
| OPLS-AA/L | 21.26 | 25.60 | 1.86 | 97 | 1.8 | 100 | 0.4 | 95.6 | 6.6 | ||
| + DEE | 21.65 | 26.04 | −242 | 1.41 | 100 | 1.3 | 100 | 0.8 | 95.6 | 2.2 | |
| AMOEBA | 20.75 | 25.19 | 2.07 | 94 | 2.3 | 100 | 0.0 | 96.4 | 12.3 | ||
| + DEE | 20.98 | 25.59 | −351 | 1.28 | 100 | 0.5 | 100 | 0.0 | 96.4 | 3.1 | |
| 3L0W | PDB_REDO | 28.58 | 32.45 | 3.14 | 53 | 30.1 | 70 | 4.0 | 86.7 | 5.4 | |
| OPLS-AA/L | 30.07 | 31.50 | 1.99 | 99 | 1.0 | 100 | 1.2 | 93.2 | 10.1 | ||
| + DEE | 28.17 | 30.80 | −316 | 1.86 | 99 | 0.8 | 100 | 1.9 | 92.3 | 7.1 | |
| AMOEBA | 27.09 | 29.71 | 2.29 | 96 | 2.9 | 100 | 1.2 | 92.3 | 9.4 | ||
| + DEE | 27.12 | 29.60 | −260 | 1.17 | 100 | 1.0 | 100 | 0.6 | 94.7 | 1.7 | |
| 3L0X | PDB_REDO | 22.26 | 23.53 | 2.71 | 89 | 8.2 | 97 | 1.6 | 91.2 | 9.6 | |
| OPLS-AA/L | 21.33 | 24.27 | 2.05 | 99 | 1.3 | 100 | 1.6 | 93.6 | 10.9 | ||
| + DEE | 20.47 | 24.78 | −334 | 1.51 | 100 | 0.8 | 100 | 1.6 | 92.8 | 2.6 | |
| AMOEBA | 20.92 | 24.10 | 2.22 | 98 | 3.0 | 100 | 0.8 | 92.4 | 7.4 | ||
| + DEE | 20.98 | 24.43 | −125 | 1.25 | 100 | 1.8 | 100 | 1.6 | 92.8 | 0.4 | |
| 3L10 | PDB_REDO | 28.01 | 32.04 | 3.35 | 37 | 43.8 | 33 | 5.0 | 84.2 | 5.4 | |
| OPLS-AA/L | 30.41 | 33.08 | 1.79 | 100 | 0.4 | 100 | 2.2 | 92.9 | 8.4 | ||
| + DEE | 27.04 | 30.65 | −281 | 1.67 | 100 | 0.6 | 100 | 1.6 | 91.0 | 4.0 | |
| AMOEBA | 27.58 | 30.20 | 2.25 | 97 | 3.3 | 100 | 0.6 | 92.0 | 7.1 | ||
| + DEE | 26.82 | 29.56 | −360 | 1.59 | 100 | 1.0 | 100 | 0.6 | 91.6 | 3.4 | |
| WT | PDB_REDO | 28.71 | 30.15 | 3.06 | 68 | 14.6 | 96 | 0.8 | 92.1 | 15.6 | |
| OPLS-AA/L | 23.89 | 27.22 | 1.69 | 100 | 0.5 | 100 | 1.6 | 92.9 | 5.6 | ||
| + DEE | 23.21 | 25.57 | −664 | 1.69 | 100 | 1.5 | 100 | 2.0 | 93.3 | 3.0 | |
| AMOEBA | 22.64 | 25.89 | 1.73 | 100 | 2.0 | 100 | 0.8 | 94.9 | 3.5 | ||
| + DEE | 21.63 | 24.24 | −760 | 1.09 | 100 | 0.7 | 100 | 1.2 | 94.9 | 0.9 | |
| Mean | PDB_REDO | 26.90 | 29.53 | 2.71 | 77 | 16.0 | 85 | 2.4 | 90.1 | 7.9 | |
| OPLS-AA/L | 25.30 | 28.11 | 1.87 | 99 | 0.9 | 100 | 1.8 | 92.3 | 7.6 | ||
| + DEE | 24.06 | 27.20 | −238 | 1.62 | 100 | 0.9 | 100 | 1.7 | 92.0 | 3.5 | |
| AMOEBA | 24.35 | 27.38 | 2.01 | 98 | 2.2 | 100 | 1.3 | 92.8 | 7.2 | ||
| + DEE | 23.44 | 26.70 | −299 | 1.25 | 100 | 0.9 | 100 | 1.3 | 93.2 | 1.7 | |
All R/Rfree values were calculated in FFX for consistency. Potential energy (Eff) after DEE repacking is reported relative to the energy after local minimization (kcal/mol).
