Abstract
Contact-mode atomic force microscopy (AFM) has been shown to reveal cortical actin structures. Using live endothelial cells, we visualized cortical actin dynamics simultaneously by AFM and confocal fluorescence microscopy. We present a method that quantifies dynamic changes in the mechanical ultrastructure of the cortical actin web. We argue that the commonly used, so-called error signal imaging in AFM allows a qualitative, but not quantitative, analysis of cortical actin dynamics. The approach we used comprises fast force-curve-based topography imaging and subsequent image processing that enhances local height differences. Dynamic changes in the organization of the cytoskeleton network can be observed and quantified by surface roughness calculations and automated morphometrics. Upon treatment with low concentrations of the actin-destabilizing agent cytochalasin D, the cortical cytoskeleton network is thinned out and the average mesh size increases. In contrast, jasplakinolide, a drug that enhances actin polymerization, consolidates the cytoskeleton network and reduces the average mesh area. In conclusion, cortical actin dynamics can be quantified in live cells. To our knowledge, this opens a new pathway for conducting quantitative structure-function analyses of the endothelial actin web just beneath the apical plasma membrane.
Introduction
Actin, in its globular and polymerized forms as G- and F-actin, respectively, constitutes 5–15% of the total protein in endothelial cells (1). In addition to intermediate filaments and microtubules, it is a central component of the cytoskeleton. Hence, it determines the cell shape and is involved in a multitude of cellular processes, including motility, division, and intracellular trafficking. A large number of actin-binding proteins are involved in controlling actin’s polymerization and architecture, linking it to other filaments, and anchoring it in the membrane (2). It is embedded in cellular signaling pathways, e.g., by proteins from the family of Rho-GTPases (3).
The endothelium is constantly subjected to hemodynamic forces in the form of shear stress, vessel wall tension, and hydrostatic pressure. An endothelial cell’s mechanical properties determine how the cell will resist and respond to these forces (4). Actin plays an important role in controlling endothelial barrier permeability (5). The cortical actin network in particular is assumed to underlie a cell’s stiffness and thereby mediate physiological effects in the endothelium, such as the release of nitric oxide after changes in electrolyte concentrations or inflammatory stimuli (6–8). Considering that the ultrastructural architecture of polymeric networks such as actin determines their mechanical characteristics (9), a method to enable high-resolution visualization of the cortical cytoskeleton network and its dynamic behavior is needed.
Atomic force microscopy (AFM) is an important tool in biological and biophysical research, and enables a multitude of structural, micromechanical, and molecular investigations (10). It can be used to image surfaces on scales ranging from cells to single DNA strands. Because it offers a higher resolution than conventional optical microscopes but, unlike electron microscopy, operates under physiological conditions and requires no complex sample preparation, AFM has proven particularly useful for live-cell investigations (11). Moreover, in live cells, AFM can actually be used to probe the submembranous cellular micromechanical organization and thereby image the cytoskeleton (12–14).
A number of AFM studies have provided views of the mechanical organization of the cytoskeleton using contact-mode error data or elasticity-mapping procedures (7,14–16). However, to our best knowledge, a procedure for imaging the cortical cytoskeleton in living cells and assessing quantitative density changes has not been described to date.
Here, we performed high-resolution visualization and quantification of the cortical actin cytoskeleton and its remodeling in live endothelial cells using AFM-based methods. Contact-mode AFM showed a cortical cytoskeleton network with mesh sizes on two different scales. Simultaneous live-cell imaging with AFM and confocal fluorescence microscopy of Lifeact identified part of the network as F-actin and provided a detailed view of remodeling processes in time-lapse experiments. To obtain better resolution and more readily quantifiable data, we developed a procedure to image the cortical cytoskeleton network that combines fast force-curve-based topography imaging with subsequent image processing. Pharmacological treatments with either low concentrations of the actin-disrupting agent cytochalasin D or the actin-stabilizing drug jasplakinolide correspondingly affected the cytoskeleton network morphology, validating our method and revealing the pivotal role of actin in the mechanical architecture of the endothelial cortical cytoskeleton.
Materials and Methods
Solutions and reagents
All chemicals and reagents were purchased from Sigma-Aldrich (Steinheim, Germany) unless mentioned otherwise. AFM experiments were performed at room temperature with live cells in HEPES buffer (in mM: 140 NaCl, 5 KCl, 1 MgCl2, 1 CaCl2, 5 glucose, 10 HEPES adjusted to pH 7.4). For force-mapping-mode imaging experiments, cytochalasin D was added to a concentration of 100 nM (stock 100 μM in ethanol) and jasplakinolide was added to a concentration of 1 μM (stock 8 mM in DMSO). All control buffers were supplemented with the appropriate concentration of solvent (ethanol or DMSO).
Cell culture
Bovine aortic endothelial cells (GM7373; DSMZ GmbH, Braunschweig, Germany) (17) were cultured in minimal essential medium (GE Healthcare/PAA Laboratories, Pasching, Austria) supplemented with 20% fetal calf serum (FCS; PAA), 1% nonessential amino acids (Life Technologies, Grand Island, NY), 1% minimum essential medium (MEM) vitamins (Life Technologies), and 1% penicillin and streptomycin (Biochrom, Berlin, Germany). Cells were kept in T25 culture flasks at 37°C, 5% CO2, and 100% humidity.
For all imaging experiments, cells were seeded on glass-bottom dishes (50 mm diameter from WillCo Wells, Amsterdam, The Netherlands, or 35 mm diameter from WPI, Sarasota, FL) and grown until they reached ≈90% confluence.
To enable fluorescent imaging of F-actin in living cells concurrently with contact-mode AFM, GM7373 cells were transfected with Lifeact-eGFP as described previously (18). Lifeact, a short peptide (19) that neither interferes with actin dynamics nor alters actin expression, was chosen as the most suitable means of labeling F-actin in live cells (20). Briefly, the cells were trypsinized, centrifuged, and resuspended in OptiMEM medium (Life Technologies) supplemented with 10% FCS. Then, 8 μl of FuGENE6 transfection reagent (Promega, Madison, WI) was added to 230 μl of FCS-free OptiMEM medium and incubated for 5 min at room temperature. Subsequently, 4 μg Lifeact-eGFP DNA (in pEGFP-N1 vector) was added and the solution was again incubated for 10 min. The cells in suspension were then added and mixed with the solution before they were seeded onto glass-bottom dishes. For the first 30 min in the cell culture incubator at 37°C, 5% CO2, the cells were placed on a shaker at 180 rpm. The next day, the medium was replaced with normal growth medium.
For experiments with force-mapping-mode AFM, GM7373 cells stably transfected with Lifeact-mKate2 were used. The cells were transfected as described above with a plasmid containing Lifeact-mKate2 and, in an IRES segment, hygromycin-B-phosphotransferase. Stable transfections were then selected with 40 μg/mL hygromycin B in regular growth medium (21). Polyclonal cells were further kept under selective conditions and seeded for experiments without hygromycin B. mKate2 was excited at a 633 nm wavelength.
AFM imaging
AFM imaging was done using a JPK Nanowizard 3 (JPK Instruments, Berlin, Germany). All measurements were performed in liquid with B-probes on MSCT-UC chips (Bruker, Santa Barbara, CA), which feature sharpened tips (10 nm radius of curvature) on soft silicon nitride cantilevers with a nominal spring constant of 0.02 N/m. They possess no backside coating to reduce the force drift during measurements (22). We have estimated the optimal imaging force to be in the range of ≈0.5 nN using deflection sensitivity and thermal noise cantilever calibration. However, variations of the measured optimal imaging force, which we observed, can be attributed to differences in tip sharpness and tip contamination, resulting in different contact areas, as well as to errors inherent in the cantilever calibration. Thus, imaging setpoints were generally set as low as possible and appropriate values were determined empirically at the beginning of each experiment. The probes were cleaned after each experiment by rinsing in deionized water and acetone (23), and subsequent plasma treatment (PDC 32G; Harrick Plasma, Ithaca, NY) for 10 min.
