Abstract
Mitochondria are energy-producing organelles in eukaryotic cells considered to be of bacterial origin. The mitochondrial genome has evolved under selection for minimization of gene content, yet it is not known why not all mitochondrial genes have been transferred to the nuclear genome. Here, we predict that hydrophobic membrane proteins encoded by the mitochondrial genomes would be recognized by the signal recognition particle and targeted to the endoplasmic reticulum if they were nuclear-encoded and translated in the cytoplasm. Expression of the mitochondrially encoded proteins Cytochrome oxidase subunit 1, Apocytochrome b, and ATP synthase subunit 6 in the cytoplasm of HeLa cells confirms export to the endoplasmic reticulum. To examine the extent to which the mitochondrial proteome is driven by selective constraints within the eukaryotic cell, we investigated the occurrence of mitochondrial protein domains in bacteria and eukaryotes. The accessory protein domains of the oxidative phosphorylation system are unique to mitochondria, indicating the evolution of new protein folds. Most of the identified domains in the accessory proteins of the ribosome are also found in eukaryotic proteins of other functions and locations. Overall, one-third of the protein domains identified in mitochondrial proteins are only rarely found in bacteria. We conclude that the mitochondrial genome has been maintained to ensure the correct localization of highly hydrophobic membrane proteins. Taken together, the results suggest that selective constraints on the eukaryotic cell have played a major role in modulating the evolution of the mitochondrial genome and proteome.
Keywords: mitochondria, protein domain, evolution, endoplasmatic reticulum, signal recognition particle
Mitochondria are organelles that produce energy by oxidative phosphorylation (OXPHOS), channeling electrons through the respiratory chain complexes to generate ATP, the energy currency of the cell. A unique feature of mitochondria is that they possess a distinct genome of their own. Mitochondrial (mt) genomes vary dramatically in size, but gene content is limited and remarkably similar in different organisms (1–4). The human mitochondrial genome contains only 13 protein coding genes, whereas the jakobid mitochondrial genomes contain up to 70 genes, mostly coding for subunits of the OXPHOS system complexes and ribosomal proteins (5, 6). Having two separate genomes is a costly arrangement for the cell. Approximately 250 proteins encoded in the nuclear (nu) genome are needed just to maintain and express the few remaining mitochondrial genes (7). Mitochondrial sequences are frequently copied to the nuclear genomes (8–10), confirming that mechanisms for the transfer of mitochondrial genes to the nuclear genome are in place. A major, unresolved question in evolutionary biology is why not all mitochondrial genes have been transferred to the nuclear genome, and thus why the mitochondrial genome has been retained.
Over the years, many competing hypotheses have been put forward to explain the retention of organelle genomes. Some argue that gene transfer is an on-going process and that all mitochondrial genes will eventually end up in the nuclear genome (11). Strictly speaking, organelles such as mitosomes and hydrogenosomes have lost their genomes entirely, as has also a plastid from a nonphotosynthetic species of the genus Polytomella (3). This does not however explain the universal retention of a genome in aerobic mitochondria and photosynthetic chloroplasts. Others suggest that the transfer of genes have been halted, either because of barriers against gene transfer or because the mitochondrial genome confers benefits. One such proposed barrier to functional gene transfers is codon reassignments (12). This explanation has been dismissed because it is not applicable to all mitochondrial genomes. Another suggested barrier is the extreme hydrophobicity of the mitochondrial proteins, which has been claimed to prevent their import from the cytosol (13). However, this hypothesis has also been rejected because hydrophobic proteins can be imported across the mitochondrial membrane and because some membrane-spanning proteins are nu-encoded, such as the Lhca and Lhcb proteins in chloroplasts (ct) (14). Instead, models based on beneficial functions, such as the colocation for redox regulation hypothesis (15, 16), have gained popularity during the past decade. However, no mitochondrial genes involved in redox regulation have been identified (17). Thus, all hypotheses that have been put forward are controversial and none have been experimentally verified.
In striking contrast to the few proteins encoded by the mitochondrial genome, it is estimated that more than 1,000 mitochondrial proteins are encoded by the nuclear genome (18). Many of these have no bacterial homologs. For example, the mitochondrial ATP/ADP translocase that exports the ATP produced in the mitochondrion to the cytoplasm shows no sequence similarity to the bacterial type of ATP/ADP translocase. Rather, the mitochondrial ATP/ADP translocase has evolved from a family of eukaryotic phosphate transporters (19). Studies of the yeast mitochondrial proteome have indicated that about 40% of all mitochondrial proteins have no homologos in bacteria, and might thus have originated within the eukaryotic genome (20, 21). Specifically, it has been suggested that new proteins have been added to the OXPHOS complex and the mito-ribosome before the diversification of the eukaryotic lineages (22, 23). This pattern contrasts with the core components of the mitochondrial OXPHOS system and the mito-ribosome, which are highly conserved and show strong sequence similarity to their bacterial homologs.
In this study, we revisit a hypothesis proposed 30 years ago but since long forgotten: namely that a mitochondrial genome is needed to prevent the export of highly hydrophobic mitochondrial membrane proteins to the endoplasmic reticulum (ER) (24). We use a combination of bioinformatics methods to predict protein localization and cell biological methods to verify our predictions experimentally. In addition, we analyze the nu-encoded fraction of the mitochondrial proteome for which structure, and hence function, can be assigned. Taking these data together, we are able to gain a comprehensive insight into the role that the two different genomes play in orchestrating mitochondrial functions.
Results
Mitochondrial Proteins are Potential Targets for Recognition by Signal Recognition Particle.
Revisiting the hydrophobicity hypothesis in its original formulation is motivated by recent insights into the mechanism whereby the signal recognition particle (SRP) exports proteins to the ER. This is a cotranslational pathway, where SRP binds to a hydrophobic domain, either in the form of a transmembrane domain (TMD) or a signal sequence, which leads to arrest of the nascent peptide chain. Importantly, it is the hydrophobicity rather than the signal sequence per se that is recognized by SRP. Next, the ribosome with the arrested peptide chain is transported to the ER (25). Soluble proteins shorter than 100–120 amino acids are missed by SRP, whereas proteins of 120–160 amino acids can be captured, although quite inefficiently (26). Thus, the hydrophobicity of the TMD, as well as the length of the C-terminal tail following the first hydrophobic domain, is critical for protein recognition by SRP (27).
