Significance
A key problem in biology is whether the same processes underlie morphological variation between and within species. Here, we show that the causes of leaf shape diversity at these two evolutionary scales can be divergent. Some species have simple leaves, whereas others bear complex leaves comprising leaflets. Previous work indicated that these interspecific differences result mostly from variation in local tissue growth and patterning. Now we find that a different process, age-dependent shape progression, underlies within-species variation in complex leaf morphology. Specifically, in plants with accelerated aging and early flowering, leaves progress to adult shapes with more leaflets, faster than their slower-aging counterparts. This mechanism coordinates leaf development with reproductive timing and may influence resource allocation to seeds.
Keywords: Cardamine hirsuta, natural variation, compound leaf, heterochrony, Flowering Locus C
Abstract
A key problem in biology is whether the same processes underlie morphological variation between and within species. Here, by using plant leaves as an example, we show that the causes of diversity at these two evolutionary scales can be divergent. Some species like the model plant Arabidopsis thaliana have simple leaves, whereas others like the A. thaliana relative Cardamine hirsuta bear complex leaves comprising leaflets. Previous work has shown that these interspecific differences result mostly from variation in local tissue growth and patterning. Now, by cloning and characterizing a quantitative trait locus (QTL) for C. hirsuta leaf shape, we find that a different process, age-dependent progression of leaf form, underlies variation in this trait within species. This QTL effect is caused by cis-regulatory variation in the floral repressor ChFLC, such that genotypes with low-expressing ChFLC alleles show both early flowering and accelerated age-dependent changes in leaf form, including faster leaflet production. We provide evidence that this mechanism coordinates leaf development with reproductive timing and may help to optimize resource allocation to the next generation.
Leaves of seed plants present an attractive model to address the genetic basis for morphological diversity at different scales because they show considerable variation between and within species, and their morphology also differs according to developmental age in a phenomenon known as heteroblasty (1) (Fig. 1). Leaves can be classified into two broad morphological classes: simple, where the blade is entire, or dissected (also referred to as compound), where the blade comprises individual leaflets (Fig. 1A). Both simple and dissected leaves emerge from a pluripotent structure called the shoot apical meristem. Previous work has identified two processes that underlie such interspecies diversification of leaf shape. The first is the generation of lateral cell proliferation axes that give rise to leaflets. This process typically involves reactivation of meristem genes in leaves such as class I Knotted1-like homeobox (KNOX1) and CUP-SHAPED COTYLEDON (CUC) genes, which influence the patterning of peaks of auxin activity that are required for leaflet formation (2–6). The second is the action of local growth repressors at the flanks of emerging leaflet primordia that promote leaflet separation. This process involves the leaf-specific homeobox gene REDUCED COMPLEXITY (RCO) (7). Furthermore, in dissected leafed species, leaf complexity is regulated by the activity of TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) genes, which modulate the competence of the leaf margin to respond to organogenic signals (8, 9).
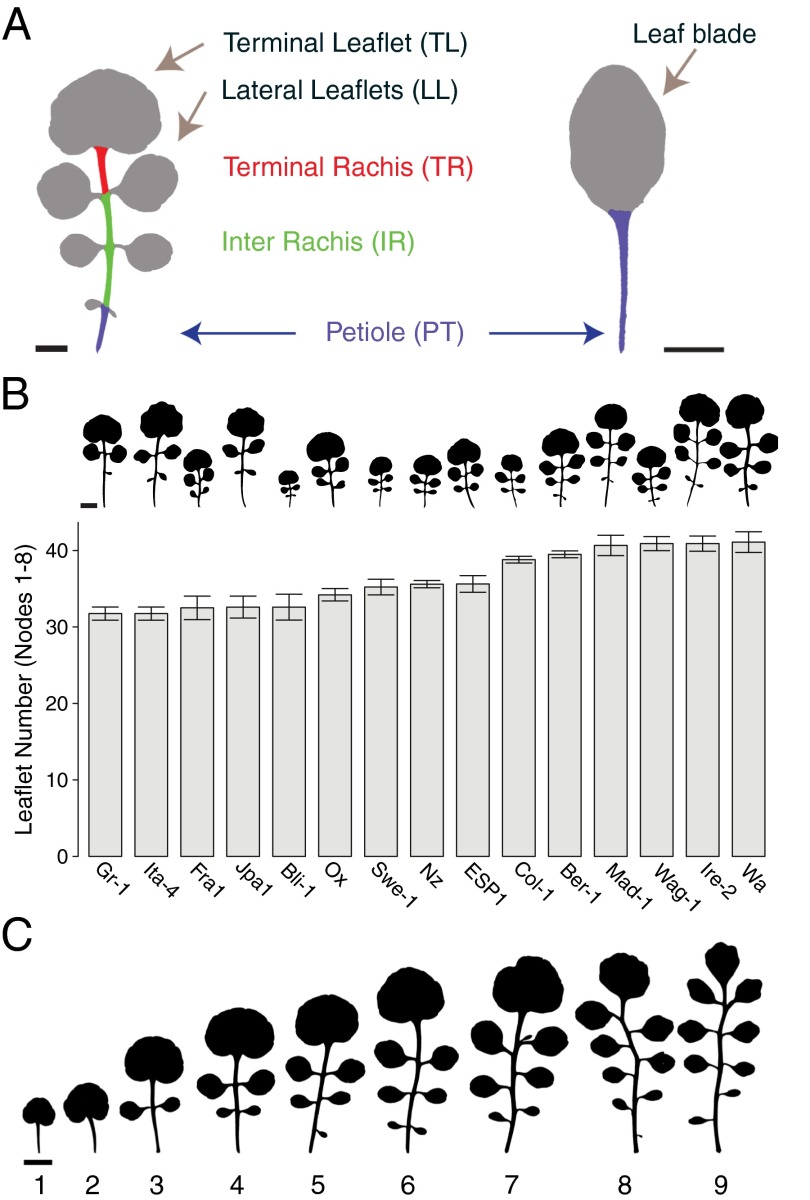
Fig. 1.
Morphological diversity of leaves across different scales. (A) Morphology of C. hirsuta (Ox) and A. thaliana (Col-0) rosette leaves; different parts of the leaf are indicated. (B, Upper) Silhouettes of the fifth rosette leaf illustrate diversity in leaf morphology of natural C. hirsuta strains. (B, Lower) Quantification of total leaflet number in the first eight rosette leaves is shown in the bar chart. Data are reported as means ± SD. (C) Silhouettes of the first nine rosette leaves from a typical heteroblastic series in C. hirsuta (Ox).
Results and Discussion
Despite progress in understanding the genetic pathways controlling leaflet development and leaf shape diversity between species (10, 11), their contribution to intraspecific differences in leaf morphology remains unclear. To study this problem, we exploited natural variation in leaf form of Cardamine hirsuta (Fig. 1B), which shows marked age-dependent variation (12) (Fig. 1C and Fig. S1 A and B). We performed quantitative trait locus (QTL) analysis in an F8 recombinant inbred line (RIL) mapping population descended from a cross between the Oxford (Ox) and Washington (Wa) strains (13) for the following traits: leaflet number; leaflet shape, quantified by extended eigenshape analysis (14); and leaf size (Fig. 2A, Fig. S1 C–M, and Table S1). Notably, no QTL for the analyzed leaf traits mapped to loci known or thought to underlie interspecific differences in leaf shape between simple and dissected leafed species, such as the KNOX1 genes SHOOT MERISTEMLESS (STM) and BREVIPEDICELLUS (BP) (6, 15), RCO (7, 16), TCP4 (8, 9), CUC3, or the microRNA MIR164A and its targets CUC1–2 (5, 17, 18) (Fig. 2A). Therefore, the genetic basis for diversity in leaf form between and within species may be distinct in this instance.
Fig. S1.
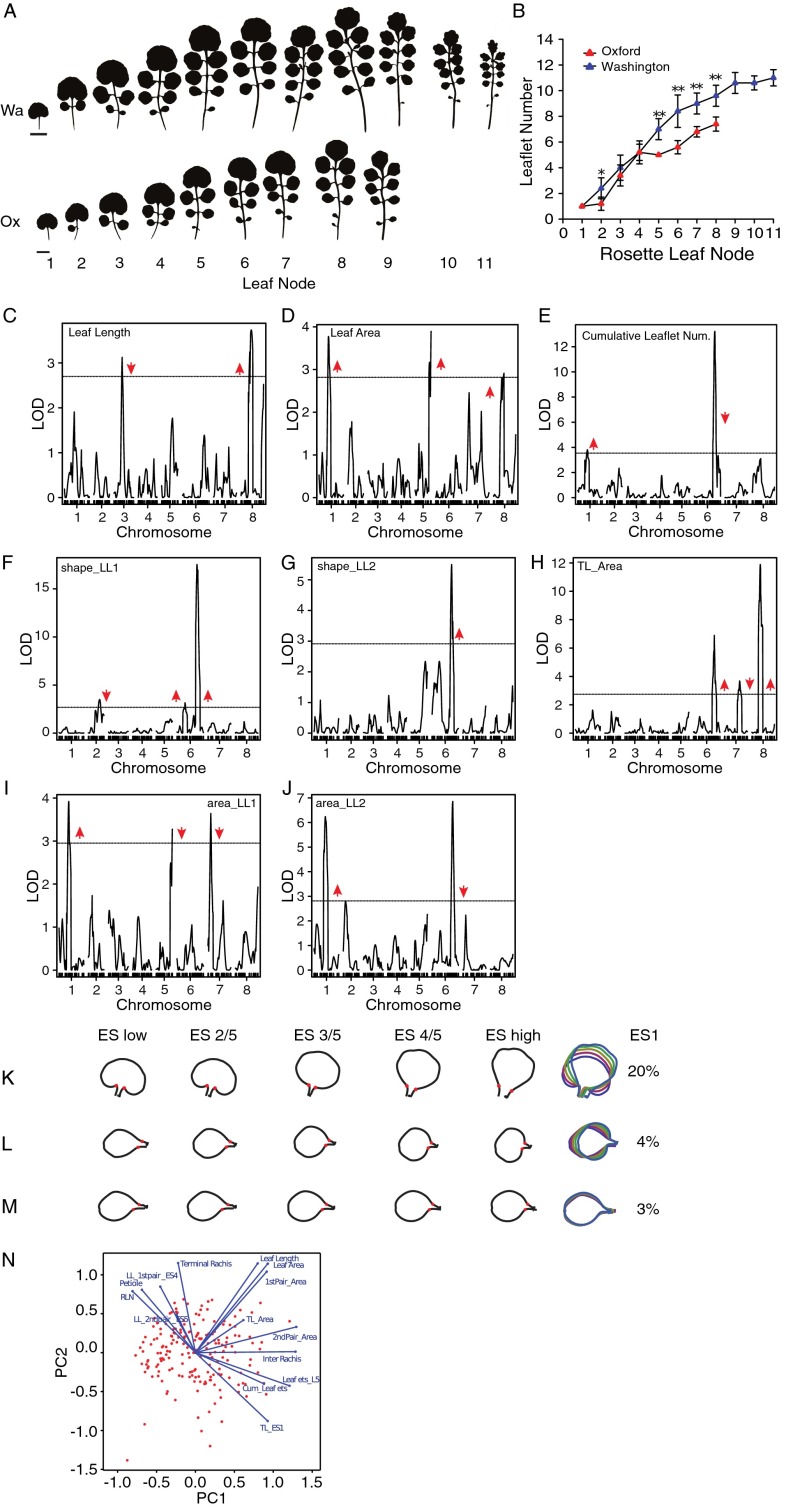
Further genetic and phenotypic characterization of the effects of QTL-LG6 on the heteroblastic progression. (A) Heteroblastic series of Wa (Upper) and Ox (Lower) parental strains. (B) Leaflet number quantification of the Ox and Wa parental strains along the heteroblastic series. Mean values are presented with SDs. Comparison of means was performed with a Student’s t test. *P < 0.05; **P < 0.01. (C–E) In addition to traits discussed in Fig. 2A, we performed QTL analysis for leaf length (C) and leaf area (D) of the fifth rosette leaf and for cumulative leaflet number produced by the first seven rosette leaves (E). (F–J) For the fifth rosette leaf, we also mapped QTL for the eigenscores ES1 of the first (F) and second (G) leaflet pairs, the area of the terminal leaflet (H), and the first (I) and second (J) leaflet pair. Horizontal black lines indicate the significance threshold corresponding to P = 0.05. Red arrowheads show the direction of the QTL effect and point upward or downward when the Wa allele at the QTL increases or decreases the trait value, respectively. (K–M) Shape models describing morphological variation along the ES1 of the terminal leaflet (K) and the first (L) and second (M) pairs of leaflets generated by phenotyping leaflet shape of rosette leaf 5 in the Ox × Wa F8 RIL population. The variance explained by ES1 is shown for each leaflet dataset. (N) Diagnostic biplot of the first two principal components generated by multivariate analysis of the 14 traits quantified in the RILs. Blue vectors represent traits, and red dots indicate individual RILs. The plot reveals how traits are correlated—e.g., blue vectors closest together or 180° apart represent traits that are most closely correlated; there is no correlation between traits if the angle between traits equals 90°.