To image cortical cytoskeleton structures, we used two different procedures: contact-mode imaging and force mapping. Contact-mode imaging was performed with a (10 μm)2 scan area, 256 × 256 pixels, 2 Hz line rate (resulting in ∼2.5 min acquisition time per image), and manually optimized feedback gains. We carried out force mapping using the JPK quantitative imaging (QI) mode with the following settings: (6 × 6 μm) scan area, 256 × 256 pixels, z range = 50 nm, extend and retract time = 1.8 ms (refers to the actual approach-retract cycle and measurement, with a resulting tip velocity of 27.8 μm/s); additional retract = 50 nm, motion time = 0.5 ms, and acceleration = 0.5 ms (parameters that control the trajectory and velocity of the tip during the lateral movement phase; ≈10 min/image). For each experiment, a single cell was selected and then imaged continuously.
We added chemicals to the cells by completely replacing the buffer in the culture dish via attached tubings while leaving the AFM head in place and briefly retracting the z-piezo.
Confocal fluorescence imaging
In the contact-mode AFM experiments, simultaneous fluorescence microscopy of F-actin in Lifeact-eGFP-transfected cells was conducted with an inverted confocal microscope (TCS SP8, Leica Microsystems, Wetzlar, Germany) equipped with a 63× NA 1.4 objective. Z stacks of the entire cell or the apical part were recorded and later visualized by maximum intensity projection. Laser scanning was performed at the same time as AFM imaging, which improved the time resolution but caused certain line artifacts in the AFM images. We established an approximate overlay with the AFM images during experiments by imaging both the fluorescent cells and the AFM probe tip in wide-field illumination using JPK’s direct overlay feature.
To check for membrane integrity and mechanosensitive stimulation of the endothelial cells during AFM scanning, we recorded the intracellular calcium concentration simultaneously with AFM-based image acquisitions by means of fluorescence microscopy. Cells were loaded with the intracellular calcium indicator Fluo4 (Life Technologies) according to the manufacturer’s protocol. Ionomycin was added to a concentration of 2.5 μM at the end of the experiment. Fluo4 was excited at 488 nm, fluorescence emission was measured at 500–550 nm, and measurements were performed every 30–60 s.
Image processing and analysis
In the contact-mode time-lapse experiments, the AFM height channel indicated the general cell topography and the error-channel data that displayed cytoskeleton network structures were exported using JPK data-processing software (v. 4.4.33+). Then, in Fiji/ImageJ (v. 2.0.0-rc-14/1.49f) (24), trace and retrace error images were overlaid by addition for better visualization, and the corresponding fluorescence images were scaled and manually matched to the AFM data. In the case of force-mapping-mode imaging, to obtain information about the fine cytoskeleton network structures, the height data were processed in a manner similar to that described in Kienberger et al. (25). Using MATLAB R2014a (The MathWorks, Natick, MA), we performed line-wise subtraction of a second-order polynomial fit, followed by difference-of-Gaussians filtering (subtraction of a smoothed version (standard deviation (SD) of the Gaussian kernel: σ = 5 pixels) of the image and Gaussian smoothing (σ = 1 px) of the image itself). We then obtained adapted root mean-square (RMS) roughness values by calculating the SD of all image values after removing outliers (values > 4 SDs distant from the mean) caused by imaging artifacts.
For the morphological analysis, the processed images were segmented using ilastik 1.1 (26). The software was manually trained to separate cytoskeleton network structures from meshwork holes with one set of images and then used to segment all shown data with the same training. Subsequently, MATLAB was used to obtain statistical measures regarding the number of holes, the mean area of holes, and the overall surface coverage of holes in each image.
With regard to the adapted RMS roughness values and morphological measures, the absolute values for each cell and thus for each experiment are offset from each other (see Fig. S5 in the Supporting Material), so all values are expressed as the difference from the control phase average for each experiment in this study.
Height data are depicted in a brown color scale, error images are shown in grayscale, and keys are shown for each image except for overlaid trace and retrace error images. A brown color scale is also used for processed height images, although since the processing makes the absolute height values arbitrary, a key is not shown. Relative differences, however, are faithfully reproduced, as the intensities of all images belonging to one experiment are adjusted identically.
Statistics
Grouped measurements were normally distributed, with a few exceptions, and therefore we compared groups using both Student’s t-test and the Mann-Whitney test. Data were considered significantly different and marked with an asterisk (∗) if p ≤ 0.05 in both tests. In diagrams, distributions are displayed by the mean, with error bars showing the mean ± standard error (SE).
Results
Contact-mode AFM imaging shows a cortical cytoskeleton network
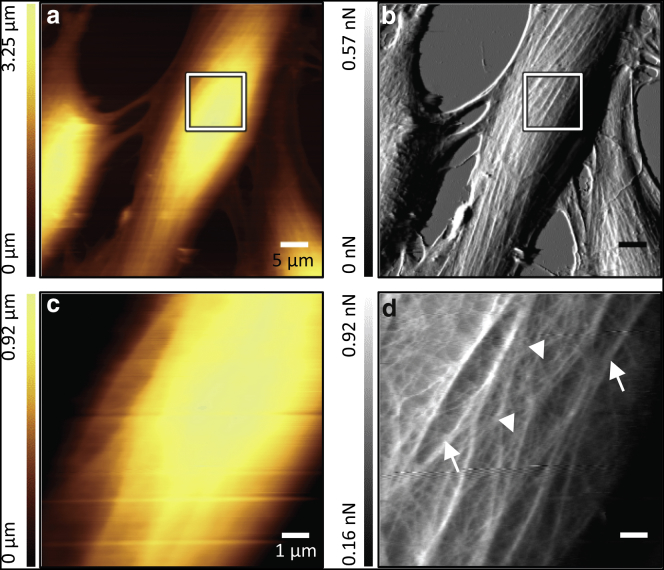
We employed standard contact-mode AFM scanning in this work. Uncoated cantilevers were used to minimize force drift and obtain stable feedback conditions over a long time, and low imaging force setpoints of ∼0.5 nN allowed us to visualize the general cell topography in the height channel (Fig. 1, a and c) and a network representing the cytoskeleton in the error channel (Fig. 1, b and d). It is composed of a coarse mesh and a fine mesh similar to those previously described in endothelial cells (14). To capture the cortical part of the cells’ cytoskeleton in a reproducible way, we selected small square areas with a 6–10 μm side length on top of the nuclei for high-resolution imaging. We estimated the spatial resolution of contact-mode AFM applied in this way to visualize cytoskeleton structures in the error-channel data to be ≈150 nm or better along the fast-scan axis by inspecting the spacing between filament structures (Fig. S4, a–c).
Figure 1.
Live GM7373 endothelial cells imaged by AFM in contact mode. (a and b) Overall topography of a cell; the region chosen for further high-resolution imaging is marked. (c and d) High-resolution scan acquired by contact-mode imaging; the coarse mesh (arrows) and fine mesh (arrowheads) can be discerned. (a and c) Height images. (b and d) Corresponding error-trace images. Scale bars are 5 μm (a and b) and 1 μm (c and d). To see this figure in color, go online.