To predict the potential of nu- and organelle-encoded mitochondrial proteins to be targets for recognition by SRP, we calculated the free insertion energy (ΔG, kcal/mol) of the TMDs (28), and categorized the TMDs as either hydrophobic or marginally hydrophobic. The proteins were then classified as arrested by SRP if they contained a hydrophobic TMD and a tail that, together with the TMD, was longer than 120 amino acids. Proteins with only marginally hydrophobic TMDs were considered to avoid recognition by SRP.
The human mitochondrial OXPHOS system consists of five membrane-spanning complexes composed of 96 proteins in total, of which 13 are encoded by the mitochondrial genome and 83 by the nuclear genome (SI Appendix, Table S1). We classified 10 of the 13 mt-encoded proteins as targets for SRP, whereas only 1 of the 83 nu-encoded proteins was predicted to be an SRP target (Fisher’s Exact χ2 test, P < 0.01) (Fig. 1A). Similarly, only 2 of the 51 nu-encoded mitochondrial membrane proteins with known structures were classified as arrested by SRP (SI Appendix, Table S2). Consistent with these results, 251 of 281 nu-encoded membrane proteins localized to mitochondria with the aid of green fluorescent protein tags in yeast and humans (29) were predicted to avoid recognition by SRP (SI Appendix, Fig. S1 and Table S3).
Fig. 1.
Biophysical characteristics of membrane proteins involved in aerobic respiration and photosynthesis. The figure shows the insertion-free energy (ΔG, dG) and the length of the TMD and the following C-terminal tail for proteins involved in (A) the OXPHOS system complexes, and (B) the photosynthetic apparatus. The insertion-free energy (kcal/mol) was estimated for either the first TMD with a calculated ΔG value below zero or if no such segment was found, the most hydropohobic segment in the protein. Red dots correspond to proteins encoded by the organelle genome, and blue dots to proteins encoded by the nuclear genome. Characteristic features of proteins that are putative targets for recognition by SRP are shown in the gray area.
We also estimated the potential of chloroplast proteins in the photosynthetic apparatus of Arabidopsis thaliana, (SI Appendix, Table S4), to be recognized by SRP. Of the 14 ct-encoded proteins longer than 100 amino acids, 11 would be arrested by SRP, compared with only 1 of the 24 nu-encoded proteins with a length of more than 100 amino acids (Fisher’s Exact χ2 test, P < 0.01) (Fig. 1B). Importantly, the nu-encoded light-harvesting (Lh) proteins Lhca and Lhcb would not be recognized by SRP according to our calculations. As much as one-third of the 30 ct-encoded membrane proteins are shorter than 50 amino acids (SI Appendix, Fig. S2). We suggest that these extremely short single-spanning membrane proteins, which integrate randomly into the thylakoid membrane in plants (30), would be difficult to transport to the thylakoid membrane if not encoded in the chloroplast genome.
Mistargeting of Mitochondrial Proteins to the ER.
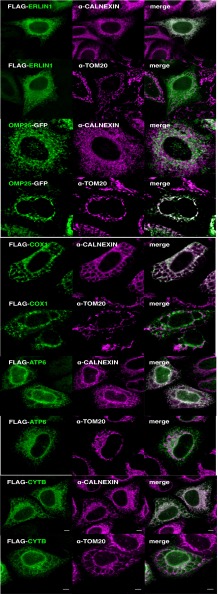
To experimentally test our predictions, mt-encoded proteins Cytochrome oxidase subunit 1 (Cox1), Apocytchrome b (Cytb), and ATP synthase subunit 6 (ATP6) were expressed in the cytoplasm of HeLa cells (Fig. 2). The proteins were FLAG-tagged at the N terminus and visualized by immunofluorescence. The FLAG-tag was placed at the N terminus rather than at the C terminus to avoid extending the length of the C-terminal sequence following the first hydrophobic domain. We used FLAG-tagged ERLIN1 and OMP25-GFP constructs as positive localization controls (for ER and mitochondria, respectively) and immunostained with antibodies marking the ER (calnexin) and mitochondria (TOM20) (Fig. 2A). As predicted, Cox1, Cytb, and ATP6 colocalized with the ER (Fig. 2). Mistargeting of these mt-encoded proteins to the ER resulted in the formation of aberrant honeycomb structures, as previously observed during viral infections (31). This finding suggests that mistargeting of mitochondrial proteins to the ER affects the morphology of the cell. We conclude that genes for hydrophobic membrane proteins of more than 120 amino acids are likely retained in distinct organelle genomes to ensure a correct localization of these proteins and avoid transport to the ER.
Fig. 2.
Mitochondrial membrane proteins atypically expressed in the cytoplasm localize to the ER. Amino acid sequences of COX1, ATP6, and CYB were reverse-translated to nuclear codon use and FLAG-tagged at their amino termini. HeLa cells were transfected with plasmids containing these synthetic genes and subjected to immunofluorescence analysis. From the top, FLAG-tagged ERLIN1 and OMP25-GFP constructs were used as positive localization controls and immunostained with antibodies marking the ER (calnexin) or mitochondria (TOM20). Below this, localization analyses of the synthetic mitochondrial membrane proteins COX1, ATP6, and CYB. (Scale bars, 5 µm.)
Phyletic Distribution Patterns of Mitochondrial Protein Folds in Eukaryotes.
It is clear that the mitochondrial genome and the supporting genetic apparatus, whether encoded by the mitochondrial or nuclear genome, are dedicated to synthesize and assemble the OXPHOS complex, which is central to energy metabolism. It is less clear to what extent selective constraints acting on the eukaryotic cell, like targeting proteins to their correct locations or regulating the activities of the two genomes, have influenced the mitochondrial proteome. To learn more about the evolutionary pressures acting on mitochondrial proteins, we surveyed a broad taxonomic range of organisms for the presence or absence patterns of proteins that are critical to mitochondrial functions. Because of the difficulty of assigning proteins to clusters of orthologous groups for highly divergent proteins and multidomain proteins (32), we used a protein domain-centric approach for this analysis. In brief, we assigned protein domains at the superfamily (SF) level of the SCOP (Structural Classification of Proteins) hierarchy (33) using hidden Markov model libraries of protein domains in the SUPERFAMILY database (E < 0.0001) (34). SCOP-SF domains are inferred to be homologous based on similarity of sequence, structure, and function.
