Abstract
The structure and composition of a biological membrane can severely influence the activity of membrane-embedded proteins. Here, we show that the E. coli aquaglyceroporin GlpF has only little activity in lipid bilayers formed from native E. coli lipids. Thus, at first glance, GlpF appears to not be optimized for its natural membrane environment. In fact, we found that GlpF activity was severely affected by negatively charged lipids regardless of the exact chemical nature of the lipid headgroup, whereas GlpF was not sensitive to changes in the lateral membrane pressure. These observations illustrate a potential mechanism by which the activity of an α-helical membrane protein is modulated by the negative charge density around the protein.
Introduction
Membrane proteins fold, assemble, and are active in lipid bilayers. Besides distinct protein-protein interactions, nonspecific as well as specific interactions between bilayer lipids and transmembrane (TM) proteins can modulate the structure and activity of TM proteins (1). Certain lipid-protein interactions can be necessary for proper protein folding and oligomerization (1–4). Some lipids are intrinsic parts of TM protein structures, whereas others act as nonprotein molecular chaperones that are crucial for proper folding of a polytopic TM protein (5,6). Furthermore, global bilayer properties, such as the bilayer thickness and the intrinsic lipid curvature, can nonspecifically influence a TM protein’s structure and activity. The hydrophobic region of TM proteins has to match the thickness of the hydrophobic bilayer core (7), and although in some cases hydrophobic mismatch can result in destabilization and inactivation of TM proteins (8,9), in other cases the thickness of the membrane adjusts locally to the thickness of the hydrophobic protein surface (10–12).
All cellular membranes contain negatively charged lipids, and TM proteins are frequently anchored in a membrane by an electrostatic interaction of the negatively charged lipid headgroups with positively charged residues of TM proteins at the bilayer-water interface. Such electrostatic interactions can also determine the orientation of a TM helix and the overall protein topology (13–16). Furthermore, many cellular membranes contain a large fraction of nonbilayer lipids, such as phosphatidylethanolamine (PE), which alone form inverted hexagonal (HII) phases and significantly affect the lateral pressure profile of membranes. Modulating the elastic properties of a biological membrane by such lipids was found to modulate the structure and activity of a TM protein in some cases (17). For example, the water conductance of the E. coli aquaglyceroporin GlpF was simulated to increase by ∼20% in palmitoyloleoyl PE (POPE) bilayers compared with palmitoyloleoyl phosphatidylcholine (POPC) (18,19). More specifically, the radius of the GlpF water/glycerol channel was calculated to be increased by 10% in POPC bilayers, and it was suggested that during water translocation, the successive exchange of hydrogen bonds is not as stringent in the translocation pore because of its increased radius. This result suggests that the activity of GlpF is modulated by changes in the lipid composition of a membrane. Therefore, we systematically investigated the effect of increasing acyl-chain lengths as well as the impact of phospholipid headgroup chemistry on the tetramer stability and activity of a TM protein. The well-characterized E. coli aquaglyceroporin GlpF belongs to the family of aquaporins (AQPs), which are polytopic TM channels that selectively facilitate the flux of water across cellular membranes in all domains of life (20,21). More specifically, GlpF is a member of the aquaglyceroporin subfamily, which additionally enables the diffusion of small, polar solutes across cellular membranes. All AQPs assemble as homotetramers in membranes (22,23), and the oligomeric state of AQPs appears to affect channel activity, with the tetramer being the most active state (24,25). In vivo, GlpF is located in the inner E. coli membrane, which contains three major phospholipid species with different headgroups and various acyl-chain lengths. The zwitterionic PE represents the major membrane lipid (70–75%), and phosphatidylglycerol (PG) (15–20%) is the major anionic phospholipid found in the E. coli inner membrane. A minor constituent (representing ∼5–10%) of the cytoplasmic E. coli membrane is the double-negatively charged lipid cardiolipin (CL) (26,27). The average length of the E. coli lipid acyl-chains is 16 carbon atoms, and approximately half of the acyl-chains are unsaturated (27). Since only little information about the activity of AQPs in different lipid bilayer environments is currently available, we decided to use GlpF to study how the activity of a complex TM protein adjusts to a natural lipid bilayer environment. As no tightly associated lipids are found in the vast majority of AQP structures, it is assumed that AQPs in general do not have defined lipid-binding sites and do not require specific lipids for activity (28).
Here, we show that the activity and stability of the GlpF tetramer, as measured by SDS-induced unfolding, do not depend on a specific lipid environment. Whereas the lateral pressure in the acyl-chain region does not affect GlpF channel function, electrostatic interactions between GlpF and negatively charged lipid headgroups severely influence the activity, but not the stability, of the GlpF tetramer. In fact, the activity of GlpF was found to be significantly reduced in a lipid bilayer system, which most closely mimics the natural E. coli membrane. The results of this study demonstrate how the lipid environment and protein-lipid interactions can directly modulate the activity of a polytopic, oligomeric TM protein.
Materials and Methods
Expression and purification of E. coli GlpF
Expression and purification of GlpF were performed as described in detail previously (24). The protein concentration was determined by measuring the absorption at 280 nm, using a calculated extinction coefficient of 37,930 M−1cm−1 (ExPasy, ProtParam tool).