Lifeact-based fluorescence microscopy identifies coarse mesh as actin
To identify actin-based structures in the AFM images, we performed live-cell fluorescence microscopy of Lifeact-eGFP-labeled F-actin simultaneously with contact-mode AFM. The endothelial cells exhibit a characteristic pattern of actin distribution comprising basal stress fibers and a thin cortical layer spanning the apical surface. In the topmost cortical parts of the cells, the Lifeact fluorescence precisely matches the coarse mesh seen in the AFM images (Fig. 2, a–d), which can thus be identified as F-actin. This agrees with previous observations made on fixed and stained samples (14). In spaces corresponding to the fine mesh, only diffuse fluorescence could be measured. Whether this lack of congruence points to a nonactin makeup of the fine mesh or must be attributed to the optical microscope’s resolution limit remains unclear.
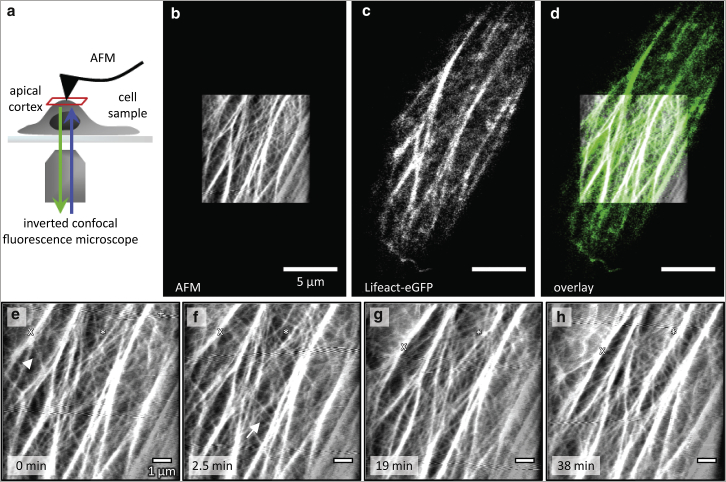
Figure 2.
(a) Illustration of combined Lifeact/AFM imaging of a living endothelial cell’s apical cortex. (b) Cortical area visualized by contact-mode AFM (error signal, trace and retrace overlaid by addition). (c) Lifeact-eGFP detected by confocal fluorescence microscopy simultaneously in the same region (maximum intensity projection of the most apical focal planes). (d) Overlay of (b) and (c). (e–h) Cytoskeleton meshwork dynamics as observed by contact-mode imaging. Coarse-mesh fibers can be tracked while moving (x) and some emerge from lighter ones (asterisk). In contrast, fine-mesh structures may disappear (arrowhead) or appear (arrow) from one frame to the next, representing a faster reorganization. In (e) a line artifact caused by confocal scanning is marked (plus sign). Scale bars are 5 μm (b–d) and 1 μm (e–h). See Movie S1 for a time-lapse visualization of the entire experiment. To see this figure in color, go online.
An optical superresolution technique called stochastic optical reconstruction microscopy (STORM) was recently demonstrated to resolve individual actin filaments in three dimensions (27). We applied the specified protocol for fixation and staining while using a custom-built STORM setup with the appropriate characteristics for imaging. However, we were not able to achieve a labeling of actin structures sufficient to visualize more detailed features compared with confocal views (data not shown).
Coarse-mesh dynamics can be tracked by time-lapse imaging
Repeated contact-mode imaging AFM and actin/Lifeact fluorescence microscopy of the same area at an interval of ∼2.5 min yielded time-lapse series that allowed us to study the dynamic behavior of cortical cytoskeleton structures (Fig. 2, e and f; Movie S1). The coarse-mesh fibers can be continuously tracked in these series. They exhibit uniform movement and reorganization in terms of splitting and joining of fibers, and in some cases the gradual formation of a coarse-mesh fiber with high contrast from a lighter structure can be observed (Fig. 2, e–h, asterisk). The fine mesh, however, appears to undergo faster remodeling, because its components cannot be tracked from one frame to the next.
An in-depth look at error-channel data
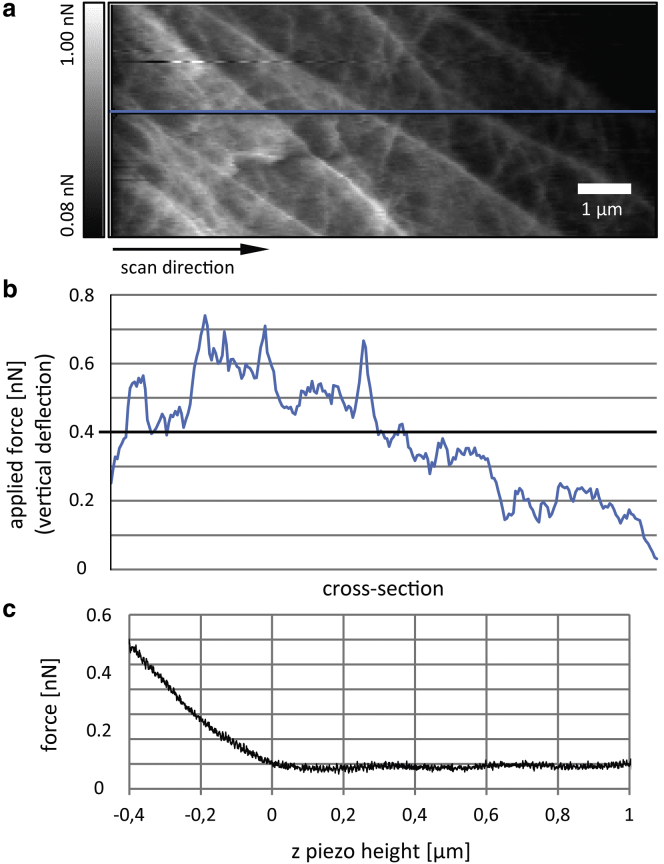
AFM deflection or error-signal data (28) have been used extensively to visualize small surface features on soft biological samples such as cells (14,16,29,30), just as they were used for the contact-mode experiments in this work. During contact-mode scanning, a feedback loop is used to constantly reposition the z-piezo (thus generating the height data) so that the cantilever deflection will stay at a certain setpoint value; its deviation from this setpoint value constitutes the error signal. The error image has a shaded appearance as it shows the slope of the height image in the fast-scan direction. As pointed out by Putman et al. (28), the feedback loop can serve as a filter separating low- and high-frequency information into height and error images, respectively. It is assumed that the applied imaging force must stay nearly constant so as not to alter the actual surface topography. In our contact-mode imaging experiments, however, we could not support this assumption. Even with optimized feedback settings, the actual imaging force varies significantly, caused on a wider scale by the convex shape of the cell surface (which is most striking in a comparison of trace and retrace error images; see Fig. S1, c and d), and also on a smaller scale caused by cytoskeleton structures (Fig. 3, a and b). This can be slightly alleviated by slowing down the scanning; however, there is a limit to the minimum scan speed, considering that dynamic structures in living cells are being imaged. A comparison with a typical force-indentation curve (Fig. 3 c) reveals that force deviations in this range considerably change the apparent cell surface as it is probed by the AFM tip. Also, it is striking that most of the filaments are revealed in the error images only by one bright ridge as the tip hits the edge of a fiber, as opposed to a bright ridge accompanied by a dark ridge, which one would expect to see if the elevation caused by a cytoskeleton fiber were properly tracked by the AFM tip. We recognize that error data acquired in this manner do not show the cytoskeleton meshwork in its entirety and hence are unsuitable for quantitatively estimating the network density, as we aimed to do in this work.
Figure 3.
Force errors in contact-mode scanning. (a) High-resolution contact-mode scan (error image); scale bar, 1 μm. (b) Cross section of (a) showing the magnitude of the force error; the thick line at 0.4 nN indicates the actual feedback setpoint force. (c) Force-height curve measured on the same cell as in (a) using the same cantilever. At 0.4 nN, the height difference is ∼400 nm. To see this figure in color, go online.