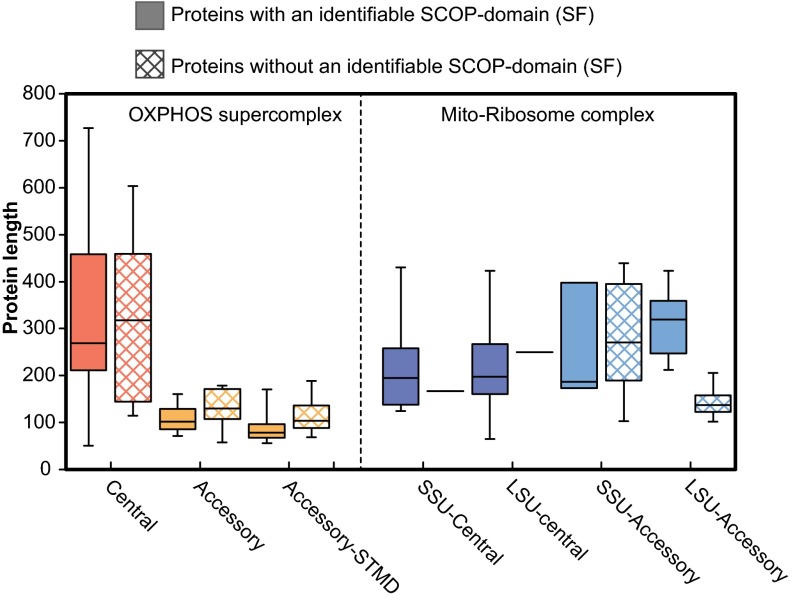
The obvious start for such a survey was the OXPHOS system itself. Thus, we predicted protein folds in the OXPHOS system in human mitochondria, which consists of a central set of 14 proteins that are conserved in both eukaryotes and bacteria, and an accessory set of 30 protein subunits that have solely been identified in mitochondria (35). In total, we assigned 39 distinct SF-domains to the central set of proteins and another 20 distinct SF-domains to the accessory proteins. A length comparison revealed a marked difference between the central and the accessory proteins in the mitochondrial OXPHOS system, with median lengths of about 300 and 100 amino acids, respectively (Fig. 3). The size difference was observed irrespectively of whether the proteins contained recognizable domains or not.
Fig. 3.
Comparison of protein lengths of the core and accessory proteins. Length distribution of the core and accessory proteins in the OXPHOS supercomplex and the mitochondrial ribosomal complex were compared between proteins for which a SF-domain could be assigned and proteins in which a SF-domain could not be identified. For each distribution center, the lines show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend to extreme data points.
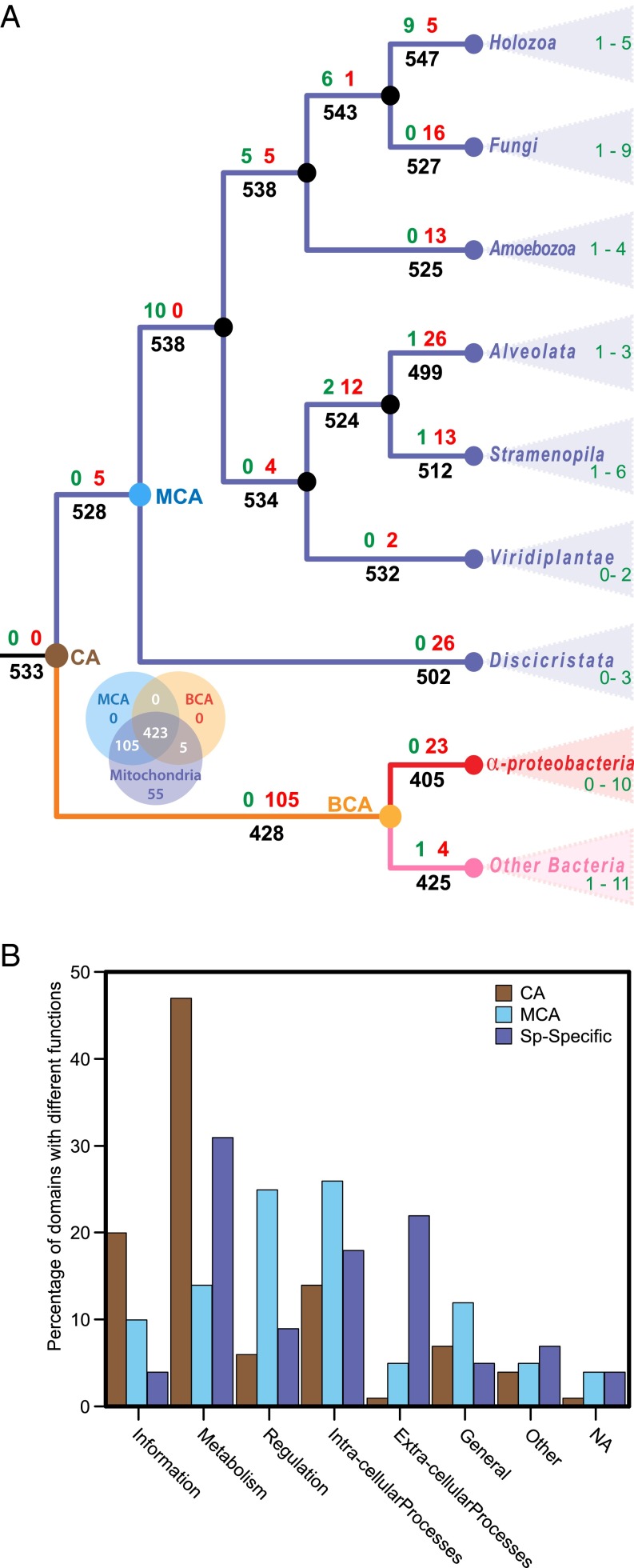
The phyletic distribution of the identified SFs was examined in 43 eukaryotes from 7 major eukaryotic lineages for which a published multigene phylogeny is available that shows their internal relationships (36). In the phylogeny, 13 bacterial species were used as outgroups, 6 of which are from the Alphaproteobacteria. The analysis showed that of the 39 SF-domains in the central set of proteins, 34 were also present in bacteria (Fig. 4). The remaining five SF-domains represent novel protein extensions located in proteins for which no homologs are present in bacteria. These additional domains include a transmembrane anchor protein domain in two proteins of the cytochrome reductase and oxidase complexes, respectively, and a nonglobular α/β subunit solely identified in human mitochondria.
Fig. 4.