Fig. 2.
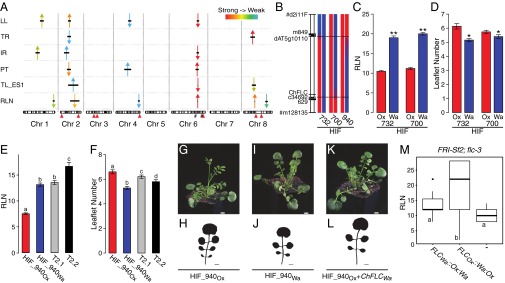
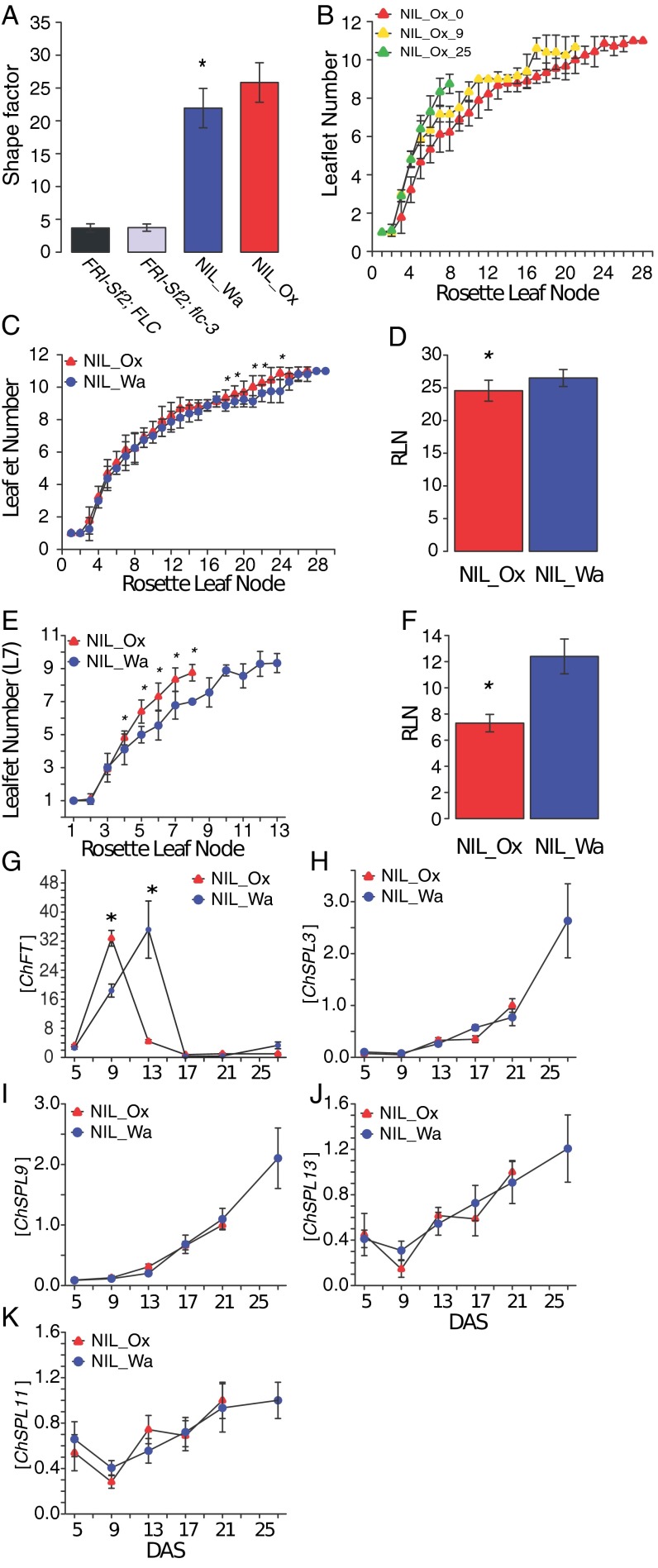
Natural variation in the regulatory sequences of ChFLC underlies phenotypic diversity in leaf morphology and flowering time in C. hirsuta. (A) Schematic representation of QTL mapped in Ox × Wa RILs using composite interval mapping for six traits. Shown from top to bottom are leaflet number (LL), terminal rachis length (TR), inter rachis length (IR), petiole length (PT), terminal leaflet shape (TL-ES1), and rosette leaf number (RLN). Leaf traits were quantified on the fifth rosette leaf. Horizontal black lines indicate the 1.5 logarithm of odds support interval for QTL. The magnitude of QTL effects is indicated by a color gradient ranging from red (strong effect) to light blue (weak effect). Arrowheads indicate the direction of the QTL effect by pointing either upward or downward according to whether the Wa allele at the QTL increases or decreases the trait value when substituted for the Ox allele. Red arrowheads below the chromosomes indicate the position of characterized leaf patterning genes. From left to right, these are: STM, CUC3, CUC1, TCP4, MIR164A, RCO, BP, and CUC2. Hashtags on chromosome 6 indicate markers used to validate QTL-LG6. (B) Schematic diagram of QTL fine mapping using HIFs. Genetic markers are shown on the left, and Ox (red) and Wa (blue) genotypes are indicated on chromosomes of three different HIFs. Black dotted lines mark the boundaries of the fine mapped region. (C and D) Validation of ChFLC allelic effects using HIFs; the Ox allele (red) reduces RLN (C) and increases leaflet number (D) compared with the Wa allele (blue) (n = 15). *P < 0.05; **P < 0.01 (Student’s t test). (E and F) Transgenic complementation of HIF940 carrying the ChFLCOx allele (HIF940Ox; red bar) with the ChFLCWa genomic locus (two independent transgenic lines shown; gray and black bars). (E) RLN is fully complemented to HIF940Wa value (blue bar) in T2.1 (gray bar) and further increased in T2.2 (black bar). (F) Leaflet number is partially complemented to HIF940Wa value (blue bar) in T2.1 (gray bar) and T2.2 (black bar). Significant differences between genotypes determined by ANOVA and post hoc Tukey’s test are indicated by letters; P < 0.05 (n = 15 per genotype). Leaflet number was quantified in rosette leaf five. Data are reported as means ± SD. (G–L) Photographs of plants (G, I, and K) and silhouettes of rosette leaf five (H, J, and L) of transgenic lines and HIFs described in E and F. (M) Boxplot of RLN in A. thaliana FRI-Sf2;flc-3 untransformed (−) or transformed with constructs containing the ChFLC genomic locus from Wa and Ox where the first intron is exchanged (FLCWa::Ox:Wa and FLCOx::Wa:Ox). Significant differences between genotypes determined by ANOVA and post hoc Tukey’s test are indicated by letters; P < 0.01 (n = 30 per genotype). (Scale bars: 1 cm.)
Table S1.
Summary table of the QTL analysis performed in the Ox × Wa F8 RIL mapping population, including heritabilities, map positions, and allelic effects for a single allele substitution of Ox alleles with Wa alleles
| Trait | Heritability, % | QTL | Linkage group | 1.5 LOD interval | % Variance | Additive allele effect |
| Rosette leaf no. | 87 | RLN_1 | 1 | 128.51–137.48 | 3.06 | −0.34 |
| RLN_2 | 2 | 48.60–83.40 | 2.82 | −0.35 | ||
| RLN_3 | 4 | 96.56–105.07 | 1.73 | −0.25 | ||
| RLN_4 | 6 | 114.802–116.66 | 36.05 | 1.21 | ||
| RLN_5 | 8 | 42.46–48.19 | 26.75 | 1.02 | ||
| RLN_6 | 8 | 94–103.8 | 2.79 | −0.32 | ||
| Leaflet no. | 55 | LL_1 | 1 | 57.32–73.19 | 7.97 | 0.42 |
| LL_2 | 2 | 32.45–45.04 | 10.82 | 0.48 | ||
| LL_3 | 4 | 52.05–66.57 | 5.54 | −0.38 | ||
| LL_4 | 6 | 109.58–120.37 | 14.98 | −0.60 | ||
| Cum. leaflet no. | 54 | CL_1 | 1 | 36.61–64.80 | 7.4 | 0.76 |
| CL_2 | 6 | 109.6–116.66 | 21.8 | −1.36 | ||
| Terminal rachis | 45 | TR_1 | 2 | 29.16–45.04 | 6.93 | −0.93 |
| TR_2 | 8 | 28.97–49.02 | 13.33 | 1.36 | ||
| Inter rachis | 43 | IR_1 | 1 | 52.26–70.73 | 9.44 | 4.98 |
| IR_2 | 2 | 14.71–37.47 | 6.61 | 3.91 | ||
| IR_3 | 6 | 109.58–120.37 | 13.9 | −6.13 | ||
| Petiole | 53 | PT_1 | 2 | 27.36–45.04 | 7.86 | −3.12 |
| PT_2 | 4 | 36.22–66.57 | 6.86 | 2.97 | ||
| PT_3 | 6 | 105.5–120.37 | 6.9 | 3.25 | ||
| Terminal leaflet eigenshape | 73 | TLES1_1 | 2 | 32.45–78.17 | 5.02 | 0.06 |
| TLES1_2 | 6 | 114.8–116.66 | 14.79 | −0.13 | ||
| TLES1_3 | 8 | 35.24–53.20 | 6.39 | −0.08 | ||
| LL_1stpairEigenshape | 77 | LL1stES4_1 | 2 | 36.61–83.39 | 5.30 | −0.02 |
| LL1stES4_2 | 6 | 28.69–56.59 | 4.83 | 0.01 | ||
| LL1stES4_3 | 6 | 105.51–116.66 | 33.91 | 0.04 | ||
| LL_2ndpairEigenshape | 58 | LL2ndES5_1 | 6 | 101.59–113.71 | 13.04 | 0.02 |
| TL_Area | 51 | TL_A1 | 6 | 109.58–120.37 | 11.55 | 26.38 |
| TL_A2 | 7 | 60.40–101.13 | 4.15 | −18.70 | ||
| TL_A3 | 8 | 42.46–49.02 | 11.81 | 33.46 | ||
| LL_2ndpairArea | 51 | LL_2ndA1 | 1 | 43.83–70.73 | 7.94 | 6.86 |
| LL_2ndA2 | 5 | 74.23–90.66 | 6.66 | −6.20 | ||
| LL_2ndA3 | 7 | 2.74–20.75 | 7.31 | −6.55 | ||
| LL_3rdpairArea | 58 | LL_3rdA1 | 1 | 52.26–73.20 | 6.45 | 6.58 |
| LL_3rdA2 | 6 | 109.58–120.37 | 1.02 | −2.64 | ||
| Leaf length | 27 | Llength_1 | 3 | 36.9–54.4 | 0.0015 | −0.01 |
| Llength_2 | 8 | 42.5–124.97 | 10.83 | 0.78 | ||
| Leaf area | 37 | LA_1 | 1 | 43.83–64.81 | 0.31 | 0.031 |
| LA_2 | 5 | 74.23–90.66 | 0.0007 | 0.001 | ||
| LA_3 | 8 | 35.24–76.58 | 1.67 | 0.09 |
LOD, logarithm of odds.