Functional reconstitution of GlpF
For GlpF reconstitution, the following Δ9-cis monounsaturated lipids were purchased from Avanti Polar Lipids (Alabaster, AL) in chloroform solutions: 1,2-dimyristoleoyl-sn-glycero-3-phosphocholine (diC14:1-PC), 1,2-dipalmitoleoyl-sn-glycero-3-phosphocholine (diC16:1-PC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (diC18:1-PC), 1,2-dieicosenoyl-sn-glycero-3-phosphocholine (diC20:1-PC) and 1,2-dierucoyl-sn-glycero-3-phosphocholine (diC22:1-PC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (diC18:1-PE), 1-(9Z-octadecenyl)-sn-glycero-3-phosphocholine (C18:1 LysoPC), and E. coli polar lipid (EPL) extract, as well as sodium salts of the negatively charged lipids 1,2-dioleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (diC18:1-PG), 1′,3′-bis[1,2-dioleoyl-sn-glycero-3-phospho]-sn-glycerol (tetrC18:1-CL), and 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (diC18:1-PS). The purchased EPL extract consists of 67% PE, 23.2% PG, and 9.8% CL (27). For functional reconstitution, a molar lipid/GlpF ratio of 400:1 was used. We added 5 mM of pure lipid or lipid mixtures dissolved in chloroform to 1.5 mL polypropylene tubes (Sarstedt, Nümbrecht, Germany), and removed the organic solvent under a gentle stream of nitrogen gas. Any remaining traces of the organic solvent were removed by vacuum desiccation overnight. The lipid film was rehydrated at 37°C for 45 min in 30 mM n-octyl β-D-glucopyranoside (OG) (Roth, Karlsruhe, Germany), 50 mM MOPS (pH 7.5; Sigma-Aldrich, Munich, Germany), 150 mM N-methyl D-glucamine (Acros Organics, Morris Plains, NJ), and 50 mM NaCl (Roth, Karlsruhe, Germany) (29). GlpF (in 50 mM of OG, 50 mM phosphate, 300 mM NaCl, and 10% glycerol) was added to the rehydrated lipids at a final concentration of 12 μM. The volume was then adjusted to 0.5 mL with 50 mM MOPS (pH 7.5), 150 mM N-methyl D-glucamine, and 50 mM NaCl (dialysis buffer). The final OG concentration was 30 mM. The GlpF/lipid mixture was dialyzed for at least 48 h at 4°C against 100 sample volumes of dialysis buffer with three buffer exchanges (SpectraPor 12–14 kDa cutoff dialysis membrane; Spectrumlabs, Breda, The Netherlands). The amount of liposome-integrated protein was determined via a semi-native SDS-PAGE analysis. In this analysis, no additional SDS is added to the sample buffer and the native tetrameric oligomerization state of GlpF is preserved (23,30). For subsequent analysis, proteoliposomes were incubated in SDS-free sample buffer (50 mM Tris-HCl (pH 6.8; Roth, Karlsruhe, Germany), 10% (v/v) glycerol (AppliChem, Darmstadt, Germany), and 0.04% (w/v) Bromphenol blue (Sigma-Aldrich, Munich, Germany) for 15 min at room temperature and separated on a 10% SDS-PAGE gel. SDS-PAGE gels were stained with Coomassie blue and the amount of liposome-incorporated GlpF (tetrameric, dimeric, and monomeric) was quantified using ImageJ software (31). The relative amount of incorporated GlpF (cGlpF,rel.) was calculated by normalizing the amount of incorporated GlpF to the amount of GlpF incorporated into pure diC18:1-PC liposomes.
GlpF activity measurements
GlpF activity was assessed by measuring the flux of the substrate ribitol across the lipid bilayer of the liposomes. Activity measurements were performed using a SX20 Stopped Flow Spectrometer (Applied Photophysics, Leatherhead, UK). Proteoliposomes were rapidly mixed with an equal volume of dialysis buffer containing 600 mM ribitol (AlfaAesar, Heysham, UK). Light scattering was followed at a wavelength of 600 nm at a 90° angle at room temperature. The scattering intensity rose quickly due to water efflux and then decayed due to the influx of ribitol. This decay was analyzed using a double exponential decay function. To compare the decay times under different conditions, a weighted rate constant (kw) was calculated according to the following equation (32):
| (1) |
where A1 and A2 are the amplitudes, and k1 and k2 are the respective rate constants.
The weighted rate constants (kw) were then normalized with respect to the relative amount of incorporated protein (cGlpF,rel.) to obtain the relative weighted rate constants (kw,rel.). Moreover, to be able to properly compare all conducted experiments, we calculated a normalized weighted rate constant (kw,norm.). For this purpose, we additionally normalized the relative weighted rate constant to the relative weighted rate constant of GlpF in diC18:1-PC liposomes.
Results
GlpF activity requires a minimal acyl-chain length
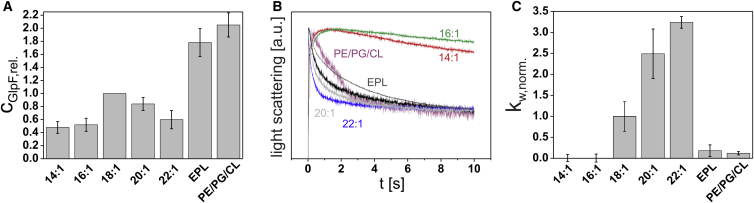
To evaluate how the lipid acyl-chain length might influence a membrane protein’s oligomerization and activity, we incorporated GlpF into liposomes formed from Δ9-cis monounsaturated PC lipids of increasing acyl-chain length (diC14:1-PC to diC22:1-PC). The thickness of the hydrophobic bilayer core increases in lipid bilayers from 29.6 to 45.5 Å (dP-P) (Table S1 in the Supporting Material) (33), and simultaneously with an increasing acyl-chain length, lipid bilayer properties, such as fluidity and the bilayer elastic bending modulus, are altered (34,35). We assessed the impact of the phospholipid headgroup chemistry by monitoring the influence of the lipids diC18:1-PE, diC18:1-PG, tetrC18:1-CL, and diC18:1-PS on the oligomeric state and activity of GlpF. For comparison, GlpF was also incorporated into liposomes formed from EPL extract and a ternary mixture of 70 mol % diC18:1-PE, 20 mol % diC18:1-PG, and 10 mol % tetrC18:1-CL (hereafter, this mixture is referred to as PE/PG/CL). Whereas the EPL liposomes mimic the lipid headgroup as well as the acyl-chain composition of the native E. coli membrane, the PE/PG/CL ternary lipid mixture only mimics the lipid headgroup composition of the native E. coli membrane, because all lipids have identical acyl-chain lengths. The average radius of all liposomes used in this study was 37 ± 11 nm, as measured by dynamic light scattering (Zetasizer-Nano-S, Malvern Instruments, Malvern, UK). Importantly, no relation between liposome size and the amount of incorporated GlpF was found. After incorporation of GlpF into the liposomes, the GlpF oligomeric state and activity were analyzed. Analyzing the GlpF oligomeric state provides information about the stability of GlpF in different lipid environments, as GlpF assembles in vivo into stable tetramers.
To investigate the role of the lipid environment, we assessed the GlpF oligomeric state by conducting a semi-native SDS-PAGE analysis. During such an analysis, the tetrameric state of GlpF remains intact (23,24,30). This remarkable feature of GlpF was utilized to assess the stability of the tetrameric GlpF complex in different lipid environments, as determined by SDS-induced unfolding of the tetramer to monomeric, dimeric, and potentially trimeric GlpF (23). Although the semi-native SDS-PAGE analysis does not visualize the oligomeric state in the bilayer of the liposomes, it nicely reflects the stability of the GlpF tetramer (23).
Moreover, the SDS-PAGE analysis enabled us to evaluate the efficiency of GlpF incorporation into liposomes with different lipid compositions, since only the incorporated and nonaggregated protein exhibited the distinct migration behavior characteristic of the monomeric, dimeric, and tetrameric GlpF states. GlpF incorporation was determined to allow the ribitol flux rate constants to be corrected, as outlined further below.
