Abstract
Aberrant self-assembly, induced by structural misfolding of the prion proteins, leads to a number of neurodegenerative disorders. In particular, misfolding of the mostly α-helical cellular prion protein (PrPC) into a β-sheet-rich disease-causing isoform (PrPSc) is the key molecular event in the formation of PrPSc aggregates. The molecular mechanisms underlying the PrPC-to-PrPSc conversion and subsequent aggregation remain to be elucidated. However, in persistently prion-infected cell-culture models, it was shown that treatment with monoclonal antibodies against defined regions of the prion protein (PrP) led to the clearing of PrPSc in cultured cells. To gain more insight into this process, we characterized PrP-antibody complexes in solution using a fast protein liquid chromatography coupled with small-angle x-ray scattering (FPLC-SAXS) procedure. High-quality SAXS data were collected for full-length recombinant mouse PrP [denoted recPrP(23–230)] and N-terminally truncated recPrP(89–230), as well as their complexes with each of two Fab fragments (HuM-P and HuM-R1), which recognize N- and C-terminal epitopes of PrP, respectively. In-line measurements by fast protein liquid chromatography coupled with SAXS minimized data artifacts caused by a non-monodispersed sample, allowing structural analysis of PrP alone and in complex with Fab antibodies. The resulting structural models suggest two mechanisms for how these Fabs may prevent the conversion of PrPC into PrPSc.
Introduction
Prion diseases are caused by proteins in an alternatively folded, self-replicating conformation of a host-encoded protein (1). As soon as these proteins misfold, they start to recruit more proteins into the same conformation through conformational templating (2). Misfolding of the prion precursor protein occurs spontaneously but may be hastened by prion protein (PrP) mutations or, rarely, via exposure to exogenous prions through infection. Irrespective of the etiology, misfolding results in multiplication of the disease-causing isoform (PrPSc), which spreads throughout the brain and other organs (3,4). Recent findings have demonstrated that many neurodegenerative diseases, such as Creutzfeldt-Jakob, Alzheimer’s, and Parkinson’s diseases, as well as the frontotemporal dementias, are all caused by proteins that become prions, accumulate, and cause disease (5). To date, no treatment is available to revert, stop, or even delay the clinical course of any of these disorders (6).
Mammalian PrP was the first protein to be discovered that can convert into a prion conformation (7). Its alternatively folded isoform, designated PrPSc, causes Creutzfeldt-Jakob disease in humans, bovine spongiform encephalopathy in cattle, and scrapie in sheep (5). PrPSc is formed from its cellular PrP precursor (PrPC) by a profound conformational change (8,9). PrPSc is insoluble and partially resistant to proteases, whereas PrPC is soluble and sensitive to protease digestion (9,10). The detailed mechanisms underlying the conversion of PrPC into PrPSc and subsequent aggregation into higher-order aggregates are still unknown.
PrPC is membrane-bound and seems likely to be involved in cellular adhesion and signaling (11). PrPC consists of two domains, a disordered N-terminal domain (residues 23–124) and a globular C-terminal domain composed of three α-helices and two short antiparallel β-strands (residues 125–230). Structural studies using solution NMR spectroscopy and x-ray crystallography revealed the structure of the C-terminal domain (12–17), but neither the structure of the flexible N-terminal domain of PrPC nor the structure of PrPSc is known (18).
Although no treatments exist for PrP prion diseases, studies have shown that antibodies directed against certain regions (including residues 95–110 and 221–230) are effective in clearing PrPSc from chronically infected cultured cells, whereas antibodies against more N-terminal residues were largely ineffective (19,20). In one study, treatment of mice with these antibodies triggered neuronal apoptosis. This result was believed to be due to antibody-induced cross-linking (21), but could not be recapitulated in follow-up studies (20). Therefore, passive immunization was suggested to be a potential treatment for prion diseases (20). Even though antibodies were able to clear cell culture systems of prion infection successfully, the underlying mechanism remains to be determined.
To obtain more information about the structure of PrP in complex with these antibodies, we structurally characterized these complexes using small-angle x-ray scattering (SAXS). In particular, we focused on potential antibody-induced structural transitions that might prevent or modify the PrPC-to-PrPSc conversion. SAXS provides comprehensive structural information about macromolecules in solution, including molecular shape and structural variability (22–24). SAXS exploits the elastic scattering of protein atoms in solution to produce a one-dimensional intensity profile that is a function of spatial frequency. The profile can be converted into an approximate distribution of pairwise electron distances in the macromolecule (i.e., the pair-distribution function) via a back-calculated Fourier transform (25). A major advantage of SAXS is that it can be applied to samples in solution, removing the requirement for crystals and thus artifacts frequently produced by crystal packing. As a result, SAXS can be used to study flexible and disordered proteins and protein fragments that are not amenable to x-ray crystallography. Interpretation of SAXS data usually involves computing theoretical SAXS profiles (26–28) for one or more atomic structures derived from x-ray crystallography, NMR spectroscopy, comparative modeling, or ab initio structure prediction. The computed profiles are compared to the experimental SAXS profiles to find a single or multiple structural models that are consistent with the data (29–34).
One of the main difficulties in applying SAXS analysis is obtaining a homogenous sample; even a low level of non-monodispersity can interfere with the accuracy of SAXS data interpretation and may be difficult to detect. This problem is even more pronounced for amyloidogenic proteins that often display aggregation in the highly concentrated samples needed for SAXS data collection. One way proteins can be purified to a suitably high level of monodispersity is by size-exclusion chromatography (SEC) via fast protein liquid chromatography (FPLC), where the macromolecule is separated into fractions based on size and shape. Therefore, an SEC-FPLC system coupled directly with a SAXS beamline (FPLC-SAXS) has several advantages over traditional data-collection strategies: a high level of monodispersity can be obtained, monodispersity can be easily verified using the ultraviolet (UV) trace, and small differences in buffer constitution are averaged out when the sample flows through the column (35,36). For the work reported here, difficulties in obtaining pure monodisperse samples suitable for measurement using the autosampler setup motivated the development of an FPLC-SAXS system at Stanford Synchrotron Radiation Lightsource (SSRL) Beamline BL4-2 (37). FPLC-SAXS has now proved helpful for many unrelated projects and is now among the core protocols on the beamline (38–40).
Here, we present an FPLC-SAXS pipeline in which the SAXS profiles of fractionated PrP could be collected immediately after SEC separation. This pipeline, in combination with integrative atomic modeling of complexes (27,28,31), was used to characterize monomers of full-length and truncated recombinant PrP (recPrP(23–230) and recPrP(89–230), respectively) and their complexes with two Fab antibodies (P and R1). Models of the recPrP–Fab complexes provided insight into the mechanism of action of each Fab. Specifically, Fab-P binds to a region that is affected by structural reorganization (residues 95–105), and thus may prevent the PrPC-to-PrPSc transition. Our model of the recPrP-Fab-R1 complex identified an additional interaction site (residues 165–175) that is known to influence the misfolding of PrP (41–43); R1 binding to this site may inhibit misfolding of PrP.
Materials and Methods
Sample preparation and FPLC-SAXS measurements
Truncated recPrP(89–230) and recPrP(23–230), as well as Fab fragments of HuM-R1 and HuM-P, were overexpressed and purified as previously described (44,45) (Fig. S1 in the Supporting Material). Lyophilized recPrP(89–230) and recPrP(23–230) pellets were dissolved in distilled water and dialyzed two times against sodium acetate buffer (20 mM sodium acetate, pH 5.1, and 150 mM NaCl) at 4°C (46). PrP-Fab complexes were made by mixing recPrP with the antibodies and leaving them for 30 min at room temperature to equilibrate. For quality control, aliquots of the PrP samples or the PrP-Fab complexes were adjusted to 1× LDS sample buffer (nonreducing conditions) and incubated for 5 min at 95°C, after which sodium dodecyl sulfate polyacrylamide gel electrophoresis was performed using 4–12% Bis-Tris gradient gels (Invitrogen, Carlsbad, CA) followed by Coomassie stain (Invitrogen) (Fig. S1).