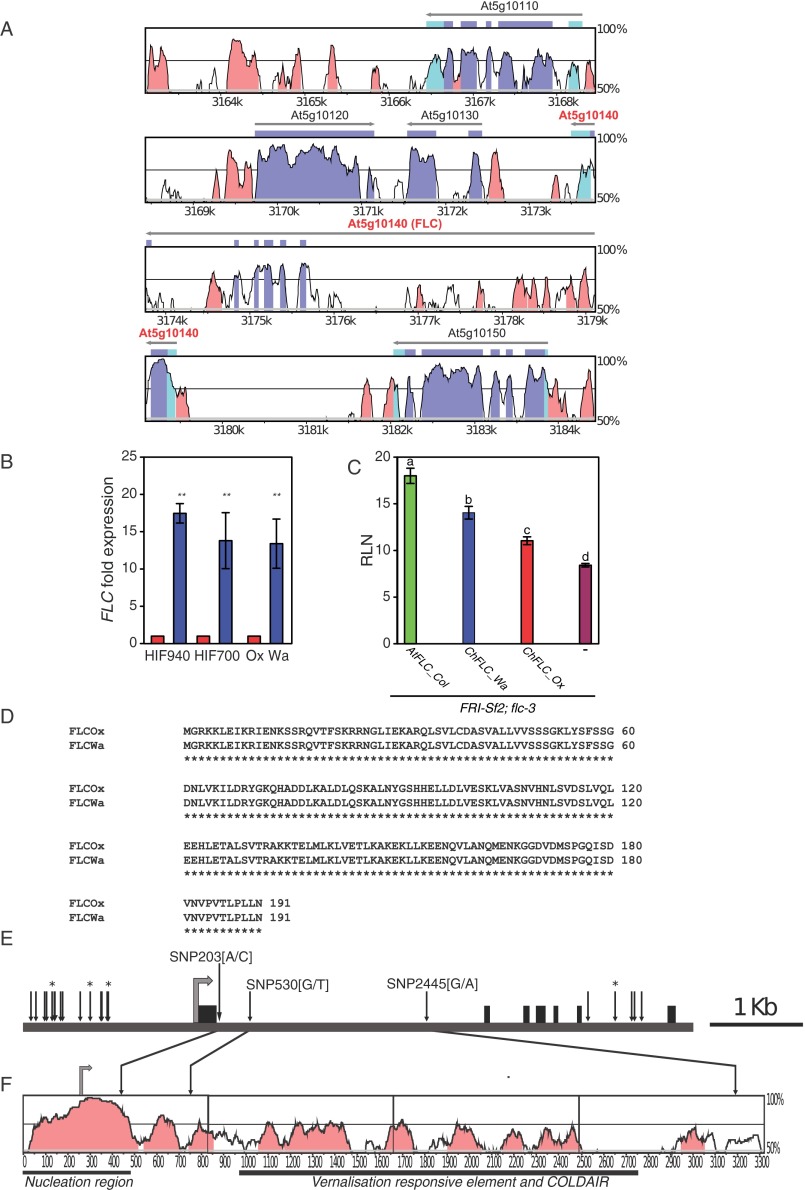
In the RIL population, phenotypic variation for leaf size, complexity, and leaflet shape correlated with flowering time, suggesting that these traits may be under common genetic control (Fig. S1N). Consistent with this result, we found that a single QTL on linkage group 6 (QTL-LG6) explained most of the phenotypic variation for leaflet number on leaf 5, as well as cumulatively over the first seven leaf nodes; leaflet shape; leaflet area; and also flowering time (Fig. 2A, Fig. S1 C–M, and Table S1). The direction of the allelic effects of this QTL on leaflet number were opposite from the difference between the founding strains because the Wa strain showed more leaflets than Ox (Fig. S1 A and B), whereas the Wa allele at QTL-LG6 reduced leaflet number. This allelic behavior is consistent with a genetic architecture of multiple loci with both negative and positive effects (19). We validated QTL-LG6 for the number of leaflets produced on the fifth rosette leaf and flowering time using heterogeneous inbred families (HIFs) (20, 21) and fine-mapped it to a 13-kb interval on chromosome 6 (Fig. 2 B–D), which contains five ORFs, including the C. hirsuta ortholog of FLOWERING LOCUS C (ChFLC) (Fig. S2A). FLC is a well-studied MCM1/AGAMOUS/DEFICIENS/SRF (MADS) box gene that represses flowering (22–24). We found that ChFLC expression was reduced in the HIF homozygous for the Ox allele (HIFOx), compared with those carrying the Wa allele (HIFWa) (Fig. S2B), suggesting that natural allelic variation at ChFLC underlies QTL-LG6 and pleiotropically influences leaf form and flowering time. For complementation analysis, we transformed HIFOx with the genomic ChFLCWa allele and found that the introduction of the transgene decreased leaflet number and increased flowering time in the direction predicted by the QTL analysis (Fig. 2 A and E–L). Similar allelic effects for flowering time were observed when we complemented the Arabidopsis thaliana flc-3 mutant [FRI-Sf2; flc-3 (24, 25)] with ChFLCWa and ChFLCOx alleles, confirming that ChFLCOx is a weaker allele compared with ChFLCWa (Fig. S2C). Because the amino acid sequences of both alleles are identical but their expression levels differed, we concluded that regulatory changes at ChFLC underlie the QTL-LG6 effect (Fig. S2D). Consistent with this view, analysis of transgenic FRI-Sf2 flc-3 plants harboring chimeric constructs between the ChFLC Wa and Ox alleles indicated that the delayed flowering conferred by the Wa allele is attributable to the first intron (Fig. 2M), which was previously shown to influence the expression of natural FLC alleles in A. thaliana (26–32). Only three SNPs in this intron differentiate the Ox from the Wa allele, making them strong candidates for contributing to QTL-LG6, one of which, SNP203, lies in the conserved nucleation region (Fig. S2 E and F and Table S2). In conclusion, natural allelic variation in the floral repressor ChFLC influences leaf morphology. Specifically, reduced expression of ChFLC results in both early flowering and increased leaflet number at leaf 5 and cumulatively over the first seven leaf nodes.
Fig. S2.
QTL-LG6 Validation and cloning of ChFLC. (A) Vista plot (genome.lbl.gov/vista/index.shtml) showing DNA sequence conservation of the C. hirsuta QTL interval within the A. thaliana genome around the FLC gene. The 19,801-bp C. hirsuta sequence that was aligned spanned 21,308-bp of the A. thaliana genome. Conserved sequences are indicated by color as follows: noncoding sequence (pink), 5′ and 3′ UTRs (light blue), and the coding sequence (purple). (B) ChFLC expression in segregating HIF940 and HIF700 and parental strains showing five visible rosette leaves is significantly higher in genotypes carrying the ChFLCWa allele (blue) compared with genotypes with the ChFLCOx allele (red). **P < 0.01 (Student’s t test). (C) Rosette leaf numbers (RLN) of transgenic A. thaliana FRISf2; flc-3 lines complemented with AtFLCCol0, ChFLCWa, and ChFLCOx alleles and untransformed controls (−) show that the ChFLCWa allele is stronger compared with ChFLCOx. Letters indicate significant differences between groups as indicated by ANOVA and post-hoc Tukey’s test (P < 0.01). Bar charts in B and C show the mean values and SDs. (D) Amino acid sequence alignment of ChFLCOx- and ChFLCWa-encoded proteins revealed no amino acid changes between alleles. (E) Diagram of the ChFLC locus. Black boxes indicate exons, and the gray arrow shows the translation start codon. Vertical arrows indicate the positions of polymorphisms between the Ox and the Wa allele. Arrows marked with * indicate InDels and those without SNPs (see Table S2 for further details). The position, relative to the adenine in the start codon, and nucleotide change are given of the three SNPs within the first intron. (F) Vista plot showing DNA sequence conservation between A. thaliana and C. hirsuta FLC genes, including 245 nt of the promoter, the first exon, and the first intron. The gray arrow indicates the FLC start codon, and the following functional regions of A. thaliana FLC intron 1 are underlined: nucleation region (27, 28), vernalisation responsive element (VRE; ref. 30), and COLDAIR long ncRNA (the transcription start site of COLDAIR is within the VRE region; ref. 29). Vertical arrows indicate the position of three SNPs that differentiate the Ox from the Wa allele in the ∼3-Kb fragment that is sufficient to recapitulate functional differences between the two ChFLC alleles. Note how SNP [A/C] at position 203 is located within a region of high-sequence homology with the nucleation region in A. thaliana where FLC silencing is initiated by the Polycomb Repressive Complex 2 (PHD-PRC2) during cold exposure.
Table S2.
The physical position of polymorphisms in ChFLC differentiating Ox from Wa
| Position | Pos.gene | Size | Ox | Wa |
| −1,840 | Promoter | 0 | A | G |
| −1,783 | Promoter | 0 | G | A |
| −1,691 | Promoter | 0 | A | T |
| −1,671 | Promoter | 0 | A | C |
| −1,614 | Promoter | 45 | — | TTAATATGTAAAATCAAACTTAGAACATTCAGAGATGCCTCTGAA |
| −1,584 | Promoter | 0 | T | A |
| −1,583 | Promoter | 0 | T | A |
| −1,523 | Promoter | 0 | T | A |
| −1,499 | Promoter | 0 | C | T |
| −1,305 | Promoter | 0 | G | A |
| −1,198 | Promoter | 0 | A | T |
| −1,085 | Promoter | −1 | T | — |
| −1,077 | Promoter | 0 | T | G |
| −1,021 | Promoter | 0 | C | A |
| −1,008 | Promoter | −1 | T | — |
| 203 | First intron | 0 | A | C |
| 530 | First intron | 0 | G | T |
| 2,445 | First intron | 0 | G | A |
| 4,186 | Sixth intron | 0 | T | A |
| 4,482 | Sixth intron | −1 | A | — |
| 4,658 | Sixth intron | 0 | G | T |
| 4,689 | Sixth intron | 0 | T | G |
| 4,771 | Sixth intron | 0 | C | A |
To understand how ChFLC influences leaf development, we focused on heterochrony (33, 34). FLC influences developmental timing in A. thaliana (35, 36); therefore, we hypothesized that its effect on leaf form in C. hirsuta might be heterochronic, such that ChFLC influences age-related changes in leaf development. C. hirsuta heteroblasty is characterized by pronounced but gradual age-dependent changes in leaflet number, area, and shape (Figs. 1C and 3 A and B; Fig. S3 A–E). The allelic state of ChFLC influenced this progression as follows: Leaflet number increased more rapidly after leaf 4 along the heteroblastic progression in early flowering HIFOx and reached the maximum leaflet number at an earlier node than HIFWa plants, with the maximum leaflet number being identical for both genotypes (Fig. 3C and Fig. S3F). Moreover, the shape of the terminal leaflet in early flowering HIFOx progressed more rapidly from a kidney-shaped to a wedge-shaped morphology, compared with late-flowering HIFWa (Fig. 3D and Fig. S3F). These results indicate that ChFLC affects leaf form by influencing the rate of age-dependent progression of development. The shape of the last rosette leaf before flowering (LLBF) differed between ChFLC genotypes, such that HIFOx had a more wedge-shaped terminal leaflet and higher perimeter-to-surface ratio for the whole leaf than HIFWa (Fig. S3 G and H). Leaf geometry also differed between ChFLC genotypes at other leaf nodes, which can be seen by comparing leaflet number between HIFOx and HIFWa leaves with equivalent terminal leaflet shapes (Fig. 3E). Specifically, we identified five nodes in HIFWa plants, where an identical terminal leaflet shape did not correspond to an identical leaflet number at any node in HIFOx; two nodes in HIFWa plants had terminal leaflet shapes unique to this ChFLC genotype and not observed in HIFOx (Fig. 3E). Finally, distinct combinations of leaflet number and terminal leaflet shape were specific to each ChFLC allele, such that each trait was a significant and independent predictor of ChFLC genotype in the HIFs (P < 0.05; Wald test; Fig. S3I). This finding highlights how distinct dissected leaf forms can be attained by combining separate contributing features—e.g., leaflet shape and number—which themselves show different rates of age-dependent variation (Fig. 3 C and D). These shape differences are not indirect effects of differences in the timing of leaf initiation, because initiation rate does not differ between the two ChFLC genotypes before flowering (Fig. 3F). In conclusion, the allelic state of ChFLC affects leaf morphology in three ways. First, leaflet shape and number progress to adult trait values more rapidly, and cumulative leaflet number over seven nodes is higher in plants carrying the Ox allele compared with the Wa allele. Second, the form of the LLBF differs between the two genotypes. Finally, different combinations of distinct age-dependent shape elements create distinct adult leaf geometries.
