Abstract
Mitochondrial and plastid genomes show a wide array of architectures, varying immensely in size, structure, and content. Some organelle DNAs have even developed elaborate eccentricities, such as scrambled coding regions, nonstandard genetic codes, and convoluted modes of posttranscriptional modification and editing. Here, we compare and contrast the breadth of genomic complexity between mitochondrial and plastid chromosomes. Both organelle genomes have independently evolved many of the same features and taken on similar genomic embellishments, often within the same species or lineage. This trend is most likely because the nuclear-encoded proteins mediating these processes eventually leak from one organelle into the other, leading to a high likelihood of processes appearing in both compartments in parallel. However, the complexity and intensity of genomic embellishments are consistently more pronounced for mitochondria than for plastids, even when they are found in both compartments. We explore the evolutionary forces responsible for these patterns and argue that organelle DNA repair processes, mutation rates, and population genetic landscapes are all important factors leading to the observed convergence and divergence in organelle genome architecture.
Keywords: endosymbiosis, chloroplast, mitochondrion, mitochondrial genome, plastid genome
Endosymbiosis can dramatically impact cellular and genomic architectures. Mitochondria and plastids exemplify this point. Each of these two types of energy-producing eukaryotic organelle independently arose from the endosymbiosis, retention, and integration of a free-living bacterium into a host cell more than 1.4 billion years ago. Mitochondria came first, evolving from an alphaproteobacterial endosymbiont in an ancestor of all known living eukaryotes, and still exist, in one form or another (1), in all its descendants (2). Plastids came later, via the “primary” endosymbiosis of a cyanobacterium by the eukaryotic ancestor of the Archaeplastida, and then spread laterally to disparate groups through eukaryote-eukaryote endosymbioses (3). Consequently, a significant proportion of the identified eukaryotic diversity has a plastid.
With few exceptions (1, 4), mitochondria and plastids contain genomes—chromosomal relics of the bacterial endosymbionts from which they evolved (2, 5). Mitochondrial and plastid DNAs (mtDNAs and ptDNAs) have many traits in common, which is not surprising given their similar evolutionary histories. Both are highly reduced relative to the genomes of extant, free-living alphaproteobacteria and cyanobacteria (2, 5), and both have transferred most of their genes to the host nuclear genome and are therefore reliant on nuclear-encoded, organelle-targeted proteins for the preservation of crucial biochemical pathways and many repair-, replication-, and expression-related functions (6). Both also show a wide and perplexing assortment of genomic architectures (7, 8), which has spurred various evolutionary explanations, ranging from ancient inheritance from the RNA world to selection for greater “evolvability” (9, 10).
At first glance, genomic complexity within mitochondria mirrors that of plastids (9, 11). Indeed, both organelle genomes have, in many instances, taken parallel evolutionary roads, independently adopting almost identical architectures (12), as well as similar mutational patterns (13), replication and gene expression strategies (14, 15), and modes of inheritance (16). There are, however, some major differences between mitochondrial and plastid genomes, including structures and/or embellishments present in one but not the other. But genomic architectural diversity has rarely been directly compared between plastids and mitochondria, and only with the recent characterization of organelle genomes from remote eukaryotic lineages (Fig. 1) can their similarities and differences be adequately addressed and fully appreciated.
Fig. 1.
Number and taxonomic distribution of all complete mitochondrial and plastid DNA (mtDNA and ptDNA) sequences at the National Center for Biotechnology Information (NCBI) Database. (A) Annual number deposited from 2003 to 2013. (B) Total number of sequences (4,965) as of 1 August 2014. Statistics taken from the NCBI Organelle Genome Resources site: www.ncbi.nlm.nih.gov/genomes/GenomesHome.cgi?taxid=2759&hopt=html.
Here, we compare the architectural diversity of mitochondrial genomes with those of plastids. First, we survey the available organelle genome sequence data across the eukaryotic domain, showing that for plastid-bearing taxa ptDNA diversity is as well or better sampled than that of mtDNA. We then assess the range of genomic complexity within mitochondria and plastids, highlighting examples of convergent evolution between these two genetic compartments, as well as illuminating features unique to one or the other. Ultimately, we argue that mitochondrial genomes harbor a greater breadth of complexity and consistently more pronounced eccentricities than plastid genomes and evaluate the potential evolutionary forces responsible for these patterns.
Three Decades of Organelle Genomics
Not surprisingly, the human and mouse mitochondrial genomes were the first organelle DNAs, and the first nonviral chromosomes, to be completely sequenced (1981) (17, 18). Five years later (1986), the first plastid genomes were deciphered—those of Marchantia polymorpha (19) and tobacco (20). In the approximately three decades following these molecular milestones, thousands of other organelle genomes were sequenced. As of 1 August 2014, there were ∼5,000 complete mtDNA and ptDNA sequences in GenBank (Fig. 1), making organelle genomes among the most highly sequenced types of chromosome. Moreover, the rate of organelle genome sequencing is increasing (Fig. 1), with nearly 400 new organelle genomes deposited in GenBank in the first half of 2014.
This sample, however, is highly biased: ∼82% of sequenced organelle genomes are metazoan mtDNAs, and only 11% are ptDNAs (Fig. 1). Nevertheless, in terms of diversity, ptDNAs are equally or better sampled than mtDNAs for many major eukaryotic lineages, including land plants, green algae, rhodophytes, diatoms, and other plastid-bearing protists (Fig. 1). Therefore, although mtDNA sequence data outnumber those of plastids by almost an order of magnitude, our understanding of plastid genomes is as good as, and in some cases better than, that of mitochondrial genomes (3, 5, 21). Thus, given the data at hand, it is possible to make meaningful and broad-level comparisons between mitochondrial and plastid genomes across the eukaryotic tree.
In addition to sequence information, there are large amounts of data on other aspects of organelle chromosomes, including their conformations (22, 23), replication strategies and inheritance patterns (15, 16), transcriptional and translational architectures (24, 25), mutational and population genetic landscapes (26, 27), and proclivity for horizontal gene transfer (6, 28).
Together, these data have helped shape our understanding of eukaryotic evolution (3, 29), and been pivotal in the fields of archaeology (30), forensics (31), industry (32), and medicine (33). Above all, mitochondria and plastids have been an endless reservoir for interesting and unconventional genomes: genomes that have broken or changed established rules in genetics (34–36), contributed to leading contemporary hypotheses on molecular evolution (26, 37), and initiated discussions about the roles of adaptive vs. nonadaptive processes in shaping cellular and genomic complexity (38).