Based on the SDS-PAGE analysis, GlpF incorporates only with very low efficiencies into diC14:1-PC and diC16:1-PC liposomes, i.e., into liposomes with a thin lipid bilayer (Figs. 1 A and S1 A). In the liposomes formed from Δ9-cis monounsaturated PC lipids, incorporation was most efficient when diC18:1-PC liposomes were used, and decreased again when liposomes were formed from diC20:1-PC and diC22:1-PC. Whereas in diC14:1-PC and diC16:1-PC the proportion of monomeric and dimeric GlpF exceeded the amount of tetrameric GlpF, in diC18:1-PC liposomes and liposomes with even longer acyl-chains, the amount of tetrameric GlpF exceeded the amount of monomeric and dimeric GlpF (diC18:1-PC: 0.78, diC20:1-PC: 0.77, and diC22:1-PC: 0.65) (Figs. S1 A and S2 A). Thus, whereas the GlpF native tetrameric state is well stabilized in EPL and PE/PG/CL liposomes (proportion of tetramer: 0.77 and 0.8, respectively), which most closely mimic the native E. coli membrane, the GlpF tetramer is also stable in nonphysiological PC lipid environments when the lipids have a sufficient chain length.
Figure 1.
GlpF incorporation and activity in liposomes of increasing bilayer thickness. (A) GlpF incorporation in the liposomes was monitored via semi-native SDS-PAGE analysis. Bars show GlpF incorporation relative to its incorporation into diC18:1-PC liposomes (cGlpF,rel.). Error bars: SD (N = 3–5). (B) GlpF activities in the various lipid liposomes were assessed by measuring the flux of the substrate ribitol across the lipid bilayer. Each curve is the average of five measurements. Representative light-scattering curves of GlpF activity measurements in liposomes of increasing acyl-chain length and EPL extract are depicted: diC14:1-PC (red), diC16:1-PC (green), diC18:1-PC (black), diC20:1-PC (light gray), diC22:1-PC (blue), EPL extract (dark gray), and ternary mixture PE/PG/CL (70 mol % diC18:1-PE, 20 mol % diC18:1-PG, 10 mol % tetrC18:1-CL) (purple). (C) Normalized weighted rate constant of ribitol conductance in the different lipid environments. The weighted rate constants (kw) of ribitol flux via GlpF were normalized to the amount of incorporated GlpF relative to the amount of GlpF incorporated into diC18:1-PC liposomes. The obtained relative weighted rate constants (kw,rel.) were additionally normalized to the relative weighted rate constant in diC18:1-PC liposomes to obtain the normalized weighted rate constant (kw,norm.). The SD of GlpF activity in diC18:1-PC liposomes was calculated from the relative weighted rate constants (kw,rel.), and the SD of GlpF activity in the other liposomal systems was calculated from the normalized weighted rate constants (kw,norm.). Error bars: SD (N = 3–5).
We evaluated the activity of liposome-incorporated GlpF by measuring the flux of a substrate across the lipid bilayer. Although the rates of glycerol and ribitol conductance via GlpF are similar, membranes have a higher intrinsic permeability for glycerol compared with ribitol (22). Thus, to minimize the background of the measurements, we chose to use the linear polyalcohol ribitol as the substrate in our measurements. In this assay, proteoliposomes were mixed rapidly with a hypertonic ribitol solution, which causes an immediate water efflux, resulting in liposome shrinkage. When ribitol subsequently diffuses into the liposomes via GlpF, water flows back into the liposomes, resulting in reswelling of the liposomes. It is worth mentioning that, in the kinetic measurements, the substrate ribitol was present in large excess (300 mM) compared with GlpF (<12.5 μM), and under the condition of substrate excess, the ribitol influx does not depend on the GlpF orientation within the lipid bilayer (19,36). We monitored the accompanying changes in the liposome size by following light scattering over time. To exclude the possibility that any OG that remained in the liposomal membranes after dialysis might somehow affect the activity measurements, we also assessed the influence of increasing OG concentrations in the liposomal membrane on GlpF activity, on the background flux in GlpF-free liposomes, and on the stability of the GlpF tetramer. For further details, see Figs. S3–S5 and the Supporting Discussion. These controls revealed that neither the GlpF activity nor the stability of the GlpF tetramer against SDS-induced unfolding was affected by any potentially remaining trace amounts of OG.
Whereas the decay in light scattering was basically absent when liposomes were formed from diC14:1-PC and diC16:1-PC, GlpF incorporated into diC18:1-PC, diC20:1-PC, and diC22:1-PC liposomes was highly active (Fig. 1 B). To quantitatively compare the rate of ribitol flux for different lipid compositions, the amount of incorporated GlpF was quantified and the relative weighted rate constants (kw,rel.) were determined and normalized to kw,rel. in diC18:1-PC liposomes to obtain the normalized weighted rate constant (kw,norm.). The underlying linear correlation between the incorporated amount of GlpF and GlpF activity is shown in Fig. S6. It is worth mentioning that, the background flux in the absence of GlpF was extremely low compared with the GlpF-specific ribitol flux and could not be fitted exponentially. Plotting these normalized rate constants as a function of the acyl-chain length revealed that GlpF is inactive in diC14:1-PC and diC16:-PC liposomes and that GlpF activity increases constantly from diC18:1-PC to diC22:1-PC liposomes (Fig. 1 C). Importantly, GlpF was remarkably tolerant toward the non-E. coli lipid PC, as both the native tetrameric state and the GlpF activity were well preserved. Thus, the results indicate that GlpF might not require the specific lipid headgroup environment found in the native E. coli membrane. In fact, GlpF activity was even strongly impaired in EPL liposomes, which most closely mimic the native E. coli membrane lipid composition.
Since both the lipid headgroups and the acyl-chains of the phospholipid species vary significantly in the EPL extract, the observed differences may simply be due to the heterogeneity of the fatty acids. However, GlpF activity was also strongly impaired in the PE/PG/CL ternary mixture, as was observed in EPL liposomes before (Fig. 1, B and C). Since the GlpF activity was significantly reduced in both EPL and PE/PG/CL lipid bilayers, we can exclude the possibility that GlpF is inactivated solely by a heterogeneous acyl-chain composition, and particularly the presence of fully saturated acyl-chains.