Fig. 3.
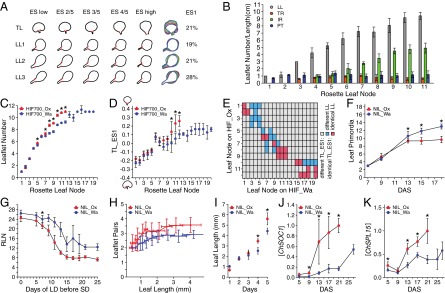
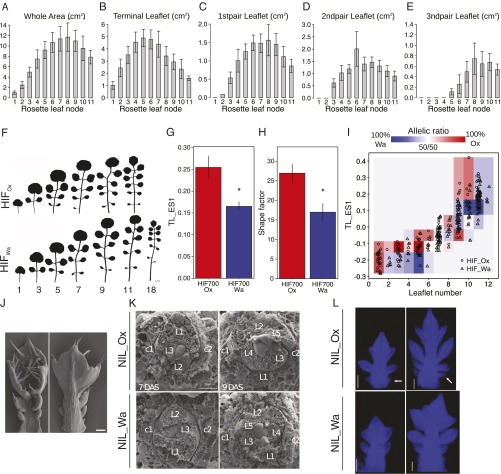
Heterochronic variation caused by allelic diversity at ChFLC underlies natural variation in C. hirsuta leaf morphology. (A and B) C. hirsuta heteroblasty is characterized by continuous changes in leaf shape and size. (A) Extended eigenshape analysis was performed on the terminal leaflet (TL) and first (LL1), second (LL2), and third (LL3) pairs of lateral leaflets along the heteroblastic series of Ox. Models describing shape variation along the first eigenshape axis (ES1) and the variance explained by ES1 are shown. (B) Quantification of leaflet number (LL), terminal rachis (TR), inter rachis (IR), and petiole (PT) length in successive rosette leaves of Ox. (C and D) Leaflet number (C) and terminal leaflet shape (TL-ES1; D) in successive rosette leaves of HIF700 segregating for ChFLC [n = 15 per genotype (C); n = 5 per genotype (D)]. (E) Comparison of leaflet number between nodes of HIFOx and HIFWa with identical terminal leaflet shape (TL_ES1). Pairs of nodes with different TL_ES1 are shown in gray, and those with identical TL_ES1 in either blue or red depending on whether the leaflet number differed significantly between these nodes or not, respectively. Results were obtained by analyzing HIF732 and HIF700 jointly. (F) Leaf primordia visible at the shoot apical meristem plotted against DAS (n = 15 per genotype). No significant differences in leaf initiation rate are observed before 13 DAS when ChFLCOx has flowered. (G) Photoperiod sensing shift experiment: rosette leaf number (RLN) of NILs plotted against the number of LD experienced before transfer to SD. Asterisk indicates the point on the x axis when both genotypes showed a significant response to floral induction (P < 0.05 by Student’s t test). (H) Average number of leaflet pairs of NILs plotted against a moving average of leaf length. (I) Length of the ninth rosette leaf measured for five consecutive days during early ontogenesis (G and H; n = 15 per genotype). (J and K) Expression levels of ChSOC1 (J) and ChSPL15 (K), measured by quantitative RT-PCR (qRT-PCR), increase significantly faster in NIL_Ox than NIL_Wa during development. Mean values and SDs are shown. *P < 0.01 (Student’s t test).
Fig. S3.
Quantification of heteroblasty in C. hirsuta and the effect of ChFLC on leaf shape. Area measurements of the whole leaf (A), the terminal leaflet (B), and the first (C), second (D), and third (E) pair of leaflets. Mean values with standard deviations are shown. (F) The pace of heteroblastic progression is increased in early flowering lines carrying the ChFLCOx allele. Heteroblastic series of HIF700_Ox (Top) and HIF700_Wa (Bottom). Rosette leaf number is indicated below leaves. (G) Terminal leaflet shape of the last rosette leaf to develop before flowering differs significantly between HIFs bearing either the Ox or the Wa ChFLC allele. Bar chart shows the mean values for TL_ES1, which is the eigenshape explaining most of the variation in shape of the terminal leaflet. Error bars indicate standard errors of the means; asterisk indicates that the difference is significant (P < 0.05 by Student's t test). (H) The last rosette leaves to develop before flowering on HIFs bearing the Ox allele have a higher shape factor [leaf perimeter2 / (leaf area * 4π)] than those bearing the Wa allele. Bar chart shows mean differences in shape factor; error bars show standard errors of the means; asterisk indicates that the difference is significant (P < 0.05 by Student's t test). (I) Relative abundance of leaves from HIFs 700 and 732 in the shape space of leaflet number and terminal leaflet shape. All samples were divided into 6 bins of TL_ES1 for each possible number of leaflets such that each bin contained as close as possible to an equal number of leaves. Colored bins indicate a higher relative ratio of leaves from Ox (red) and Wa (blue). The intensity of the color indicates how strong the respective allele dominated the ratio over the other allele. For example, leaves with 5 leaflets from plants with the Wa allele tended to have a more juvenile shape than those of plants with the Ox allele, which is shown by the dark blue color in the bin for low TL_ES1, while the bins above it are red. Actual values of TL_ES1 are plotted against leaflet number for plants with the Ox allele (circles) and the Wa allele (triangles); some jitter was applied to separate the data points in the figure. The HIFs 700 and 732 were not found to differ from each other for these traits so they were analyzed together [P < 0.05 by Student's t test (TL_ES1) or Kruskal–Wallis test (leaflet number)]. (J) Scanning electron micrographs (SEM) show rosette leaf 5 lacking abaxial trichomes. (Scale bar: 100 μm.) (K) SEM of vegetative apices of NIL_Ox and NIL_Wa at 7 and 9 DAS revealed the initiation of leaf 3 and 5, respectively. Stipules are indicated with a *. Cotyledons are indicated with the letter C. Rosette leaves are indicated with the letter L. (Scale bar: 100 μm.) (L) Confocal laser scanning micrographs of rosette leaf 9 show that more leaflets are initiated per unit leaf length in the early flowering NIL_Ox compared to NIL_Wa. White arrows point to developing leaflets in NIL_Ox. (Scale bar: 100 μm.)
To understand how the allelic status of ChFLC influences heteroblasty, we studied phase transitions. Vegetative shoot development progresses from a juvenile to an adult vegetative phase, and the transition is closely related to the competence to respond to flower-inducing signals (37, 38). The timing of this transition and the relative length of the juvenile and adult vegetative phases often influence age-dependent changes in leaf form (39). On this basis, we investigated whether ChFLC affects leaf shape by modulating the timing of the juvenile-to-adult phase change in C. hirsuta. The first leaf that produces abaxial trichomes marks the time point of the juvenile-to-adult transition in A. thaliana (40). C. hirsuta leaves lack abaxial trichomes, and we could not identify another obvious morphological feature that discretely marks this transition (Fig. S3 A–E and J). Therefore, we identified the time point of the juvenile-to-adult phase change in C. hirsuta near isogenic lines (NILs) at the ChFLC locus by performing a photoperiod-shift experiment, where we used failure to respond to photoperiodic floral induction to identify juvenile plants (SI Materials and Methods) (38). By these criteria, we placed the end of juvenility between 5 and 7 DAS, when both genotypes responded to the photoperiodic induction of flowering (Fig. 3G). At this stage, both genotypes had produced on average three leaves at the shoot apical meristem (Fig. 3F and Fig. S3K). Therefore, allelic variation at ChFLC does not affect the onset of the juvenile-to-adult transition but, rather, influences the rate at which adult features are acquired. Growth analysis of developing leaves revealed that leaves of the early flowering NIL_Ox expanded faster and produced more leaflets per unit leaf length than NIL_Wa (Fig. 3 H and I and Fig. S3L). These observations indicate an acceleration of the leaf development program, including leaflet initiation, in the early flowering genotypes. From these results, we conclude that natural variation in a heterochronic pathway, which affects the length of the adult vegetative phase, influences heteroblastic progression in C. hirsuta by modulating the rate at which leaves mature and produce leaflets. Although FLC affects heteroblasty in A. thaliana (36), quantitative shape comparisons show that the consequences of heterochronic variation on leaf geometry are more pronounced in the complex leaf of C. hirsuta (Fig. S4A).
Fig. S4.
Allelic variation at ChFLC affects the coordination between leaf morphology and flowering time in C. hirsuta. The consequences of FLC variation on leaf morphology are more pronounced in C. hirsuta than A. thaliana. (A) The shape factor [perimeter2/(4π*area)] for leaf 8 is shown to compare the effects of distinct FLC alleles on leaf shape in C. hirsuta and A. thaliana. (B) Heteroblastic increase of leaflet number in NIL_Ox plants grown in LD for 0 d (red), 9 d (blue), and 25 d (green) and then transferred to SD (n = 15 per genotype). Leaflet number of all rosette leaves produced before flowering is shown. (C and D) Leaflet number (C) and total rosette leaf number (D) of NILs grown in SD. (E and F) Leaflet number (E) and total rosette leaf number (F) of NILs grown in LD. (G and K) qRT-PCR analysis of the expression levels of ChFT (G), ChSPL3 (H), ChSPL9 (I), ChSPL13 (J), and ChSPL11 (K) along a developmental time series. *P < 0.01 (Student’s t test). Data are represented as mean values with SD.
Next, we tested whether accelerated acquisition of adult leaf morphology in early flowering genotypes reflected a developmental feedback of the flowering state to developing vegetative leaves. We hypothesized that such a feedback might exist because accelerated flowering in response to inductive photoperiod also increased the rate of heteroblastic progression, in a manner reminiscent of the early flowering ChFLCOx allele (Fig. S4B). We observed that the ChFLC allelic status still affected leaflet number and flowering time, even when plants were grown under short-day conditions that delay flowering in C. hirsuta. However, in this case, the effect was shifted to later leaves in the heteroblastic series (Fig. S4 C and D). That the effect of ChFLC on leaf complexity cannot be uncoupled from flowering, even in noninductive conditions, suggests that ChFLC influences leaf shape, concomitant with its effect on the floral transition. Consistent with this view, we first detected differences in the expression of the floral integrators and likely targets of FLC, ChFLOWERING LOCUS T (FT) and ChSUPPRESSOR OF OVEREXPRESSION OF CO 1 (ChSOC1) (41, 42), between NIL_Ox and NIL_Wa at 9 days after sowing (DAS) (Fig. 3J and Fig. S4G). At this stage, long-day-grown plants have initiated four or five leaves (Fig. 3F and Fig. S3K), which is coincident with the first phenotypic differences in the heteroblastic progression between the two genotypes (Fig. 3C and Fig. S4 E and F). ChSQUAMOSA PROMOTER BINDING PROTEIN-LIKE 15 (ChSPL15) is another potential target of ChFLC (35), for which transcript levels rise faster in NIL_Ox than in NIL_Wa (Fig. 3K and Fig. S4 H–K). SPL15 contributes to both flowering- and vegetative-phase transitions in A. thaliana (38, 43), which makes ChSPL15 a strong candidate for mediating the effects of ChFLC on C. hirsuta leaf shape.