A Multiplicity of Mitochondrial and Plastid Genome Architectures
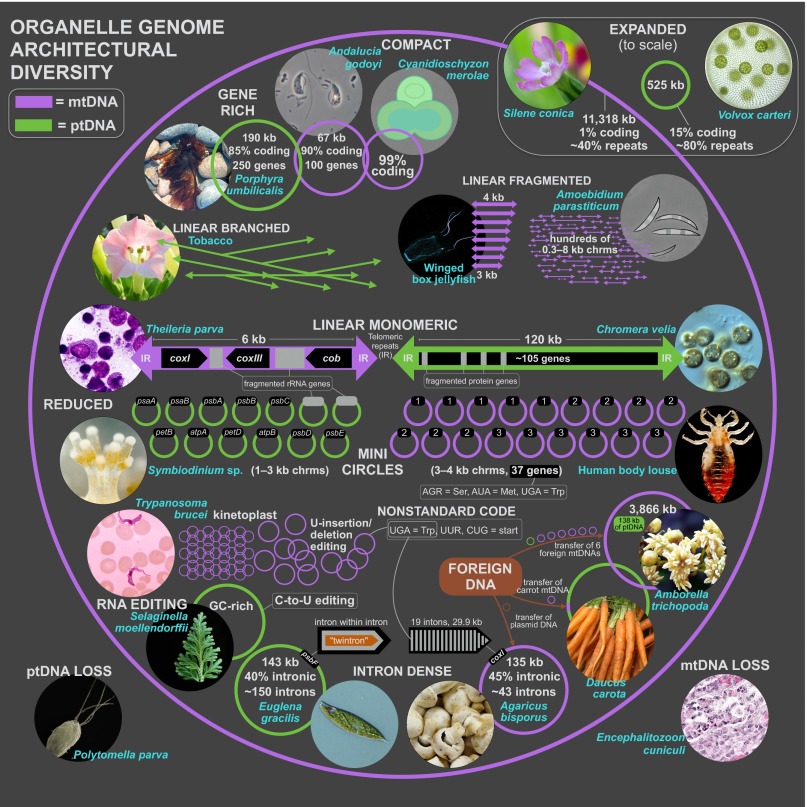
Organelle genomes were once thought to be simple, circular molecules with homogeneous size classes, contents, and organizations (11, 39), but they have proven to be anything but ordinary (7). From the enormous, multichromosomal mtDNAs of various angiosperms (40) to the baffling miniature fragmented ptDNAs of dinoflagellates (8), many organelle DNAs are complex and dynamic, making the genome hard to define and leading to intense debates on the evolutionary explanations for such tremendous genomic variance (37, 38). Although it is infeasible to describe the full extent of this complexity in a single review, the following paragraphs summarize the main architectural themes and extremes of mitochondrial and plastid chromosomes, underlining parallels between both systems, but also the greater tendency for things to go awry in mitochondria than in plastids (Fig. 2 and Table S1).
Fig. 2.
Organelle genome architectural diversity. Mitochondrial and plastid genome/gene maps are shown in pink and green, respectively. Genome maps are not to scale, unless stated otherwise on figure. Alantina image courtesy of Dr. Andre Seale (Hawaii Institute of Marine Biology, Kaneohe, HI); Amborella image courtesy of Wikimedia Commons/bff; Andalucia © Alastair Simpson; Chromera image courtesy of the National Center for Marine Algae and Microbiota (NCMA); Daucus image courtesy of Jeremy Keith on Flickr; E. cuniculi image courtesy of The Joint Pathology Center; Head louse image courtesy of the CDC; Porphyra image courtesy of Nicolas Blouin; Selaginella image courtesy of Jing-Ke Weng (Purdue University, West Lafayette, IN); Symbiodinium image by Eiichi Shoguchi, reprinted with permission of OIST; Theileria © 2008 Jan Votýpka; Trypanosoma image courtesy of the CDC.
Nucleotide Composition.
There is a near-universal adenine (A) and thymine (T) bias in organelle genomes throughout the eukaryotic domain (41), the most extreme of which (∼90% AT) is in the mitochondria of yeasts and arthropods. Plastid genomes can also be very AT rich, reaching 87% in Plasmodium falciparum, but overall, they have less severe nucleotide biases than mtDNAs (41). Although rare, guanine and cytosine (GC)-rich organelle DNAs do exist, occurring in euglenozoans, green plants, animals, and fungi. The highest documented mitochondrial GC content (68%) is from the lycophyte Selaginella moellendorffii (42) and is significantly greater than the record for plastids (58%), belonging to a trebouxiophyte green alga. Remarkably, all known species with GC-biased ptDNA also have GC-rich mtDNA. The reverse, however, is not true (41).
Structure.
Although widely depicted as circular chromosomes, organelle DNAs usually have more multifarious structures. Land plant mitochondrial and plastid genomes and yeast mtDNAs typically assemble as circles but are thought to exist in vivo as multigenomic, linear-branched structures (23). Linear-mapping mitochondrial genomes with defined telomeres are found within various green algae, protists, animals, and fungi (22, 43). The telomeres of linear mtDNAs can have ornate conformations (22), such as hairpin loops (44), single-stranded overhangs (45), and/or covalently attached proteins (46), which likely preserve the chromosome ends independent of telomerase (22). The alveolate Chromera velia boasts the only known linear-mapping plastid genome, which has a telomeric arrangement mirroring those of many linear mitochondrial chromosomes (47).
Chromosome Number.
The shift from a single to a multipartite chromosomal architecture has happened many times in mtDNA evolution but is a surprisingly rare event for ptDNAs (12). No fewer than 12 eukaryotic lineages are known to contain fragmented mitochondrial genomes, and mtDNA splintering has transpired more than once within certain groups, including cnidarians (48), chlamydomonadalean algae (44), and vascular plants (13, 49). Sometimes the levels of fragmentation are minor: the mtDNA of the colorless green alga Polytomella piriformis is divided into two small (<15 kb) linear chromosomes (44). In other instances, the fragmentation is extensive: human head louse mtDNA is broken into 20 miniature (1.5–3 kb) circular chromosomes, each encoding one to three genes (50), and the euglenozoan Diplonema papillatum mtDNA comprises >75 small (6–7 kb) circular DNAs with single-gene modules (51). Trypanosome mtDNAs form giant, intertwined networks (kinetoplasts) comprising thousands of small (0.5–10 kb) and a few dozen large (20–40 kb) circular chromosomes (52), and the Amoebidium parasiticum mtDNA is made up of hundreds of 0.3- to 8.3-kb linear pieces (53). Fragmented plastid genomes are currently restricted to dinoflagellates (8, 54), including peridinin-containing dinoflagellates, whose ptDNAs have shattered into ∼2- to 3-kb minicircles with one to a few genes apiece (8).
Genome Size.
Mitochondrial and plastid genome size can vary by orders of magnitude, but mtDNAs exhibit a wider range of sizes, both among and within lineages, than ptDNAs (40, 55). At one extreme are the giant mtDNAs of seed plants, like cucumber (∼1.6 Mb) (49) and Silene conica (∼11 Mb) (40), as well as those of diplonemids, which can exceed 500 kb (51). At the other end of the spectrum are the 6-kb mtDNAs of certain apicomplexans (56) and the fragmented mtDNA of their close relative C. velia, which is even smaller (57). Diminutive mtDNAs (<13 kb) have also been uncovered in green algae (44), ctenophores (58), and fungi (59). Although narrower than that of mtDNAs, the size range for ptDNAs is nonetheless astounding. Prodigious ptDNAs have been sequenced from the chlorophycean algae Volvox carteri (∼525 kb) (27) and Floydiella terrestris (∼520 kb) (60) and the ulvophyte Acetabularia acetabulum (≥1 Mb) (61). The smallest plastid genomes are found in peridinin dinoflagellates, whose fragmented ptDNAs are ∼30 kb (e.g., Symbiodinium) (62), as well as in nonphotosynthetic algae, such as Plasmodium species (∼35 kb) (63) and Helicosporidium sp. (37.5 kb) (64).
Noncoding Nucleotides.