Phyletic distribution of SF-domains of the human mitochondrial OXPHOS complex. The number of central and accessory SF-domains identified in each species of eukaryotes and bacteria are compared along the phylogeny. Columns correspond to the species in the phylogenetic tree and rows are individual SF-domains of the OXPHOS supercomplex. The colors indicate the number of domains in each species if present and absences are shown as white. Descriptions of each row include the complex in which a SF-domain is identified in, followed by the SCOP classification of the SF-domain. The corresponding SF-domains descriptions are listed in SI Appendix, Table S7. SF-domains encoded in the nuclear genome are shown for all species. The SFs encoded by the human mitochondrial genome are indicated in bold. The number of domains in eukaryotes only include nu-encoded SFs.
All of the 20 SF-domains identified in the accessory proteins, one-third of which are single transmembrane-spanning domains (STMD), were markedly absent from bacteria. The functions of the accessory proteins are largely unknown, although a few have been implicated in the biogenesis of the OXPHOS complex (35, 37). Some of the short STMDs are quite hydrophobic, and as such might help stabilize the respiratory protein complexes in the membrane. The most prominent candidates are UQCR10 and COX6A, which have a high hydrophobicity and a number of positive charges flanking this segment, making them well anchored in the membrane. Our phyletic analysis showed a highly scattered distribution pattern of these domains in the eukaryotic lineages and most domains were also not associated with proteins of other functions. This finding suggests that the identified domains represent cases of de novo evolution of protein folds.
However, these domains are all located in membrane-associated proteins. For comparison, we also examined protein folds of the human mitochondrial ribosome, which is composed of a central set of soluble proteins, 21 and 32 for the small and large ribosomal subunit, respectively, plus 12 and 18 soluble accessory proteins that are unique to mitochondria. We predicted 34 SFs for the 53 central proteins and 10 SFs for the 30 accessory proteins in the mitochondrial ribosome. Unlike the accessory proteins of the OXPHOS complex, the accessory proteins of the ribosome are of similar sizes as the central proteins (Fig. 3). A study of the phyletic distribution patterns of the protein folds in the accessory ribosomal proteins showed that they are broadly present in our reference set of bacteria and eukaryotes (SI Appendix, Fig. S3). The protein domains in the central mitochondrial ribosomal proteins were also identified in the cytosolic ribosomal proteins, which explain why these domains are present in more than one copy in most eukaryotes. Unlike the accessory proteins of the OXPHOS system, which are unique to mitochondria, only 1 of the 11 identified SF-domains in the accessory ribosomal proteins was solely present in eukaryotic proteins, the peptidyl-tRNA hydrolase domain in MRLP58. Thus, the mitochondrial ribosome seems to have expanded in complexity by reusing SF-domains for multiple functions.
Protein Folds in the Mitochondrial Common Ancestor.
To study the history of mitochondrial SF-domain evolution, we performed ancestral reconstruction analyses. Inferring protein-fold cohorts in the mitochondrial common ancestor (MCA) can indicate selection at the level of the eukaryotic cell. We mapped SF-domains onto the reference phylogeny based on the most parsimonious reconstructions (SI Appendix, SI Methods). Here, the simple assumption is that proteins for which no bacterial homologs are available have evolved within the eukaryotic cell. For this analysis, we used two datasets: one consists of mitochondrial protein folds encoded by the mitochondrial genome, and the other of mitochondrial protein folds inferred from experimentally determined mitochondrial proteomes.
For the first analysis, we used the previously published “MitoCOG” dataset, which consists of 34,751 proteins from more than 2,000 species that have been clustered into 140 clusters of orthologous groups (MitoCOGs) (38). We assigned 67 distinct SF-domains to 82 of the 140 MitoCOGs (SI Appendix, Table S5). These include 16 of the 25 MitoCOGs in the OXPHOS pathway and all 30 MitoCOGs for translation functions. Several lineage-specific MitoCOGs previously annotated as hypotheticals were identified as ribosomal proteins in our analysis, including ribosomal proteins S3, S7, and L6 in ciliates, S3 in Amoebozoa, and L10 in Stramenophiles. The 67 SF-domains are, with the exception of a few mobile elements, also present in bacterial proteins. Importantly, all folds identified in the proteins encoded by the mitochondrial genomes are also present in bacteria, indicating that none is the result of selective constraints acting specifically on the eukaryotic cell.
For the second analysis, we investigated the occurrence patterns of SFs in 58 large-scale proteomics datasets in the mitochondrial proteomes of 12 organisms compiled in the MitoMiner resource (29), (SI Appendix, Table S6). Included in this resource are proteins that have been identified in at least one study with GFP tags or three or more independent mass spectrometry studies, and should thus be reliable. However, it cannot be excluded that a few nonmitochondrial proteins are included in the datasets, particularly for the 25% of proteins assigned to mitochondria in a single species. Some domains may also represent mis-assignments. The mitochondrial proteomes of yeast, humans, mouse, and Drosophila contain a large number of proteins deduced from several different experimental approaches. Other taxa are less well covered, and “the mitochondrial proteome” is thus biased toward the model organisms. However, it should be noted that the subsequent identification of homologous SF-domains in the reference set of taxa is not.
The experimental dataset contained ∼13,000 proteins in total, which we assigned to 588 distinct SF-domains (i.e., about 10-times as many as the SFs identified in the mt-encoded proteome). The distribution patterns of these SFs were surveyed in the reference phylogeny of eukaryotes and bacteria (36), and most parsimonious scenarios of their gains and losses were estimated using a penalty of one for loss and two for gains.
About two-thirds of the SFs associated with the experimentally determined mitochondrial proteomes were broadly present in all species and thus assigned to the common ancestor (CA) of eukaryotes and bacteria, and 105 SFs were assigned to the MCA (Fig. 5A). However, no more than 10% of the SF-domains in MCA were uniquely associated with mitochondrial proteins, whereas the rest of the SF domains were also identified in proteins with other cellular locations (SI Appendix, Table S8). The mitochondrial proteins include the peptidyl-tRNA hydrolase domain of mitochondrial protein L58 in the large ribosomal subunit, a mitochondria-specific protein domain in a mitochondrial carrier protein, and two of the extra domains in the central proteins and three of the domains in the accessory proteins of the OXPHOS complex.
Fig. 5.