Different phospholipids have been shown to modulate the activity of certain membrane proteins (17,37,38), and GlpF was found to be significantly less active in EPL and PE/PG/CL liposomes compared with PC liposomes. To understand the basis of the decreased GlpF activity in E. coli membrane lipids, we next analyzed the influence of the lipid headgroups, which naturally occur in the E. coli inner membrane, on the oligomeric structure and activity of GlpF. By gradually adding PE, PG, and CL to PC liposomes, which were used as the basic lipid system, we were able to analyze the effect of each E. coli phospholipid on GlpF activity and tetramerization. Importantly, the total concentration of lipids was kept constant. Since incorporation of GlpF was most efficient in diC18:1-PC liposomes and GlpF activity was only moderate in those liposomes, we used diC18:1 acyl-chain-containing lipids in all further experiments, which allowed us to monitor potential activity decreases as well as increases.
GlpF activity is not altered by changes in the lateral membrane pressure in the acyl-chain region
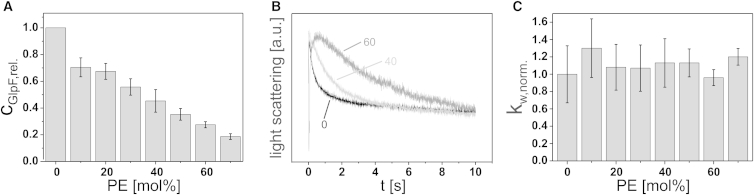
GlpF was found to have a reduced activity in EPL and PE/PG/CL liposomes compared with PC liposomes of the appropriate acyl-chain length. Therefore, we first determined the effect of PE, the major lipid in the E. coli membrane, on the GlpF tetrameric state and activity. Whereas PC and the major E. coli lipid, PE, are both zwitterionic, the PE headgroup is smaller in size than the PC headgroup. In PE, the cross-sectional area of the headgroup is smaller than the cross-sectional area of the acyl-chains, and thus it is a nonbilayer-forming lipid that causes intrinsic membrane negative curvature stress (39,40). In fact, pure PE forms inverted hexagonal phases (HII) rather than lamellar bilayer structures in aqueous environments (41), and integration of PE into lipid bilayers will induce an elastic stress in the bilayer by increasing the lateral pressure in the lipid acyl-chain region of the membrane (42–44). Thus, the reduced GlpF activity in EPL and PE/PG/CL membranes might be due to the increased lateral pressure in the acyl-chain region in these membranes.
The efficiency of GlpF incorporation into liposomes with a diC18:1-PC background and increasing diC18:1-PE concentrations decreased continuously (Figs. 2 A and S1 B). Up to a concentration of 50 mol % PE, the tetramer level was maintained at ∼0.80, whereas PE contents higher than 50 mol % caused a decrease in the proportion of tetrameric GlpF (60 mol % PE: 0.62; 70 mol % PE: 0.54) (Figs. S1 B and S2 B). The changes in the proportion of tetrameric GlpF were associated with an increase in the proportion of monomeric/dimeric GlpF (Fig. S2 B). Although the GlpF activity decreased constantly with an increasing PE content in the liposomes (Fig. 2 B), this could be attributed to the reduced incorporation efficiency, since the rate constants relative to the incorporated protein remained unaffected by the PE content (Fig. 2 C). Therefore, no relation between the GlpF activity and the oligomeric state, as determined via semi-native SDS-PAGE analysis, was found in diC18:1-PE-containing diC18:1-PC liposomes. Thus, the major E. coli lipid, PE, does not appear to affect GlpF activity in lipid bilayers, and therefore changes in the lateral membrane pressure in the acyl-chain region do not influence the GlpF channel activity. It is worth mentioning that addition of up to 16 mol % 1-(9Z-octadecenyl)-sn-glycero-3-phosphocholine (C18:1 LysoPC), which in contrast to PE lowers the lateral pressure in the acyl-chain region, as it causes a positive intrinsic membrane curvature stress, also did not affect GlpF tetramerization or activity (data not shown).
Figure 2.
GlpF incorporation and activity in diC18:1-PC liposomes with increasing diC18:1-PE content. (A) Relative GlpF incorporation. Error bars: SD (N = 6). (B) Representative light-scattering curves of GlpF activity measurements at 0 (black), 40 (light gray), and 60 (gray) mol % diC18:1-PE. Each curve is the average of five measurements. (C) Normalized weighted rate constants of ribitol flux in liposomes of increasing mol % diC18:1-PE. Error bars: SD (N = 6). The SD of GlpF activity in diC18:1-PC liposomes was calculated from the relative weighted rate constants (kw,rel.), and the SD of GlpF activity in the other liposomes was calculated from the normalized weighted rate constants (kw,norm.). The PE concentration in the native E. coli inner membrane is ∼70–75%.
GlpF activity is modulated by negatively charged lipids
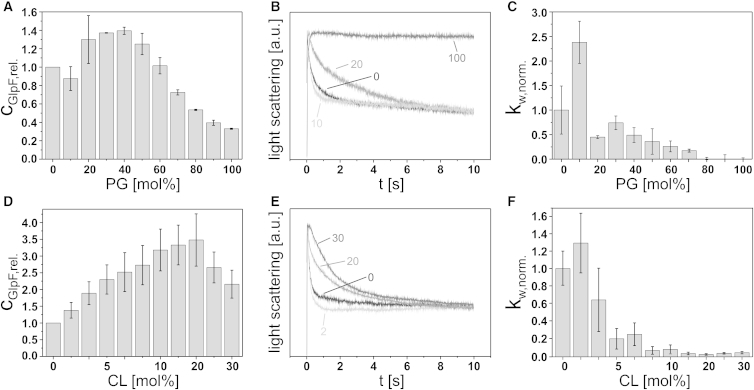
Next, we analyzed the influence of the anionic E. coli lipids PG and CL on GlpF activity. The efficiency of GlpF incorporation into liposomes containing increasing amounts of the negatively charged lipids followed a bell-shaped profile (Figs. 3, A and D, and S1, C and D).
Figure 3.
GlpF incorporation and activity in diC18:1-PC liposomes with increasing amounts of negatively charged lipids. (A and D) Relative GlpF incorporation into liposomes with increasing mol % diC18:1-PG (error bars: SD, N = 3) or tetrC18:1-CL (error bars: SD, N = 6), respectively. (B) Representative light-scattering curves of GlpF activity measurements at 0 (black), 10 (light gray), 20 (gray), and 100 (dark gray) mol % diC18:1-PG. Each curve is the average of five measurements. (C and F) Normalized weighted rate constants of ribitol flux in liposomes with increasing mol % diC18:1-PG (error bars: SD, N = 3) and tetrC18:1-CL (error bars: SD, N = 6), respectively. (E) Representative light-scattering curves of GlpF activity measurements at mole fractions of 0 (black), 2 (light gray), 20 (gray), and 30 (dark gray) mol % tetrC18:1-CL. Each curve is the average of five measurements. The SD of GlpF activity in diC18:1-PC liposomes was calculated from the relative weighted rate constants (kw,rel.), and the SD of GlpF activity in the other liposomes was calculated from the normalized weighted rate constants (kw,norm.). The PG and CL concentrations in the native E. coli inner membrane are ∼15–20% and ∼5–10%, respectively.