Most of the variation in organelle genome size is due to differences in noncoding DNA content (26), which varies from ∼1% to 99% for mtDNAs (40) and from ∼5% to 80% for ptDNAs (55). Small organelle genomes, such as vertebrate mtDNAs and apicomplexan ptDNAs, are usually coding-dense and devoid of introns, whereas large genomes, including the S. conica mtDNA and V. carteri ptDNA, are distended with noncoding nucleotides. These noncoding regions are often riddled with repeats, introns, mobile elements, and, for mitochondrial genomes, foreign DNA (6, 26, 55). The number of introns within organelle genomes can differ from none (e.g., human mtDNA) to >150 (65), and mitochondria generally display more intronic variability than plastids (26), although euglenid plastids are a notable exception. The cox1 gene of the button mushroom Agaricus bisporus has 19 introns, making it the largest (29.9 kb) and most intron-rich organelle gene from all eukaryotes (66). The types and arrangements of introns within organelle DNAs can be quite bizarre: the Euglena gracilis plastid genome contains 15 twintrons (introns within introns), which must be removed sequentially for accurate splicing (65); in the S. moellendorffii mtDNA, nad4L is located within an intron of nad1 (42).
Gene Number.
As with size and structure, mtDNAs show greater variation in gene number and organization than ptDNAs (12). The jakobid Andalucia godoyi has the largest, least-derived mitochondrial gene content (100 genes, ∼66 of which encode proteins) (67), whereas dinoflagellates, apicomplexans, and their close relatives have the most reduced: three or fewer proteins, no tRNAs, and in some instances (e.g., C. velia) even lack complete rRNAs (36, 56, 57, 68). Chlamydomonadalean mtDNAs also have diminished coding contents (10–13 genes) (44), and some land plants (e.g., S. moellendorffii), animals (e.g., the winged box jellyfish), and trypanosomes have lost all or most of their mitochondrial tRNA-coding regions (39, 42, 48). Plastid genomes are typically more gene-rich than mtDNAs (12), maxing out at ∼250 genes in red algae (69). Plants and algae that have lost photosynthetic capabilities have reduced plastid gene contents (<75) (63, 64, 70), but the absolute lowest are in the photosynthetic, peridinin plastids of dinoflagellates, which encode <20 genes (62).
Genome Loss.
Some organelles have lost their genome entirely. This loss can occur following the disappearance of oxidative phosphorylation in mitochondria or photosynthesis in plastids but is not inevitable and once again is more common in mitochondria than in plastids. Genome-lacking mitochondria-derived organelles (mitosomes and hydrogenosomes) are known from several anaerobes and microaerophiles, such as Giardia, Trichomonas, Mikrocytos, and Entamoeba (1). In contrast, nonphotosynthetic plastids are relatively common, but in almost all known cases retain a reduced genome (4), exemplified by the parasitic plant Epifagus (70) and the parasitic algae Plasmodium and Helicosporidium (63, 64). Well-supported examples of genome-lacking plastids are currently only known from the green algal genus Polytomella (3).
Gene Fragmentation.
In a number of eukaryotes, mitochondrial-encoded genes are fragmented and scrambled (12), a feature unmatched in most of the ptDNAs sequenced to date (5). Well-studied examples include the discontinuous and jumbled mitochondrial rRNA- and/or protein-coding genes of various green algae (44, 71), alveolates (36, 63), and euglenozoans (51, 52, 72). The large- and small-subunit (LSU and SSU) rRNA genes of Chlamydomonas reinhardtii, for example, have split into eight and four unordered coding segments, respectively, which come together after transcription through secondary-pairing interactions (71). RNA splintering has gone even further in the P. falciparum mtDNA, where at least 27 distinct modules encode the LSU and SSU rRNAs (73). Fragmented ptDNA-located genes are uncommon (5) but have been documented in chlamydomonadalean algae (55), peridinin dinoflagellates (74), C. velia (47), and the haptophyte-derived plastid of the dinoflagellate Karlodinium veneficum (54). Also, rps12 is transspliced in the plastids of various land plants (13).
Noncanonical Genetic Codes.
Among animal mtDNAs, there have been at least 12 unique changes to the “universal genetic code,” with up to five codon reassignments in certain groups (34, 39), and most microbial eukaryotes have experienced one or two mtDNA codon alterations (7), as well as the occasional loss of start and stop codons (36, 68). Overall, mitochondria that retain the universal code are relatively rare exceptions. In contrast, no noncanonical genetic codes have been observed in primary plastids, and there are only a few reports from other types of plastids, including some from aplicomplexans, C. velia, and the dinoflagellate Lepidodinium chlorophorum (47, 75, 76).
RNA Editing.
Mitochondrial RNA editing has evolved in diverse eukaryotes, including slime molds (77), land plants (14), and dinoflagellates (36). The most elaborate display of editing occurs in kinetoplast mitochondria, where ∼90% of codons experience uracil-insertion/deletion editing (52, 78). Plastid transcript editing has only been reported in land plants and in peridinin and fucoxanthin dinoflagellates (8, 14, 79, 80). The extent of plastid editing in each of these groups is not as widespread and elaborate as that in the mitochondrial compartment (12).
Foreign DNA.
Many mitochondrial genomes abound with horizontally acquired sequences, whereas plastid genomes are conspicuously lacking in foreign DNA (6, 81). For instance, ∼13% (∼130 kb) of the zucchini mtDNA is represented by plastid- and nuclear-derived sequences, almost all of which are noncoding (82). Other angiosperms, like Plantago and ginger species, contain mitochondrial gene mosaics, formed by gene conversion between native and foreign homologs (83). Even more impressive is the mtDNA of the shrub Amborella trichopoda, which harbors the equivalent of six horizontally acquired mitochondrial genomes (84). Until recently, it was thought that plastids were impenetrable to foreign DNA (85, 86). However, new data from various angiosperms uncovered mitochondrion-to-plastid (87, 88) and nucleus-to-plastid DNA migration events (89), and there are also examples of plastids, including those of some diatoms and the red alga Gracilaria tenuistipitata, acquiring genes from plasmid or bacterial genomes (69, 90). Foreign DNA can also come in the form of extrachromosomal elements. Plasmids have been characterized from the mitochondria of land plants, fungi, and various protists (43, 91), but are near absent from plastids, with some notable exceptions (92).
Convergent Organelle Genome Evolution
As touched on in the previous section, convergent evolution can be observed between mtDNAs and ptDNAs of distantly related species, between those within the same lineages, and even the same cell (Fig. 2 and Table S1). Below we examine in more detail parallels in mitochondrial and plastid genomic architecture, emphasizing that among and within species, mtDNAs tend to attain greater levels of complexity than ptDNAs.
Selaginella moellendorffii is an excellent illustration of how mitochondrial and plastid genomes can arrive at similar extremes in a single organism (Table S2). Both the mtDNA and ptDNA of this seedless vascular plant have some of the highest observed organelle GC contents, severely reduced gene (particularly tRNA) repertoires, inflated numbers of introns, record levels of C-to-U RNA editing, and accelerated rates of structural rearrangements (42, 93). However, for each of these different traits, the mtDNA has been pushed to a greater extreme than the ptDNA (Table S2). It has a stronger GC bias (68% vs. 52%), fewer tRNA regions (0 vs. 13), more introns (37 vs. 11), and a larger number and ratio of RNA-editing sites (thousands vs. hundreds). Moreover, the mtDNA has peculiarities not found in the ptDNA, such as standard genes situated within introns, repetitive elements, and a recombinant structure (42). Not all land plant organelle genomes are as unconventional as those of S. moellendorffii, but across land plants, there is a general tendency toward higher levels of RNA editing, higher intron densities, and larger, more complex genomic structures in mtDNA vs. ptDNA (21, 26, 40).