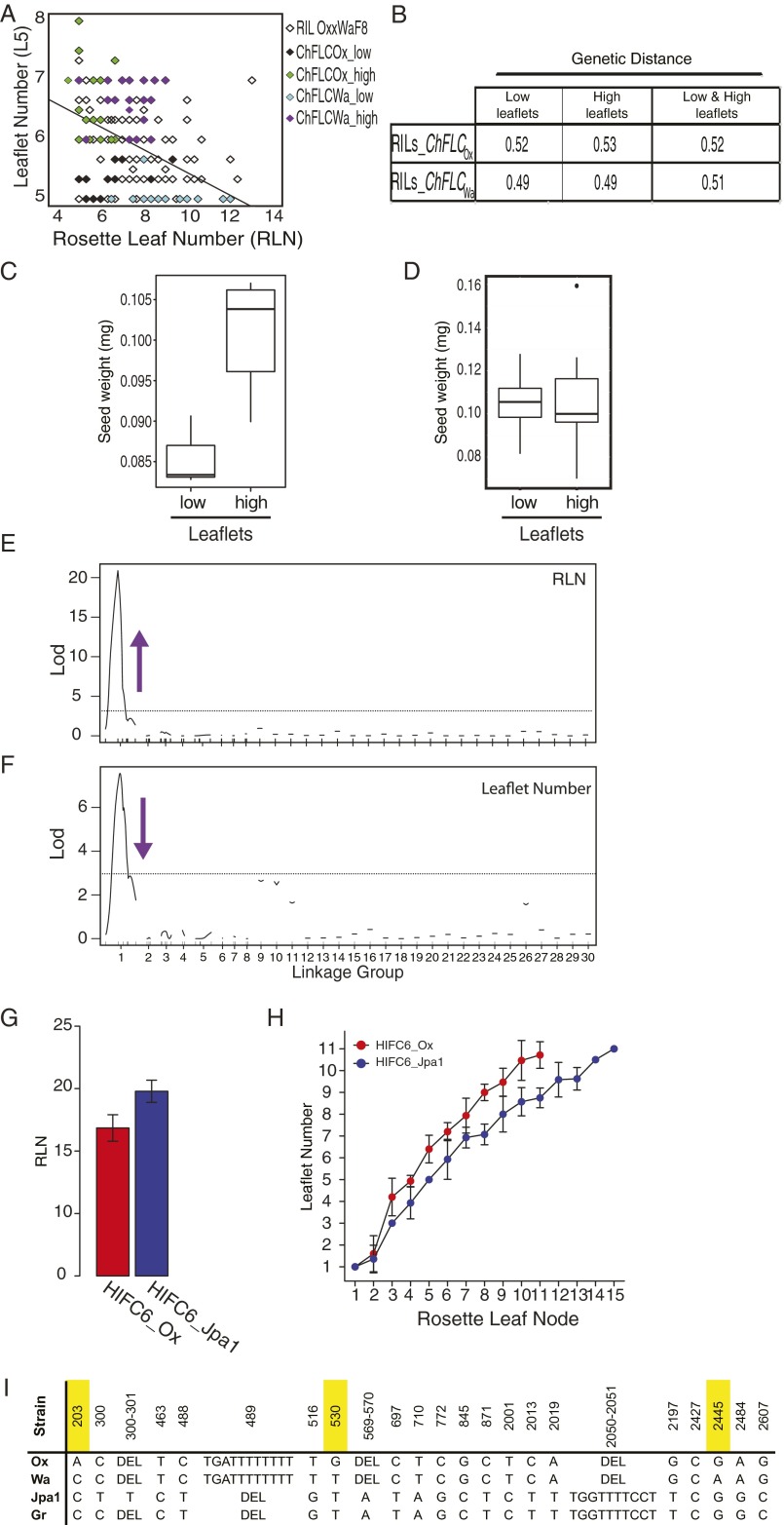
We have shown that ChFLC influences heteroblasty by regulating the adult vegetative phase, such that early flowering plants produce more complex adult leaf shapes at an earlier leaf node than their late-flowering counterparts (Fig. 4A). We reasoned that if this coordination between heteroblasty and reproductive timing is physiologically relevant, then genotypes lacking it should show perturbations in aspects of development that are influenced by it. Because flowering time has been suggested to influence seed yield (44, 45), we hypothesized that coordination of leaf growth with flowering may be part of a reproductive strategy to optimize resource allocation for the next generation. To test this idea, we evaluated the consequence of disrupting the coordination between leaf heteroblasty and flowering time on seed weight, by studying exceptional RIL families where this correlation is not found. To identify these lines, we first grouped RILs depending on their ChFLC allele, and then compared seed weight of lines belonging to the extreme tails of the distribution in leaflet number in each group (Fig. S5 A and B). Mean seed weight was significantly lower in early flowering ChFLCOx RILs that had low leaflet number compared with the ChFLCOx RILs with high leaflet number (Fig. 4B and Fig. S5 C and D). We did not detect any significant effect of flowering time on seed weight (ANOVA: RLN, P = 0.67; RLN × Leaflet, P = 0.54), which excludes the possibility that residual genetic variation for flowering time segregating in the RILs underlies differences in seed weight between RILs with low and high leaflet number. Thus, early flowering ChFLCOx RIL plants that failed to acquire high leaflet number showed reduced seed weight. A plausible interpretation of these results is that early flowering plants optimize their leaf growth program to support seed set in the context of a shorter life cycle. One possibility is that leaves with increased adult character and more leaflets have a higher capacity to produce and/or transfer photosynthetic metabolites to floral tissues (46).
Fig. 4.
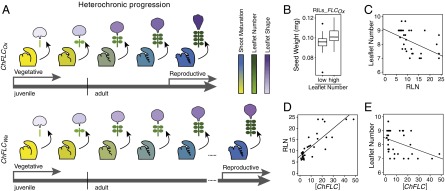
The impact of ChFLC on heteroblasty, the correlation with seed weight, and the prevalence of ChFLC-dependent leaflet number variation in C. hirsuta. (A) Cartoon summarizing how ChFLC-dependent shoot maturation influences heteroblasty in C. hirsuta. The weaker ChFLCOx allele causes increased leaflet number and rapid terminal leaflet shape changes at equivalent nodes compared with the stronger ChFLCWa allele, owing to faster progression through the adult vegetative phase of the shoot. Differential age-dependent variation of leaflet number and terminal leaflet shape results in adult leaves with divergent morphologies. In addition, the weaker allele results in earlier flowering. Gray arrows mark the juvenile, adult vegetative, and reproductive phases. Meristems with developing leaves are colored following a gradient from yellow (not competent to flower) to dark blue (flowering), which reflects shoot maturation. Schematic representations of heteroblastic leaf traits are colored following a gradient from low leaflet number and juvenile leaflet shape (light green and purple, respectively) to high leaflet number and adult leaflet shape (dark green and purple, respectively). Leaf primordia and lateral and terminal leaflets are not drawn to scale. (B) Boxplot showing the median and interquartile range of seed weight of RILs selected for their leaflet number and for having the Ox allele at the ChFLC locus. Mean seed weight of low leaflet number (5–5.6) and high leaflet number (6–8) RILs is significantly different (P < 0.05 by ANOVA). (C) Differences in RLN (x axis) can explain variation in leaflet number (y axis) in a sample of 33 C. hirsuta strains (R2 = 0.25; P < 0.03). (D and E) Variation in ChFLC expression measured by qRT-PCR (x axis) can explain differences in RLN (D; y axis; R2 = 0.68; P < 0.001; n = 35) and leaflet number on leaf 7 (E; y axis; R2 = 0.15; P < 0.01; n = 33). Transcript abundance is shown relative to that of the Ber-1 strain, which had the lowest ChFLC expression levels in our sample.
Fig. S5.
FLC-dependent heterochrony is widespread in C. hirsuta and affects seed weight in the context of rapid cycling. (A) Selection of RILs based on the total number of rosette leaves produced, their allele at ChFLC, and leaflet number on the fifth rosette leaf. Leaflet number of the Ox × Wa F8 RILs is plotted against RLN, and the line shows the negative correlation that exists between them (R2 = 0.21; P < 0.01; also see Fig. S1L). Lines with extremely high and low numbers of leaflets were selected from early flowering RILs carrying the ChFLCOx allele (green and black circles, respectively) and from late-flowering RILs carrying the ChFLCWa allele (purple and cyan circles, respectively). (B) Genetic distances estimated within and between low and high leaflet groups show that RILs are equally genetically related and thus satisfy the assumption underlying the Student’s t test used for comparing mean seed weight. (C and D) A repeat of the experiment shown in Fig. 4B with a subset of the RILs (SI Materials and Methods). The box plots show the median seed weight and associated interquartile range of RILs selected for their leaflet number and for having the Ox allele at the ChFLC locus. Mean seed weight between low and high leaflet number groups is significantly different in lines selected for having the Ox allele at ChFLC (C) (ANOVA; P < 0.05). In contrast, there is no significant difference in seed weight in RILs selected for their leaflet number and for having the Wa allele at the ChFLC locus (D). (E and F) QTL mapping for flowering time (E) and leaflet number (F) in the F6 generation of the Ox × Gr-1 RIL population. At present, the genetic map consists of eight small linkage groups (two to seven markers) and 20 unmapped markers. Horizontal dotted lines correspond to a significance threshold at P < 0.05. The purple arrows indicate the direction of the QTL effects by pointing either upward or downward according to whether the Gr-1 allele at the QTL increases or decreases the trait value when substituted for the Ox allele. For both traits, a single QTL is detected at marker 424 of linkage group 1, which is closely linked to the ChFLC locus in the Ox × Wa linkage map (SI Materials and Methods). (G and H) Ox × Jpa1 HIFs segregating at the ChFLC locus differ significantly for flowering time and leaflet number (n = 15 per genotype). Mean values are shown, and error bars indicate SD of the mean. (I) Two of the three SNPs in the first intron of ChFLC that are polymorphic between the Ox and Wa strains in the sequence interval causal for phenotypic variation between them (Fig. S2E) are shared between the Wa, Gr-1, and Jpa1 strains, making them candidates for also contributing to phenotypic variation revealed in corresponding crosses. Column headers indicate the positions of the polymorphisms in base pairs, with respect to the adenine in the translational start codon of ChFLCOx. SNP203, SNP530, and SNP2445, which are referred to in the text and are indicated in Fig. S2E, are highlighted in yellow. DEL, deletion of the alternative allele at the respective position.
By investigating leaf shape in additional C. hirsuta accessions, we showed that the contribution of ChFLC to C. hirsuta leaflet number diversity is broadly informative for diversification patterns within the species, and not confined to the Ox and Wa genotypes. A major QTL controlling both flowering time and leaflet number was detected at the ChFLC locus in a different Ox × Gr-1 RIL population derived from a Greek founder strain (Fig. S5 E, F, and I). Additionally, both flowering time and leaflet number cosegregated with markers linked to the ChFLC locus in an Ox × Jpa1 HIF derived from a founder strain from Japan (Fig. S5 G–I). Finally, in a panel of 33 natural strains, 25% of the variation in leaflet number could be explained by flowering-time variance (Fig. 4C), whereas ChFLC expression levels explained 15% of the variation in leaflet number and 68% of the variation in flowering time (Fig. 4 D and E). From these results, we conclude that allelic diversity at ChFLC is a key contributor to natural variation in C. hirsuta leaf form. Natural variation in the FLC gene has been widely described to contribute to differences in flowering time within the Brassicaceae (23, 47–50). Given that FLC also acts as an environmental sensor (30, 51), it is possible that evolutionary changes in its regulation provide an opportunity for concerted modulation of leaf growth with flowering time in response to changing environments, making this gene a “hot spot” for evolutionary diversification. This view is also consistent with the hypothesis that regulatory divergence in pleiotropic genes provides a favorable path for morphological evolution to occur (52, 53).