To mitigate aggregation, an FPLC-SAXS system was developed on SSRL Beamline BL4-2, utilizing an Ettan FPLC system (GE Healthcare, Little Chalfont, United Kingdom) connected to an autosampler via PEEK tubing (37). A Superdex PC3.2/200 SEC column was used to purify the protein, with a flow rate of 0.05 or 0.08 mL, depending on the protein’s sensitivity to radiation damage. The column was equilibrated with one and one-half column volumes of running buffer (20 mM sodium acetate, pH 5.1, and 150 mM NaCl). A 100-μL sample loop was used, and for each data collection, a slight excess of sample (PrP alone, antibodies alone, and PrP-Fab complexes) was injected into the system. An x-ray scattering image was taken every second during most of the FPLC run. All SAXS data were collected with a nominal 1.7 m sample-to-detector distance, at 11 keV, with an exposure time of 1 s per image (Table 1).
Table 1.
Summary of SAXS experiments
| Data Collection | |
|---|---|
| Beamline | SSRL Beamline BL4-2 |
| Defining slit size (mm) | 0.3(H) × 0.3 (V) |
| Beam energy (keV) | 11 |
| Sample-detector distance (m) | 1.7 |
| Detector | Rayonix MX225-HE |
| Pixel binning | 8 × 8 |
| Pixel size (μm) | 292 |
| Exposure time (s) | 1 |
| Images | images taken for duration of run =∼700 images |
| Type of sample cell | quartz capillary (diameter 1.5 mm) |
| Temperature (K) | 283 |
| Calibrant | AgBe |
| Final q range (Å−1) | 0.01 to 0.5 |
| Data Analysis | |
| Programs | SASTOOL (47), PRIMUS (48) |
| Buffer | 20 mM Tris, 50 mM NaCl, pH 7.5 |
| Injected protein concentrations (mg/mL) | see Table 2 |
| Points used for Guinier analysis | see Table 2 |
| Guinier qRg limits | see Table 2 |
| Guinier Rg (Å) | see Table 2 |
| GNOM q range (Å−1) | see Table 2 |
| Dmax (Å) | see Table 2 |
| Rg (real) (Å) | see Table 2 |
| Rg (reciprocal) (Å) | see Table 2 |
| Modeling | |
| Programs | IMP (rrt_sample, foxs, multi_foxs), Pymol |
| Data plotting | GNUPLOT |
FPLC-SAXS data processing and analysis
All data were collected using a Mar225 CCD detector. For each image, subtracted intensities were obtained using SasTool (47). For the autosampler data, the intensities from the buffer data were subtracted from the sample data to produce the final scattering profile (37). For the FPLC data, the first 100 fraction images were used to create a combined buffer scattering profile, followed by subtracting the profile from each of the remaining images to produce the final scattering profile for each exposure. Data were initially analyzed using the ATSAS package (29). The scattering profiles for samples around peaks in UV absorbance (determined by UV-visible spectroscopy) (see Fig. 2) were selectively merged to produce the final SAXS profiles (Table 2). First, we selected a range of relevant SAXS profiles that were in the vicinity of the UV peak with low variance in both Rg and I0 traces. Depending on the data, this range included 10–50 consecutive SAXS profiles. Second, we merged each non-overlapping window of 10 profiles in this range using Primus (48) and estimated the molecular weight based on the merged profile using SAXS MOW (49). Finally, we selected a merged profile with the best match to the molecular weight calculated from the sequence.
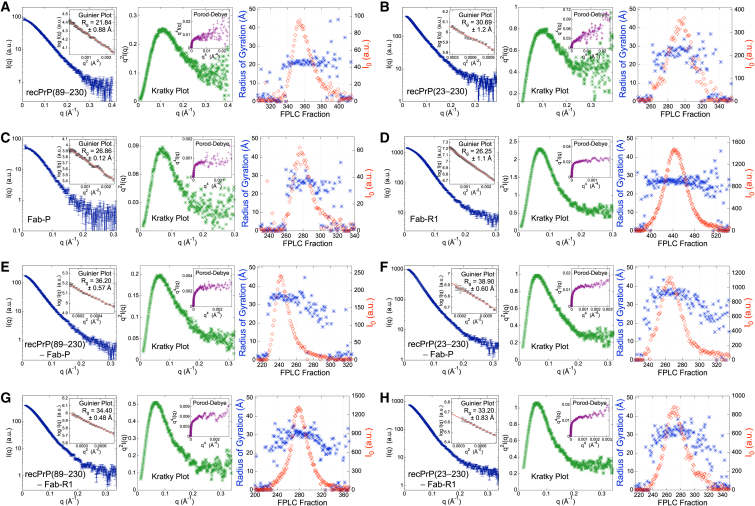
Figure 2.
Summary of FPLC-SAXS data analysis. FPLC-SAXS data were obtained for (A) recPrP(89–230), (B) recPrP(23–230), (C) Fab-P, (D) Fab-R1, (E) the recPrP(89–230)-Fab-P complex, (F) the recPrP(23–230)-Fab-P complex, (G) the recPrP(89–230)-Fab-R1 complex, and (H) the recPrP(23–230)-Fab-R1 complex. For each sample, SAXS profiles (left), Kratky plots (middle), and FPLC traces (right) are shown. The SAXS profiles (dark blue) and the accompanying Guinier plot (left inset, black dots) are shown together. The radius of gyration (Rg) value was determined from a linear fit in the Guinier plot (left inset, red line). Porod-Debye plots (middle inset, purple) are shown for the corresponding SAXS profile, visually depicting the level of flexibility for the corresponding sample in solution. The FPLC traces of Rg (blue) and I0 (red) are shown around the peak in UV absorbance for the corresponding sample (right). To see this figure in color, go online.
Table 2.
Summary of data analysis of the experimental SAXS profiles
| Samples | Injected Concentrationa (mg/mL) | FPLC Flow Rate (mL/min) | Rg (Real ± SD) (Å) | Guinier Points | q∗Rg | Dmax (Å) | Native Molecular Weight (kDa) | Molecular Weight Estimationb (kDa) | Porod Volumec (Å3) (q-range (Å−1)) |
|---|---|---|---|---|---|---|---|---|---|
| recPrP(89–230) | 3.6 | 0.05 | 21.84 ± 0.88 | 13–44 | 1.081 | 76.5 | 16.2 | 15.4 | 20,548 (0.010–0.255) |
| recPrP(23–230) | 4.1 | 0.08 | 30.69 ± 1.20 | 1–18 | 0.495 | 104.4 | 22.9 | 26.0 | 39,832 (0.017–0.258) |
| Fab-P | 5.3 | 0.05 | 26.86 ± 0.12 | 9–43 | 1.305 | 84.8 | 47.4 | 48.4 | 67,280 (0.010–0.150) |
| Fab-R1 | 4.1 | 0.05 | 26.25 ± 1.10 | 20–45 | 1.301 | 96.8 | 47.5 | 48.8 | 68,292 (0.010–0.142) |
| recPrP(89–230)-Fab-P | 1.0 | 0.05 | 36.20 ± 0.57 | 3–15 | 0.850 | 121.1 | 63.6 | 68.6 | 84,955 (0.010–0.191) |
| 2.6 | |||||||||
| recPrP(23–230)-Fab-P | 1.0 | 0.08 | 38.90 ± 0.60 | 1–15 | 0.986 | 136.1 | 70.3 | 73.8 | 104,183 (0.013–0.217) |
| 3.7 | |||||||||
| recPrP(89–230)-Fab-R1 | 1.0 | 0.05 | 34.40 ± 0.48 | 1–16 | 0.914 | 120.4 | 63.7 | 62.8 | 78,474 (0.013–0.197) |
| 3.7 | |||||||||
| recPrP(23–230)-Fab-R1 | 1.0 | 0.08 | 33.20 ± 0.83 | 1–10 | 0.904 | 113.6 | 70.4 | 62.2 | 79,039 (0.019–0.205) |
| 2.9 |
We estimate that the eluted concentration at the beam capillary is ∼67% of the corresponding injected concentration, according to UV spectroscopy (data not shown).
Molecular weights were estimated using SAXS MOW (49) with a threshold of Qmax = 0.25 Å−1.