Similar trends emerge from dinoflagellates. The genomes within their mitochondria and peridinin plastids are remarkable examples of convergent evolution (Table S2). Both organelle DNAs have complicated and fragmented organizations, among the lowest gene contents of all eukaryotes, aberrant gene sequences, and undergo large amounts of posttranscriptional modification, which can include A-to-G editing and 5′ and/or 3′ processing (8, 36, 68). However, although the mitochondrial and plastid genomes have followed similar evolutionary paths, the mtDNA has consistently gone further (Table S2). It has more severe gene losses, more widespread and elaborate forms of posttranscriptional editing and processing, more gene isoforms, and more extensive gene fragmentations. Unlike their plastid counterparts, dinoflagellate mtDNAs can contain noncanonical start and stop codons (or none at all), transspliced genes, and oligonucleotide caps at both the 5′ and 3′ ends of transcripts (36, 68). Many of these tendencies are consistent across other major alveolate groups as well (47, 56, 63, 73).
The mtDNAs and ptDNAs of green algae are also paragons of parallel evolution. In Dunaliella salina and Volvox carteri, both the mitochondrial and plastid genomes have undergone massive expansions, resulting in uncharacteristically long intergenic regions and large amounts of repetitive DNA (Table S2) (27, 55). What’s more, similar repeats exist in the mtDNA and ptDNA of each alga (27, 55). In the prasinophyte Ostreococcus tauri the reverse has occurred: both organelle genomes have contracted, and consequently they have very little noncoding DNA (94). However, the degree of genome reduction within the mitochondrion (zero introns; ∼92% coding) surpasses that of the plastid (one intron; ∼80% coding) (55, 94). The coexpansion/cocontraction of mitochondrial and plastid genomes is observed in a variety of other species (55) (Table S2), and in most instances, the ptDNA is outdone by the mtDNA.
Evolution of Characteristics Mediated by Intrinsic vs. Extrinsic Factors
Similar genomic eccentricities in both the plastid and mitochondrial genomes of the same species may at first seem to be remarkable parallelisms; however, it may rather be that one system evolved from the other. Unlike endosymbionts or parasites, the evolution of specific host-to-organelle protein targeting systems means that organelle genetic information is partitioned between two genomes. Factors that govern organelle processes can, therefore, be considered “intrinsic” if they reside within the organelle genome or “extrinsic” if they reside within the nuclear genome, and the processes themselves may be dominated by one or the other type of factor. For example, RNA editing in trypanosome mitochondria is governed by intrinsic factors: the editing process is mediated by mitochondrial-encoded guide RNAs (52). In contrast, mitochondrial RNA editing in land plants, and probably dinoflagellates, is governed by extrinsic factors: nuclear-encoded proteins identify the editing sites (14, 35, 37). Noncanonical genetic codes can similarly be intrinsic (if the tRNAs in question are organelle-encoded) or extrinsic (if the tRNAs or release factors are imported). This is an important distinction because targeting can be “leaky,” and both dual-targeted and retargeted proteins have led to the specific import of new proteins into organelles (95, 96). Consequently, predominantly extrinsic processes that evolved in one compartment could spread if key proteins found their way into another compartment.
The appearance of rare and apparently complex genomic eccentricities in both the plastid and mitochondrion of the same species is, therefore, not that surprising if they are mediated by extrinsic factors. To continue with the example of RNA editing, even though essentially all of the enzymes involved in editing processes are nuclear encoded, the fact that trypanosome editing is mediated by intrinsic information and plant editing by extrinsic information means that plant editing is more likely to spread to other genetic systems within the same cell, as it apparently has in plants and dinoflagellates (12, 14). Unfortunately, we do not know the direction, and as trypanosomes lack a plastid, the corollary is also not testable. A spatial spread of genomic characteristics might explain many of the other shared tendencies of plastids and mitochondria within the same lineages described in the proceeding section, and might also explain the recent description of RNA editing in the tertiary plastid of a dinoflagellate (97). Finally, there is increasing evidence for molecular crosstalk between mitochondria and plastids (98), so certain mtDNA-ptDNA correlations could be due to a direct link between the organelles. Some mitochondrial mutations, for instance, can have major effects on chloroplast properties (98).
Understanding Mitochondrial and Plastid Genome Evolution
Many explanations have been put forward to explain the strange nature of plastid and especially mitochondrial genomes (26), but in most cases, they focus on a specific trait and do not take into account the similarities and differences of both kinds of organelle genome. Even when mitochondrial and plastid genomes acquire similar traits in a given group or species, the eccentricities tend to be more pronounced in mtDNAs than in ptDNAs. Accordingly, if there is a root cause of these characteristics, it must reflect the similarities and differences of both organelles. One could argue that the range of plastid genomic complexity might be equivalent to or greater than that of mitochondria but that it has not yet been sufficiently explored and documented. However, as discussed earlier, ptDNAs are equally or better sampled than mtDNAs for many major lineages (Fig. 1). Nevertheless, the oversampling of mitochondrial genomes within some clades has contributed to the range of observed organelle genome diversity. For example, an excess of arthropod mitochondrial genome sequencing resulted in the discovery of the abnormal mtDNAs from lice (50). There are, of course, many eukaryotic lineages that harbor mitochondria but not plastids, so even if plastids genomes are well sampled in the lineages that contain plastids, there are still more mitochondrial lineages to sample (52).
It could also be argued that because the endosymbiotic event that led to mitochondria occurred hundreds of millions of years before that which gave rise to plastids, mtDNAs have had more time than ptDNAs to become complex. However, many genomic eccentricities observed within mtDNAs and ptDNAs are believed to have arisen at approximately the same time in both compartments. For example, within the green lineage, mitochondrial and plastid RNA editing is thought to have evolved concurrently in the common ancestor of land plants (14), yet editing is still more elaborate in mitochondria than in plastids. Similar cases can be made for the GC biases within Selaginella mtDNA and ptDNA or organelle genome compaction within Ostreococcus. Besides, many of the most peculiar mitochondrial genomic features, such as the fragmented, scrambled rRNA-coding genes of Chlamydomonadalean algae, arose relatively recently in evolutionary time, long after the establishment and diversification of plastids. Thus, there appears to be a genuine tendency for mitochondrial genomes to reach greater extremes than those of plastids.
One of the many reasons for this could be linked to the fact that mitochondria contain fewer genes than plastids. For example, changes to the genetic code are expected to occur more frequently in genomes with a small amount of protein-coding sequence (99). Likewise, a smaller gene complement could also allow for larger fluctuations in mutation rates because selection against a mutator allele would be proportional to its effect on the rate of functionally deleterious mutations per genome (not per nucleotide).
Structural and physiological differences between the organelles could also cause extreme architectures. Unlike plastids, mitochondria regularly fuse in vivo, potentially exposing themselves to foreign elements. The fusion between domestic and foreign mitochondria is likely how Amborella acquired mtDNA sequences from mosses, green algae, and other angiosperms (84). Another possible reason for the presence of exogenous sequences in mtDNAs and their absence from ptDNAs is that mitochondria (at least those of land plants and mammals) have an active DNA import system (85, 100, 101), whereas no such system has been identified in plastids. However, these explanations focus on only a few aspects of mitochondrial vs. plastid genome complexity. A broader understanding of organelle genome evolution requires knowledge of the underlying processes that fashion chromosomes, namely mutation, recombination, natural selection, and random genetic drift (10).