Force-mapping-mode imaging enables visualization of the fine mesh using height data
To address the above issues associated with standard contact-mode imaging, we performed force-mapping-mode imaging (termed QI by JPK Instruments) in a manner that allowed us to image the cortical cytoskeleton network with high resolution. In this fast force-curve-based mode, a complete vertical approach-retract cycle is carried out at every pixel, with lateral movement of the tip only in between vertical cycles. With the settings we optimized to attain a reasonable time resolution, the tip remains in continuous contact with the cell. For analysis, we used the height at the setpoint force, which was optimized to be as low as possible while achieving good image quality (≈0.5 nN). In principle, it is also possible to calculate elasticity data from the force curves (31); however, the data acquired with our experimental setup are unsuitable for this kind of measurement because they contain only partial force curves.
In contrast to contact-mode imaging, with force-mapping-mode imaging we were able to visualize cortical cytoskeleton mesh-like structures in the height data (Fig. 4 a). We attribute this to smaller deviations of the applied imaging force and to a reduction in lateral forces compared with contact-mode scanning. To extract the cytoskeleton network morphology and quantify its density, we employed an image-processing procedure that emphasizes small features on large-scale objects (25). First, to eliminate line-to-line offsets caused by cantilever force drift and sample drift in z, we carried out line-wise subtraction of a fitted second-order polynomial. We chose a second-order polynomial so as to approximate the general shape of the cell surface. Then, most importantly, we applied a difference-of-Gaussians filter, which subtracts a smoothed copy of the image from the original and thereby enhances features smaller than the smoothing range. In the resulting images, both coarse and fine meshes are clearly visible, and the underlying cell shape has been removed (Fig. 4 b).
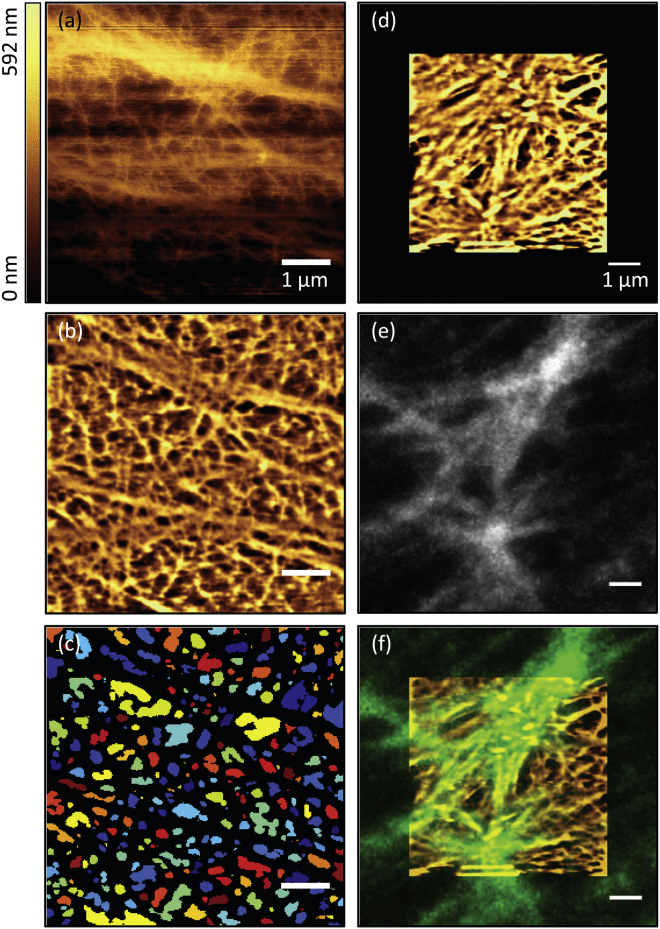
Figure 4.
High-resolution force-mapping-mode imaging of the cortical cytoskeleton, subsequent image processing, and analysis. (a–c) Different views of the same data: (a) raw height data, (b) processed image used to calculate the adapted RMS roughness and to further quantify filament densities, and (c) detected holes for the morphological quantification of mesh sizes, each displayed in a randomly chosen color. (d–f) Correlation with Lifeact imaging: (d) processed force-mapping-mode image, (e) Lifeact-mKate2 detected by confocal fluorescence microscopy simultaneously in the same region (maximum intensity projection of the most apical focal planes), and (f) overlay of (d) and (e), adjusted in brightness and contrast. Scale bars are 1 μm. To see this figure in color, go online.
When the same processing is conducted with height data acquired in contact mode, it becomes apparent that a lower resolution compared with the force-mapping-mode data is achieved, and the level of detail is not as good as in the corresponding error images (Fig. S1). This supports the notion that force-mapping-mode imaging ensures better tracking of the apparent cell surface by reducing the lateral forces and improving the force precision.
This AFM modality also allows correlations with live-cell Lifeact fluorescence views (Fig. 4, d–f). Again, only the coarse-mesh structures (although they are harder to differentiate in this case) can be correlated with the corresponding F-actin fluorescence. Precise overlays are more prone to be affected by possible motion artifacts in the AFM images (Fig. S3) compared with the faster contact-mode AFM scans (Fig. 2).
The resolution of force-mapping-mode imaging (as determined by assessing the smallest visible filament structures) is ∼120 nm or better along the fast-scan direction (Fig. S4, d–f).
Pharmacological manipulation of actin dynamics impacts the cortical cytoskeleton network’s morphology characteristically
To evaluate our imaging procedure and to gain further insight into the composition of the cortical cytoskeleton network, we monitored the effects of pharmacological manipulation of actin polymerization dynamics with cytochalasin D and jasplakinolide using AFM force-mapping-mode imaging. Cytochalasin D, a fungal toxin, caps the barbed ends of actin filaments and thus disrupts the organization of the actin cytoskeleton (32). It has been shown to selectively soften the cortex of endothelial cells in low concentrations (e.g., 100 nM), thereby triggering physiologically relevant changes in cell signaling (18,33). The cyclic peptide jasplakinolide promotes actin polymerization by binding to F-actin competitively with phalloidin while being cell permeable (34). It increases the density of actin filaments when applied to cells in the short term, whereas after longer incubations, the regular cytoskeleton architecture is destroyed as amorphous aggregates of actin develop (35).
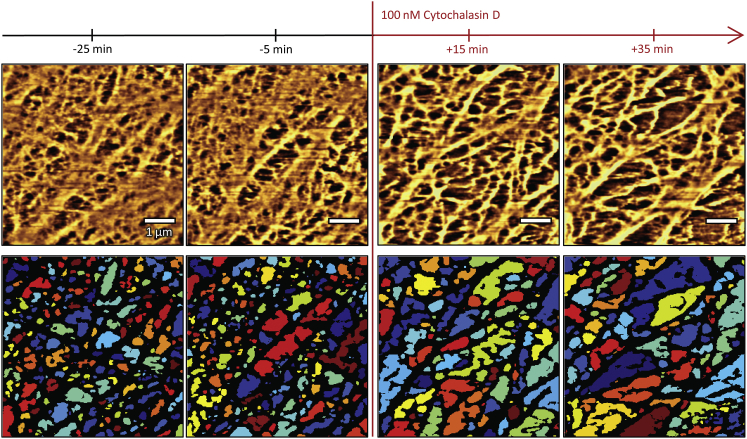
Upon treatment with cytochalasin D (n = 8; corresponding control experiments: n = 7), the cortical cytoskeleton meshwork thins out, the holes between filaments become larger, and the contrast of the processed images increases (Fig. 5). Predominantly the fine mesh appears to be affected.
Figure 5.
The cortical cytoskeleton network is thinned out by actin depolymerization with cytochalasin D, as observed by force-mapping-mode imaging. Exemplary images from one experiment are shown. Top row: filtered height data; bottom row: detected meshwork holes. Scale bars are 1 μm. To see this figure in color, go online.