Computational modeling
Representation and scoring function
Atomic-resolution structural models were used for interpretation of the samples. The models were compared to the SAXS profiles using the FoXS method, which optimizes hydration layer density (c2) and excluded volume (c1) to improve the fit of a model to the experimental SAXS profile (27,28). The quality of the fit is assessed by the χ score (26):
where Iexp(q) is the experimental profile, σ(q) is the experimental error of the measured profile, and S is the number of points in the profile. I(q, c1, c2) is the computed profile, given by the Debye formula (27), with c, c1, and c2 optimized to minimize the score.
Here, we are also considering a possibility of a mixture of states contributing to the observed single SAXS profile. In such a case, the score of a multi-state model (a model that specifies two or more coexisting structural states and their population weights) is given by
where In(q, c1, c2) and wn are the computed profile and the corresponding weight, respectively, for each of the N states in the model; this equation minimizes data overfitting by using a single set of c1 and c2 values for all N states.
Sampling of the disordered regions in recPrP
Initial models for the disordered regions were built using MODELER 9.13 (50). The disordered region of PrP (residues 23–125) was sampled using rapidly exploring random trees (RRTs) (51,52). The RRT algorithm samples the conformational space by leveraging an iteratively constructed nearest-neighbor linked tree. This iterative strategy expands the tree toward unexplored regions of the conformational space and significantly improves the sampling efficiency compared to random sampling. The φ and ψ angles of the disordered regions were sampled independently. We generated 10,000 models for each disordered region. To increase the confidence in the sampling protocol, we generated 50,000 models and validated that no other models with lower χ scores were produced in the ensemble.
Sampling of the Fab-P and Fab-R1 antibodies
The Fab models were generated using MODELER 9.13 (50) using different templates to account for 28 different Fab elbow angles (range 130–180°) (53). For each template, 10 models were generated and fitted to the experimental SAXS profile of the Fab using FoXS (27,28).
Sampling of the recPrP-Fab complexes
The computational modeling of the recPrP-Fab complexes was performed using an integrative docking protocol (54). To account for the flexibility of the C-terminal region, we used 20 conformations of recPrP from the solution NMR spectroscopy (Protein Data Bank (PDB) code 2L39) (16). Over 400,000 models were generated using the rigid-body docking program PatchDock with antibody-antigen protocol (55). The disordered N-terminal regions of recPrP(89–230) and recPrP(23–230) were not used in the docking stage, but subsequently were sampled using RRT and added for fitting to SAXS profiles using FoXS (27,28). The interface between the Fab-R1 and recPrP was scored with the SOAP-PP statistical potential (56). Each docking model was ranked by the sum of the Z-scores for the SAXS χ and SOAP-PP scores.
Multi-state model enumeration
Given N input conformations and their computed SAXS profiles, our goal was to find multi-state models of size n (n << N), such that the corresponding sum of weighted SAXS profiles fitted the experimental SAXS profile. Multi-state model enumeration was performed by MultiFoXS iteratively using the branch-and-bound method (57). In each branch step, we extended K (K = 10,000) best-scoring models of size n to KN models of size n + 1 by addition of each of the N input conformations. In the bound step, we selected K best-scoring models out of the total KN models for the next iteration. Therefore, generation of K multi-state models of size n + 1 from K multi-state models of size n required KN SAXS score calculations. This greedy approach avoided the exponential growth in scale of enumeration while still producing the good-scoring multi-state models.
Results
Solution structures of full-length and N-terminally truncated recPrP
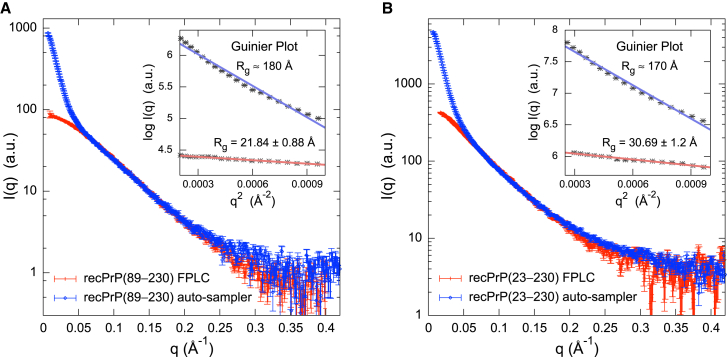
Previously, only residues 119–230 (mouse sequence) of full-length (residues 23–230) recPrP were structurally characterized by solution NMR spectroscopy and x-ray crystallography (12–14,16,17). Initial SAXS data collection was attempted on recPrP(89–230) and recPrP(23–230) at SSRL Beamline BL4-2 using an autosampler (37) (Fig. 1). The initial SAXS profiles indicated that both recPrP samples suffered from severe aggregation effects; all attempts to reduce aggregation via filtering, centrifugation, and ultracentrifugation failed (Fig. 1, blue). In comparison, FLPC-SAXS runs of recPrP(89–230) and recPrP(23–230) showed linearity in the Guinier region of the SAXS profiles (Fig. 1, red).
Figure 1.