Mutations, Populations, and Nonadaptive Hypotheses
There is growing evidence that many aspects of organelle genome complexity are not the direct product of natural selection but instead have their roots in nonadaptive processes (10), particularly neutral evolutionary ratchets (37). Lynch put forward a universal hypothesis, called the mutational hazard hypothesis (MHH), explaining organelle genomic architecture (10, 26). The hypothesis proposed that genomic embellishments, such as introns, fragmented genes, and RNA editing sites, are mutationally burdensome and therefore have a greater tendency to accumulate in systems with low mutation rates and small effective population sizes, where random genetic drift can overpower selection (10, 26). This theory is supported by the exceptionally low mutation rate estimates from the massive, highly embellished mitochondrial genomes of many land plants (26). Similarly, the giant plastid genomes of some volvocine green algae, like V. carteri, harbor very little genetic diversity, suggesting that their inflated, repeat-rich structures might be made possible by their low mutation rates and/or increased random genetic drift (27). Other studies, however, have found the opposite trend: the enormous mtDNAs of the angiosperms Silene noctiflora and S. conica appear to have unprecedentedly high mutation rates and no obvious reductions in effective population size relative to species with smaller mtDNAs, which contradicts the MHH (40). It is thought that changes in mtDNA recombinational processes rather than increased genetic drift are driving the evolution of Silene mtDNAs (40). There is little doubt, however, that the irremediable complexity of some organelle genetic systems is the result of neutral processes. For example, mutational ratchets, which can be exacerbated by increased genetic drift, eloquently explain the gratuitous levels of posttranscriptional editing in mitochondria and plastids (37) and could equally be applied to a variety of other complex embellishments.
Whether neutral or not, investigations on mtDNA and ptDNA evolution have also shown that there is a strong link between extreme organelle genomic architectures and extreme rates of mutation and recombination, be they high or low. For a range of different eukaryotes, high predicted mutation rates and elevated levels of recombination and gene conversion correlate with structural and sequence upheaval of organelle chromosomes (43, 51, 102). Embellished genomic features, such as extensive posttranscriptional modification and/or dramatic departures in genome organization, content, and structure, also occur alongside extraordinarily low rates of organelle mutation (103, 104). The tulip tree has one of the most mutationally quiescent but most heavily RNA-edited mitochondrial genomes of any plastid-bearing eukaryote (104), and the unprecedentedly low mitochondrial mutation rates of corals concurs with intron accumulation, enlarged intergenic regions, and a paucity of tRNA genes (103). Once again, these patterns are sometimes observed in the mtDNA and ptDNA of a single species. For example, the extraordinarily high mitochondrial mutation rates of S. noctiflora and S. conica coincide with accelerated ptDNA sequence and structural evolution (13). For V. carteri, low plastid genetic diversity and a distended ptDNA structure is accompanied by equally low mtDNA diversity and genomic expansion (27).
Organelle mutation and recombination rates are a reflection of the underlying organelle DNA maintenance pathways, virtually all of which are nuclear encoded (95). In land plants and algae, many of the DNA replication, recombination, and repair proteins targeted to the mitochondrion have plastid-targeted paralogs (95), and a significant proportion is targeted to both organelles (96). The extrinsic nature of these factors could therefore explain why some species have similar mitochondrial and plastid mutation rates (13, 105) and genomic architectures. That said, the proficiency of organelle DNA maintenance can vary substantially between species and compartments (95), which is not surprising because the evolution of these processes involves a complex history of gene transfer, co-option, duplication, and replacement events, leading to patterns of general tendencies rather than absolutes.
Indeed, organelle repair mechanisms can even differ across a single chromosome, a trait that has provided fundamental insights into organelle genome evolution. After discovering that the rates of mutation and modes of molecular evolution differ drastically between coding and noncoding regions in the Arabidopsis thaliana mitochondrial genome, Christensen (106) proposed that land plants use two types of mtDNA repair, each of which has shaped mitochondrial genomic architecture: “Within genes, a bias toward gene conversion would keep measured mutation rates low, whereas in noncoding regions, break-induced replication (BIR) explains the expansion[s] and rearrangements. Both processes are types of double-strand break repair, but enhanced second-strand capture in transcribed regions versus BIR in non-transcribed regions can explain the two seemingly contradictory features of plant mitochondrial genome evolution—the low mutation rates in genes and the striking expansions of noncoding sequences” (106). Differences in DNA repair between and within mitochondria and plastid lineages might also account for much broader patterns in organelle genome evolution (107), including why mtDNAs are more architecturally varied than ptDNAs. If repair processes are impacting organelle genomic diversity, there should be observable differences between mitochondrial and plastid mutational spectrums, with mitochondria exhibiting more severe and wide-ranging mutational features than plastids. Likewise, mitochondrial and plastid genomes with similar degrees of complexity should also show similar mutational trends, especially when housed in the same cell.
Mitochondrial Mutation Rates Are More Variable and Often Much Higher or Lower Than Those of Plastids
Data on mitochondrial and plastid mutation rates are slowly accumulating for diverse species, and a general trend is emerging: the mtDNA mutational spectrum is broader and more variable than that of ptDNA (26). Mitochondria boast some of the highest (108) and lowest (104) mutation rate estimates from any eukaryotic or bacterial genome. For angiosperms alone there is a 5,000-fold range in the absolute rate of synonymous site substitutions (dS) among explored mitochondrial genomes (104), and up to a 340-fold range among genes within a single mtDNA (109). The mitochondrial synonymous substitution rate range is almost as staggering for cnidarians (103), sponges (110), and other animals (111), as well as for various protists, including alveolates (112, 113) and amoebozoans (114). Plastid genome mutation rate estimates, on the other hand, are less variable within and across lineages and do not display such extreme values as those for mtDNAs (115, 116). Even for the few lineages with notably high or low rates of ptDNA synonymous site substitution, such as the olive tree, Silene, legumes, and the dinoflagellate Symbiodinium, the rate increases/decreases are generally localized to a subset of genes (13, 117–119), and the levels of synonymous site divergence in the mitochondrial compartment are often equally as high or low (13, 118, 119) (Fig. 3).
Fig. 3.
Synonymous substitution rates in the organelle and nuclear genomes from various plastid-bearing lineages. Plastid DNA (ptDNA) is green, mitochondrial DNA (mtDNA) is pink, and nuclear DNA (nucDNA) is orange. The Archaeplastida (i.e., Plantae) comprises glaucophytes, red algae, green algae, and land plants, all of which have primary plastids. The haptophyte Phaeocystis and the dinoflagellate Symbiodinium have secondary, red algal-derived plastid. Synonymous site substitution rates come from ref. 113 and references therein. Data for Symbiodinium come from ref. 118 and were calculated by comparing species A2 to species C90.