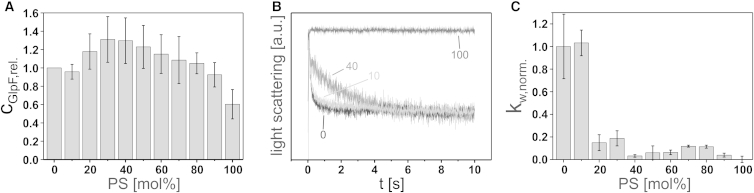
Maximal GlpF incorporation was achieved at 40 mol % PG and 20 mol % CL, respectively. Strikingly, the best incorporation was reached when the same amount of negative lipid charges were present on the liposomes. However, the relative increase of incorporated protein differed between PC/PG and PC/CL lipid mixtures. Whereas in mixed PC/PG liposomes the maximal amount of incorporated GlpF was increased by a factor of 1.3 compared with pure PC liposomes (Fig. 3 A), in PC/CL liposomes the incorporation efficiency increased by a factor of 3.2 relative to pure PC liposomes (Fig. 3 D). Importantly, the GlpF tetramer was quite stable in all tested PC/PG and PC/CL lipid mixtures. However, a slight increase of monomeric and dimeric GlpF from ∼0.18 to 0.35 was detected at high PG contents (≥70 mol %; Figs. S1 C and S2 C) and the stability of tetrameric GlpF appeared to increase slightly at CL concentrations of ≥4 mol % (Figs. S1 D and S2 D), associated with a decrease of monomeric/dimeric GlpF. Nevertheless, these minor changes cannot explain the rather drastic effect of low mol % CL on the GlpF activity (Fig. 3 F). In general, GlpF activity was highest in the absence or at very low concentrations of negatively charged lipids. The highest rates of substrate flux per relative amount of incorporated protein were measured at 10 mol % PG and 2 mol % CL (Fig. 3, B, C, E, and F). It is worth mentioning that these levels are below the PG and CL concentrations found in native E. coli membranes. With increasing PG and CL contents, the ribitol flux across the liposomal membrane decreased significantly, and thus the channel activity was severely impaired by negatively charged lipid headgroups. To test whether negatively charged lipids per se influence GlpF oligomerization and activity, we additionally analyzed the impact of the negatively charged lipid phosphatidylserine (PS), which is not a major E. coli membrane constituent. In liposomes containing increasing amounts of diC18:1-PS, up to a concentration of 90 mol %, no influence on the incorporation of GlpF was detected within the range of the standard deviation (SD) (Figs. 4 A, S1 E, and S2 E). However, in pure diC18:1-PS liposomes, the amount of incorporated GlpF was slightly decreased. The influence of PS on the GlpF activity was found to be similar to that observed in liposomes containing PG and CL: the relative GlpF activity was highest at low mol % PS and drastically decreased thereafter, starting at a PS concentration of 20 mol % (Fig. 4, B and C). Based on these findings, it can be concluded that the negative charge itself, and not the PG and CL headgroup chemistry, had caused inactivation of GlpF.
Figure 4.
GlpF incorporation and activity in diC18:1-PC liposomes containing increasing amounts of diC18:1-PS. (A) Relative GlpF incorporation. Error bars: SD (N = 3). (B) Representative light-scattering curves of GlpF activity measurements at 0 (black), 10 (light gray), 40 (gray), and 100 (dark gray) mol % diC18:1-PS. Each curve is the average of five measurements. (C) Normalized weighted rate constants of ribitol flux in liposomes of increasing mol % diC18:1-PS. Error bars: SD (N = 3). The SD of GlpF activity in diC18:1-PC liposomes was calculated from the relative weighted rate constants (kw,rel.), and the SD of GlpF activity in the other liposomes was calculated from the normalized weighted rate constants (kw,norm.).
Discussion
The lipid environment and lipid bilayer properties can affect the structure and function of membrane-integrated proteins; however, only a few studies have analyzed this in great detail (1,37). Recently, it was suggested that accommodation of defined lipid species in the vicinity of a membrane protein might allow the activity of the protein to be controlled (45). This is an intriguing concept, as in many cases it is not yet clear how cells regulate the activity of certain membrane proteins in vivo. In this study, we demonstrate that the lipid acyl-chain length and the lipid headgroup charge, but not the lateral membrane pressure, modulate the activity of GlpF, a polytopic, oligomeric TM channel protein.
Recent molecular-dynamics simulations indicated higher GlpF activity in a POPE bilayer compared with a POPC bilayer (18,19). A 10% decreased channel radius was calculated when GlpF was embedded in a POPE bilayer compared with a POPC bilayer. Based on this observation, it was suggested that the successive exchange of hydrogen bonds during water translocation might be less stringent in a translocation pore with an increased radius. Whereas in the previous simulations GlpF was incorporated into a POPE bilayer with one saturated (C16:0) and one unsaturated (C18:1) acyl-chain, in this study we increased the lateral membrane pressure in the acyl-chain region using DOPE containing two unsaturated acyl-chains (C18:1). Although shortening and, more importantly, increasing the saturation of the acyl-chains will increase the temperature at which the lipid bilayer changes from a lamellar crystalline to the hexagonal HII phase (46), both DOPE and POPE exert a negative curvature when incorporated into a lipid bilayer system (the spontaneous curvature (J0m) of DOPE and POPE is −0.399 and −0.316, respectively) (47), and thus will increase the lateral pressure in the acyl-chain region. Therefore, the different lipids used here and in the simulations cannot explain the discrepancy between the simulation and experimental results.
In contrast to the simulations, our study demonstrates that increasing amounts of PE do not affect the GlpF channel function at all, and thus the GlpF activity appears to be resistant to changes in the lateral membrane pressure (Fig. 2 C). This finding supports the assumption that tetrameric GlpF and, most likely, AQPs in general have a rather rigid protein structure. As the AQP channel activity does not involve major conformational changes of the protein structure, the functional properties of these rigid structures are unlikely to be affected by changes in the lateral pressure in the acyl-chain region. The resistance of GlpF to major changes in the lipid composition implies an enormous structural stability, as previously discussed for AQP0 (48). This observation additionally helps to explain the remarkably high activity of GlpF in the nonphysiological PC bilayer: whereas the lateral pressure in the acyl-chain region will be higher in native E. coli membranes than in a PC bilayer, due to the high abundance of PE, the activity of GlpF is not sensitive to changes in the lateral pressure in the acyl-chain region and thus is well preserved in PC bilayers. However, this cannot explain the considerably higher GlpF activity in PC bilayers compared with EPL and PE/PG/CL liposomes (Fig. 1, B and C). An interrelation between the acyl-chain composition and the lipid headgroup composition was recently described for the lactose permease (LacY) of E. coli (49), as LacY’s energy-dependent uphill transport of lactose was only supported in the presence of unsaturated acyl-chains (49). However, our data rule out a significant impact of the heterogeneous acyl-chain composition, since we also found significantly decreased channel activity in the ternary PE/PC/CL mixture with completely monounsaturated acyl-chains (Fig. 1).