Comparison of the SAXS profiles obtained using FPLC and the SSRL BL4-2 autosampler. The SAXS profiles of (A) recPrP(89–230) and (B) recPrP(23–230) were collected using both FPLC (red) and the SSRL BL4-2 autosampler (blue). Data obtained from the SSRL BL4-2 autosampler showed that both protein samples suffered from severe aggregation effects, and any attempts to reduce this via filtering, centrifugation, and ultracentrifugation failed. In comparison, the SAXS profiles collected using FPLC showed linearity in the Guinier region (insets). To see this figure in color, go online.
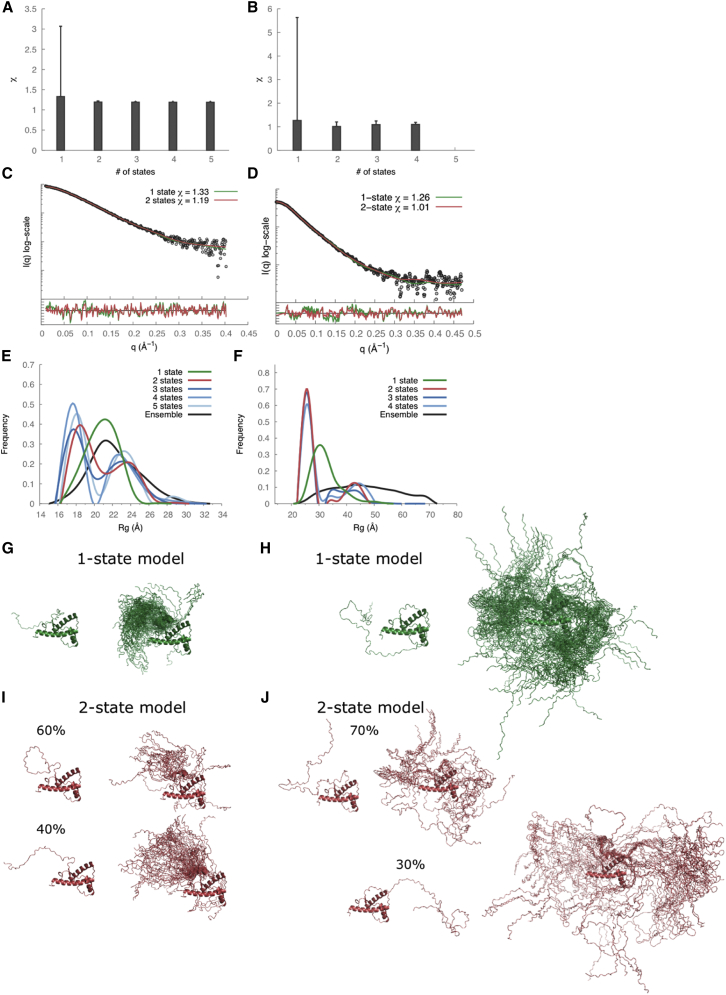
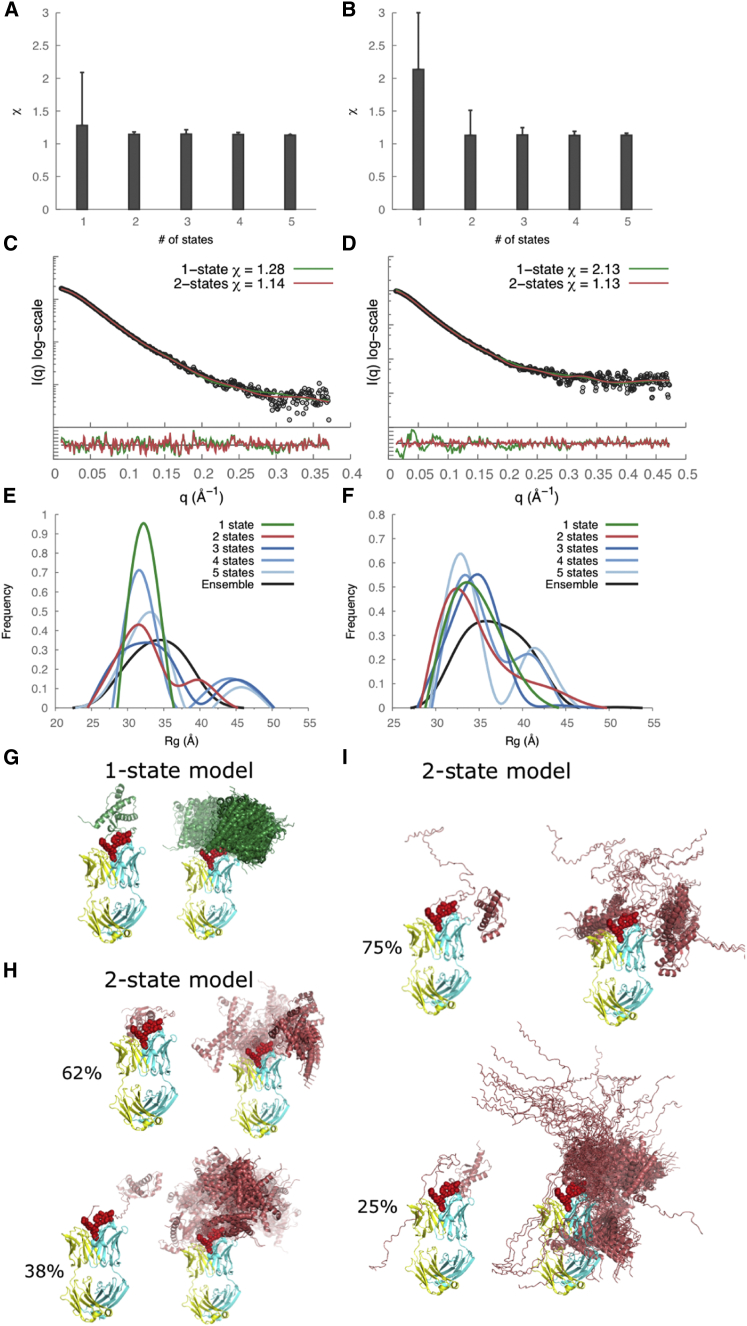
The radius of gyration (Rg) values calculated from the SAXS profiles were 21.8 ± 0.9 Å and 30.7 ± 1.2 Å for recPrP(89–230) and recPrP(23–230), respectively (Fig. 2, A and B, and Table 2). For comparison, the Rg of recPrP(120–231) calculated from the solution NMR structure (PDB code 2L39) (16) was 15.0 Å; however, the structure lacks atomic coordinates for 31 and 97 disordered N-terminal residues compared to recPrP(89–230) and recPrP(23–230), respectively. The top 1000 of the 10,000 models generated by RRT had χ values from 1.33 to 3.05 for recPrP(89–230) (Fig. 3 A) and from 1.26 to 5.70 for recPrP(23–230) (Fig. 3 B). Thus, a single structural state explains the data within the data noise. However, it is unlikely that the system actually exists in a single conformation, given the failure of both NMR spectroscopy and x-ray crystallography to determine the structure of the N-terminal fragment (residues 23–125). Therefore, we enumerated possible N-state models, with N between 2 and 5, saving the 1000 top-scoring models for each N. The multi-state models fit the data within the noise with even better scores than any single-state model (a model that specifies a single structural state), as expected (because there are more degrees of freedom in these models to fit the data). The 1000 top-scoring two-state models had χ values from 1.19 to 1.22 for recPrP(89–230) (Fig. 3 A) and from 1.01 to 1.20 for recPrP(23–230) (Fig. 3 B). There was no significant improvement in the χ scores for models of three or more states.
Figure 3.
SAXS data analysis and modeling for recPrP(89–230) (left column) and recPrP(23–230) (right column). (A and B) The lowest χ scores for each of the N-state models (N = 1–5) are shown with an error bar indicating the range of χ values for the top 1000 models. (C and D) Comparison of the experimental SAXS profiles (black) with computed SAXS profiles for the top-scoring single-state (green) and two-state (red) models. The lower plots show the residuals defined as (Iexp(q) − Icalc(q))/σexp(q), corresponding to the difference between the experimental and the computed intensities weighted by the experimental uncertainty. (E and F) Rg distributions for the entire ensemble of 10,000 conformations and the top 1000 N-state models are shown. (G–J) Conformations of the top-scoring single-state (G and H) and two-state models (I and J) are shown on the left, and the top 100 models aligned on the structured C-terminal domain of PrPC on the right. To see this figure in color, go online.
In an effort to characterize the range of conformations consistent with the SAXS data, we analyzed distributions of the Rg for the entire ensemble of 10,000 conformations as well as the 1000 best-scoring N-state models (N = 1 … 5). For recPrP(89–230), the Rg peak was 22 Å for the entire ensemble of conformations and 21 Å for the single-state models; two Rg peaks (18 Å and 24 Å) were conserved among the best-scoring N-state models for N from 2 to 5 (Fig. 3 E). Similar distributions were observed for recPrP(23–230) (Fig. 3 F). For both recPrP constructs, the first Rg peak (∼60–70% of the population) corresponded to compact closed conformations, whereas the second peak (∼30–40%) corresponded to extended open conformations (Fig. 3, I and J). In other words, each one of the two states contained many different conformations with similar Rg values.
Solution structures of humanized antibody fragments Fab-P and Fab-R1
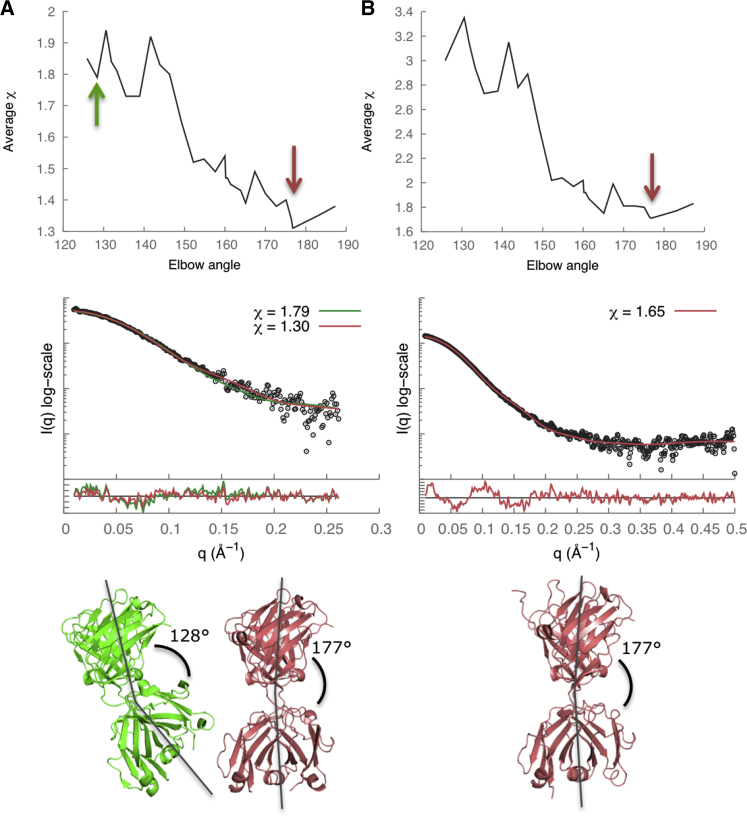
There are multiple Fab structures deposited in the PDB, including the structure of HuM-P (PDB code 2HH0) (58). A change in the elbow angle results in a different orientation between the constant and variable Fab domains, and subsequently in a significant difference in the distance distribution function. Therefore, accurate modeling of the elbow angle was necessary to obtain a good fit to the SAXS profile. The x-ray structure of HuM-P Fab (PDB code 2HH0) (58) has an elbow angle of 128° and a χ score of 1.79. Our model of Fab-P with the lowest χ score of 1.30 had an elbow angle of 177°, indicating that its solution state might be different from the crystallographic state (Fig. 4 A). Similarly, a Fab-R1 model with the lowest χ score of 1.65 also had an elbow angle of 177° (Fig. 4 B). The Rg values calculated from the SAXS profiles were 26.9 ± 0.1 Å and 26.3 ± 1.1 Å for Fab-P and Fab-R1, respectively (Fig. 2, C and D, and Table 2).