Much of our knowledge on mitochondrial vs. plastid mutations comes from studies on relative (as opposed to absolute) levels of synonymous site substitution within mtDNA and ptDNA of closely related species (120) (Fig. 3). When looking across eukaryotes, there is a propensity toward higher rates of mutation in mtDNA compared with ptDNA and nuclear DNA (nucDNA) (113, 116), with notable exceptions from seed plants, which can have exceptionally small mitochondrial-to-plastid (and mitochondrial-to-nuclear) mutation rate ratios (120). Studies on glaucophytes, rhodophytes, and streptophyte green algae have revealed a 2- to 10-fold higher mutation rate in mtDNA compared with ptDNA and in some cases nucDNA (116). A higher mutation rate in mtDNA vs. ptDNA is consistently observed in plastid-bearing taxa outside the Archaeplastida as well: diverse photosynthetic eukaryotes with red algal-derived plastids, including haptophyte and stramenopile algae, have ∼5–20 times greater mutation rates in mtDNA relative to ptDNA and nucDNA (116). A similar pattern has also been exposed in apicomplexan parasites (113). In fact, one of the few known lineages with higher levels of substitution in ptDNA relative to mtDNA and nucDNA is the peridinin dinoflagellate Symbiodinium (118), which has among the most bizarre plastid genome structures ever described (62). Some chlorophyte green algae are also exceptional in that they appear to have similar rates of mutation in all three genetic compartments (27, 105), which might explain why they can have very similar mitochondrial and plastid genomic architectures (105).
Given all of this, it is hard to ignore that mitochondrial mutation rate estimates are wider-ranging and more erratic than those of ptDNAs and that an mtDNA/ptDNA mutation rate ratio of »1 or «1 is seen in a diversity of plastid-bearing species. Ultimately, the highly variable and extreme mutation rates of mitochondrial genomes are consistent with there being major differences, incongruences, and/or inefficiencies in the underlying DNA maintenance processes among mitochondrial lineages, much more so than for ptDNA. These incongruences, in turn, could help account for the large breadth of and severities in mitochondrial genomic complexity compared with plastid genomes, but raises additional questions as to what underlying forces might have led to the evolution and maintenance of such differences.
Conclusion
Organelle genomes are truly a playground for genomic diversity, but we have yet to see if there is a unifying explanation for the bizarre array of changes to structure, function, and content that they have sustained. Alternatively, the desire to find a single explanation for a suite of related characteristics in diverse lineages might be an oversimplistic trap, and in reality, their evolution is the sum of a combination of many factors. By comprehensively comparing the architectures of mtDNAs and ptDNAs, however, we can add another piece to the puzzle of organelle genome evolution. For all their similarities, mitochondrial genomes have a greater proclivity than ptDNAs for genomic embellishments, adornments, and eccentricities of all types. A recurring narrative is the crucial role that DNA repair, replication, and recombination have in fashioning organelle chromosomes. The DNA maintenance machineries of mitochondria and plastids often have common origins and shared parts, which can help account for convergent organelle genome evolution within and among species, and the nuclear location of genes that mediate other strange processes could explain the apparent parallel distribution of other eccentric features as well. Nucleotide substitution rate data from across the eukaryotic tree suggest that DNA maintenance in mitochondrial systems is more capricious and variable than those of plastids, which could lead to mitochondrial chromosomes being more structurally varied than ptDNAs, but why this is the case remains unknown. Future studies on organelle DNA maintenance and mutation from diverse eukaryotes should help explain the outstanding array of organelle genomic architectures.
Supplementary Material
Acknowledgments
This work was supported by Discovery grants (to D.R.S. and P.J.K.) from the Natural Sciences and Engineering Research Council of Canada. P.J.K. is a fellow of the Canadian Institute for Advanced Research.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. J.P.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1422049112/-/DCSupplemental.
References
- 1.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith DR, Lee RW. A plastid without a genome: Evidence from the nonphotosynthetic green algal genus Polytomella. Plant Physiol. 2014;164(4):1812–1819. doi: 10.1104/pp.113.233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66(1):34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]
- 6.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–138. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 7.Burger G, Gray MW, Lang BF. Mitochondrial genomes: Anything goes. Trends Genet. 2003;19(12):709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 8.Howe CJ, Nisbet RER, Barbrook AC. The remarkable chloroplast genome of dinoflagellates. J Exp Bot. 2008;59(5):1035–1045. doi: 10.1093/jxb/erm292. [DOI] [PubMed] [Google Scholar]
- 9.Gray MW. Evolution of organellar genomes. Curr Opin Genet Dev. 1999;9(6):678–687. doi: 10.1016/s0959-437x(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 10.Lynch M. The Origins of Genome Architecture. Sinauer; Sunderland, MA: 2007. p. 494. [Google Scholar]
- 11.Palmer JD. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- 12.Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol Evol. 2012;4(3):294–306. doi: 10.1093/gbe/evs006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray MW. RNA editing in plant organelles: A fertile field. Proc Natl Acad Sci USA. 1996;93(16):8157–8159. doi: 10.1073/pnas.93.16.8157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186(2):299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- 16.Birky CW., Jr Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proc Natl Acad Sci USA. 1995;92(25):11331–11338. doi: 10.1073/pnas.92.25.11331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 18.Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- 19.Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322(6079):572–574. [Google Scholar]
- 20.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol Biol. 2011;76(3-5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nosek J, Tomáska L. Mitochondrial genome diversity: Evolution of the molecular architecture and replication strategy. Curr Genet. 2003;44(2):73–84. doi: 10.1007/s00294-003-0426-z. [DOI] [PubMed] [Google Scholar]
- 23.Bendich AJ. The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. BioEssays. 2007;29(5):474–483. doi: 10.1002/bies.20576. [DOI] [PubMed] [Google Scholar]
- 24.Mercer TR, et al. The human mitochondrial transcriptome. Cell. 2011;146(4):645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhelyazkova P, et al. The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell. 2012;24(1):123–136. doi: 10.1105/tpc.111.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311(5768):1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- 27.Smith DR, Lee RW. Low nucleotide diversity for the expanded organelle and nuclear genomes of Volvox carteri supports the mutational-hazard hypothesis. Mol Biol Evol. 2010;27(10):2244–2256. doi: 10.1093/molbev/msq110. [DOI] [PubMed] [Google Scholar]
- 28.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 29.Williams TA, Foster PG, Cox CJ, Embley TM. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504(7479):231–236. doi: 10.1038/nature12779. [DOI] [PubMed] [Google Scholar]
- 30.Orlando L. A 400,000-year-old mitochondrial genome questions phylogenetic relationships amongst archaic hominins: Using the latest advances in ancient genomics, the mitochondrial genome sequence of a 400,000-year-old hominin has been deciphered. BioEssays. 2014;36(6):598–605. doi: 10.1002/bies.201400018. [DOI] [PubMed] [Google Scholar]
- 31.Budowle B, Allard MW, Wilson MR, Chakraborty R. Forensics and mitochondrial DNA: Applications, debates, and foundations. Annu Rev Genomics Hum Genet. 2003;4:119–141. doi: 10.1146/annurev.genom.4.070802.110352. [DOI] [PubMed] [Google Scholar]