Ancestral reconstruction analysis of SF-domains in the mitochondrial proteome. (A) Flux of SF-domains in the mitochondrial proteome during evolution of eukaryotes and bacteria. Parsimonious reconstruction of domain flux is mapped onto a eukaryote phylogeny rooted with bacterial outgroup. Numbers at nodes show the number of SF-domains inferred to have been gained (green), lost (red), and the total number present at each node or branch (black) along the phylogeny. Only the major internal nodes are shown for clarity. Colored nodes represent the Common Ancestor of eukaryotes and bacteria (CA) (brown); Mitochondrial Common Ancestor (MCA) (blue); Bacterial Common Ancestor (BCA) (orange). Because the number of gains and losses varies between species, only a range from a minimum to a maximum in each clade is shown here. Details of flux for all species are shown in SI Appendix, Fig. S4. (B) A comparison of the functions associated to SF-domains estimated to be in the CA of eukaryotes and bacteria, MCA, and species specific SF-domains (Sp-specific). Bars represent the percent of SF-domains associated with a functional category.
Another 55 SF-domains were variably present and thus inferred to have evolved more recently (Fig. 5A and SI Appendix, Table S8). We functionally categorized the three sets of SFs (Fig. 5B). The majority of SF-domains in the shared CA were found in proteins involved in information and bioenergetic processes, consistent with previous findings. In contrast, domains in the MCA were mostly associated with regulation and intracellular processes, whereas metabolic and extracellular processes dominated the variably present (Sp-specific) protein domains. The functional breakdown of the mitochondrial protein domains with evolutionary age suggests that the emergence of the mitochondrion was a gradual process that required major adaptive changes in the regulation and assembly of the mitochondrial complexes. These results further imply that the mitochondrial proteome has expanded in complexity through the reuse of protein domains already present in other eukaryotic proteins. True innovations in the form of novel protein folds have mostly been targeted to the OXPHOS system.
Discussion
We have presented bioinformatic and experimental data suggesting that genes for hydrophobic membrane proteins of more than 120 amino acids are retained in mitochondrial and chloroplast genomes to avoid SRP recognition and subsequent mistargeting to the ER. To gain a comprehensive view of the functions and ostensible relationships of mitochondria to bacteria for proteins encoded by two distinct genomes, we also analyzed protein domains associated with mitochondria. We explored this in two different ways: (i) we analyzed proteins that are known to exclusively function in mitochondria—that is the respiratory complex and the mito-ribosome—and (ii) we assembled a nominal mitochondrial proteome based on experimentally verified proteins localized to mitochondria and analyzed flux of domains during evolution on a known phylogeny. The studies confirmed that the evolution of mitochondria has been associated with the evolution of novel protein domains that are specific for mitochondrial functions. The prime examples are the accessory proteins in the OXPHOS system, which have been suggested to be involved in the organization, regulation, and biogenesis of the complex (35, 37, 39).
The evolution of the mitochondrial ribosome has been studied in great detail previously (40), and it has been inferred that 19 accessory ribosomal proteins were present already in the mitochondrial ancestor (41). We identified SF-domains for 6 of these 19 proteins, all of which were associated with generalized functions present in many bacterial and cytosolic proteins. For example, the Nudix domain in MRPL46 is also found in decapping enzymes, ADP ribose diphosphatase, and similar proteins, and the double-stranded (ds) RNA-binding protein domain found in MRPL44 and MRPS5 is a generic RNA-binding domain found in diverse enzymes, such as RNA helicases, dsRNA-dependent protein kinase, RNase III, and so forth. The accessory proteins, which are situated on the surface of the ribosome, have been suggested to be involved in cotranslational processes and programmed cell death (42, 43), providing examples of how the eukaryotic cell controls the mitochondrial processes.
Overall, one-third of the SFs identified in mitochondrial proteins are mainly found in eukaryotic genomes and only rarely found in bacterial genomes. About 25% of the protein domains assigned to the mitochondrial ancestor was associated with regulatory functions, whereas metabolic functions dominate among the clade and species-specific mitochondrial protein domains. A large majority of these folds are generic and present in proteins with many functions and destinations within the eukaryotic cell. One of the few nu-encoded mitochondrial proteins, which was classified as arrested by SRP in our analysis, is mistargeted to the ER in the absence of a strong mitochondrial target peptide (44). Thus, some of the nu-encoded proteins with characteristics that are typical of proteins recognized by SRP might in fact be targeted to both mitochondria and the ER. Taken together, these results are fully consistent with previous gene-based studies, which have shown that about one-third of the mitochondrial proteins, mostly associated with regulatory, transport, or membrane functions, have evolved in response to the specific constraints presented by the compartmentalized eukaryotic cell (20–23).
Our results also provide an explanation for several rare cases of gene transfers of otherwise universal mitochondrial genes to the nuclear genome. As predicted by our hypothesis, the mitochondrial proteins show a reduced hydrophobicity of the first TMDs in the few species in which these genes have been successfully transferred in nature (45–47). For example, the first TMD of the nu-encoded Cox2 protein in legumes is less hydrophobic than the first TMD of the mt-encoded homolog (48). Experimental studies have confirmed that the mitochondrial variant of Cox2 could not be imported into mitochondria unless the first TMD was removed or if the sequence was changed to that of the less hydrophobic nu-encoded homolog (48) and when an exceptionally strong mitochondrial targeting peptide of 130 amino acids was added (49).
Simlarly, all three TMDs of the mt-encoded SdhC protein in Reclinomonas americana are highly hydrophobic, whereas the TMDs of the nu-encoded homologs in yeast, humans, and acanthamoeba are only marginally or not at all hydrophobic. Our findings also explain why the 3′-end of the cox2 gene could successfully be transferred to the nuclear genome in some green algae, whereas the 5′-end of the gene encoding the hydrophobic TMD remains situated in the mitochondrial genome.
Thus, successful transfers of mitochondrial genes for hydrophobic proteins have occurred in nature and have been achieved in the laboratory by modifications, such as: (i) reducing the hydrophobicity of the first TMD; (ii) reducing the length of the C-terminal sequence of the hydrophobic TMD; (iii) moving the hydrophobic TMD toward the C-terminal end; or (iv) by splitting the gene into two, leaving the 5′-end, which codes for the hydrophobic TMD in the mitochondrial genome. We suggest that the effect of these modifications is a reduced potential for recognition by SRP, enabling the protein to reach the mitochondrial destination.