See main text for explanation.
The MOLPROBITY score was improved by ∼1.0 using local minimization alone, by 1.24 using pairwise OPLS-AA/L rotamer optimization, and by 1.61 using many-body AMOEBA rotamer optimization. The latter placed all seven structures in the 100th MOLPROBITY percentile among structures of this resolution range (3.0 Å). Clash score was improved to the 100th percentile by all methods except PDB_REDO. The deposited structures averaged poor rotameric positions for 8.4% of side chains, which was not significantly improved by local minimization methods. DEE using both pairwise OPLS-AA/L and many-body AMOEBA algorithms reduced the percent of poor rotamers by 4.9 and 6.7%, respectively. Although both DEE methods yielded marked improvements in most structure quality metrics, the many-body AMOEBA improvements were greatest. Relative to OPLS-AA/L pairwise DEE, AMOEBA DEE shows mean additional improvements of a lower Rfree by 0.5, lower MOLPROBITY score by 0.37, 1.8% fewer poor rotamers, better clash score, and more favorable Ramachandran values. These additional improvements are driven by the inclusion of many-body polarization and atomic multipole electrostatics, which are critically important to capture the bifurcated hydrogen bonding that stabilizes both α-helical and β-sheet secondary structure (Fig. 3 and Fig. S1 in the Supporting Material) (46). As shown in Table 3, truncation of the many-body expansion at pairwise interactions neglects ∼1 kcal/mol/residue of interaction energy under the AMOEBA polarizable force field. Fortunately, truncation at three-body interactions neglects <0.1 kcal/mol/residue, which is a reasonable compromise between efficiency and residual error due to higher-order neglected interactions (i.e., four-body and higher). Distributions of self, pair, and three-body energies for the wild-type structure, as well as distributions of slack (i.e., the amount of energy by which the elimination criterion was exceeded), are available in Fig. S4. Ninety-percent of three-body energies by absolute magnitude are greater than only 0.04% of self-elimination slacks and none of the pair slacks. The largest individual three-body energy, however, is >10 kcal/mol. We thus expect that there exist individual fourth- and higher-order energies (at short distances) with significant impact on elimination, but calculation of fourth-order energies represents an infeasible computational cost for structures of PCNA’s size. Comparison of run times for the methods tested herein is available in Table S4.
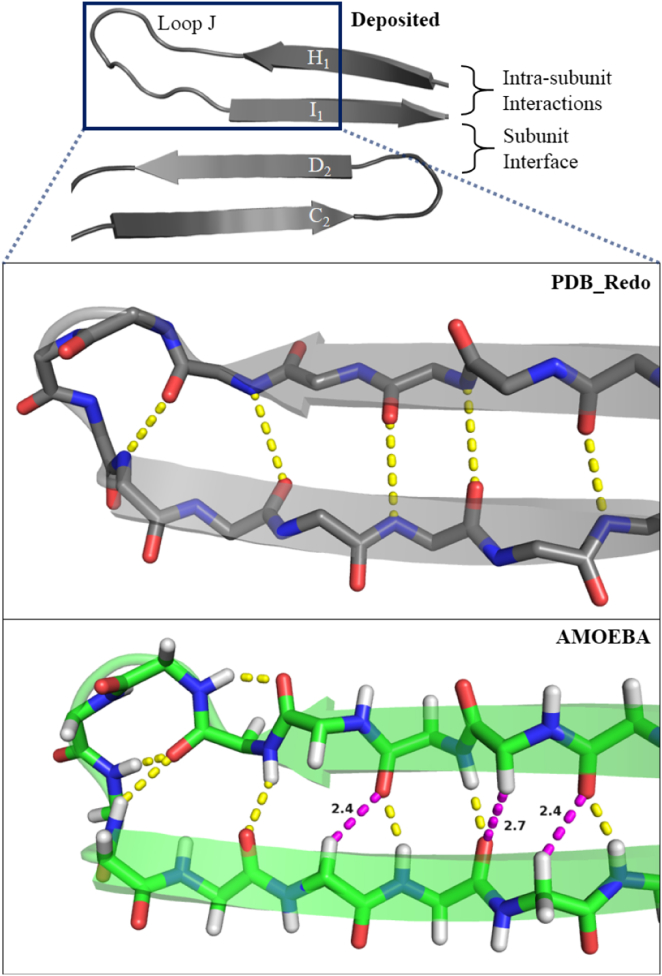
Figure 3.