Although GlpF activity was unaffected by changes in the membrane lateral pressure profile, it was found to be highly sensitive to the negative charge density of the surrounding lipid bilayer (Figs. 3, C and F, and 4 C). Low PG and CL contents initially caused an increased GlpF activity, whereas we observed a decreased GlpF activity at higher negative charge levels. Stabilizing effects of small amounts of anionic lipids on the configuration of TM helices that are flanked by cationic residues were previously described for both PG and PS (50). This effect on GlpF activity, however, seems to be lipid-headgroup specific, as PS did not increase the GlpF activity. Thus, low mol % of PG and CL stabilizes an active GlpF structure, possibly via direct, headgroup-specific interactions. Similarly, it was recently reported that negatively charged lipids have different abilities to stabilize the native tetrameric state of the E. coli AQP AqpZ, and especially CL appeared to stabilize the AqpZ tetrameric state well (45). However, in our study, a further increase in the amount of negatively charged lipids resulted in a severely reduced GlpF function in all three analyzed lipid systems, regardless of the lipid headgroup chemistry. The decreased GlpF activity observed in lipid bilayers with a negative surface charge also explains the rather low GlpF activity observed in EPL and PE/PG/CL liposomes compared with pure PC liposomes. The significantly decreased GlpF activity in liposomes from the EPL extract is most likely due to the negative surface charge density and not to the chemical nature of the lipids. In this study, the GlpF activity was already decreased at 20 mol % PG/PS or at 5 mol % CL. With concentrations of ∼20 mol % PG and ∼10 mol % CL (27), the negative charge level of the EPL and PE/PG/CL liposomes was even higher.
But how could the negative lipid headgroups affect the GlpF channel activity? A direct interference of the negatively charged lipids with substrate conduction seems unlikely, as the amino acids interacting with the permeating substrate do not interact with the surrounding lipids (Fig. 5). Previous in silico analyses indicated a voltage-dependent regulation of AQP1 and AQP4 (51). The conformation of the arginine residue in the selectivity filter of AQP1 and AQP4 differed depending on the membrane potential. The respective arginine is highly conserved within the AQP family and plays a key role in substrate conduction. When a negative membrane potential was applied (i.e., with an increased anion concentration on the intracellular side of AQP1 and AQP4), the arginine residue of the AQP selectivity filter was found to be in an orientation that reduced single-channel water permeability. In contrast, in the presence of a positive membrane potential, the different orientation of arginine appeared to facilitate rapid water flux. Direct electrostatic interactions have been proposed to cause the conformational change of arginine in the selectivity filter, and therefore a direct influence of negative lipid headgroup charges on the orientation of the arginine side chain seems possible. However, in the in silico studies, free diffusion of the anions was allowed in solution, whereas in our experiments the negative charges were localized at the lipid headgroups. This most likely constrains (or even abolishes) the possibility of direct interactions with arginine in the selectivity filter located within the GlpF channel pore.
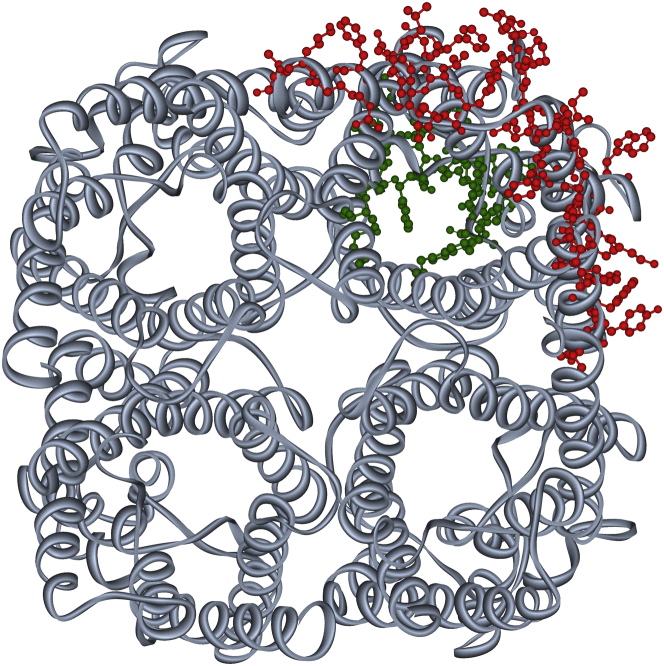
Figure 5.
GlpF crystal structure. A periplasmic view of the native tetrameric GlpF structure (PDB ID: 1FX8) (22) is shown. Amino acids interacting with glycerol are indicated in green, and those suggested by molecular-dynamics simulations to interact with lipid headgroups are shown in red. The residues that interact frequently with lipid headgroups are localized at the lipid-exposed surface of the monomeric GlpF unit (58), whereas the residues that interact with glycerol are found at the periplasmic vestibule and in the channel interior (59). To see this figure in color, go online.
In biological membranes, the ratio of negatively charged lipids and neutral lipids is carefully regulated, and positively charged amino acids are significantly overrepresented in TM proteins at the membrane-water interface region (52). Electrostatic interactions between positively charged amino acid side chains and negatively charged lipid headgroups help to anchor and position TM proteins within lipid bilayers, and they can determine the topology of membrane integral proteins (15,16). A dramatic destabilization of a TM helix bundle structure, caused by electrostatic interactions between positively charged amino acids and negative lipid headgroups, was recently observed in a simple TM helix dimer of human glycophorin A (53–55). Nevertheless, in contrast to this observation, it appears that electrostatic interactions between negatively charged lipid headgroups and GlpF do not dramatically destabilize the GlpF tetramer structure against SDS-induced unfolding, but rather inactivate the protein.
Most likely, interactions of negatively charged lipids with positively charged amino acids subtly affect the structure of GlpF, resulting in protein inactivation. An influence of the negative lipid headgroup charges on the dipole moment of the two AQP characteristic half-helices would potentially also result in an altered channel activity. However, these assumptions remain speculative, and determining the exact molecular mechanism by which GlpF is inactivated will likely require high-resolution structures in the presence of negatively charged lipids.