However, arrest by SRP and subsequent mistargeting to the ER is unlikely to explain all of the peculiarities of mitochondrial genomes in individual species. For example, genes for SdhD, ATP1, ATP3, and ATP9 in the R. americana and other jakobid protists are too short, do not contain hydrophobic TMDs, or the C-terminal tail following the TMD segment is too short to allow arrest by SRP. These mitochondrial genomes also encode several ribosomal proteins, which serve key functions in ribosomal assembly and initial rRNA binding (50). Interestingly, a similar set of ribosomal proteins is also encoded by chloroplast genomes, indicative of convergent evolution in response to similar selective constraints. This finding suggests that there is a hierarchical order of mitochondrial gene loss and that the genes that encode hydrophobic TMDs are subjected to the strongest selective constraints and therefore the last to be lost or transferred to the nuclear genomes (48).
Chloroplasts have several membranes, making the import of membrane proteins in the photosynthetic apparatus to the chloroplast a more complex process than targeting membrane proteins to mitochondria. Like mitochondria, chloroplasts have a double-membrane envelope, but additionally chloroplasts contain internal membrane structures (thylakoids) where the two photosystems are embedded. To accommodate this extra layer of complexity, higher plants contain three different SRP targeting systems (51). One is the cytosolic SRP system for transport to the ER, whereas the other two SRP systems specifically mediate protein targeting to the thylakoids. One of the latter is a cotranslational SRP system that inserts plastid-encoded membrane proteins with hydrophobic TMDs in the thylakoid membrane (51). The other SRP system acts posttranslationally (51). Thus, neither of the plastid SRP systems would be able to transport proteins with hydrophobic TMDs to the thylakoid membrane if the genes were located in the nuclear genomes.
Secondary plastids found in photosynthetic eukaryotes other than plants, like algae, are even more complicated in their membrane structures. Compared with primary plastids that have a double-membrane envelope, secondary plastids are bound by up to four membranes, further complicating targeting and transport of proteins. However, the gene content in the secondary plastids of photosynthetic algae is largely the same as in the primary plastids, although dinoflagellates have exceptionally few chloroplast genes (52). In addition to 2 rRNA genes, dinoflagellates contain 12 genes for key proteins in the photosynthetic apparatus, located on plasmid-like minicircles of 2–4 kb. Consistent with our predictions, 8 of these 12 proteins are targets for recognition by the cytosolic SRP. The mitochondrial genomes of the dinoflagellates are also highly reduced and encode Cytochrome b and Cytochrome oxidase subunits, similarly prime targets for recognition by the cytosolic SRP.
Soluble nu-encoded chloroplast proteins are transported into the secondary plastids through the ER with the aid of protein translocases, like the TIC-TOC (53, 54). However, bulky hydrophobic proteins are unlikely to fold properly in a soluble environment and cannot be transported across several layers of membranes in their nonnative state, even with the aid of a series of translocases. Furthermore, when unfolded or misfolded these proteins would be extremely prone to degradation by proteases and, even worse, are likely to aggregate in deleterious manner for the cell. Transporting membrane proteins with multiple hydrophobic TMDs to the thylakoid membranes of the secondary plastids would be a major obstacle if nu-encoded. The retention of genes for membrane proteins with highly hydrophobic TMDs in the extremely reduced mitochondrial and chloroplast genomes of dinoflagellates is thus fully consistent with our hypothesis.
An alternative hypothesis is that chloroplasts need a genome to facilitate stoichiometry adjustments and ensure similar transcription rates in all chloroplasts within the cell (15, 55). A two-component regulatory system encoded by the nuclear genome has been identified that regulates the expression of plastid-encoded proteins of photosystems (PS) I and II (56). Here, a sensor kinase (CSK) phosphorylates itself as well as sigma factor 1 (SIG1) in response to increased oxidation levels, thereby repressing transcription form the PSI promoter, but leaving the promoter of PSII unaffected. A more reduced state of the plastoquinone pool inactivates CSK, after which SIG1 becomes dephosphorylated and the repression of PS1 gene transcription is removed.
Thus, selection against mistargeting to the ER and selection for redox regulation may both pose strong selective constraints on the retention of the chloroplast genome. The two theories are nonexclusive and there are ample of support for both in plastids. The evolution of specific targeting systems for plastid-encoded proteins and the extra layer of membranes in organisms with secondary plastids provide strong barriers against gene transfers to the nuclear genomes of these organisms. Currently, we can only speculate about which of these forces was the original driver for the maintenance of the chloroplast genomes. Suffice it to conclude that many selective constraints are operating on organelle genomes in modern organisms, including recently evolved regulatory circuits and transport systems, which have limited the loss and transfer of organelle genes.
However, other types organelles derived from aerobic mitochondria and photosynthetic chloroplasts have been identified in anaerobic and heterotrophic eukaryotic lineages (3). The mt-derived organelles belong to five different classes, of which aerobic mitochondria, anaerobic mitochondria, and hydrogen-producing mitochondria contain a genome. The genomes of anaerobic mitochondria contain genes for NADH dehydrogenase subunits, a few of which are on our list of potential targets for SRP, which could explain the retention of their genomes. Mitosomes and most hydrogenosomes lack a genome; this shows that the mitochondrial genome can be lost when the OXPHOS system is no longer required. Similarly, the photosynthetic apparatus has been lost independently in many nonphotosynthetic species, and these losses are often associated with the evolution of parasitism. The plastid genomes in these organisms are severely affected by the change in lifestyle, showing signs of rearrangements, losses, and pseudogenization. Notably, the nonphotosynthetic green algal Polytomella (57) and the parasitic flowering plant Rafflesia (58) seem to have lost their genomes. This finding suggests that the plastid genome can also be lost in the absence of selection on the photosynthetic apparatus.
On the clinical side, an important goal has been to develop methods to cure human mitochondrial genetic diseases. Because no genetic system is available for the manipulation of mammalian mitochondrial genomes, gene therapy by allotopic expression of mt-encoded proteins is an attractive alternative. However, mitochondrial proteins, such as Cox1 and Cytb, could not be imported to the mitochondrion, despite the insertion of very strong mt-targeting peptides (59, 60). To date, no study has provided unequivocal evidence for functional protein import and integration of these two proteins into mitochondria when their genes have been transferred to the nuclear genome (61). The results presented in this study suggest that SRP protein targeting presents a major barrier in allotopic expression of mitochondrial proteins. This insight can explain previous studies of allotopic gene expression and have important implications for curing mitochondrial genetic diseases.