The PCNA H1 and I1β-strands and loop J backbone electrostatic network for 3GPM from PDB_REDO and from the AMOEBA DEE refinement in FFX. The intrastrand interaction is stabilized by five backbone hydrogen bonds in the PDB_REDO structure (above), while the AMOEBA model (below) contains seven. The AMOEBA model has three additional low-energy CαH···O bonds (purple). The bifurcated hydrogen bonding is driven largely by the quadrupole moment of the carbonyl oxygen. See Fig. S1 for similar improvements in α-helical structure.
Table 3.
Neglected higher-order energy when truncating at two- or three-body interactions when using the AMOEBA force field
| Data Set | Etotal | Ebackbone | ΣEself | ΣEpair | ΣEtrimer |
Eneglected |
Eneglected/Residue |
||
|---|---|---|---|---|---|---|---|---|---|
| Two-Body | Three-Body | Two-Body | Three-Body | ||||||
| 3F1W | −7303.7 | −2392.7 | −2737.2 | −2387.8 | 230.0 | 213.9 | −16.0 | 0.8 | −0.1 |
| 3GPM | −7777.1 | −2459.3 | −2886.1 | −2687.5 | 283.0 | 255.7 | −27.3 | 1.0 | −0.1 |
| 3GPN | −7618.3 | −2838.6 | −2368.5 | −2628.5 | 232.7 | 217.3 | −15.4 | 0.9 | −0.1 |
| Wild-type | −7591.7 | −2385.2 | −2685.5 | −2765.7 | 269.6 | 244.7 | −24.9 | 1.0 | −0.1 |
| Mean | 232.9 | −20.9 | 0.9 | −0.1 | |||||
Total energy neglected when truncating the expansion after pairwise interactions is ∼1 kcal/mol/residue. By contrast, truncation after three-body interactions reduces the neglected energy by an order of magnitude to <0.1 kcal/mol/residue.
Structural insights into the relative stability of PCNA mutants
The newly refined AMOEBA models provide structural and mechanistic insights that are supported by the x-ray diffraction data, but were not achieved in the original models due to limitations in available refinement algorithms. To demonstrate this, we now focus on E113G and G178S TLS-deficient separation-of-function PCNA mutants (38,53). These substitutions, E113G in β-strand I1 and G178S in β-strand D2, are at the subunit interface of PCNA, where antiparallel strand interactions between I1 and D2 stabilize the PCNA trimer (36). The original structural models demonstrated partial separation of these β-strands in both mutant proteins relative to the wild-type protein (40). In addition, biochemical studies showed that both mutant proteins have significantly reduced trimer stability relative to the wild-type protein, which is responsible for their inability to support TLS (54).
The structural basis for the separation of β I1 and β D2 was indicated by the original model of the G178S mutant protein. The side-chain hydroxyl group on substituted Ser178 (on β D2) forms a new hydrogen bond with the backbone carbonyl of Glu113 (on β I1), and this interaction alters the trajectory of β I1 in the mutant protein (40). By contrast, the structural basis for the strand separation was not clear from the original model of the E113G mutant protein. The newly refined models, however, have provided what are, to our knowledge, novel insights into how the E113G substitution alters the structure of the subunit interface.