Our work identifies heterochronic variation arising through regulatory evolution as a key driver of leaf shape diversity at the intraspecific scale, whereas developmental pathways controlling tissue patterning and local growth appear to dominate at the scale of interspecific variation in crucifers (5–7, 34). Flowering time can diversify rapidly in annuals such as C. hirsuta, so this coordinated regulation of leaf shape and flowering time by ChFLC may provide a mechanism for trait integration that anchors vegetative growth to reproduction to optimize resource allocation to seeds (48). Similar examples were reported as early as 1944 in cotton (54, 55), where early flowering genotypes developed leaves with increased complexity, and our work provides a framework to interpret these results from diverse taxa. However, heteroblasty can be uncoupled from reproduction in other examples (56), and early flowering can be associated with decreased, rather that increased, leaf complexity (57). Therefore, the timekeeping activity of common genetic modules may tend to associate flowering time with the rate of progression of vegetative development, but whether and how this tendency is expressed in different species may depend on the interplay of these timekeeping modules with lineage-specific leaf developmental programs, ecological challenges, reproductive strategies, and drift.
SI Materials and Methods
Plant Growth Conditions.
Experiments shown in Figs. 1; 2 A–L; 3 A–E, H, and I; and 4 B and D; and Figs. S1; S2; S3 A–E and G–I; and S5 were performed at the University of Oxford, Department of Plant Sciences. Plants were grown in growth chambers in long-day conditions (LD) (days: 24 °C, 16 h; nights: 22 °C, 8 h, 113 μmol·m−2·s−1 light and 70% relative humidity). Experiments shown in Figs. 2M; 3 F–G and J–K; and 4 C and E; and Figs. S3 F and J–L; and S4 were conducted at the Max Planck Institute for Plant Breeding Research in Cologne in growth cabinets in LD (days: 20 °C, 16 h; nights: 18 °C, 8 h, 200 μmol·m−2·s−1 and 70% relative humidity) and short-day conditions (SD) (days: 20 °C, 8 h; nights: 18 °C, 16 h, 175 μmol·m−2·s−1 light and 70% relative humidity).
Genetic Stocks.
The C. hirsuta ecotype Ox [specimen voucher Hay 1 (OXF)] has been described (6). All accessions used in this work are listed in Table S3. The Ox × Wa F8 mapping population was described in ref. 13. Initial QTL validation was performed by using the RIL 57 selected for being heterozygous at the QTL between the Derived Cleaved Amplified Polymorphic Sequences (dCAPS) markers m128135 and d2i11F, corresponding to a physical distance of ∼263 kb. Fine mapping was performed by genotyping 1,000 F9 progenies of RIL 57 to a region of ∼13 kb flanked by markers c34692 and snpAT5G10110. NILs were generated by back-crossing the F8 RIL_57 to the Ox ecotype for five generations. Marker-assisted selection was used to select for the Ox alleles in genomic regions outside the ChFLC locus. The introgression of the Wa genome spanning the ChFLC gene is estimated to be 1.3 Mbp. NILs were genotyped by using the Sequenom technology at the Wellcome Trust Center for Human Genetics (Oxford) as described (13) and by dCAPS markers. Ox × Jpa1 HIF was selected from a F5 mapping population by using marker 09880 located at 97 Kb from the ChFLC promoter. A complete list of the markers used for genotyping is given in Table S4.
Table S3.
List of accessions used in this work
| Accession code | Published material | Source | Collector | Year | Country | Latitude | Longitude | Major area | Minor area |
| Ber1 | Ref. 13 | ||||||||
| Bli1 | Ref. 13 | ||||||||
| Col-1 | Ref. 13 | ||||||||
| Cor-1 | Ref. 13 | ||||||||
| Cor-2 | Ref. 13 | ||||||||
| Cor-5 | MPIPZ | M.C., B.P. | 2011 | U.K. | 50.54169 | −4.940179 | Cornwall | Hayle | |
| Cor-6 | Ref. 13 | ||||||||
| Gr-1 | Ref. 13 | ||||||||
| Eng16 | Ref. 13 | ||||||||
| Epw1 | MPIPZ | B.P. | 2010 | Netherlands | 52.152271 | 6.703301 | Twente | Haaksbergen | |
| ESP1 | Ref. 13 | ||||||||
| Fra1 | Ref. 13 | ||||||||
| Fra127 | Ref. 13 | ||||||||
| Ger1 | Ref. 13 | ||||||||
| Ger2 | Ref. 13 | ||||||||
| Ger3 | Ref. 13 | ||||||||
| Ger4 | Ref. 13 | ||||||||
| Hbg-1 | Ref. 13 | ||||||||
| HOV-1 | Ref. 13 | ||||||||
| Ire2 | Ref. 13 | ||||||||
| Ire7 | Ref. 13 | ||||||||
| ITA4 | Ref. 13 | ||||||||
| jpa1 | Ref. 13 | ||||||||
| Mad1 | Ref. 13 | ||||||||
| Net5 | Ref. 13 | ||||||||
| Nmg1 | Ref. 13 | ||||||||
| Nz* | Ref. 13 | ||||||||
| Wa | Ref. 13 | ||||||||
| Ox | Ref. 6 | ||||||||
| Sch1 | MPIPZ | M. Koornneef | 2009 | Netherlands | 53.488352 | 6.15086 | Schiermonnikoog | ||
| Sch2 | MPIPZ | M. Koornneef | 2009 | Netherlands | 53.488352 | 6.15086 | Schiermonnikoog | ||
| Spa2 | Ref. 13 | ||||||||
| Swe1 | Ref. 13 | ||||||||
| Swi2* | Ref. 13 | ||||||||
| Wag1 | Ref. 13 |
Only shown in Fig. 1.
Table S4.
Details of dCAPs markers used for genetic mapping
| Experiment | Marker name | primer_F | primer_R | Restriction enzyme | Linkage group | Synteny block (6) |
| HIF_OxxWa_fine mapping | m128135 | TTCCATCCATGACTTGTAGACTGC | TAAAAATTTAGGAGTTTCCGATT | MseI | 6 | R |
| HIF_OxxWa_fine mapping | d2i11F | CACAAACTTGCCTAGACACAAGTTGA | AAACGTAGAACCTGAATCTGTTCTTACC | EcoRI | 6 | R |
| HIF_OxxWa_fine mapping | 629 | AAAAGCTGTGGAATGGGATG | GAACTGTTCGATTTTAACATTAGCTT | TaqI | 6 | R |
| HIF_OxxWa_fine mapping | c34692 | CTATACATTTATATGTTGGATTTAGC | TAGACCAACCCTGGTCTAATTATATCAAGACC | AluI | 6 | R |
| HIF_OxxWa_fine mapping | snpAT5g10110 | TTGGTTAATTTGACTAATCTTCTTCCAGCTAA | AAACTTCTCTCTCGAACATTCATAAGC | DdeI | 6 | R |
| HIF_OxxWa_fine mapping | m849 | TTGGAGGAAGGAGGAAAGCTCTCTG | GAAGAAAAGAAGCAAGAGAAGCA | DdeI | 6 | R |
| HIF_OxxJpa1_validation | dcaps09880 | CTGCGGTTTGCATCGGTGCAGGCTGC | GGCTTGGCTTTGGCTTCATATTCATGAGG | PstI | 6 | R |
| Production of NIL | SNP_135 | AGCAAAGCAAAATACTACTTGG | GAAGCCTCTGATGCTTCAGTTA | MseI | 1 | A |
| Production of NIL | SNP_1972 | TTACATATTAGTTCAATAAAAGTG | GAGGCTCAACACCCTTTAAACTCAATAGC | AluI | 1 | B |
| Production of NIL | SNP_4501 | CAATCACCATCTCGACCTTCATCGTC | CGTCTTGTTTATCTTCAACGGGAAGGCTA | DdeI | 2 | D |
| Production of NIL | SNP_468 | AGGAGATGAGAAACGGAG | AAGCTGTTCTTGATAATGTA | RsaI | 2 | E |
| Production of NIL | SNP_74 | ACAAAGAATCGGGAGCATGT | ACAATATAAGTAATGTTAGTGGC | DdeI | 3 | F |
| Production of NIL | SNP_1154 | AAAGTCTTCCTTGTGAGTAGAGAGACC | GGCTTGACGTTGCTCGTCAGAAGCTTGTTGT | EcoRI | 5 | L |
| Production of NIL | SNP_13146 | AAATCGCGATATATGAGACCTGAATCAGC | AGGATACAAAGAGAGAACAATGCTCCTGC | DdeI | 5 | N |
| Production of NIL | SNP_22377 | ACCATACCAAAGAACAATAACATGAG | CCGTCACGTCTCGACCTGCTCCTTTGTA | RsaI | 6 | V |
QTL Analyses.
QTL mapping was performed in R (www.r-project.org/) by using the R/qtl package (58). The description of the genetic map has been provided in ref. 13. QTL mapping was performed with the Composite Interval Mapping method using 1,000 permutations for determining the significance threshold. For the phenotypic analysis, 195 RILs of the Ox × Wa F8 population were grown in three replicates. Phenotyping of the fifth rosette leaf morphology and flowering time was performed at the University of Oxford. Analysis of the cumulative leaflet number was performed in Cologne. To QTL map in the Ox × Gr-1 F6 population, we used 182 RILs. Genotyping of the RILs was carried out by the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics by using Sequenom technology. The linkage map was generated in Joinmap (Version 4.0; Kyazma B.V.), and it consists of a total of 50 markers distributed in nine small linkage groups and 22 singletons. The position of the QTL-LG6 locus was compared by using six markers (636_1, 535_3, 211_1, 424_1, 851_1, and 369_1), which were mapped in both populations (13).
Further Statistical Analyses.
The diagnostic biplot shown in Fig. S1N was generated in GenStat (16th Edition; ref. 59). All other statistical analyses were performed in R (Version 3.1.0; www.r-project.org). Logistic regression (glm) was used to predict the allele in HIFs 700 and 732 combined from the nominal variable leaflet number and the continuous variable terminal leaflet shape. The Wald statistic was used to test for the individual contribution of both traits to the prediction of allele. For visualization of the results, the terminal leaflet shape data for the HIF were binned into six bins with breakpoints determined by the quantiles. The relative ratios between the number of leaves from the HIF with the Ox and Wa alleles were calculated for each bin per leaflet combination. A regulatory constant of 2/n, where n is the number of bins, was added to the nominator and denominator. These ratios were then plotted according to a color gradient indicating the relative representation of both alleles.
Phenotyping.
To quantify flowering time, we counted the total number of rosette leaves produced by the plants. Leaflet number was counted for each rosette leaf after the plants had fully flowered. To obtain leaf silhouettes, fully developed rosette leaves were adhered to white paper by using adhesive spray and digitally scanned. Photoshop (Adobe) and a script developed in MATLAB were used to quantify the size of the rosette leaves. For rosette leaf 9 growth analysis, seedlings were mounted in 24-h intervals starting from 2.5 wk after germination onwards and observed in water without fixation. Confocal imaging was performed by using a Leica TCS SP8 microscope and a 10× objective (N.A. 0.30) or a 20× water immersion objective (N.A. 0.5). A 488-nm argon laser was used for excitation and images of the chlorophyll autofluorescence were collected at 660–749 nm. The resulting confocal z-stacks (15–20 sections) were then processed by using the Leica LAS AF lite software to generate maximum projections.