Figure 4.
SAXS data analysis and modeling for (A) Fab-P and (B) Fab-R1. The Fab elbow angle versus χ score (upper), the SAXS profile fits and the residual plots (middle), and the best-scoring structural models (lower, red) are shown. The lowest χ scores were 1.30 for Fab-P and 1.65 for Fab-R1. In (A, lower), the x-ray crystal structure of Fab-P (green; PDB code 2HH0; χ = 1.79; elbow angle 128°) (58) was compared to the best-scoring model of Fab-P (red; χ = 1.30; elbow angle 177°). To see this figure in color, go online.
Solution structures of recPrP(89–230)-Fab-P and recPrP(23–230)-Fab-P complexes
The SAXS profiles of recPrP(89–230)-Fab-P and recPrP(23–230)-Fab-P were subtly different, with Rg values of 36.2 ± 0.6 Å and 38.9 ± 0.6 Å, respectively (Fig. 2, E and F, and Table 2). An initial model of the complex was obtained using a PDB structure of Fab-P associated with the epitope peptide from PrP (PDB code 2HH0) (58), adding the missing linker residues (either 89–94 or 23–94, and 105–124) to the prion structure (using MODELER 9.13 (50)). The elbow angle of the Fab was set to 177° to be consistent with the solution Fab state.
The top 1000 of the 10,000 models generated using RRT had χ values from 1.28 to 2.11 for the recPrP(89–230)-Fab-P complex (Fig. 5 A) and from 2.13 to 6.00 for the recPrP(23–230)-Fab-P complex (Fig. 5 B). As with recPrP alone, we explored a single-state model as well as N-state models (N = 2–5) for both complexes. A single-state model fit the data within the noise only for the recPrP(89–230)-Fab-P complex, but not for the recPrP(23–230)-Fab-P complex (Fig. 5, C and D). For the two-state models, both recPrP(89–230)-Fab-P and recPrP(23–230)-Fab-P complexes fit the data within the noise. The 1000 top-scoring two-state models had χ values from 1.14 to 1.18 for recPrP(89–230)-Fab-P (Fig. 5 A) and from 1.13 to 1.51 for recPrP(23–230)-Fab-P (Fig. 5 B). Again, there was no significant improvement in the χ score for models of three or more states (Fig. 5, A and B).
Figure 5.
SAXS data analysis and modeling for the recPrP(89–230)-Fab-P (left column) and recPrP(23–230)-Fab-P complexes (right column). (A and B) The lowest χ scores for each of the N-state models (N = 1–5) are shown with error bars indicating the range of χ values for the top 1000 models. (C and D) Comparison of the experimental SAXS profiles (black) with the computed SAXS profiles for the top-scoring single-state (green) and two-state (red) models. The lower plots show the residuals defined as (Iexp(q) − Icalc(q))/σexp(q), corresponding to the difference between the experimental and computed intensities weighted by the experimental uncertainty. (E and F) Rg distributions for the entire ensemble of 10,000 conformations and the top 1000 N-state models are shown. (G–I) Conformations of the top-scoring single-state (G) and two-state models (H and I) are shown on the left and the top 100 models aligned on the Fab structure on the right. The heavy chain of Fab is shown in cyan, the light chain in yellow, and the epitope residues in red spacefill. To see this figure in color, go online.
In an effort to characterize the range of conformations consistent with the SAXS data, we analyzed distributions of the Rg for the entire ensemble of 10,000 conformations as well as the 1000 best-scoring N-state models (N = 1 … 5). For the recPrP(89–230)-Fab-P complex, the Rg peak was 35 Å for the entire ensemble of conformations and 32 Å for the single-state models (Fig. 5 E). For the best-scoring N-state models (N = 2 … 5), two Rg peaks at 31 Å and 43 Å were conserved (Fig. 5 E). Similar distributions were observed for the recPrP(23–230)-Fab-P complex (Fig. 5 F). For both recPrP-Fab-P complexes, the first Rg peak (∼60–75% of the population) corresponded to compact closed conformations, whereas the second peak (25–40%) corresponded to extended open conformations (Fig. 5, H and I). Similar to recPrP alone, each one of the two states for these recPrP-Fab complexes contains many different conformations with similar Rg.
Solution structures of recPrP(89–230)-Fab-R1 and recPrP(23–230)-Fab-R1 complexes
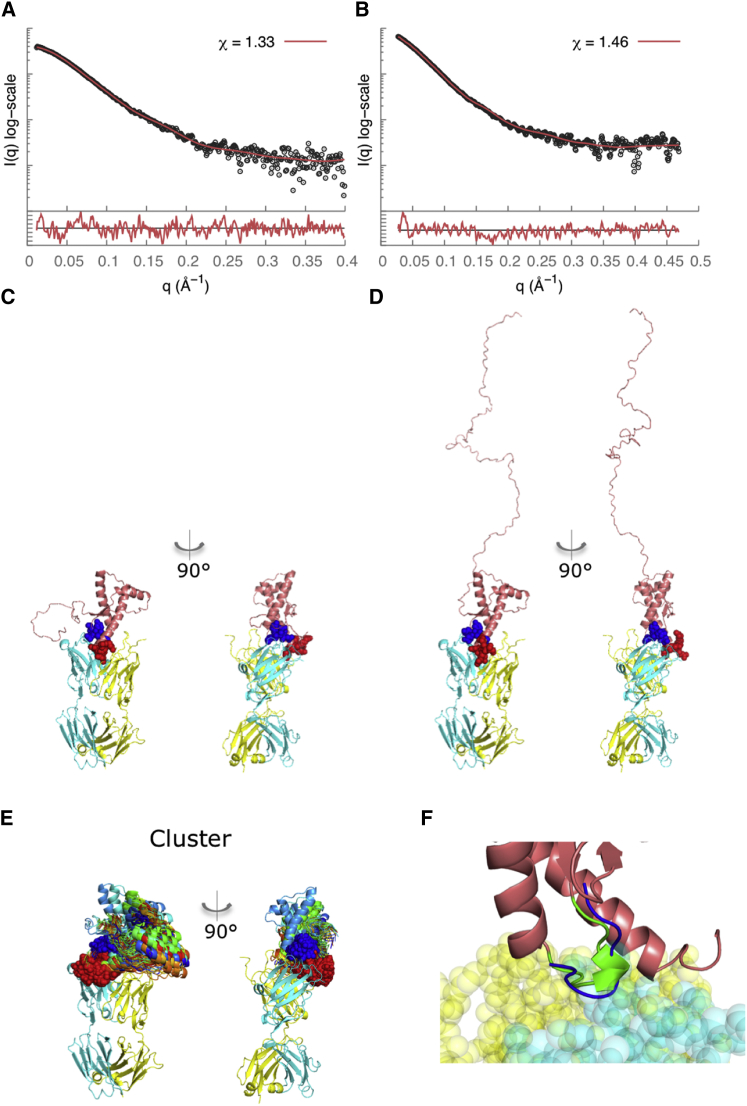
In contrast to Fab-P, which binds in the disordered linker region in the N-terminus, Fab-R1 binds at the C-terminal structured region (residues 223–230). Thus, the resulting complexes were more compact, with Rg values of ∼34.0 Å for both recPrP(89–230)-Fab-R1 and recPrP(23–230)-Fab-R1 complexes (Fig. 2, G and H, and Table 2). The samples used for SAXS experiments remained intact, with no degradation products found under denaturing conditions by sodium dodecyl sulfate polyacrylamide gel electrophoresis (Fig. S1). However, for the recPrP(23–230)-Fab-R1 complex, the Rg value was determined using only 10 data points (qRg < 0.9 (Table 2)), potentially explaining why we did not observe a significant difference between the Rg values for the two complexes. The epitope residues 223–230 were used as the site for computational docking, resulting in a single top-scoring cluster with an average Cα root mean-square deviation of 2.1 Å between each model in the cluster (Fig. 6 E); we use this variability as a measure of model precision. The model in each cluster with the best fit to the SAXS profile had a χ score of 1.33 for the recPrP(89–230)-Fab-R1 Fab complex (Fig. 6 A) and 1.46 for the recPrP(23–230)-Fab-R1 complex (Fig. 6 B). Of the recPrP interface residues, 75% corresponded to the known Fab-R1 epitope site (residues 221–230), whereas the remaining interface residues came from the α2-β2 loop (residues 165–175).