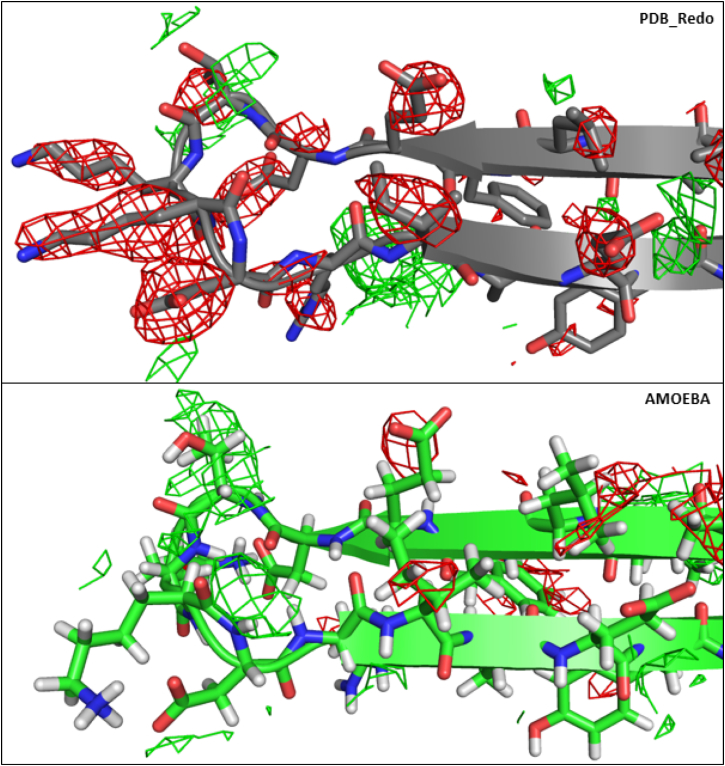
In the AMOEBA side-chain optimized model of the E113G mutant, the interaction between β I1 and β H1 is stabilized by increased hydrogen bonding. Comparing the structures of the wild-type and E113G mutant protein, we see that β H1 is extended by one residue (position 105) and that β I1 is extended by two residues (positions 109 and 110) in the mutant protein. In addition, the loop between β H1 and β I1 (loop J) appears to be in a more energetically favorable conformation in the mutant proteins. There are three intrastrand backbone hydrogen bonds in the mutant protein that are not present in AMOEBA side-chain optimized wild-type protein. A similar mechanism was proposed for a loss-of-flexibility S115P mutant (36) that caused trimer instability due to loss of interstrand hydrogen bonds. In wild-type PCNA, the trimeric form is more stable than the monomeric form by 1667 kcal/mol of AMOEBA energy; this stabilization drops to 1424 kcal/mol in the E113G mutant. Our results suggest a similar mechanism for the gain-of-flexibility E113G mutant based on an energetic tradeoff between the intermolecular interactions of β D2 and β I1 at the subunit interface and intramolecular interactions between β H1 and β I1 and within the backbone of loop J. The greater flexibility of β I1 due to introduction of glycine at position 113 has shifted the balance in favor of stronger intramolecular β H1-β I1 interactions and loop-J hydrogen bonds (see Fig. 3 and Table S1). This is a possible explanation for the observed separation of the subunit interface and is consistent with reduced trimer stability. The AMOEBA PCNA electrostatic networks at the subunit interface are supported not only by lower MOLPROBITY score and lower R/Rfree values, but also by dramatically cleaner σA-weighted Fo-Fc electron density maps (Fig. 4).
Figure 4.
The β-strands H1 and I1 of PCNA G178S mutant (3F1W) are shown with Fo-Fc maps contoured at 2σ (green) and −2σ (red) for PDB_REDO (above) and AMOEBA (below).
Conclusions
Biomolecular x-ray refinement strategies that place side chains, such as PDB_REDO (34) and RINGER (55), have achieved some success in improving the quality and interpretation of x-ray diffraction experiments. However, protein structure refinement methods have been limited by their assumption of side-chain independence and/or the absence of rigorous electrostatic interactions. For example, PDB_REDO is based on choosing a rotameric state for one residue at a time (56), which is reflected by a mean poor rotamer percentage of 6.6% for the PCNA structures examined here. On the other hand, many-body DEE using AMOEBA reduced the percentage of poor rotamers to 1.7% while simultaneously improving overall MOLPROBITY score and lowering both Rfree and AMOEBA potential energy.
Model bias is an important consideration for any refinement procedure that optimizes atomic coordinates to a target function that depends on calculated phases. Neither systematic removal of backbone model bias nor optimization of backbone conformation beyond what is achieved by local minimization was considered in this work. However, several methods have been proposed for considering limited backbone flexibility during repacking, which could be coupled to many-body DEE in the future. For example, generation of a discrete set of backbone conformations to include during DEE has been described in Su and Mayo (57). Alternatively, deterministic DEE has been extended to find a flexible-backbone rigid-rotamer GMEC by calculating bounds on rotameric interaction energies given a limited range of backbone dihedral movements imposed by per-residue restraining boxes (58).