- 32.Hannon M, Gimpel J, Tran M, Rasala B, Mayfield S. Biofuels from algae: Challenges and potential. Biofuels. 2010;1(5):763–784. doi: 10.4155/bfs.10.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dahl EL, Rosenthal PJ. Apicoplast translation, transcription and genome replication: Targets for antimalarial antibiotics. Trends Parasitol. 2008;24(6):279–284. doi: 10.1016/j.pt.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Jukes TH, Osawa S. The genetic code in mitochondria and chloroplasts. Experientia. 1990;46(11-12):1117–1126. doi: 10.1007/BF01936921. [DOI] [PubMed] [Google Scholar]
- 35.Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993;9(8):265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- 36.Waller RF, Jackson CJ. Dinoflagellate mitochondrial genomes: Stretching the rules of molecular biology. BioEssays. 2009;31(2):237–245. doi: 10.1002/bies.200800164. [DOI] [PubMed] [Google Scholar]
- 37.Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Cell biology. Irremediable complexity? Science. 2010;330(6006):920–921. doi: 10.1126/science.1198594. [DOI] [PubMed] [Google Scholar]
- 38.Speijer D. Does constructive neutral evolution play an important role in the origin of cellular complexity? Making sense of the origins and uses of biological complexity. BioEssays. 2011;33(5):344–349. doi: 10.1002/bies.201100010. [DOI] [PubMed] [Google Scholar]
- 39.Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27(8):1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1):e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith DR. Updating our view of organelle genome nucleotide landscape. Front Genet. 2012;3:175. doi: 10.3389/fgene.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith DR, Keeling PJ. Gene conversion shapes linear mitochondrial genome architecture. Genome Biol Evol. 2013;5(5):905–912. doi: 10.1093/gbe/evt059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith DR, Hua J, Lee RW. Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr Genet. 2010;56(5):427–438. doi: 10.1007/s00294-010-0311-5. [DOI] [PubMed] [Google Scholar]
- 45.Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. Mitochondrial DNA of Chlamydomonas reinhardtii: The structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr Genet. 1993;24(3):241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- 46.Fricova D, et al. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology. 2010;156(Pt 7):2153–2163. doi: 10.1099/mic.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Janouskovec J, et al. Split photosystem protein, linear-mapping topology, and growth of structural complexity in the plastid genome of Chromera velia. Mol Biol Evol. 2013;30(11):2447–2462. doi: 10.1093/molbev/mst144. [DOI] [PubMed] [Google Scholar]
- 48.Smith DR, et al. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 2012;4(1):52–58. doi: 10.1093/gbe/evr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011;23(7):2499–2513. doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao R, Zhu XQ, Barker SC, Herd K. Evolution of extensively fragmented mitochondrial genomes in the lice of humans. Genome Biol Evol. 2012;4(11):1088–1101. doi: 10.1093/gbe/evs088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vlcek C, Marande W, Teijeiro S, Lukes J, Burger G. Systematically fragmented genes in a multipartite mitochondrial genome. Nucleic Acids Res. 2011;39(3):979–988. doi: 10.1093/nar/gkq883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lukes J, et al. Kinetoplast DNA network: Evolution of an improbable structure. Eukaryot Cell. 2002;1(4):495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci USA. 2003;100(3):892–897. doi: 10.1073/pnas.0336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Espelund M, et al. Genome fragmentation is not confined to the peridinin plastid in dinoflagellates. PLoS ONE. 2012;7(6):e38809. doi: 10.1371/journal.pone.0038809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith DR, et al. The Dunaliella salina organelle genomes: Large sequences, inflated with intronic and intergenic DNA. BMC Plant Biol. 2010;10:83. doi: 10.1186/1471-2229-10-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hikosaka K, et al. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol. 2010;27(5):1107–1116. doi: 10.1093/molbev/msp320. [DOI] [PubMed] [Google Scholar]
- 57.Petersen J, et al. Chromera velia, endosymbioses and the rhodoplex hypothesis—Plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages) Genome Biol Evol. 2014;6(3):666–684. doi: 10.1093/gbe/evu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pett W, et al. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA. 2011;22(4):130–142. doi: 10.3109/19401736.2011.624611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pramateftaki PV, Kouvelis VN, Lanaridis P, Typas MA. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: A unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 2006;6(1):77–90. doi: 10.1111/j.1567-1364.2005.00018.x. [DOI] [PubMed] [Google Scholar]
- 60.Brouard JS, Otis C, Lemieux C, Turmel M. The exceptionally large chloroplast genome of the green alga Floydiella terrestris illuminates the evolutionary history of the Chlorophyceae. Genome Biol Evol. 2010;2:240–256. doi: 10.1093/gbe/evq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Vries J, et al. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 2013;5(12):2540–2548. doi: 10.1093/gbe/evt205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barbrook AC, Voolstra CR, Howe CJ. The chloroplast genome of a Symbiodinium sp. clade C3 isolate. Protist. 2014;165(1):1–13. doi: 10.1016/j.protis.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 63.Arisue N, et al. The Plasmodium apicoplast genome: Conserved structure and close relationship of P. ovale to rodent malaria parasites. Mol Biol Evol. 2012;29(9):2095–2099. doi: 10.1093/molbev/mss082. [DOI] [PubMed] [Google Scholar]
- 64.de Koning AP, Keeling PJ. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006;4:12. doi: 10.1186/1741-7007-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hallick RB, et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21(15):3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Férandon C, Xu J, Barroso G. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal Genet Biol. 2013;55:85–91. doi: 10.1016/j.fgb.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 67.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5(2):418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slamovits CH, Saldarriaga JF, Larocque A, Keeling PJ. The highly reduced and fragmented mitochondrial genome of the early-branching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J Mol Biol. 2007;372(2):356–368. doi: 10.1016/j.jmb.2007.06.085. [DOI] [PubMed] [Google Scholar]
- 69.Janouškovec J, et al. Evolution of red algal plastid genomes: Ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS ONE. 2013;8(3):e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krause K. Plastid genomes of parasitic plants: a trail of reductions and losses. In: Bullerwell C, editor. Organelle Genetics. Springer; Berlin: 2012. pp. 79–103. [Google Scholar]
- 71.Boer PH, Gray MW. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell. 1988;55(3):399–411. doi: 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 72.Spencer DF, Gray MW. Ribosomal RNA genes in Euglena gracilis mitochondrial DNA: Fragmented genes in a seemingly fragmented genome. Mol Genet Genomics. 2011;285(1):19–31. doi: 10.1007/s00438-010-0585-9. [DOI] [PubMed] [Google Scholar]
- 73.Feagin JE, et al. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS ONE. 2012;7(6):e38320. doi: 10.1371/journal.pone.0038320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dang Y, Green BR. Substitutional editing of Heterocapsa triquetra chloroplast transcripts and a folding model for its divergent chloroplast 16S rRNA. Gene. 2009;442(1-2):73–80. doi: 10.1016/j.gene.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 75.Lang-Unnasch N, Aiello DP. Sequence evidence for an altered genetic code in the Neospora caninum plastid. Int J Parasitol. 1999;29(10):1557–1562. doi: 10.1016/s0020-7519(99)00119-8. [DOI] [PubMed] [Google Scholar]