Figure 6.
SAXS data analysis and modeling for the recPrP(89–230)-Fab-R1 (left column) and recPrP(23–230)-Fab-R1 (right column) complexes. The SAXS profile fits (A and B) and the corresponding best-scoring structural models (C and D) are shown for each of the samples. The lower plots in (A) and (B) show the residuals defined as (Iexp(q) − Icalc(q))/σexp(q), corresponding to the difference between the experimental and computed intensities weighted by the experimental uncertainty. (E) The cluster of the top-scoring models is shown. The heavy chain of the Fab is shown in cyan, the light chain in yellow, and the epitope residues in red spacefill; the rigid loop is shown in blue. (F) Our structural model reveals interactions of the α2-β2 loop of PrPC (residues 165–175) with Fab-R1 (spacefill model) (41–43). This loop region in murine PrPC exists in two structural states, a highly populated state that forms a 310-helical turn (green) and a minor state that forms a type-I β-turn (blue) (41,43). To see this figure in color, go online.
Discussion
The FPLC-SAXS enables measurement of a non-monodispersed sample
PrPC has been difficult to study due to the difficulty of obtaining large amounts of purified protein from mammalian sources (10,59). Furthermore, natural PrP is anchored to the neuronal membranes via a glycosylphosphatidylinositol anchor (10,60), usually requiring preparations with lipids or detergents that render them inaccessible to SAXS-based studies (10,59). Structural comparison between natural and recombinant PrP did not reveal major structural differences; therefore, recombinant PrP can be used to study the PrPC isoform in vitro (59,61).
Here, we demonstrate that an FPLC-SAXS system enabled collection of SAXS data on challenging protein samples (Figs. 1 and 2), and this system facilitate further work on recombinant multimerization-prone proteins, or indeed any system that can be purified to homogeneity with FPLC. In addition, FPLC-SAXS has the advantage of minimizing differences between the protein buffer and buffer measurements due to the continuous FPLC flow, helping to minimize potential radiation damage. These advantages allowed us to collect and interpret SAXS data for recPrP(89–230)-antibody and recPrP(23–230)-antibody complexes.
Although FPLC-SAXS allows collection of high-quality SAXS data sets that do not suffer from aggregation and compositional heterogeneity, the sample can still be conformationally heterogeneous, making data interpretation challenging. Usually, the best explanation of the data is obtained by minimizing the number of conformations that resulted in the data (Occam’s razor) (33). For most of our samples, we found that a single conformation explains the SAXS data within its noise (Figs. 3 and 5). However, we know that PrPC is almost certain to be conformationally heterogeneous in solution (thus, Occam’s razor as expressed above does not apply here). To explicitly model this heterogeneity, we analyzed the data with multi-state models. Here, a multi-state model consists of multiple conformations and their weights; thus, the data are interpreted through the weighted combination of all states in a model, not any single state. We overcame the challenge of the resulting overfitting by computing ensembles of multi-state models that fit the data, using MultiFoXS (see Materials and Methods), and highlighting only the conserved features of these ensembles. Other similar approaches include EOM (62,63), MaxOcc (64–67), ASTEROIDS (68–70), SES (71), and EROS (72). A parsimonious explanation of the SAXS profiles for each of the recPrP and recPrP-Fab-P samples is provided by the open and closed states, corresponding to the two Rg peaks that were conserved for N-state models (for N = 2 … 5; Figs. 3 and 5). Each of the two states consists of multiple different conformations with similar Rg values.
Possible inhibitory mechanisms of prion conversion by Fab-P and Fab-R1
Both Fab-P and Fab-R1 inhibit conversion from PrPC to PrPSc but bind to different sites on PrP (19). Fab-P binds a region of PrP that is structurally accessible in PrPC but not in PrPSc (73), whereas the epitope of Fab-R1 is accessible in both isoforms (74). Structural models of the recPrP-Fab-P and recPrP-Fab-R1 complexes were built computationally by integrating information from SAXS profiles, known epitopes (45), structures from NMR spectroscopy and x-ray crystallography (16,58), as well as statistical potentials (56).
Based on our structural models, we speculate how each antibody prevents the conversion, as follows. Fab-P binds to the N-terminal region (residues 95–105) that is known to undergo structural conversion from PrPC to PrPSc. The Fab-P might prevent the PrPC-PrPSc interaction by acting as a structural clamp on the epitope, thus preventing its conversion from the disordered conformation in PrPC to the ordered β-sheet conformation in PrPSc. Given the size of Fab-P relative to PrP, the Fab-P-PrP interaction could also interfere with the refolding of PrPC into PrPSc outside of the epitope.
In contrast to Fab-P, Fab-R1 binds to residues 225–230, whose conformation probably does not change during the conversion, allowing for binding of both PrPC and PrPSc. The interaction of Fab-R1 with both isoforms of PrPC and PrPSc might sterically inhibit a crucial interaction between PrPC and PrPSc. In addition to the known Fab-R1 epitope of PrPC, our structural model revealed putative contacts between Fab-R1 and PrP residues 165–175, the α2-β2 loop of PrPC (41–43). This loop region in murine PrP exists in two structural states, a major state that forms a 310-helical turn and a minor state that forms a type-I β-turn (41,43) (Fig. 6 F). Interestingly, the sequence of this loop region has also been shown to be strongly associated with the rate of prion infection (41–43). Our structural model rationalizes this observation, suggesting that Fab-R1 prevents or slows down the conversion from PrPC to PrPSc by influencing the structure and/or dynamics of the α2-β2 loop.
Alternatively, the binding of either Fab-P or Fab-R1 may be sufficient to divert PrPC from conversion into PrPSc by accelerating its degradation (75,76). It may be possible to determine whether this explanation applies to either or both of the PrPC-Fab complexes described here. Radiolabeling of nascent PrPC and measuring its turnover may help extend our understanding of the mechanism by which anti-PrP antibodies inhibit PrPSc formation (77,78).
Conclusions
The FPLC-SAXS pipeline presented here minimizes data artifacts caused by a non-monodispersed sample, enabling high-quality data collection on challenging macromolecular systems that are prone to aggregation. We applied this approach toward structural characterization of recPrP, both on its own and in complex with two different antibodies. Based on the resulting structural models, we propose two different mechanisms for antibody-mediated inhibition of the PrPC-to-PrPSc conversion: 1) direct inhibition by Fab-P binding to a PrPC region (residues 95–105) known to undergo rearrangement during PrPC-to-PrPSc conversion (74); and 2) indirect inhibition by Fab-R1 that is predicted here to bind to the α2-β2 loop structure (residues 165–175), which is known to impact the conversion rate (41–43). As noted above, antibody-mediated inhibition of PrPSc formation may also occur through accelerating the degradation of PrPC.
Although anti-Aβ antibody therapies for Alzheimer’s disease have been widely tested, none have been successful. Because <0.1% of systemically administered antibodies cross the blood-brain barrier, the use of anti-Aβ antibodies to treat or prevent Alzheimer’s disease is probably a poor strategy. The same problems apply for antibody therapeutics for PrP prion diseases: Anti-PrP antibodies have not extended the lives of mice inoculated intracerebrally (79), but they have prolonged the lives of mice inoculated intraperitoneally (80). Whether the structural insights reported here prove useful in facilitating the discovery of therapeutically effective small molecules remains to be determined.