The side-chain repacking algorithm presented here, to our knowledge, is the first deterministic method compatible with many-body potential energy functions. This opens the door to using polarizable force fields, Poisson-Boltzmann electrostatics, and quantum mechanical potential energy functions alone or in combination with experimental data to improve protein structural models. In this work, a hybrid target function has shown success in improving MOLPROBITY score and lowering both Rfree and AMOEBA potential energy based on a series of mid- to low-resolution PCNA x-ray diffraction data sets. In this case, electrostatic networks from coupled side-chain reorientations, which are difficult or impossible to refine by hand, revealed intramolecular stabilization of PNCA monomers at the expense of intermolecular hydrogen-bonding and destabilization of the active PNCA trimer.
In addition to x-ray structure determination, this work sets the foundation for application of many-body potential energy functions to computational protein design, homology modeling, and design of protein-ligand interactions. The advantage of many-body over pairwise DEE is of greatest importance for driving molecular forces that are inherently many-body in nature, including polarizable electrostatic interactions and the hydrophobic effect. For example, it has been suggested that the inherent many-body nature of the hydrophobic effect has made computational protein design a challenge for implicit solvents (59). Future applications of many-body DEE may help determine whether the use of polarizable force fields (26) and self-consistent reaction-field implicit solvents (28–30) can overcome the limitations of previous generation pairwise force fields (14) and pairwise implicit solvents (21–23) for computational protein design (15–17).
Author Contributions
S.D.L., S.G., W.T.A.T., and M.J.S. conceived the theory; S.D.L. performed the experiments; S.D.L., J.M.L., K.T.P., M.T.W., and M.J.S. analyzed the data; S.D.L., J.M.L., K.T.P., A.M.L., W.T.A.T., T.D.F., M.T.W., and M.J.S. contributed code/tools/structures; and S.D.L., J.M.L., K.T.P., M.T.W., and M.J.S. wrote the article.
Acknowledgments
The authors thank Lokesh Gakhar and Lynne Dieckman for helpful discussions. All computations were performed on The University of Iowa NEON cluster with support and guidance from Glenn Johnson and Ben Rogers.
M.J.S. was supported by National Science Foundation award No. CHE-1404147 and National Institutes of Health award No. R01 DC002842 from The National Institute on Deafness and Other Communication Disorders. M.T.W. was supported by National Institutes of Health award No. 01-GM081433 from the National Institute of General Medical Sciences. S.D.L. acknowledges a National Institutes of Health fellowship from award No. T32-GM067795. J.M.L. acknowledges a National Institutes of Health fellowship from award No. T32-GM008365 and a University of Iowa Presidential Fellowship. S.G. acknowledges support from a University of Iowa Biochemistry Summer Undergraduate Research Fellowship. A.M.L. and W.T.A.T. were partially supported by fellowships from The University of Iowa Center for Research by Undergraduates.
Editor: Nathan Baker.
Footnotes
Supporting Materials and Methods, three figures, four tables, and derivations of many-body dead-end elimination criteria are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00676-1.
Supporting Material
References
- 1.Berman H.M., Westbrook J., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams P.D., Afonine P.V., Zwart P.H. PHENIX: a comprehensive PYTHON-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winn M.D., Ballard C.C., Wilson K.S. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen V.B., Arendall W.B., 3rd, Richardson D.C. MOLPROBITY: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponder J.W., Case D.A. Vol. 66. Academic Press; London, UK: 2003. Force fields for protein simulations; pp. 27–85. (Advances in Protein Chemistry). [DOI] [PubMed] [Google Scholar]