- 76.Matsumoto T, Ishikawa SA, Hashimoto T, Inagaki Y. A deviant genetic code in the green alga-derived plastid in the dinoflagellate Lepidodinium chlorophorum. Mol Phylogenet Evol. 2011;60(1):68–72. doi: 10.1016/j.ympev.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 77.Byrne EM, Gott JM. Unexpectedly complex editing patterns at dinucleotide insertion sites in Physarum mitochondria. Mol Cell Biol. 2004;24(18):7821–7828. doi: 10.1128/MCB.24.18.7821-7828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Simpson AGB, Stevens JR, Lukeš J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;22(4):168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 79.Lin S, Zhang H, Spencer DF, Norman JE, Gray MW. Widespread and extensive editing of mitochondrial mRNAS in dinoflagellates. J Mol Biol. 2002;320(4):727–739. doi: 10.1016/s0022-2836(02)00468-0. [DOI] [PubMed] [Google Scholar]
- 80.Jackson CJ, Gornik SG, Waller RF. A tertiary plastid gains RNA editing in its new host. Mol Biol Evol. 2013;30(4):788–792. doi: 10.1093/molbev/mss270. [DOI] [PubMed] [Google Scholar]
- 81.Smith DR. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol. 2011;3:743–748. doi: 10.1093/gbe/evr068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Alverson AJ, et al. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae) Mol Biol Evol. 2010;27(6):1436–1448. doi: 10.1093/molbev/msq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hao W, Richardson AO, Zheng Y, Palmer JD. Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc Natl Acad Sci USA. 2010;107(50):21576–21581. doi: 10.1073/pnas.1016295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 85.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58(1):1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 86.Smith DR, Asmail SR. Next-generation sequencing data suggest that certain nonphotosynthetic green plants have lost their plastid genomes. New Phytol. 2014;204(1):7–11. doi: 10.1111/nph.12919. [DOI] [PubMed] [Google Scholar]
- 87.Iorizzo M, et al. De novo assembly of the carrot mitochondrial genome using next generation sequencing of whole genomic DNA provides first evidence of DNA transfer into an angiosperm plastid genome. BMC Plant Biol. 2012;12:61. doi: 10.1186/1471-2229-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Straub SCK, Cronn RC, Edwards C, Fishbein M, Liston A. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae) Genome Biol Evol. 2013;5(10):1872–1885. doi: 10.1093/gbe/evt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Knox EB. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci USA. 2014;111(30):11097–11102. doi: 10.1073/pnas.1403363111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ruck EC, Nakov T, Jansen RK, Theriot EC, Alverson AJ. Serial gene losses and foreign DNA underlie size and sequence variation in the plastid genomes of diatoms. Genome Biol Evol. 2014;6(3):644–654. doi: 10.1093/gbe/evu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Handa H. Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions? Mitochondrion. 2008;8(1):15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 92.La Claire JW, Wang J. Localization of plasmid like DNA in giant-celled marine green algae. Protoplasma. 2000;213:157–164. [Google Scholar]
- 93.Smith DR. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 2009;71(6):627–639. doi: 10.1007/s11103-009-9545-3. [DOI] [PubMed] [Google Scholar]
- 94.Robbens S, et al. The complete chloroplast and mitochondrial DNA sequence of Ostreococcus tauri: Organelle genomes of the smallest eukaryote are examples of compaction. Mol Biol Evol. 2007;24(4):956–968. doi: 10.1093/molbev/msm012. [DOI] [PubMed] [Google Scholar]
- 95.Sloan DB, Taylor DR. Evolutionary rate variation in organelle genomes: the role of mutational processes. In: Bullerwell C, editor. Organelle Genetics. Springer; Berlin: 2012. pp. 123–146. [Google Scholar]
- 96.Carrie C, Giraud E, Whelan J. Protein transport in organelles: Dual targeting of proteins to mitochondria and chloroplasts. FEBS J. 2009;276(5):1187–1195. doi: 10.1111/j.1742-4658.2009.06876.x. [DOI] [PubMed] [Google Scholar]
- 97.Dorrell RG, Howe CJ. Functional remodeling of RNA processing in replacement chloroplasts by pathways retained from their predecessors. Proc Natl Acad Sci USA. 2012;109(46):18879–18884. doi: 10.1073/pnas.1212270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Leister D. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene. 2005;354:110–116. doi: 10.1016/j.gene.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 99.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5(7):e1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22(6):1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum Mol Genet. 2006;15(1):143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 102.Lavrov DV, et al. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): Six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol Biol Evol. 2013;30(4):865–880. doi: 10.1093/molbev/mss274. [DOI] [PubMed] [Google Scholar]
- 103.Hellberg ME. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol Biol. 2006;6:24. doi: 10.1186/1471-2148-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The “fossilized” mitochondrial genome of Liriodendron tulipifera: ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11:29. doi: 10.1186/1741-7007-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hua J, Smith DR, Borza T, Lee RW. Similar relative mutation rates in the three genetic compartments of Mesostigma and Chlamydomonas. Protist. 2012;163(1):105–115. doi: 10.1016/j.protis.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 106.Christensen AC. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol Evol. 2013;5(6):1079–1086. doi: 10.1093/gbe/evt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oliveira DCSG, Raychoudhury R, Lavrov DV, Werren JH. Rapidly evolving mitochondrial genome and directional selection in mitochondrial genes in the parasitic wasp nasonia (Hymenoptera: Pteromalidae) Mol Biol Evol. 2008;25(10):2167–2180. doi: 10.1093/molbev/msn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhu A, Guo W, Jain K, Mower JP. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 2014;31(5):1228–1236. doi: 10.1093/molbev/msu079. [DOI] [PubMed] [Google Scholar]
- 110.Huang D, Meier R, Todd PA, Chou LM. Slow mitochondrial COI sequence evolution at the base of the metazoan tree and its implications for DNA barcoding. J Mol Evol. 2008;66(2):167–174. doi: 10.1007/s00239-008-9069-5. [DOI] [PubMed] [Google Scholar]
- 111.Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312(5773):570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- 112.Strüder-Kypke MC, Lynn DH. Comparative analysis of the mitochondrial cytochrome coxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst Biodivers. 2010;8(1):131–148. [Google Scholar]
- 113.Smith DR, Keeling PJ. Twenty-fold difference in evolutionary rates between the mitochondrial and plastid genomes of species with secondary red plastids. J Eukaryot Microbiol. 2012;59(2):181–184. doi: 10.1111/j.1550-7408.2011.00601.x. [DOI] [PubMed] [Google Scholar]
- 114.Heger TJ, Mitchell EAD, Leander BS. Holarctic phylogeography of the testate amoeba Hyalosphenia papilio (Amoebozoa: Arcellinida) reveals extensive genetic diversity explained more by environment than dispersal limitation. Mol Ecol. 2013;22(20):5172–5184. doi: 10.1111/mec.12449. [DOI] [PubMed] [Google Scholar]
- 115.Duchene D, Bromham L. Rates of molecular evolution and diversification in plants: Chloroplast substitution rates correlate with species-richness in the Proteaceae. BMC Evol Biol. 2013;13:65. doi: 10.1186/1471-2148-13-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Smith DR, Jackson CJ, Reyes-Prieto A. Nucleotide substitution analyses of the glaucophyte Cyanophora suggest an ancestrally lower mutation rate in plastid vs mitochondrial DNA for the Archaeplastida. Mol Phylogenet Evol. 2014;79:380–384. doi: 10.1016/j.ympev.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 117.Magee AM, et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010;20(12):1700–1710. doi: 10.1101/gr.111955.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pochon X, Putnam HM, Gates RD. Multi-gene analysis of Symbiodinium dinoflagellates: A perspective on rarity, symbiosis, and evolution. PeerJ. 2014;2:e394. doi: 10.7717/peerj.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Besnard G, Hernández P, Khadari B, Dorado G, Savolainen V. Genomic profiling of plastid DNA variation in the Mediterranean olive tree. BMC Plant Biol. 2011;11:80. doi: 10.1186/1471-2229-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84(24):9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.