Conclusions
GlpF has a very high structural stability, which renders the protein resistant to major changes in the lateral membrane pressure. However, we found that GlpF activity was greatly influenced by negatively charged lipid headgroups, although the observed effects arose from negative surface charges and not from the specific headgroup chemistry. Most likely, electrostatic interactions between basic amino acids and the negative headgroup charge induce subtle conformational changes in the GlpF TM domain, which affect the GlpF channel function. In a living cell, membrane-binding molecules (e.g., peripheral proteins and sugars) modulate the surface chemistry and also the surface charge density of a membrane (56,57). Thus, shielding lipid headgroup charges by membrane-binding components might guarantee proper GlpF activity in vivo, in living E. coli cells. On the other hand, if the activity of GlpF is regulated in E. coli, the selective enrichment of negatively charged lipids in the direct vicinity of GlpF might allow a cell to reversibly control the activity of the glycerol channel. Thus, our work illustrates a mechanism by which a cell could control the activity of an α-helical membrane protein merely by varying the negative charge density around the protein.
Author Contributions
N.K. and D.S. designed research. N.K and N.H. performed research. N.K., N.H., and D.S. analyzed data and wrote the manuscript.
Acknowledgments
This work was supported by grants from the Stiftung Rheinland-Pfalz für Innovation, the Research Center for Complex Materials (COMATT), and the German Chemical Industry fund.
Editor: Andreas Engel.
Footnotes
Supporting Discussion, six figures, and one table are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)00677-3.
Supporting Citations
Reference (60) appears in the Supporting Material.
Supporting Material
References
- 1.Lee A.G. How lipids affect the activities of integral membrane proteins. Biochim. Biophys. Acta. 2004;1666:62–87. doi: 10.1016/j.bbamem.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 2.Debnath D.K., Basaiawmoit R.V., Otzen D.E. The role of membrane properties in Mistic folding and dimerisation. Protein Eng. Des. Sel. 2011;24:89–97. doi: 10.1093/protein/gzq095. [DOI] [PubMed] [Google Scholar]
- 3.Neumann J., Klein N., Schneider D. Folding energetics and oligomerization of polytopic α-helical transmembrane proteins. Arch. Biochem. Biophys. 2014;564:281–296. doi: 10.1016/j.abb.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Raja M., Spelbrink R.E., Killian J.A. Phosphatidic acid plays a special role in stabilizing and folding of the tetrameric potassium channel KcsA. FEBS Lett. 2007;581:5715–5722. doi: 10.1016/j.febslet.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 5.Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 6.Bogdanov M., Dowhan W. Phospholipid-assisted protein folding: phosphatidylethanolamine is required at a late step of the conformational maturation of the polytopic membrane protein lactose permease. EMBO J. 1998;17:5255–5264. doi: 10.1093/emboj/17.18.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Killian J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta. 1998;1376:401–415. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 8.Williamson I.M., Alvis S.J., Lee A.G. Interactions of phospholipids with the potassium channel KcsA. Biophys. J. 2002;83:2026–2038. doi: 10.1016/S0006-3495(02)73964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A.G. How lipids interact with an intrinsic membrane protein: the case of the calcium pump. Biochim. Biophys. Acta. 1998;1376:381–390. doi: 10.1016/s0304-4157(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 10.Fattal D.R., Ben-Shaul A. A molecular model for lipid-protein interaction in membranes: the role of hydrophobic mismatch. Biophys. J. 1993;65:1795–1809. doi: 10.1016/S0006-3495(93)81249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen C., Goulian M., Andersen O.S. Energetics of inclusion-induced bilayer deformations. Biophys. J. 1998;74:1966–1983. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogdanov M., Heacock P.N., Dowhan W. A polytopic membrane protein displays a reversible topology dependent on membrane lipid composition. EMBO J. 2002;21:2107–2116. doi: 10.1093/emboj/21.9.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogdanov M., Xie J., Dowhan W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 2008;182:925–935. doi: 10.1083/jcb.200803097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seppälä S., Slusky J.S., von Heijne G. Control of membrane protein topology by a single C-terminal residue. Science. 2010;328:1698–1700. doi: 10.1126/science.1188950. [DOI] [PubMed] [Google Scholar]
- 17.van den Brink-van der Laan E., Killian J.A., de Kruijff B. Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim. Biophys. Acta. 2004;1666:275–288. doi: 10.1016/j.bbamem.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 18.Jensen M.O., Mouritsen O.G. Lipids do influence protein function—the hydrophobic matching hypothesis revisited. Biochim. Biophys. Acta. 2004;1666:205–226. doi: 10.1016/j.bbamem.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 19.Jensen M.O., Mouritsen O.G. Single-channel water permeabilities of Escherichia coli aquaporins AqpZ and GlpF. Biophys. J. 2006;90:2270–2284. doi: 10.1529/biophysj.105.073965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preston G.M., Carroll T.P., Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- 21.Fujiyoshi Y., Mitsuoka K., Engel A. Structure and function of water channels. Curr. Opin. Struct. Biol. 2002;12:509–515. doi: 10.1016/s0959-440x(02)00355-x. [DOI] [PubMed] [Google Scholar]
- 22.Fu D., Libson A., Stroud R.M. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 23.Veerappan A., Cymer F., Schneider D. The tetrameric α-helical membrane protein GlpF unfolds via a dimeric folding intermediate. Biochemistry. 2011;50:10223–10230. doi: 10.1021/bi201266m. [DOI] [PubMed] [Google Scholar]
- 24.Cymer F., Schneider D. A single glutamate residue controls the oligomerization, function, and stability of the aquaglyceroporin GlpF. Biochemistry. 2010;49:279–286. doi: 10.1021/bi901660t. [DOI] [PubMed] [Google Scholar]
- 25.Klein N., Neumann J., Schneider D. Folding and stability of the aquaglyceroporin GlpF: Implications for human aqua(glycero)porin diseases. Biochim. Biophys. Acta. 2015;1848:622–633. doi: 10.1016/j.bbamem.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 26.Lugtenberg E.J., Peters R. Distribution of lipids in cytoplasmic and outer membranes of Escherichia coli K12. Biochim. Biophys. Acta. 1976;441:38–47. doi: 10.1016/0005-2760(76)90279-4. [DOI] [PubMed] [Google Scholar]