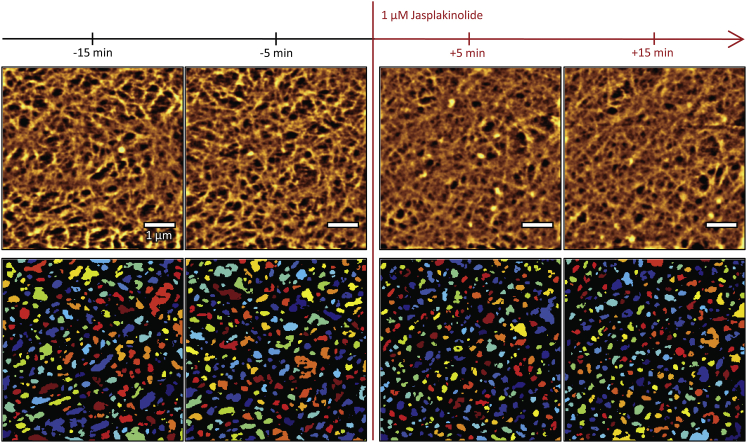
In contrast, jasplakinolide increases the meshwork’s density, shrinking the holes and reducing the image contrast (Fig. 6) in our experiments (n = 6; corresponding control experiments: n = 5). After a short period of ∼15 min, most cells lose their mechanical integrity. In some cases, we note that under the influence of jasplakinolide, the cortical cytoskeleton network structures stay fixed in their configuration and are continuously imaged, and only a slight shift of the image can be attributed to thermal drift of the microscope setup. Based on this observation, we reason that AFM imaging by itself does not mechanically disturb the imaged cytoskeleton structures.
Figure 6.
Jasplakinolide increases the density of the cortical cytoskeleton network. Representative images from one force-mapping-mode imaging experiment are shown. Top row: filtered height data; bottom row: detected holes. Scale bars are 1 μm. To see this figure in color, go online.
Quantification of changes in cytoskeleton network density
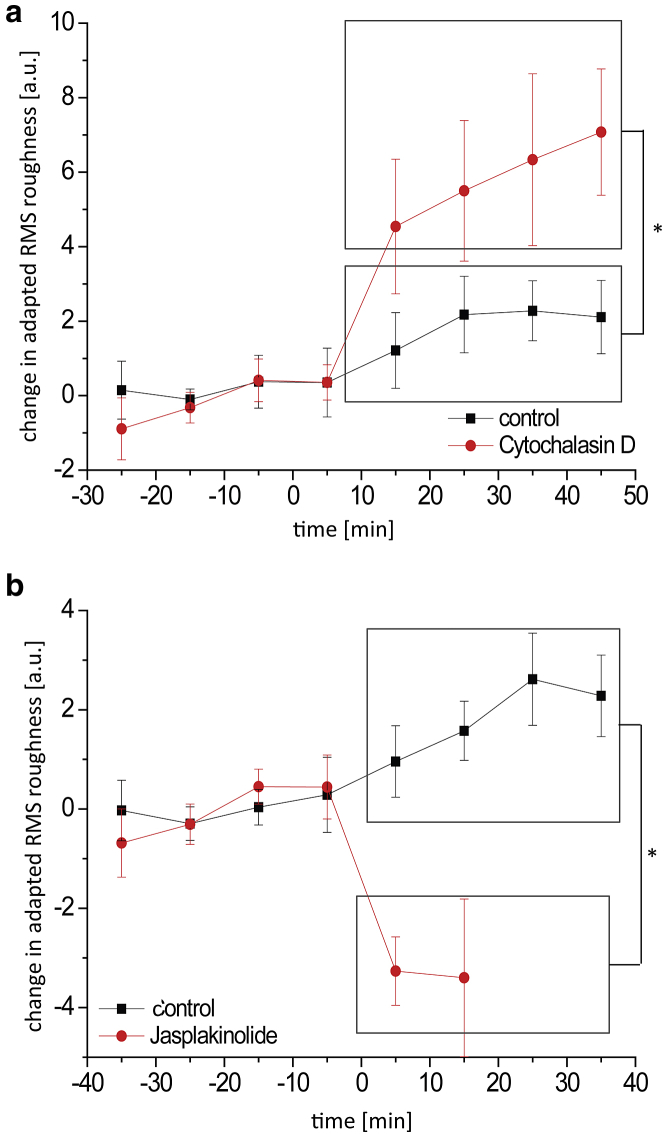
We calculated the RMS roughness of the processed height images (adapted roughness) to estimate the density of the cytoskeleton network. Although absolute values for RMS roughness are generally given in nanometers, the adapted roughness is given in arbitrary units (a.u.) due to changes in the absolute values during image processing. As shown in Fig. 7, a and b, the roughness of the scanned cells’ apical surface increases slightly over time. This increase, however, appears to not be significant. Upon treatment with cytochalasin D, the adapted roughness increases significantly by 6.8 ± 2.2 a.u. (Fig. 7 a). This increase in contrast is due to the disruption (or reduced polymerization) of fine-mesh filaments in the cell cortex. Larger filament bundles are not affected by concentrations of cytochalasin D as low as we applied, so the AFM cantilever indents deeper, larger valleys between the coarse-mesh structures, resulting in an increase in contrast and thus in adapted roughness. Filament stabilization results in the opposite effect, as the apical surface roughness decreases by 3.3 ± 0.9 a.u. after jasplakinolide treatment (Fig. 7 b).
Figure 7.
(a and b) Adapted RMS roughness measurements (calculated using force-mapping-mode imaging data) show the effects of 100 nM cytochalasin D (a) and 1 μM jasplakinolide (b). Values are expressed as differences from the respective control phase average and shown as mean ± SE. Sample sizes (individual experiments and the number of cells studied): cytochalasin D, n = 8 (treatment), n = 7 (control); jasplakinolide n = 6 (treatment) and n = 5 (control). Time = 0 signifies replacement with control or treatment buffer. To see this figure in color, go online.
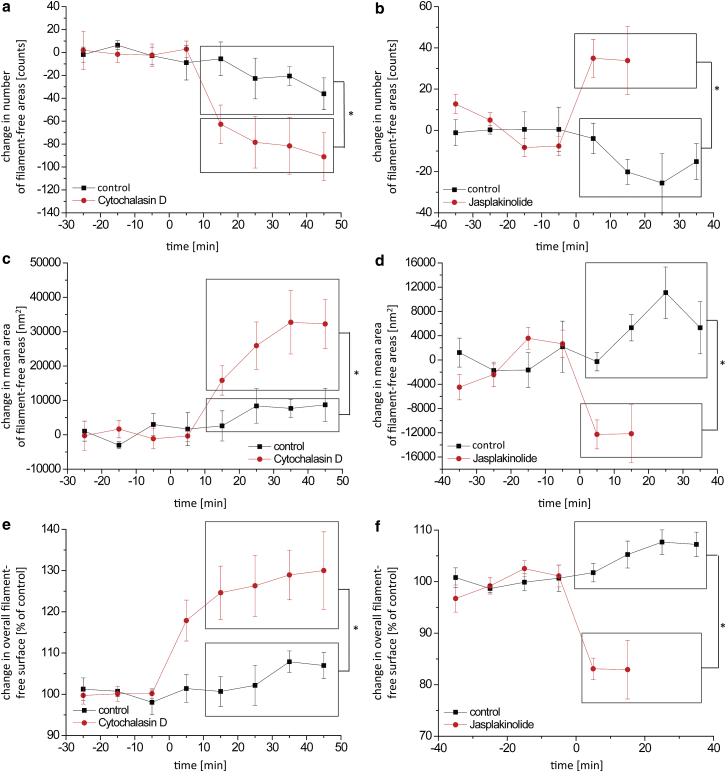
To obtain a more direct and intuitive assessment of the cortical actin cytoskeleton’s dynamic behavior, we estimated morphological changes in our experiments by characterizing filament-free regions using a more sophisticated approach. We segmented the processed images into foreground network structures and background filament-free holes (Fig. 4 c) using the software ilastik, an interactively trainable random forest classifier that relies on multiple mathematical functions specifically designed to detect features such as intensities, gradients, edges, and orientations. We trained the software by manually providing labels for the foreground and background in one series of images until the segmentation approximately matched our visual perception; all further images could be processed automatically. Subsequently, for each image we determined the number of filament-free holes, the mean area of all holes in an image, and the overall surface coverage of holes as a percentage of the imaging area. For all measurements except the overall surface coverage and the total number, holes touching the border of the image were excluded from the analysis because their actual full dimensions could not be determined. In our experiments, apical cortical cytoskeleton exhibited an average number of 271 ± 27.7 holes per 6 × 6 μm surface area. Application of cytochalasin D reduced the average number of holes by 99 ± 31.3 (Fig. 7 a), whereas the area of each hole increased (from basal 42,045 ± 5553.2 nm2) by 37,886 ± 10,898.1 nm2 (Fig. 7 c). Hence, the relative surface coverage of the filament-free area rose by 30% (Fig. 7 e). Jasplakinolide at a concentration of 1 μM increased the number of holes (+33 ± 16.4) and reduced the mean hole area (–12,160 ± 4786.4 nm2) and overall coverage of the filament-free area (−18%; Fig. 8, b, d, and f).