Author Contributions
L.C., S.J.K., D.S.-D., J.S., T.M.W., H.T., S.B.P., and A.S. designed the research; J.S., G.P.-M., and S.B.P. prepared samples for data collection; L.C., S.J.K., T.M.W., and H.T. collected SAXS data; L.C., S.J.K., D.S.-D., and J.S. analyzed the data; D.S.-D., S.J.K., and A.S. performed computational modeling; and L.C., S.J.K., D.S.-D., J.S., S.B.P., and A.S. wrote the article.
Acknowledgments
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Funding for this work was provided by National Institutes of Health grants R01 GM083960 and P41 GM109824. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of the SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the U.S. Department of Energy Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program (P41 RR001209).
Editor: Lois Pollack.
Footnotes
Hiro Tsuruta died on August 25, 2011.
Lester Carter, Seung Joong Kim, Dina Schneidman-Duhovny, and Jan Stöhr contributed equally to this work.
Guillaume Poncet-Montange’s present address is Department of Genomic Medicine, University of Texas, MD Anderson Cancer Center, 1515 Holcombe Blvd. Unit 1000, Houston, TX 77030.
One figure is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00708-0.
Contributor Information
Stanley B. Prusiner, Email: stanley@ind.ucsf.edu.
Andrej Sali, Email: sali@salilab.org.
Supporting Material
References
- 1.Prusiner S.B. Cell biology. A unifying role for prions in neurodegenerative diseases. Science. 2012;336:1511–1513. doi: 10.1126/science.1222951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colby D.W., Prusiner S.B. Prions. Cold Spring Harb. Perspect. Biol. 2011;3:a006833. doi: 10.1101/cshperspect.a006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walker L.C., Diamond M.I., Hyman B.T. Mechanisms of protein seeding in neurodegenerative diseases. JAMA Neurol. 2013;70:304–310. doi: 10.1001/jamaneurol.2013.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker L.C., Jucker M. Seeds of dementia. Sci. Am. 2013;308:52–57. doi: 10.1038/scientificamerican0513-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prusiner S.B. Biology and genetics of prions causing neurodegeneration. Annu. Rev. Genet. 2013;47:601–623. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissmann C., Aguzzi A. Approaches to therapy of prion diseases. Annu. Rev. Med. 2005;56:321–344. doi: 10.1146/annurev.med.56.062404.172936. [DOI] [PubMed] [Google Scholar]
- 7.Prusiner S.B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 8.Pan K.-M., Baldwin M., Prusiner S.B. Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pergami P., Jaffe H., Safar J. Semipreparative chromatographic method to purify the normal cellular isoform of the prion protein in nondenatured form. Anal. Biochem. 1996;236:63–73. doi: 10.1006/abio.1996.0132. [DOI] [PubMed] [Google Scholar]
- 10.Pan K.M., Stahl N., Prusiner S.B. Purification and properties of the cellular prion protein from Syrian hamster brain. Protein Sci. 1992;1:1343–1352. doi: 10.1002/pro.5560011014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas O., Gavín R., del Río J.A. New insights into cellular prion protein (PrPc) functions: the “ying and yang” of a relevant protein. Brain Res. Brain Res. Rev. 2009;61:170–184. doi: 10.1016/j.brainresrev.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Riek R., Hornemann S., Wüthrich K. NMR structure of the mouse prion protein domain PrP(121–231) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 13.James T.L., Liu H., Cohen F.E. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl. Acad. Sci. USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donne D.G., Viles J.H., Dyson H.J. Structure of the recombinant full-length hamster prion protein PrP(29–231): the N terminus is highly flexible. Proc. Natl. Acad. Sci. USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knaus K.J., Morillas M., Yee V.C. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 2001;8:770–774. doi: 10.1038/nsb0901-770. [DOI] [PubMed] [Google Scholar]
- 16.Damberger F.F., Christen B., Wüthrich K. Cellular prion protein conformation and function. Proc. Natl. Acad. Sci. USA. 2011;108:17308–17313. doi: 10.1073/pnas.1106325108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baral P.K., Swayampakula M., James M.N. Structural basis of prion inhibition by phenothiazine compounds. Structure. 2014;22:291–303. doi: 10.1016/j.str.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 18.Wille H., Bian W., Stubbs G. Natural and synthetic prion structure from x-ray fiber diffraction. Proc. Natl. Acad. Sci. USA. 2009;106:16990–16995. doi: 10.1073/pnas.0909006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peretz D., Williamson R.A., Prusiner S.B. Antibodies inhibit prion formation and abolish prion infectivity. In: Lehmann S., editor. Transmissible Spongiform Encephalopathies: New Perspectives for Prion Therapeutics. Editions de Condé; Paris, France: 2002. pp. 107–118. [Google Scholar]
- 20.Klöhn P.C., Farmer M., Collinge J. PrP antibodies do not trigger mouse hippocampal neuron apoptosis. Science. 2012;335:52. doi: 10.1126/science.1215579. [DOI] [PubMed] [Google Scholar]
- 21.Solforosi L., Criado J.R., Williamson R.A. Cross-linking cellular prion protein triggers neuronal apoptosis in vivo. Science. 2004;303:1514–1516. doi: 10.1126/science.1094273. [DOI] [PubMed] [Google Scholar]
- 22.Putnam C.D., Hammel M., Tainer J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 23.Mertens H.D., Svergun D.I. Structural characterization of proteins and complexes using small-angle x-ray solution scattering. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Jacques D.A., Trewhella J. Small-angle scattering for structural biology—expanding the frontier while avoiding the pitfalls. Protein science. 2010;19:642–657. doi: 10.1002/pro.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 26.Svergun D., Barberato C., Koch M.H.J. CRYSOL—a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 27.Schneidman-Duhovny D., Hammel M., Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 2010;38:W540–W544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneidman-Duhovny D., Hammel M., Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petoukhov M.V., Konarev P.V., Svergun D.I. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. J. Appl. Crystallogr. 2007;40:S223–S228. [Google Scholar]
- 30.Pons C., D’Abramo M., Fernández-Recio J. Structural characterization of protein-protein complexes by integrating computational docking with small-angle scattering data. J. Mol. Biol. 2010;403:217–230. doi: 10.1016/j.jmb.2010.08.029. [DOI] [PubMed] [Google Scholar]
- 31.Schneidman-Duhovny D., Hammel M., Sali A. Macromolecular docking restrained by a small angle x-ray scattering profile. J. Struct. Biol. 2011;173:461–471. doi: 10.1016/j.jsb.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernadó P., Pérez Y., Pons M. Structural characterization of the active and inactive states of Src kinase in solution by small-angle x-ray scattering. J. Mol. Biol. 2008;376:492–505. doi: 10.1016/j.jmb.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 33.Pelikan M., Hura G.L., Hammel M. Structure and flexibility within proteins as identified through small angle x-ray scattering. Gen. Physiol. Biophys. 2009;28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hammel M. Validation of macromolecular flexibility in solution by small-angle x-ray scattering (SAXS) Eur. Biophys. J. 2012;41:789–799. doi: 10.1007/s00249-012-0820-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathew E., Mirza A., Menhart N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron Radiat. 2004;11:314–318. doi: 10.1107/S0909049504014086. [DOI] [PubMed] [Google Scholar]
- 36.Pérez J., Nishino Y. Advances in x-ray scattering: from solution SAXS to achievements with coherent beams. Curr. Opin. Struct. Biol. 2012;22:670–678. doi: 10.1016/j.sbi.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Martel A., Liu P., Tsuruta H. An integrated high-throughput data acquisition system for biological solution x-ray scattering studies. J. Synchrotron Radiat. 2012;19:431–434. doi: 10.1107/S0909049512008072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trastoy B., Lomino J.V., Sundberg E.J. Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies. Proc. Natl. Acad. Sci. USA. 2014;111:6714–6719. doi: 10.1073/pnas.1322908111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho M.C., Wilczek C., Shechter D. Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One. 2013;8:e57008. doi: 10.1371/journal.pone.0057008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.West A.L., Evans S.E., Michel S.L. Ni(II) coordination to mixed sites modulates DNA binding of HpNikR via a long-range effect. Proc. Natl. Acad. Sci. USA. 2012;109:5633–5638. doi: 10.1073/pnas.1120283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Christen B., Damberger F.F., Wüthrich K. Structural plasticity of the cellular prion protein and implications in health and disease. Proc. Natl. Acad. Sci. USA. 2013;110:8549–8554. doi: 10.1073/pnas.1306178110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kyle L.M., John T.R., Lewis R.V. Introducing a rigid loop structure from deer into mouse prion protein increases its propensity for misfolding in vitro. PLoS One. 2013;8:e66715. doi: 10.1371/journal.pone.0066715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kurt T.D., Bett C., Sigurdson C.J. Prion transmission prevented by modifying the β2-α2 loop structure of host PrPC. J. Neurosci. 2014;34:1022–1027. doi: 10.1523/JNEUROSCI.4636-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehlhorn I., Groth D., Prusiner S.B. High-level expression and characterization of a purified 142-residue polypeptide of the prion protein. Biochemistry. 1996;35:5528–5537. doi: 10.1021/bi952965e. [DOI] [PubMed] [Google Scholar]
- 45.Williamson R.A., Peretz D., Burton D.R. Mapping the prion protein using recombinant antibodies. J. Virol. 1998;72:9413–9418. doi: 10.1128/jvi.72.11.9413-9418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poncet-Montange G., St Martin S.J., Ghaemmaghami S. A survey of antiprion compounds reveals the prevalence of non-PrP molecular targets. J. Biol. Chem. 2011;286:27718–27728. doi: 10.1074/jbc.M111.234393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SasTool. 2013. http://ssrl.slac.stanford.edu/∼saxs/analysis/sastool.htm.