- 27.Oursel D., Loutelier-Bourhis C., Lange C.M. Lipid composition of membranes of Escherichia coli by liquid chromatography/tandem mass spectrometry using negative electrospray ionization. Rapid Commun. Mass Spectrom. 2007;21:1721–1728. doi: 10.1002/rcm.3013. [DOI] [PubMed] [Google Scholar]
- 28.Gonen T., Cheng Y., Walz T. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borgnia M.J., Agre P. Reconstitution and functional comparison of purified GlpF and AqpZ, the glycerol and water channels from Escherichia coli. Proc. Natl. Acad. Sci. USA. 2001;98:2888–2893. doi: 10.1073/pnas.051628098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galka J.J., Baturin S.J., O’Neil J.D. Stability of the glycerol facilitator in detergent solutions. Biochemistry. 2008;47:3513–3524. doi: 10.1021/bi7021409. [DOI] [PubMed] [Google Scholar]
- 31.Bearer E.L. Overview of image analysis, image importing, and image processing using freeware. Curr. Protoc. Mol. Biol. 2003;Chapter 14 doi: 10.1002/0471142727.mb1415s63. Unit 14.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu H., Xu T.L. The general anesthetic pentobarbital slows desensitization and deactivation of the glycine receptor in the rat spinal dorsal horn neurons. J. Biol. Chem. 2002;277:41369–41378. doi: 10.1074/jbc.M206768200. [DOI] [PubMed] [Google Scholar]
- 33.Kucerka N., Gallová J., Katsaras J. Areas of monounsaturated diacylphosphatidylcholines. Biophys. J. 2009;97:1926–1932. doi: 10.1016/j.bpj.2009.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anbazhagan V., Schneider D. The membrane environment modulates self-association of the human GpA TM domain--implications for membrane protein folding and transmembrane signaling. Biochim. Biophys. Acta. 2010;1798:1899–1907. doi: 10.1016/j.bbamem.2010.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Rawicz W., Olbrich K.C., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jensen M.O., Park S., Schulten K. Energetics of glycerol conduction through aquaglyceroporin GlpF. Proc. Natl. Acad. Sci. USA. 2002;99:6731–6736. doi: 10.1073/pnas.102649299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen O.S., Koeppe R.E., 2nd Bilayer thickness and membrane protein function: an energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007;36:107–130. doi: 10.1146/annurev.biophys.36.040306.132643. [DOI] [PubMed] [Google Scholar]
- 38.Charalambous K., Miller D., Booth P.J. Lipid bilayer composition influences small multidrug transporters. BMC Biochem. 2008;9:31. doi: 10.1186/1471-2091-9-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cullis P.R., de Kruijff B. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 40.Cullis P.R., Hope M.J., Tilcock C.P. Lipid polymorphism and the roles of lipids in membranes. Chem. Phys. Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- 41.Seddon J.M. Structure of the inverted hexagonal (HII) phase, and non-lamellar phase transitions of lipids. Biochim. Biophys. Acta. 1990;1031:1–69. doi: 10.1016/0304-4157(90)90002-t. [DOI] [PubMed] [Google Scholar]
- 42.Cantor R.S. Lateral pressures in cell membranes: a mechanism for modulation of protein function. J. Phys. Chem. B. 1997;101:1723–1725. [Google Scholar]
- 43.Cantor R.S. The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry. 1997;36:2339–2344. doi: 10.1021/bi9627323. [DOI] [PubMed] [Google Scholar]
- 44.Epand R.M. Lipid polymorphism and protein-lipid interactions. Biochim. Biophys. Acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 45.Laganowsky A., Reading E., Robinson C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koynova R., Caffrey M. Phases and phase transitions of the hydrated phosphatidylethanolamines. Chem. Phys. Lipids. 1994;69:1–34. doi: 10.1016/0009-3084(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 47.Kollmitzer B., Heftberger P., Pabst G. Monolayer spontaneous curvature of raft-forming membrane lipids. Soft Matter. 2013;9:10877–10884. doi: 10.1039/C3SM51829A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sanders C.R., Mittendorf K.F. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50:7858–7867. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vitrac H., Bogdanov M., Dowhan W. Proper fatty acid composition rather than an ionizable lipid amine is required for full transport function of lactose permease from Escherichia coli. J. Biol. Chem. 2013;288:5873–5885. doi: 10.1074/jbc.M112.442988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shahidullah K., London E. Effect of lipid composition on the topography of membrane-associated hydrophobic helices: stabilization of transmembrane topography by anionic lipids. J. Mol. Biol. 2008;379:704–718. doi: 10.1016/j.jmb.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hub J.S., Aponte-Santamaría C., de Groot B.L. Voltage-regulated water flux through aquaporin channels in silico. Biophys. J. 2010;99:L97–L99. doi: 10.1016/j.bpj.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ulmschneider M.B., Sansom M.S., Di Nola A. Properties of integral membrane protein structures: derivation of an implicit membrane potential. Proteins. 2005;59:252–265. doi: 10.1002/prot.20334. [DOI] [PubMed] [Google Scholar]
- 53.Hong H., Bowie J.U. Dramatic destabilization of transmembrane helix interactions by features of natural membrane environments. J. Am. Chem. Soc. 2011;133:11389–11398. doi: 10.1021/ja204524c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russ W.P., Engelman D.M. The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 55.Cymer F., Veerappan A., Schneider D. Transmembrane helix-helix interactions are modulated by the sequence context and by lipid bilayer properties. Biochim. Biophys. Acta. 2012;1818:963–973. doi: 10.1016/j.bbamem.2011.07.035. [DOI] [PubMed] [Google Scholar]
- 56.Murray D., Arbuzova A., McLaughlin S. The role of electrostatic and nonpolar interactions in the association of peripheral proteins with membranes. Curr. Top. Membr. 2002;52:277–307. [Google Scholar]
- 57.Goldenberg N.M., Steinberg B.E. Surface charge: a key determinant of protein localization and function. Cancer Res. 2010;70:1277–1280. doi: 10.1158/0008-5472.CAN-09-2905. [DOI] [PubMed] [Google Scholar]
- 58.Stansfeld P.J., Jefferys E.E., Sansom M.S. Multiscale simulations reveal conserved patterns of lipid interactions with aquaporins. Structure. 2013;21:810–819. doi: 10.1016/j.str.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen M.O., Tajkhorshid E., Schulten K. The mechanism of glycerol conduction in aquaglyceroporins. Structure. 2001;9:1083–1093. doi: 10.1016/s0969-2126(01)00668-2. [DOI] [PubMed] [Google Scholar]
- 60.Urbani A., Warne T. A colorimetric determination for glycosidic and bile salt-based detergents: applications in membrane protein research. Anal. Biochem. 2005;336:117–124. doi: 10.1016/j.ab.2004.09.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.