In conclusion, highly hydrophobic membrane proteins in mitochondria are recognized by SRP according to our bionformatic predictions, and exported to the ER when expressed in the cytoplasm, as verified by our experimental studies. These results resolve the long-standing question about why aerobic mitochondria and photosynthetic chloroplasts need a distinct compartmental genome, by and large, although other factors may also be involved. En passant, we note that the results also imply that the SRP targeting system for transport of proteins to the ER was likely in place before the evolution of mitochondria.
Methods
SRP-prediction.
To predict if a protein is a target of SRP, we use a program called DGpred that calculates the free insertion energy (ΔG, kcal/mol) of a TMD (28). If a TMD gets a score of zero or less the TMD is considered to be hydrophobic and if it has a higher score it is considered to be marginally hydrophobic. If one TMD is considered to be hydrophobic and the length of that TMD and its tail is longer than 120 amino acids we predict it to be arrested by SRP.
Experimental Analyses.
Synthetic genes were constructed by LifeTechnologies based on the amino acid sequences of mitochondrial proteins following reverse-translation to nuclear codon usage. The synthetic genes were cloned into a plasmid after the addition of restriction sites, FLAG tags, and linker sequences. HeLa cells were transfected with the plasmids and subjected to immunofluorescence staining with antibodies.
Bioinformatic Analyses.
Transmembrane domains were predicted and their hydrophobicity was calculated (28). SCOP SF-domain assignments were obtained from the SUPERFAMILY (ver 1.75) database (32). Proteins for which SUPERFAMILY assignments were unavailable, SF-domain assignments were carried out as described in (32). To estimate ancestral domain content and to analyze domain flux during evolution, the most parsimonious scenarios of mitochondrial SF-domain content evolution were reconstructed by using generalized parsimony based ancestral-state reconstruction methods.
Supplementary Material
Acknowledgments
We thank Karl Dyrhage for assisting with data analyses. This work was supported by Swedish Research Council Grant 621-2011-4669 (to S.G.E.A.) and the Knut and Alice Wallenberg Foundation Grant 2011.148 (to S.G.E.A.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. P.J.K. is a guest editor invited by the Editorial Board.
1P.B. and A.H. contributed equally to this work.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421372112/-/DCSupplemental.
References
- 1.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283(5407):1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW. Mitochondrial genes on the move. Nature. 2000;408(6810):302–303, 305. doi: 10.1038/35042663. [DOI] [PubMed] [Google Scholar]
- 3.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Recurring themes, but significant differences at the extremes. Proc Natl Acad Sci USA. 2015;112:10177–10184. doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MW. Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci USA. 2015;112:10133–10138. doi: 10.1073/pnas.1421379112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387(6632):493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 6.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5(2):418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park CB, Larsson NG. Mitochondrial DNA mutations in disease and aging. J Cell Biol. 2011;193(5):809–818. doi: 10.1083/jcb.201010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams KL, Daley DO, Qiu YL, Whelan J, Palmer JD. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature. 2000;408(6810):354–357. doi: 10.1038/35042567. [DOI] [PubMed] [Google Scholar]
- 9.Stupar RM, et al. Complex mtDNA constitutes an approximate 620-kb insertion on Arabidopsis thaliana chromosome 2: Implication of potential sequencing errors caused by large-unit repeats. Proc Natl Acad Sci USA. 2001;98(9):5099–5103. doi: 10.1073/pnas.091110398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazkani-Covo E, Zeller RM, Martin W. Molecular poltergeists: Mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6(2):e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer JD. Organelle genomes: Going, going, gone! Science. 1997;275(5301):790–791. doi: 10.1126/science.275.5301.790. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs HT. Structural similarities between a mitochondrially encoded polypeptide and a family of prokaryotic respiratory toxins involved in plasmid maintenance suggest a novel mechanism for the evolutionary maintenance of mitochondrial DNA. J Mol Evol. 1991;32(4):333–339. doi: 10.1007/BF02102192. [DOI] [PubMed] [Google Scholar]
- 13.Popot JL, de Vitry C. On the microassembly of integral membrane proteins. Annu Rev Biophys Biophys Chem. 1990;19:369–403. doi: 10.1146/annurev.bb.19.060190.002101. [DOI] [PubMed] [Google Scholar]
- 14.Allen JF. Why chloroplasts and mitochondria contain genomes. Comp Funct Genomics. 2003;4(1):31–36. doi: 10.1002/cfg.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol. 1993;165(4):609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- 16.Race HL, Herrmann RG, Martin W. Why have organelles retained genomes? Trends Genet. 1999;15(9):364–370. doi: 10.1016/s0168-9525(99)01766-7. [DOI] [PubMed] [Google Scholar]
- 17.de Grey ADJN. Forces maintaining organellar genomes: Is any as strong as genetic code disparity or hydrophobicity? BioEssays. 2005;27(4):436–446. doi: 10.1002/bies.20209. [DOI] [PubMed] [Google Scholar]
- 18.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 19.Amiri H, Karlberg O, Andersson SGE. Deep origin of plastid/parasite ATP/ADP translocases. J Mol Evol. 2003;56(2):137–150. doi: 10.1007/s00239-002-2387-0. [DOI] [PubMed] [Google Scholar]
- 20.Kurland CG, Andersson SGE. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64(4):786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlberg O, Canbäck B, Kurland CG, Andersson SGE. The dual origin of the yeast mitochondrial proteome. Yeast. 2000;17(3):170–187. doi: 10.1002/1097-0061(20000930)17:3<170::AID-YEA25>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gabaldón T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301(5633):609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- 23.Huynen MA, Duarte I, Szklarczyk R. Loss, replacement and gain of proteins at the origin of the mitochondria. Biochim Biophys Acta. 2013;1827(2):224–231. doi: 10.1016/j.bbabio.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 24.von Heijne G. Why mitochondria need a genome. FEBS Lett. 1986;198(1):1–4. doi: 10.1016/0014-5793(86)81172-3. [DOI] [PubMed] [Google Scholar]