- 48.Konarev P.V., Volkov V.V., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 49.Fischer H., Neto M.D., Craievich A.F. Determination of the molecular weight of proteins in solution from a single small-angle x-ray scattering measurement on a relative scale. J. Appl. Crystallogr. 2010;43:101–109. [Google Scholar]
- 50.Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 51.Amato N.M., Song G. Using motion planning to study protein folding pathways. J. Comput. Biol. 2002;9:149–168. doi: 10.1089/10665270252935395. [DOI] [PubMed] [Google Scholar]
- 52.Raveh B., Enosh A., Halperin D. Rapid sampling of molecular motions with prior information constraints. PLOS Comput. Biol. 2009;5:e1000295. doi: 10.1371/journal.pcbi.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanfield R.L., Zemla A., Rupp B. Antibody elbow angles are influenced by their light chain class. J. Mol. Biol. 2006;357:1566–1574. doi: 10.1016/j.jmb.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 54.Schneidman-Duhovny D., Rossi A., Sali A. A method for integrative structure determination of protein-protein complexes. Bioinformatics. 2012;28:3282–3289. doi: 10.1093/bioinformatics/bts628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schneidman-Duhovny D., Inbar Y., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dong G.Q., Fan H., Sali A. Optimized atomic statistical potentials: assessment of protein interfaces and loops. Bioinformatics. 2013;29:3158–3166. doi: 10.1093/bioinformatics/btt560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lawler E.L., Wood D.E. Branch-and-bound methods: a survey. Oper. Res. 1966;14:699–719. [Google Scholar]
- 58.Luginbühl B., Kanyo Z., Plückthun A. Directed evolution of an anti-prion protein scFv fragment to an affinity of 1 pM and its structural interpretation. J. Mol. Biol. 2006;363:75–97. doi: 10.1016/j.jmb.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 59.Elfrink K., Ollesch J., Gerwert K. Structural changes of membrane-anchored native PrPC. Proc. Natl. Acad. Sci. USA. 2008;105:10815–10819. doi: 10.1073/pnas.0804721105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baldwin M.A., Stahl N., Prusiner S.B. Glycosylinositol phospholipid anchors of prion proteins. In: Prusiner S.B., Collinge J., Powell J., Anderton B., editors. Prion Diseases of Humans and Animals. Ellis Horwood; London: 1992. pp. 380–397. [Google Scholar]
- 61.Hornemann S., Schorn C., Wüthrich K. NMR structure of the bovine prion protein isolated from healthy calf brains. EMBO Rep. 2004;5:1159–1164. doi: 10.1038/sj.embor.7400297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernadó P., Mylonas E., Svergun D.I. Structural characterization of flexible proteins using small-angle x-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 63.Bernadó P., Svergun D.I. Structural analysis of intrinsically disordered proteins by small-angle x-ray scattering. Mol. Biosyst. 2012;8:151–167. doi: 10.1039/c1mb05275f. [DOI] [PubMed] [Google Scholar]
- 64.Bertini I., Giachetti A., Svergun D.I. Conformational space of flexible biological macromolecules from average data. J. Am. Chem. Soc. 2010;132:13553–13558. doi: 10.1021/ja1063923. [DOI] [PubMed] [Google Scholar]
- 65.Bertini I., Ferella L., Svergun D.I. MaxOcc: a web portal for maximum occurrence analysis. J. Biomol. NMR. 2012;53:271–280. doi: 10.1007/s10858-012-9638-1. [DOI] [PubMed] [Google Scholar]
- 66.Cerofolini L., Fields G.B., Teixeira J.M. Examination of matrix metalloproteinase-1 in solution: a preference for the pre-collagenolysis state. J. Biol. Chem. 2013;288:30659–30671. doi: 10.1074/jbc.M113.477240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andrałojć W., Luchinat C., Ravera E. Exploring regions of conformational space occupied by two-domain proteins. J. Phys. Chem. B. 2014;118:10576–10587. doi: 10.1021/jp504820w. [DOI] [PubMed] [Google Scholar]
- 68.Nodet G., Salmon L., Blackledge M. Quantitative description of backbone conformational sampling of unfolded proteins at amino acid resolution from NMR residual dipolar couplings. J. Am. Chem. Soc. 2009;131:17908–17918. doi: 10.1021/ja9069024. [DOI] [PubMed] [Google Scholar]
- 69.Salmon L., Nodet G., Blackledge M. NMR characterization of long-range order in intrinsically disordered proteins. J. Am. Chem. Soc. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 70.Fisher C.K., Stultz C.M. Constructing ensembles for intrinsically disordered proteins. Curr. Opin. Struct. Biol. 2011;21:426–431. doi: 10.1016/j.sbi.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berlin K., Castañeda C.A., Fushman D. Recovering a representative conformational ensemble from underdetermined macromolecular structural data. J. Am. Chem. Soc. 2013;135:16595–16609. doi: 10.1021/ja4083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Różycki B., Kim Y.C., Hummer G. SAXS ensemble refinement of ESCRT-III CHMP3 conformational transitions. Structure. 2011;19:109–116. doi: 10.1016/j.str.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peretz D., Williamson R.A., Burton D.R. A conformational transition at the N terminus of the prion protein features in formation of the scrapie isoform. J. Mol. Biol. 1997;273:614–622. doi: 10.1006/jmbi.1997.1328. [DOI] [PubMed] [Google Scholar]
- 74.Govaerts C., Wille H., Cohen F.E. Evidence for assembly of prions with left-handed β-helices into trimers. Proc. Natl. Acad. Sci. USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Enari M., Flechsig E., Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc. Natl. Acad. Sci. USA. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peretz D., Williamson R.A., Prusiner S.B. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 77.Caughey B., Race R.E., Chesebro B. Prion protein biosynthesis in scrapie-infected and uninfected neuroblastoma cells. J. Virol. 1989;63:175–181. doi: 10.1128/jvi.63.1.175-181.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Borchelt D.R., Scott M., Prusiner S.B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ohsawa N., Song C.-H., Horiuchi M. Therapeutic effect of peripheral administration of an anti-prion protein antibody on mice infected with prions. Microbiol. Immunol. 2013;57:288–297. doi: 10.1111/1348-0421.12037. [DOI] [PubMed] [Google Scholar]
- 80.White A.R., Enever P., Hawke S. Monoclonal antibodies inhibit prion replication and delay the development of prion disease. Nature. 2003;422:80–83. doi: 10.1038/nature01457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.