- 6.Tama F., Miyashita O., Brooks C.L., 3rd Flexible multi-scale fitting of atomic structures into low-resolution electron density maps with elastic network normal mode analysis. J. Mol. Biol. 2004;337:985–999. doi: 10.1016/j.jmb.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 7.Schröder G.F., Levitt M., Brunger A.T. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponder J.W., Richards F.M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 9.Lovell S.C., Word J.M., Richardson D.C. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 10.Shapovalov M.V., Dunbrack R.L., Jr. A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desmet J., De Maeyer M., Lasters I. The dead-end elimination theorem and its use in protein side-chain positioning. Nature. 1992;356:539–542. doi: 10.1038/356539a0. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein R.F. Efficient rotamer elimination applied to protein side-chains and related spin glasses. Biophys. J. 1994;66:1335–1340. doi: 10.1016/S0006-3495(94)80923-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahiyat B.I., Mayo S.L. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 14.Boas F.E., Harbury P.B. Potential energy functions for protein design. Curr. Opin. Struct. Biol. 2007;17:199–204. doi: 10.1016/j.sbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Das R., Baker D. Macromolecular modeling with ROSETTA. Annu. Rev. Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 16.Gainza P., Roberts K.E., Donald B.R. OSPREY: protein design with ensembles, flexibility, and provable algorithms. Methods Enzymol. 2013;523:87–107. doi: 10.1016/B978-0-12-394292-0.00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simonson T., Gaillard T., Archontis G. Computational protein design: the PROTEUS software and selected applications. J. Comput. Chem. 2013;34:2472–2484. doi: 10.1002/jcc.23418. [DOI] [PubMed] [Google Scholar]
- 18.Kaminski G.A., Friesner R.A., Jorgensen W.L. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J. Phys. Chem. B. 2001;105:6474–6487. [Google Scholar]
- 19.Cornell W.D., Cieplak P., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1996;118:2309. [Google Scholar]
- 20.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 21.Lazaridis T., Karplus M. Effective energy function for proteins in solution. Proteins. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 22.Marshall S.A., Vizcarra C.L., Mayo S.L. One- and two-body decomposable Poisson-Boltzmann methods for protein design calculations. Protein Sci. 2005;14:1293–1304. doi: 10.1110/ps.041259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaillard T., Simonson T. Pairwise decomposition of an MMGBSA energy function for computational protein design. J. Comput. Chem. 2014;35:1371–1387. doi: 10.1002/jcc.23637. [DOI] [PubMed] [Google Scholar]
- 24.Kauzmann W. Some factors in the interpretation of protein denaturation. In: Anfinsen C.B. Jr., Anson M.L., Bailey K., Edsall J.T., editors. Vol. 14. Academic Press; London, UK: 1959. (Advances in Protein Chemistry). [DOI] [PubMed] [Google Scholar]
- 25.Ponder J.W., Wu C., Head-Gordon T. Current status of the AMOEBA polarizable force field. J. Phys. Chem. B. 2010;114:2549–2564. doi: 10.1021/jp910674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopes P.E.M., Roux B., MacKerell A.D., Jr. Molecular modeling and dynamics studies with explicit inclusion of electronic polarizability. Theory and applications. Theor. Chem. Acc. 2009;124:11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomasi J., Mennucci B., Cammi R. Quantum mechanical continuum solvation models. Chem. Rev. 2005;105:2999–3093. doi: 10.1021/cr9904009. [DOI] [PubMed] [Google Scholar]
- 28.Maple J.R., Cao Y.X., Friesner R.A. A polarizable force field and continuum solvation methodology for modeling of protein-ligand interactions. J. Chem. Theory Comput. 2005;1:694–715. doi: 10.1021/ct049855i. [DOI] [PubMed] [Google Scholar]
- 29.Schnieders M.J., Ponder J.W. Polarizable atomic multipole solutes in a generalized Kirkwood continuum. J. Chem. Theory Comput. 2007;3:2083–2097. doi: 10.1021/ct7001336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnieders M.J., Baker N.A., Ponder J.W. Polarizable atomic multipole solutes in a Poisson-Boltzmann continuum. J. Chem. Phys. 2007;126:124114. doi: 10.1063/1.2714528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schnieders M.J., Fenn T.D., Pande V.S. Polarizable atomic multipole x-ray refinement: particle mesh Ewald electrostatics for macromolecular crystals. J. Chem. Theory Comput. 2011;7:1141–1156. doi: 10.1021/ct100506d. [DOI] [PubMed] [Google Scholar]
- 32.Fenn T.D., Schnieders M.J., Brunger A.T. A smooth and differentiable bulk-solvent model for macromolecular diffraction. Acta Crystallogr. D Biol. Crystallogr. 2010;66:1024–1031. doi: 10.1107/S0907444910031045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenn T.D., Schnieders M.J. Polarizable atomic multipole x-ray refinement: weighting schemes for macromolecular diffraction. Acta Crystallogr. D Biol. Crystallogr. 2011;67:957–965. doi: 10.1107/S0907444911039060. [DOI] [PubMed] [Google Scholar]