Morphometric Analysis.
To quantify variation in shape of the terminal and lateral leaflets, outline curves were digitized in tpsDig (life.bio.sunysb.edu/morph/) by locating a series of coordinate points along the margin of the leaflets. To enhance the topological correspondence of the digitized leaflets, two landmarks were manually added at the maximum inception point located between the rachis and the leaflet lamina. We then performed Extended Eigenshape analysis (www.morpho-tools.net; ref. 14), which converts the Cartesian coordinates used to describe the position of the coordinate points along the outline into the ϕ form of the shape function (60). Finally, multivariate decomposition returned a series of orthogonal eigenshape axes that capture the principal component of variance among the outlines. ANOVA was used to identify the eigenshape axes significantly different among the RILs, and shape models were then generated to visualize the nature of the morphological variation described by each axis. Eigenscores of each selected eigenshape axis were then used as phenotypic traits for the QTL mapping. For HIF validation and quantification of heteroblasty, the leaflet shape of the whole heteroblastic series was analyzed. In Fig. 3D, the eigenscores of the first principal axis were plotted on a Cartesian (x, y) plane.
Transgenic Plant Construction.
To construct the Col-0 AtFLC::FLC locus, a 9.1-Kb fragment was amplified from the T13P16 (NASC) BAC clone. Then, 7.7-Kb fragments of the Wa ChFLC:FLC locus were amplified from genomic DNA. PCR products were amplified by using the Platinum Taq polymerase (Invitrogen). Primer sequences are listed in Table S5. The amplified products were cloned in the pGEM-T Easy vector System (Promega) and dideoxy-sequenced (pGChFLCWa). To obtain the Ox ChFLC::FLC locus, a BAC library of the Ox C. hirsuta genomic clones was screened (13), and an 8.8-kb SalI–BamHI fragment was cloned into pBJ36 (pBChFLCOx). To test for the presence of causal variants in the ChFLC first intron, 3-Kb fragments containing the first exon and the intron were obtained by digesting both pBChFLCOx and pGChFLCWa with AflII and BspeI restriction enzymes and swapped. AtFLC::FLC, pGChFLCWa, pBChFLCOx, ChFLCWa::FLCOx:FLCWa, and ChFLCOx::FLCWa:FLCOx were transferred to pMLBART by using NotI restriction enzyme, transformed into Agrobacterium tumefaciens strain GV3101, and used to floral dip C. hirsuta and A. thaliana plants as described (13). Transgenic analyses in Fig. 2 E and F and Fig. S2C were performed by phenotyping T2 lines. To assess the contribution of allelic variation at the first intron, we phenotyped RLN in 30 independent T1 lines generated from each construct.
Table S5.
List of primers used for gene cloning, qRT-PCR analysis, and dideoxy sequencing
| Experiment | Gene | Primer_F | Primer_R |
| Gene cloning | FLCCol-0 | TTCCTTAGTGTCGGCTGGAT | ATGCCGTACATTCACCAAT |
| Gene cloning | FLCWa | CACTCGTTCACGAGAAAAGG | CACGACGCGTTTTGAAATAA |
| qRT | ChGADPH | TGACCACCGTCCACTCCATCAC | GCTCTTCCACCTCTCCAGTCCTTC |
| qRT | ChFT | AGGTCCTCTCCACCAATCTCAACTC | TGTAAGAGACCCTCTTCTGGTCAGC |
| qRT | ChSOC1 | TCAATCGAGGAGCTGCAACA | TCCGAACTTGGGCTACTCTC |
| qRT | ChFLC | AACCCAAACCTCACCTCAGGATCAAATTAGG | CGGCTACTTTTGTTCTCAATTCGC |
| qRT | ChSPL3 | GAAGGAGAAGAAGATTCTGAGGACACT | GTTGGTGAAGACCAGAGATCCGA |
| qRT | ChSPL9 | GTGGAATGAACAATGGAAGC | ATGAAAAGCTTGTCCCTCCT |
| qRT | ChSPL13 | CTTCATCCTCTGTCCCGCAT | GTGGAAGAGCAACAGCCTCA |
| qRT | ChSPL11 | GTCCAAGTTTCAACTTCCTGGCG | AGAATAGAGTAGAGAAAGCGGCTGC |
| qRT | ChSPL15 | ATATGGCGAGCATCTTCAGG | TGTGGTGCGATTTCTCATGT |
| Sequencing | ChFLC_F1/R1 | TAGCGGCAGCATTATCGAATCGG | TATCACTCTGGTAGAGATTCC |
| Sequencing | ChFLC_F1seq | AAACGTTATTAGTCATTCATTAGCGGG | |
| Sequencing | ChFLC_F2/R2 | TAGAGTTCAAAGACAAATGAAAATGG | TAAAGTAGAGATCACAATATGTAGC |
| Sequencing | ChFLC_F3/R3 | ACTAAATTAATACTCCTAGACTCC | AACTAAATCGCTATTACAACTAACC |
| Sequencing | ChFLC_F4/R4 | ATCTTTATGATTTGTTTGTTTTAGGTCC | TAGTTAAGAAACTAGATACAATACAAGAGG |
| Sequencing | ChFLC_F5/R5 | TCTAGAATCAGTTTGGGTTTCGTTTCG | ACGTGTACCGCATGACTAATTCCCAGG |
| Sequencing | ChFLC_F6*/R6 | AGTATAAATTTGAGACTAAAAGAGG | TTGTCGGCTACTTTTGTTCTCAATTCG |
| Sequencing | ChFLC_F7/R7* | AACGCTCAGTATCTCCGGCGACTTGAACCC | TTTAGCTCATCACGACATAGTTCTTGC |
| Sequencing | ChFLC_F8*/R8 | TTCCTTATATGGGCTTTCATTTATTTCCC | TAGATCTCTCGGAAGTACACTGCATTGC |
| Sequencing | ChFLC_F9/R9* | AAGTTGCATGTCCTTCAAGATCTTTTCG | AAGATGTTTATATATACACACATCAACTGC |
| Sequencing | ChFLC_F10/R10 | ATCCAAATTATGGTTTCTGATTGATTCC | AATTAAGGTCGTGTTGTAAATTTCC |
| Sequencing | ChFLC_F11/R11 | AAATGTATGCTACATTGTGCAACTATTGC | TTTGCACGAAAATCAAGGATGCACAAACG |
| Sequencing | ChFLC_F12*/R12* | ATTTGTTCTTGCCTTATATTGAATCC | AAACGACTTCCCCAGTAGTCTTCCACTTTCC |
| Sequencing | ChFLC_F13/R13 | TTAACTTTCAGTGGCAGAAAAATCTCTACC | TATATTTAAACCCGAAAATCAAACAATCAAGCC |
| Sequencing | ChFLC_F14*/R14* | TAACCACTGGCTCTAATATCACAATTTGTGG | AAAGGAGATCTAATATTTACCAAGTATTGC |
| Sequencing | ChFLC_F15/R15 | AATTGATAGGTTACAACATGTACTGG | ATCTGGAACTCAGGCCTTAAGTAAATTTTGG |
| Sequencing | ChFLC_F16/R16 | TTCAGAGTTAGCTAGTAGCTTTGATCC | AAGGTCTAGTAGCTCGTGGTGTGAACC |
| Sequencing | ChFLC_F17/R17 | ACCTTGACATTGTGGAGATCTAGATCC | TACTTTAAACTTGGAGGCAAATATCC |
| Sequencing | ChFLC_F18/R18 | TAACTAGAGCCAAGAAGGTAAGTTGATTCC | AATGCTAATTAGTGAGTGCTTAGGGTGC |
| Sequencing | ChFLC_F19/R19 | TTGGCTAACCAGGTAATTAACGAAAGC | TTGTTTTTGCTAAAACCCTGACTTCATCGG |
| Sequencing | ChFLC_F20/R20 | TTTTATTGATAAAACTATACATACACACATAC | AACCCAGGGAATATTCTGTTCATACAACC |
| Sequencing | ChFLC_F21/R21 | ACATGCTCAAGGTGGACAAGGTTACATGC | CAGGTGACCGACGTAGGCCCATAGAAACC |
| Sequencing | ChFLC_F21seq | AGTACACACAACGTCTCATTTTTGC |
These primers were used to sequence parts of the first intron of ChFLC in the Gr-1 and Jpa1 strains.
qRT-PCR Analysis.
For ChFLC expression analysis, seedlings bearing four visible leaves were collected. Of the same experiment, three plants per genotype were left to grow, and leaflet number and flowering time were scored. To analyze temporal dynamics of gene expression in the NILs, apical regions with the oldest leaf being <5 mm in length were collected at intervals of 2 d until flower buds became visible. Total RNA was isolated by using the Qiagen RNeasy kit (Qiagen) with extra DNaseI treatment (Qiagen). cDNA was synthesized by using the SuperScript VILO kit (Invitrogen) and amplified by using the ViiA7 Real-Time PCR system (Life Technologies). The amplification reactions were prepared by using the Power SYBR green PCR master mix (Applied Biosystems) using 5 µL of cDNA (diluted 1:100) and 0.8 µM primers in 13-µL reactions. Each experiment was performed in two biological replicates and three technical replicates. Primer efficiency was estimated according to the manufacturer’s instruction (Applied Biosystems). Expression levels were estimated by using the Comparative Ct method and were normalized against values obtained for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). All primers used are listed in Table S5.
Dideoxy Sequencing.
The entire ORF and the 5′ intergenic region of ChFLCWa were dideoxy-sequenced on both strands by using the primers listed in Table S5. Two regions of ChFLC that spanned the locations of the three SNPs discovered between Ox and Wa in the first intron were dideoxy-sequenced on both strands in C. hirsuta strains Gr-1 and Jpa1 (Table S3) by using the indicated primer pairs listed in Table S5.
Photoperiod-Shift Sensing Experiment.
To investigate the effect of ChFLC on vegetative phase change, we performed a photoperiod-shift experiment. NILs grown in LD were transferred to SD condition at regular interval of times (0, 5, 7, 9, 11, 13, 15, 17, 19, 21, and 25 DAS). Plants were left in SD to flower, and RLN and leaflet number was scored when the inflorescence bolt reached ∼15 cm in length. The onset of the juvenile-to-adult phase change (i.e., the end of the photoperiod-insensitive phase) was determined by Student’s t test comparing RLN of NILs shifted plants to a SD grown control. To determine the number of leaf primordia produced by the NILs entering the adult phase, seedlings were collected at each time point and analyzed by scanning electron microscopy (SEM).
SEM.
In preparation for SEM, seedlings were cut a few millimeters below the cotyledonary node and stored in FAA fixative [3.7% (vol/vol) formaldehyde, 50% (vol/vol) ethanol, and 5% (vol/vol) acetic acid] for up to several weeks at 4 °C in a refrigerator. Subsequently, the fixed material was dehydrated by passing through a series of increasing ethanol concentrations [75% (vol/vol), 85% (vol/vol), and 96% (vol/vol)] with a final transfer to dimethoxymethane (formaldehyde dimethyl acetal; Sigma-Aldrich Chemie) and stored overnight before critical-point drying in liquid CO2 in a Leica CPD300 (Leica Microsystems). After mounting the dried samples in an upright position on SEM stubs by using LeitC, cotelydons and immature leaves were dissected manually by using a fine needle to expose the shoot apical meristem. After gold/palladium-coating the prepared samples in a Polaron SC7640 sputtercoater (Quorum Technologies), SEM imaging was performed on a Zeiss Supra 40VP (Carl Zeiss Microscopy).
Analysis of Seed Weight.
Seed weight was quantified by weighting ∼100 mg of seeds, counting the corresponding seed number, and dividing it by their weighed value. Student’s t test was used to analyze the difference in mean seed weight between the low and high leaflet number group. RILs within and between groups were estimated as equally genetically distant by calculating the pairwise genetic distance in GGT (Version 2.0). A two-way ANOVA factorial design was performed to test for the effect of leaflet number and residual flowering time (Leaflets × RLN) on seed weight.