Figure 8.
(a–f) Morphological analysis of force-mapping-mode imaging data shows a decrease in network density induced by treatment with 100 nM cytochalasin D (a, c, and e) and an increase in network density induced by treatment with 1 μM jasplakinolide (b, d, and f). (a and b) Total number of detected holes. (c and d) Mean area of all holes in one image. (e and f) Overall area covered by holes. Values are expressed as the difference from the respective control phase average and shown as mean ± SE. Cytochalasin D: n = 8 (treatment), n = 7 (control); jasplakinolide: n = 6 (treatment) and n = 5 (control). Time = 0 signifies replacement with control or treatment buffer. To see this figure in color, go online.
Intracellular calcium concentration is unaffected by force-mapping-mode imaging
We cannot exclude the possibility that the cells were mechanically stimulated by the continuous and rapid indentation during scanning. To verify that force-mapping-mode imaging does not disrupt the membrane integrity or stimulate mechanosensitive signaling mechanisms, we measured intracellular calcium levels simultaneously with AFM imaging. Fluo4 fluorescence intensity was measured for 3 min without force mapping to obtain control values corresponding to an unstimulated cell. Subsequently, AFM scanning (force-mapping-mode imaging) was performed for 20 min. Finally, ionomycin was added to a concentration of 2.5 μM to elicit an increase in the intracellular calcium concentration as a positive control. As shown in Fig. S2, indentation of the plasma membrane during force-mapping-mode imaging did not affect the intracellular calcium concentration, and therefore appears to leave the plasma membrane intact and not activate any mechanosensitive signaling mechanism.
Discussion
As has been widely reported, contact-mode AFM imaging of living cells reveals filamentous structures corresponding to the cytoskeleton beneath the general membrane surface topography (14–16,29). The local mechanical properties of the cell surface have been shown to reflect the organization of the underlying submembranous cytoskeleton (12,13), so AFM scanning with small indentation depths or low applied forces, respectively, can be used to selectively probe and image cytoskeleton structures in the cortical compartment of a cell. In contrast, tapping-mode AFM is mostly used to visualize the membrane surface of a cell (36); however, one can image the submembranous cytoskeleton by choosing high damping setpoints and analyzing the cantilever deflection (37). Fixed cells imaged by AFM exhibit a smooth surface without filamentous features (14), although the state of the actin cytoskeleton may still be inferred from the cell’s mechanical properties (33). Special preparations enable AFM imaging of the cytoskeleton after fixation. For example, removing the membrane by detergent treatment makes the underlying cytoskeleton accessible to AFM imaging (30), and unroofing the cells allows one to study the apical membrane and the attached cytoskeleton from the cytoplasmic side (38).
By employing contact-mode imaging, we can image the reorganization of the coarse cortical cytoskeleton meshwork in live cells and qualitatively visualize the dynamics of finer-mesh structures. However, it appears that acquisition and analysis of the AFM error signal, are not suitable for gathering quantitative data regarding the fine mesh, such as its density. Using a slower, adapted measurement and image processing technique, we show that the cortical cytoskeleton network can be imaged in height data. The force error is reduced compared with contact-mode scanning, and thus very soft filament structures are more reliably represented in the acquired data. With commonly available AFM technology, this procedure allows one to image the cortical cytoskeleton’s mechanical architecture in live cells with a resolution better than that achieved by conventional optical microscopy.
One can detect changes in the meshwork build quantitatively by calculating the RMS roughness as a statistical parameter or by morphological measurements. Surface roughness measurements by AFM are an established means to evaluate the structure of cell surfaces (39) and have also been used to quantify cytoskeletal structures (30). Here, we have shown that the adapted RMS roughness, calculated using preprocessed height data, can be used as an indicator of cortical cytoskeleton density in living cells. Cortical actin polymerization or depolymerization, however, is not the only possible determinant of surface roughness. The cell volume; the occurrence of microvilli, membrane ruffles, or filopodia; the composition of the membrane, including membrane proteins; and the activity of cytoskeletal motor proteins may also induce changes in the surface roughness. Therefore, one must exercise some caution when taking roughness as a parameter for cytoskeleton network density. An automated geometric analysis of cytoskeleton structures based on AFM data was previously described for erythrocyte preparations (40). By analyzing successive images using our morphological segmentation routine, we were able to objectively quantify changes in the actual organization of the cortical cytoskeleton in live cells, and directly acquire and observe mesh sizes on an ultrastructural scale.
The applicability of our method based on force-mapping-mode imaging and processing may be limited by its time resolution. The imaged structures rearrange significantly faster than the acquisition time for an image. Thus, morphological features such as the holes we analyzed probably do not reflect the actual network architecture. This also becomes apparent when the AFM data are compared with the fluorescence data (Fig. S3). Fast remodeling processes such as treadmilling of actin filaments may not be observed in this fashion either. However, the line-wise scanning modality of AFM needs to be taken into account. The positions of filament structures in a single (horizontal) scan line are determined in a short time and therefore are accurate. The vertical distance in an image corresponds to the time offset during acquisition. For a small number of neighboring, successively scanned lines, the time resolution is thus better than that for the entire image. Nonsquare rectangular scans could be taken to exploit this fact and improve the general time resolution of the experiment, although in return the scan area and hence the robustness of the morphological analysis would be reduced.
The validity of our imaging procedure with respect to actin cytoskeleton dynamics on a timescale of several minutes is empirically shown in the pharmacological experiments. The results obtained by quantitative image analysis are in line with theoretical expectations and previous observations concerning the effect of cytochalasin D and jasplakinolide on the actin cytoskeleton.