- 34.Joosten R.P., Salzemann J., Vriend G. PDB_REDO: automated re-refinement of x-ray structure models in the PDB. J. Appl. Cryst. 2009;42:376–384. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 36.Krishna T.S.R., Kong X.-P., Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 37.Stelter P., Ulrich H.D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 38.Amin N.S., Holm C. In vivo analysis reveals that the interdomain region of the yeast proliferating cell nuclear antigen is important for DNA replication and DNA repair. Genetics. 1996;144:479–493. doi: 10.1093/genetics/144.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau P.J., Flores-Rozas H., Kolodner R.D. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol. Cell. Biol. 2002;22:6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freudenthal B.D., Ramaswamy S., Washington M.T. Structure of a mutant form of proliferating cell nuclear antigen that blocks translesion DNA synthesis. Biochemistry. 2008;47:13354–13361. doi: 10.1021/bi8017762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Freudenthal B.D., Gakhar L., Washington M.T. Structure of monoubiquitinated PCNA and implications for translesion synthesis and DNA polymerase exchange. Nat. Struct. Mol. Biol. 2010;17:479–484. doi: 10.1038/nsmb.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dieckman L.M., Boehm E.M., Washington M.T. Distinct structural alterations in proliferating cell nuclear antigen block DNA mismatch repair. Biochemistry. 2013;52:5611–5619. doi: 10.1021/bi400378e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brünger A.T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987;235:458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 44.Moulinier L., Case D.A., Simonson T. Reintroducing electrostatics into protein x-ray structure refinement: bulk solvent treated as a dielectric continuum. Acta Crystallogr. D Biol. Crystallogr. 2003;59:2094–2103. doi: 10.1107/s090744490301833x. [DOI] [PubMed] [Google Scholar]
- 45.Brünger A.T. FREE R VALUE: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 46.Ho B.K., Curmi P.M.G. Twist and shear in β-sheets and β-ribbons. J. Mol. Biol. 2002;317:291–308. doi: 10.1006/jmbi.2001.5385. [DOI] [PubMed] [Google Scholar]
- 47.Vargas R., Garza J., Hay B.P. How strong is the Cα−H···OC hydrogen bond? J. Am. Chem. Soc. 2000;122:4750–4755. [Google Scholar]
- 48.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Ren P., Wu C., Ponder J.W. Polarizable atomic multipole-based molecular mechanics for organic molecules. J. Chem. Theory Comput. 2011;7:3143–3161. doi: 10.1021/ct200304d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi Y., Xia Z., Ren P. The polarizable atomic multipole-based AMOEBA force field for proteins. J. Chem. Theory Comput. 2013;9:4046–4063. doi: 10.1021/ct4003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Read R. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A. 1986;42:140–149. [Google Scholar]
- 52.Cowtan K. Likelihood weighting of partial structure factors using spline coefficients. J. Appl. Cryst. 2005;38:193–198. [Google Scholar]
- 53.Zhang H., Gibbs P.E.M., Lawrence C.W. The Saccharomyces cerevisiae rev6-1 mutation, which inhibits both the lesion bypass and the recombination mode of DNA damage tolerance, is an allele of POL30, encoding proliferating cell nuclear antigen. Genetics. 2006;173:1983–1989. doi: 10.1534/genetics.106.058545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dieckman L.M., Washington M.T. PCNA trimer instability inhibits translesion synthesis by DNA polymerase η and by DNA polymerase δ. DNA Repair (Amst.) 2013;12:367–376. doi: 10.1016/j.dnarep.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fraser J.S., Clarkson M.W., Alber T. Hidden alternative structures of proline isomerase essential for catalysis. Nature. 2009;462:669–673. doi: 10.1038/nature08615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Joosten R.P., Joosten K., Perrakis A. Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank. Bioinformatics. 2011;27:3392–3398. doi: 10.1093/bioinformatics/btr590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Su A., Mayo S.L. Coupling backbone flexibility and amino acid sequence selection in protein design. Protein Sci. 1997;6:1701–1707. doi: 10.1002/pro.5560060810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Georgiev I., Keedy D., Donald B.R. Algorithm for backrub motions in protein design. Bioinformatics. 2008;24:i196–i204. doi: 10.1093/bioinformatics/btn169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaramillo A., Wodak S.J. Computational protein design is a challenge for implicit solvation models. Biophys. J. 2005;88:156–171. doi: 10.1529/biophysj.104.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.