- 25.Noriega TR, Chen J, Walter P, Puglisi JD. Real-time observation of signal recognition particle binding to actively translating ribosomes. eLife. 2014;3:1–12. doi: 10.7554/eLife.04418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lakkaraju AK, et al. Efficient secretion of small proteins in mammalian cells relies on Sec62-dependent posttranslational translocation. Mol Biol Cell. 2012;23(14):2712–2722. doi: 10.1091/mbc.E12-03-0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reithinger JH, et al. A short C-terminal tail prevents mis-targeting of hydrophobic mitochondrial membrane proteins to the ER. FEBS Lett. 2013;587(21):3480–3486. doi: 10.1016/j.febslet.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 28.Hessa T, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450(7172):1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 29.Smith AC, Blackshaw JA, Robinson AJ. MitoMiner: A data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 2012;40(Database issue):D1160–D1167. doi: 10.1093/nar/gkr1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aldridge C, Cain P, Robinson C. Protein transport in organelles: Protein transport into and across the thylakoid membrane. FEBS J. 2009;276(5):1177–1186. doi: 10.1111/j.1742-4658.2009.06875.x. [DOI] [PubMed] [Google Scholar]
- 31.Moffat K, et al. Effects of foot-and-mouth disease virus nonstructural proteins on the structure and function of the early secretory pathway: 2BC but not 3A blocks endoplasmic reticulum-to-Golgi transport. J Virol. 2005;79(7):4382–4395. doi: 10.1128/JVI.79.7.4382-4395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gabaldón T, Koonin EV. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 2013;14(5):360–366. doi: 10.1038/nrg3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 34.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol. 2001;313(4):903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 35.Kmita K, Zickermann V. Accessory subunits of mitochondrial complex I. Biochem Soc Trans. 2013;41(5):1272–1279. doi: 10.1042/BST20130091. [DOI] [PubMed] [Google Scholar]
- 36.He D, et al. An alternative root for the eukaryote tree of life. Curr Biol. 2014;24(4):465–470. doi: 10.1016/j.cub.2014.01.036. [DOI] [PubMed] [Google Scholar]
- 37.Kmita K, et al. Accessory NUMM (NDUFS6) subunit harbors a Zn-binding site and is essential for biogenesis of mitochondrial complex I. Proc Natl Acad Sci USA. 2015;112(18):5685–5690. doi: 10.1073/pnas.1424353112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannan S, Rogozin IB, Koonin EV. MitoCOGs: Clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol Biol. 2014;14:237. doi: 10.1186/s12862-014-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zickermann V, Angerer H, Ding MG, Nübel E, Brandt U. Small single transmembrane domain (STMD) proteins organize the hydrophobic subunits of large membrane protein complexes. FEBS Lett. 2010;584(12):2516–2525. doi: 10.1016/j.febslet.2010.04.021. [DOI] [PubMed] [Google Scholar]
- 40.Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35(14):4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S. On the last common ancestor and early evolution of eukaryotes: Reconstructing the history of mitochondrial ribosomes. Res Microbiol. 2011;162(1):53–70. doi: 10.1016/j.resmic.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Cavdar Koc E, et al. A new face on apoptosis: Death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492(1-2):166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 43.O’Brien TW, O’Brien BJ, Norman RA. Nuclear MRP genes and mitochondrial disease. Gene. 2005;354:147–151. doi: 10.1016/j.gene.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki E, Kida Y, Mihara K, Sakaguchi M. Switching the sorting mode of membrane proteins from cotranslational endoplasmic reticulum targeting to posttranslational mitochondrial import. Mol Biol Cell. 2005;16(4):1788–1799. doi: 10.1091/mbc.E04-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Funes S, et al. The typically mitochondrial DNA-encoded ATP6 subunit of the F1F0-ATPase is encoded by a nuclear gene in Chlamydomonas reinhardtii. J Biol Chem. 2002;277(8):6051–6058. doi: 10.1074/jbc.M109993200. [DOI] [PubMed] [Google Scholar]
- 46.Déquard-Chablat M, et al. Two nuclear life cycle-regulated genes encode interchangeable subunits c of mitochondrial ATP synthase in Podospora anserina. Mol Biol Evol. 2011;28(7):2063–2075. doi: 10.1093/molbev/msr025. [DOI] [PubMed] [Google Scholar]
- 47.Bietenhader M, et al. Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution. PLoS Genet. 2012;8(8):e1002876. doi: 10.1371/journal.pgen.1002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daley DO, Clifton R, Whelan J. Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA. 2002;99(16):10510–10515. doi: 10.1073/pnas.122354399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daley DO, et al. Gene transfer from mitochondrion to nucleus: Novel mechanisms for gene activation from Cox2. Plant J. 2002;30(1):11–21. doi: 10.1046/j.1365-313x.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 50.Maier U-G, et al. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol. 2013;5(12):2318–2329. doi: 10.1093/gbe/evt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Träger C, et al. Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: The signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell. 2012;24(12):4819–4836. doi: 10.1105/tpc.112.102996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koumandou VL, Nisbet RE, Barbrook AC, Howe CJ. Dinoflagellate chloroplasts—Where have all the genes gone? Trends Genet. 2004;20(5):261–267. doi: 10.1016/j.tig.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 53.Kunau W-H, Agne B, Girzalsky W. The diversity of organelle protein import mechanisms. Trends Cell Biol. 2001;11(9):358–361. doi: 10.1016/s0962-8924(01)02083-9. [DOI] [PubMed] [Google Scholar]
- 54.Chaal BK, Green BR. Protein import pathways in ‘complex’ chloroplasts derived from secondary endosymbiosis involving a red algal ancestor. Plant Mol Biol. 2005;57(3):333–342. doi: 10.1007/s11103-004-7848-y. [DOI] [PubMed] [Google Scholar]
- 55.Allen JF, de Paula WB, Puthiyaveetil S, Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011;16(12):645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 56.Puthiyaveetil S, Ibrahim IM, Allen JF. Evolutionary rewiring: A modified prokaryotic gene-regulatory pathway in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 2013;368(1622):20120260. doi: 10.1098/rstb.2012.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164(4):1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31(4):793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Claros MG, et al. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem. 1995;228(3):762–771. [PubMed] [Google Scholar]
- 60.Oca-Cossio J, Kenyon L, Hao H, Moraes CT. Limitations of allotopic expression of mitochondrial genes in mammalian cells. Genetics. 2003;165(2):707–720. doi: 10.1093/genetics/165.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perales-Clemente E, Fernández-Silva P, Acín-Pérez R, Pérez-Martos A, Enríquez JA. Allotopic expression of mitochondrial-encoded genes in mammals: achieved goal, undemonstrated mechanism or impossible task? Nucleic Acids Res. 2011;39(1):225–234. doi: 10.1093/nar/gkq769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.