Selection of ChFLCOx F8 RILs.
ChFLCOx F8 RILs with the lowest leaflets at rosette leaf 5 were selected from the lower tail of the distribution. Out of 61 ChFLCOx F8 RILs, we found 12 having low, 5–5.7, leaflets and compared their seed weight to 25, randomly selected around the mode, ChFLCOx F8 RILs with 6–8 leaflets (Fig. 4B). The seed weight of three biological replicates was quantified. We repeated the experiment by germinating three and four RILs bearing 5–5.3 and 6–7 leaflets, respectively (Fig. S5C). The seed weight of their progeny was quantified on five biological replicates.
Selection of ChFLCWa F8 RILs.
ChFLCWa F8 RILs with the highest leaflet number at rosette leaf 5 were selected from the upper tail of the distribution. Out of 124 ChFLCWa F8 RILs, we found 21 having a high number of 6–7 leaflets and compared their seed weight to 16 randomly selected around the mode ChFLCWa F8 RILs with 5–5.6 leaflets (Fig. S5D). The seed weight of three biological replicates was quantified.
Materials and Methods
Plants were grown in controlled environment chambers under long-day (16h/8h; day/night) or short-day (8h/16h; day/night) conditions. All C. hirsuta strains used in this work are listed in Table S3 and the Ox x Wa F8 RIL population was previously described (13). Variation in shape of the terminal leaflet was quantified by Extended Eigenshape analysis (14). For the photoperiod-shift sensing experiment, plants were grown in LD conditions and transferred to SD conditions 0–25 days after sowing. Detailed methods for plant growth conditions, phenotyping, QTL and statistical analysis, morphometric analysis, transgenic plant construction, quantitative RT-PCR, DNA sequencing, and microscopy can be found in the SI Materials and Methods and Tables S4 and S5.
Acknowledgments
We thank S. Poethig and C. Alonso-Blanco for helpful discussions; G. Coupland for critical reading of the manuscript; M. Rast, J. Lamb, A. Hallab, X. Gan, E. Kirdök, J. Pietsch, S. Höhmann, B. Grosardt, I. Will, R. Pabst, N. MacLeod, and J. D. Krieger for assistance in molecular biology, phenotypic characterization, and bioinformatics; and M. Koornneef for donating C. hirsuta strains. This work was supported by Deutsche Forschungsgemeinschaft Priority Programme “Adaptomics” Grant TS 229/1-1 (to M.T. and A.H.); Biotechnology and Biological Sciences Research Council Grants BB/H011455/1 (to M.T.), BB/H006974/1 (to M.T. and A.H.); the Gatsby Charitable Foundation (M.T.); Cluster of Excellence on Plant Science (M.T.); HFSP-RGP0047/2010 (M.T.); the Minerva Programme of the Max Planck Society (A.H.) and a Max Planck Society core grant (M.T.). M.C. was supported by a Federation of European Biochemical Societies postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KM110949 and KM096577–KM096584).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419791112/-/DCSupplemental.
References
- 1.Goebel K. Organography of Plants Especially of the Archegoniatae and Spermatophyta. Clarendon; Oxford: 1900. [Google Scholar]
- 2.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40(9):1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 3.Bharathan G, et al. Homologies in leaf form inferred from KNOXI gene expression during development. Science. 2002;296(5574):1858–1860. doi: 10.1126/science.1070343. [DOI] [PubMed] [Google Scholar]
- 4.Bilsborough GD, et al. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci USA. 2011;108(8):3424–3429. doi: 10.1073/pnas.1015162108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blein T, et al. A conserved molecular framework for compound leaf development. Science. 2008;322(5909):1835–1839. doi: 10.1126/science.1166168. [DOI] [PubMed] [Google Scholar]
- 6.Hay A, Tsiantis M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat Genet. 2006;38(8):942–947. doi: 10.1038/ng1835. [DOI] [PubMed] [Google Scholar]
- 7.Vlad D, et al. Leaf shape evolution through duplication, regulatory diversification, and loss of a homeobox gene. Science. 2014;343(6172):780–783. doi: 10.1126/science.1248384. [DOI] [PubMed] [Google Scholar]
- 8.Ori N, et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet. 2007;39(6):787–791. doi: 10.1038/ng2036. [DOI] [PubMed] [Google Scholar]
- 9.Shleizer-Burko S, Burko Y, Ben-Herzel O, Ori N. Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development. 2011;138(4):695–704. doi: 10.1242/dev.056770. [DOI] [PubMed] [Google Scholar]
- 10.Chitwood DH, et al. Resolving distinct genetic regulators of tomato leaf shape within a heteroblastic and ontogenetic context. Plant Cell. 2014;26(9):3616–3629. doi: 10.1105/tpc.114.130112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Efroni I, Eshed Y, Lifschitz E. Morphogenesis of simple and compound leaves: A critical review. Plant Cell. 2010;22(4):1019–1032. doi: 10.1105/tpc.109.073601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubio-Somoza I, et al. Temporal control of leaf complexity by miRNA-regulated licensing of protein complexes. Curr Biol. 2014;24(22):2714–2719. doi: 10.1016/j.cub.2014.09.058. [DOI] [PubMed] [Google Scholar]
- 13.Hay AS, et al. Cardamine hirsuta: A versatile genetic system for comparative studies. Plant J. 2014;78(1):1–15. doi: 10.1111/tpj.12447. [DOI] [PubMed] [Google Scholar]
- 14.MacLeod N. Generalizing and extending the eigenshape method of shape space visualization and analysis. Paleobiology. 1999;25(1):107–138. [Google Scholar]
- 15.Piazza P, et al. Arabidopsis thaliana leaf form evolved via loss of KNOX expression in leaves in association with a selective sweep. Curr Biol. 2010;20(24):2223–2228. doi: 10.1016/j.cub.2010.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Sicard A, et al. Repeated evolutionary changes of leaf morphology caused by mutations to a homeobox gene. Curr Biol. 2014;24(16):1880–1886. doi: 10.1016/j.cub.2014.06.061. [DOI] [PubMed] [Google Scholar]
- 17.Hasson A, et al. Evolution and diverse roles of the CUP-SHAPED COTYLEDON genes in Arabidopsis leaf development. Plant Cell. 2011;23(1):54–68. doi: 10.1105/tpc.110.081448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Todesco M, et al. Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol. 2012;22(2):166–170. doi: 10.1016/j.cub.2011.11.060. [DOI] [PubMed] [Google Scholar]
- 19.deVicente MC, Tanksley SD. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134(2):585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alonso-Blanco C, Koornneef M, Ooijen JWv. 2006. QTL analysis. Arabidopsis Protocols, Methods in Molecular Biology, eds Salinas J, Sanchez-Serrano JJ (Humana, Totowa, NJ), pp 79–99.
- 21.Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: A method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95(5-6):1005–1011. [Google Scholar]
- 22.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 23.Lempe J, et al. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet. 2005;1(1):109–118. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11(5):949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee I, Bleecker A, Amasino R. Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol Gen Genet. 1993;237(1-2):171–176. doi: 10.1007/BF00282798. [DOI] [PubMed] [Google Scholar]
- 26.Angel A, Song J, Dean C, Howard M. A Polycomb-based switch underlying quantitative epigenetic memory. Nature. 2011;476(7358):105–108. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 27.Coustham V, et al. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science. 2012;337(6094):584–587. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- 28.Finnegan EJ, Dennis ES. Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol. 2007;17(22):1978–1983. doi: 10.1016/j.cub.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 29.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331(6013):76–79. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 30.Li P, et al. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 2014;28(15):1635–1640. doi: 10.1101/gad.245993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheldon CC, Conn AB, Dennis ES, Peacock WJ. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell. 2002;14(10):2527–2537. doi: 10.1105/tpc.004564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sung S, et al. Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet. 2006;38(6):706–710. doi: 10.1038/ng1795. [DOI] [PubMed] [Google Scholar]
- 33.Gould SJ. Ontogeny and Phylogeny. Harvard Univ Press; Cambridge, MA: 1977. p. 522. [Google Scholar]
- 34.Hudson CJ, et al. Genetic control of heterochrony in Eucalyptus globulus. Genes Genomes Genet. 2014;4(7):1235–1245. doi: 10.1534/g3.114.011916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng W, et al. FLOWERING LOCUS C (FLC) regulates development pathways throughout the life cycle of Arabidopsis. Proc Natl Acad Sci USA. 2011;108(16):6680–6685. doi: 10.1073/pnas.1103175108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Willmann MR, Poethig RS. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development. 2011;138(4):677–685. doi: 10.1242/dev.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poethig RS. 2013. Vegetative phase change and shoot maturation in plants. Current Topics in Developmental Biology (Academic, New York), Vol 105, pp 125–152.
- 38.Schwarz S, Grande AV, Bujdoso N, Saedler H, Huijser P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol Biol. 2008;67(1-2):183–195. doi: 10.1007/s11103-008-9310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang JW, et al. miRNA control of vegetative phase change in trees. PLoS Genet. 2011;7(2):e1002012. doi: 10.1371/journal.pgen.1002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124(3):645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- 41.Searle I, et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006;20(7):898–912. doi: 10.1101/gad.373506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59(1):573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- 43.Usami T, Horiguchi G, Yano S, Tsukaya H. The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development. 2009;136(6):955–964. doi: 10.1242/dev.028613. [DOI] [PubMed] [Google Scholar]
- 44.Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96(8):4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2010;107(49):21199–21204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taiz L, Zeiger E. 2010. Translocation in the phloem. Plant Physiology (Sinauer Associates Inc., Sunderland, MA), 5th Ed.
- 47.Guo Y-L, Todesco M, Hagmann J, Das S, Weigel D. Independent FLC mutations as causes of flowering-time variation in Arabidopsis thaliana and Capsella rubella. Genetics. 2012;192(2):729–739. doi: 10.1534/genetics.112.143958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Méndez-Vigo B, de Andrés MT, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C. Temporal analysis of natural variation for the rate of leaf production and its relationship with flowering initiation in Arabidopsis thaliana. J Exp Bot. 2010;61(6):1611–1623. doi: 10.1093/jxb/erq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tadege M, et al. Control of flowering time by FLC orthologues in Brassica napus. Plant J. 2001;28(5):545–553. doi: 10.1046/j.1365-313x.2001.01182.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, et al. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459(7245):423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- 51.Méndez-Vigo B, Picó FX, Ramiro M, Martínez-Zapater JM, Alonso-Blanco C. Altitudinal and climatic adaptation is mediated by flowering traits and FRI, FLC, and PHYC genes in Arabidopsis. Plant Physiol. 2011;157(4):1942–1955. doi: 10.1104/pp.111.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martin A, Orgogozo V. The Loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67(5):1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 53.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323(5915):746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashby E. Studies in the morphogenesis of leaves. New Phytol. 1948;47(2):153–176. [Google Scholar]
- 55.Stephens SG. The genetic organization of leaf-shape development in the genus Gossypium. J Genet. 1944;46(1):28–51. [Google Scholar]
- 56.Costa MMR, et al. The genetic basis for natural variation in heteroblasty in Antirrhinum. New Phytol. 2012;196(4):1251–1259. doi: 10.1111/j.1469-8137.2012.04347.x. [DOI] [PubMed] [Google Scholar]
- 57.Shalit A, et al. The flowering hormone florigen functions as a general systemic regulator of growth and termination. Proc Natl Acad Sci USA. 2009;106(20):8392–8397. doi: 10.1073/pnas.0810810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 59.International VSN. 2013. GenStat for Windows (VSN International, Hemel Hempstead, U.K.), 16th Ed.
- 60.Zahn CT, Roskies RZ. Fourier descriptors for plane closed curves. IEEE Trans Comput. 1972;21(3):269–281. [Google Scholar]