The force resolution of the AFM instrument is still limited, however, which means that not necessarily all filaments are accurately mapped. A much denser meshwork has been resolved in raster electron micrographs of the endothelial cortical cytoskeleton (41), although this method does not need to represent the mechanical organization as visualized by AFM. Also, the actual spatial resolution of the cytoskeleton imaging techniques demonstrated here is difficult to assess. From a technical point of view, AFM is capable of subnanometer resolution. However, the scanned surface is convoluted with the probe shape, which in this case includes not only the tip (with a nominal radius of curvature of ≈10 nm for the probes used here) but also the plasma membrane that lies between the tip and the cytoskeleton. Furthermore, the apical part of the cell probably does not represent a rigid sample, but slightly shifts upon exertion of lateral forces by the AFM. Therefore, a proper test sample to assess the resolution of this imaging application does not exist. To provide a rough estimate of the attained spatial resolution, we measured the smallest spacings between apparent filament structures that resulted in structures of ∼100–150 nm distance (Fig. S4). Although this lies in the same order of magnitude as the theoretical Abbe limit for optical microscopy, we must acknowledge that our measurement provides only an upper boundary of the resolution, and that in practice, the resolution of live-cell fluorescence microscopy is likely to be worse because it is limited by the achievable signal strength.
Concerning the identification of structures imaged by AFM, in principle, various procedures can be used to infer their molecular makeup. In the case of actin microfilaments, direct observations by AFM of distinctive morphological features such as the filament width and height (30) and a periodic helical ultrastructure (42) have been used for identification, but this requires removal of the membrane and fixation of the cell. The same applies for dedicated techniques such as on-stage labeling and imaging (42), and topography and recognition imaging (43). The mesh geometry of the cortical cytoskeleton in live cells was previously analyzed and found to be consistent with the notion that the cortical cytoskeleton is composed of F-actin (14). The combination of AFM and fluorescence microscopy has demonstrated that actin accounts for the majority of features present in AFM images of the cortical cytoskeleton (14,16). As samples can only be fixed and stained after AFM imaging is completed, resulting in a time lag and possible artifacts, we performed live-cell fluorescence microscopy of Lifeact simultaneously with AFM scans. The results show that the coarse part of the cortical cytoskeleton network is made up of actin.
These coarse-mesh structures may be akin to what has been described in motile fibroblasts as transverse arcs, stress fibers located on the dorsal side of the cell (44). The reorganization processes that we imaged (Fig. 2) are in line with the intact and altered boundary mode remodeling described previously based on investigations on a broader timescale (45). The observation that coarse-mesh fibers gradually emerge from smaller structures can be interpreted as the formation of stress fibers by myosin-based bundling of actin filaments, an effect based on Rho signaling (44,46).
The marked effects of cytochalasin D and jasplakinolide, both of which specifically target actin polymerization dynamics, let us arrive at the conclusion that, indeed, the fine meshwork structures affected are constituted of actin. However, it is also conceivable that a different protein, such as spectrin (5), actually constitutes the fine mesh, in which case the fact that it is supported by actin would explain our experimental results.
Based on our calcium measurements (Fig. S2) and our observation that cells subjected to AFM scanning remain viable for hours, we conclude that force-mapping-mode imaging does not appear to harm or mechanically stimulate endothelial cells. In contrast to other microscopy techniques, AFM imaging of cultured cells does not require any special sample preparation, such as fixation or staining, or expression of a genetic vector, and the risk of phototoxicity in live cells is avoided. The microscopy techniques described here thus present a very suitable alternative for live-cell imaging of the apical actin cytoskeleton.
Cytoskeletal imaging by AFM could benefit from technological advances in high-speed AFM that would result in increased feedback bandwidth (47) and especially better force precision with small (48) or uncoated/partially coated cantilevers (22,49). However, the maximum speed for cytoskeletal imaging may be limited by the viscoelastic properties of a cell (i.e., the cell hardens when indented too fast (36)). We hope that AFM-based imaging of the cortical cytoskeleton will prove useful in the field of mechanobiology for answering questions about how structure, mechanics, and function are linked.
Author Contributions
C.K. designed the project, performed experiments, and wrote the manuscript. M.S.-H. analyzed data. D.B. performed experiments. H.O. and J.F. designed the project and wrote the manuscript.
Acknowledgments
We thank Arthur Carlson (Max-Planck Institute of Biochemistry, Martinsried, Germany) and Marianne Wilhelmi (Institute of Physiology II, Münster, Germany) for their technical support and advice.
This work was supported by the Cells-in-Motion Cluster of Excellence, the Programm für Innovative Medizinische Forschung (I-FE 220904), and the Deutsche Forschungsgemeinschaft (Koselleck grant OB 63/18). It was also supported through the networking activities of COST action TD 1002.
Editor: Christopher Yip.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Five figures and one movie are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00709-2.
Supporting Material
Left: cortical area visualized by contact-mode imaging AFM (error signal, trace and retrace overlaid by addition); middle: Lifeact-eGFP detected by confocal fluorescence microscopy simultaneously in the same region (maximum intensity projection of the most apical focal planes); right: overlay. Total length of movie: 1 h 55 min.
References
- 1.Patterson C.E., Lum H. Update on pulmonary edema: the role and regulation of endothelial barrier function. Endothelium. 2001;8:75–105. doi: 10.3109/10623320109165319. [DOI] [PubMed] [Google Scholar]
- 2.Winder S.J., Ayscough K.R. Actin-binding proteins. J. Cell Sci. 2005;118:651–654. doi: 10.1242/jcs.01670. [DOI] [PubMed] [Google Scholar]
- 3.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 4.Kliche K., Jeggle P., Oberleithner H. Role of cellular mechanics in the function and life span of vascular endothelium. Pflugers Arch. 2011;462:209–217. doi: 10.1007/s00424-011-0929-2. [DOI] [PubMed] [Google Scholar]
- 5.Prasain N., Stevens T. The actin cytoskeleton in endothelial cell phenotypes. Microvasc. Res. 2009;77:53–63. doi: 10.1016/j.mvr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fels J., Jeggle P., Oberleithner H. Nanomechanics of vascular endothelium. Cell Tissue Res. 2014;355:727–737. doi: 10.1007/s00441-014-1853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birukova A.A., Arce F.T., Birukov K.G. Endothelial permeability is controlled by spatially defined cytoskeletal mechanics: atomic force microscopy force mapping of pulmonary endothelial monolayer. Nanomedicine (Lond.) 2009;5:30–41. doi: 10.1016/j.nano.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szczygiel A.M., Brzezinka G., Szymonski M. Elasticity changes anti-correlate with NO production for human endothelial cells stimulated with TNF-α. Pflugers Arch. 2012;463:487–496. doi: 10.1007/s00424-011-1051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fletcher D.A., Mullins R.D. Cell mechanics and the cytoskeleton. Nature. 2010;463:485–492. doi: 10.1038/nature08908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Müller D.J., Dufrêne Y.F. Atomic force microscopy: a nanoscopic window on the cell surface. Trends Cell Biol. 2011;21:461–469. doi: 10.1016/j.tcb.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Francis L.W., Lewis P.D., Conlan R.S. Atomic force microscopy comes of age. Biol. Cell. 2010;102:133–143. doi: 10.1042/BC20090127. [DOI] [PubMed] [Google Scholar]
- 12.Pogoda K., Jaczewska J., Lekka M. Depth-sensing analysis of cytoskeleton organization based on AFM data. Eur. Biophys. J. 2012;41:79–87. doi: 10.1007/s00249-011-0761-9. [DOI] [PubMed] [Google Scholar]
- 13.Kasas S., Wang X., Catsicas S. Superficial and deep changes of cellular mechanical properties following cytoskeleton disassembly. Cell Motil. Cytoskeleton. 2005;62:124–132. doi: 10.1002/cm.20086. [DOI] [PubMed] [Google Scholar]
- 14.Pesen D., Hoh J.H. Micromechanical architecture of the endothelial cell cortex. Biophys. J. 2005;88:670–679. doi: 10.1529/biophysj.104.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotsch C., Radmacher M. Drug-induced changes of cytoskeletal structure and mechanics in fibroblasts: an atomic force microscopy study. Biophys. J. 2000;78:520–535. doi: 10.1016/S0006-3495(00)76614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson E., Haydon P.G., Sakaguchi D.S. Actin filament dynamics in living glial cells imaged by atomic force microscopy. Science. 1992;257:1944–1946. doi: 10.1126/science.1411511. [DOI] [PubMed] [Google Scholar]
- 17.Grinspan J.B., Mueller S.N., Levine E.M. Bovine endothelial cells transformed in vitro by benzo(a)pyrene. J. Cell. Physiol. 1983;114:328–338. doi: 10.1002/jcp.1041140312. [DOI] [PubMed] [Google Scholar]
- 18.Fels J., Jeggle P., Oberleithner H. Cortical actin nanodynamics determines nitric oxide release in vascular endothelium. PLoS One. 2012;7:e41520. doi: 10.1371/journal.pone.0041520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riedl J., Crevenna A.H., Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemieux M.G., Janzen D., Knecht D.A. Visualization of the actin cytoskeleton: different F-actin-binding probes tell different stories. Cytoskeleton (Hoboken) 2014;71:157–169. doi: 10.1002/cm.21160. [DOI] [PubMed] [Google Scholar]
- 21.Mortensen R.M., Kingston R.E. Selection of transfected mammalian cells. Curr. Protoc. Mol. Biol. 2009;Chapter 9 doi: 10.1002/0471142727.mb0905s86. Unit 9.5. [DOI] [PubMed] [Google Scholar]
- 22.Churnside A.B., Sullan R.M.A., Perkins T.T. Routine and timely sub-picoNewton force stability and precision for biological applications of atomic force microscopy. Nano Lett. 2012;12:3557–3561. doi: 10.1021/nl301166w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Souza C.P., Andrade M.S., Neves B.R.A. Implementation of recycling routes for scanning probe microscopy tips. Microsc. Microanal. 2002;8:509–517. doi: 10.1017/S1431927602010449. [DOI] [PubMed] [Google Scholar]
- 24.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kienberger F., Pastushenko V.P., Hinterdorfer P. Improving the contrast of topographical AFM images by a simple averaging filter. Ultramicroscopy. 2006;106:822–828. doi: 10.1016/j.ultramic.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Sommer C., Straehle C., Hamprecht F. Ilastik: interactive learning and segmentation toolkit. Proc. 8th IEEE Int. Symp. Biomed. Imaging. 2011:230–233. [Google Scholar]
- 27.Xu K., Babcock H.P., Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat. Methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Putman C.A., van der Werf K.O., Hansma P.K. New imaging mode in atomic-force microscopy based on the error signal. Proc. SPIE. 1992;1639:198–204. [Google Scholar]
- 29.Barbee K.A., Davies P.F., Lal R. Shear stress-induced reorganization of the surface topography of living endothelial cells imaged by atomic force microscopy. Circ. Res. 1994;74:163–171. doi: 10.1161/01.res.74.1.163. [DOI] [PubMed] [Google Scholar]
- 30.Berdyyeva T., Woodworth C.D., Sokolov I. Visualization of cytoskeletal elements by the atomic force microscope. Ultramicroscopy. 2005;102:189–198. doi: 10.1016/j.ultramic.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Chopinet L., Formosa C., Dague E. Imaging living cells surface and quantifying its properties at high resolution using AFM in QI™ mode. Micron. 2013;48:26–33. doi: 10.1016/j.micron.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 32.Cooper J.A. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 1987;105:1473–1478. doi: 10.1083/jcb.105.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grimm K.B., Oberleithner H., Fels J. Fixed endothelial cells exhibit physiologically relevant nanomechanics of the cortical actin web. Nanotechnology. 2014;25:215101. doi: 10.1088/0957-4484/25/21/215101. [DOI] [PubMed] [Google Scholar]
- 34.Bubb M.R., Senderowicz A.M., Korn E.D. Jasplakinolide, a cytotoxic natural product, induces actin polymerization and competitively inhibits the binding of phalloidin to F-actin. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- 35.Bubb M.R., Spector I., Fosen K.M. Effects of jasplakinolide on the kinetics of actin polymerization. An explanation for certain in vivo observations. J. Biol. Chem. 2000;275:5163–5170. doi: 10.1074/jbc.275.7.5163. [DOI] [PubMed] [Google Scholar]
- 36.Putman C.A., van der Werf K.O., Greve J. Viscoelasticity of living cells allows high resolution imaging by tapping mode atomic force microscopy. Biophys. J. 1994;67:1749–1753. doi: 10.1016/S0006-3495(94)80649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vié V., Giocondi M.-C., Le Grimellec C. Tapping-mode atomic force microscopy on intact cells: optimal adjustment of tapping conditions by using the deflection signal. Ultramicroscopy. 2000;82:279–288. doi: 10.1016/s0304-3991(99)00137-0. [DOI] [PubMed] [Google Scholar]
- 38.Usukura J., Yoshimura A., Cho S.-J. Use of the unroofing technique for atomic force microscopic imaging of the intra-cellular cytoskeleton under aqueous conditions. J. Electron Microsc. (Tokyo) 2012;61:321–326. doi: 10.1093/jmicro/dfs055. [DOI] [PubMed] [Google Scholar]
- 39.Francis L.W., Lewis P.D., Conlan R.S. Progesterone induces nano-scale molecular modifications on endometrial epithelial cell surfaces. Biol. Cell. 2009;101:481–493. doi: 10.1042/BC20080189. [DOI] [PubMed] [Google Scholar]
- 40.Shi H., Liu Z., Lim C.T. Life cycle-dependent cytoskeletal modifications in Plasmodium falciparum infected erythrocytes. PLoS One. 2013;8:e61170. doi: 10.1371/journal.pone.0061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satcher R., Dewey C.F., Jr., Hartwig J.H. Mechanical remodeling of the endothelial surface and actin cytoskeleton induced by fluid flow. Microcirculation. 1997;4:439–453. doi: 10.3109/10739689709146808. [DOI] [PubMed] [Google Scholar]
- 42.Jung S.-H., Park J.-Y., Ha K.-S. Identification and ultrastructural imaging of photodynamic therapy-induced microfilaments by atomic force microscopy. Ultramicroscopy. 2009;109:1428–1434. doi: 10.1016/j.ultramic.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Chtcheglova L.A., Wildling L., Hinterdorfer P. AFM functional imaging on vascular endothelial cells. J. Mol. Recognit. 2010;23:589–596. doi: 10.1002/jmr.1052. [DOI] [PubMed] [Google Scholar]
- 44.Pellegrin S., Mellor H. Actin stress fibres. J. Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 45.Pesen D., Hoh J.H. Modes of remodeling in the cortical cytoskeleton of vascular endothelial cells. FEBS Lett. 2005;579:473–476. doi: 10.1016/j.febslet.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 46.Machesky L.M., Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J. Cell Biol. 1997;138:913–926. doi: 10.1083/jcb.138.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ando T. High-speed atomic force microscopy coming of age. Nanotechnology. 2012;23:062001. doi: 10.1088/0957-4484/23/6/062001. [DOI] [PubMed] [Google Scholar]
- 48.Leitner M., Fantner G.E., Hinterdorfer P. Increased imaging speed and force sensitivity for bio-applications with small cantilevers using a conventional AFM setup. Micron. 2012;43:1399–1407. doi: 10.1016/j.micron.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labuda A., Bates J.R., Grütter P.H. The noise of coated cantilevers. Nanotechnology. 2012;23:025503. doi: 10.1088/0957-4484/23/2/025503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Left: cortical area visualized by contact-mode imaging AFM (error signal, trace and retrace overlaid by addition); middle: Lifeact-eGFP detected by confocal fluorescence microscopy simultaneously in the same region (maximum intensity projection of the most apical focal planes); right: overlay. Total length of movie: 1 h 55 min.