Abstract
The endosymbiotic origin of plastids from cyanobacteria was a landmark event in the history of eukaryotic life. Subsequent to the evolution of primary plastids, photosynthesis spread from red and green algae to unrelated eukaryotes by secondary and tertiary endosymbiosis. Although the movement of cyanobacterial genes from endosymbiont to host is well studied, less is known about the migration of eukaryotic genes from one nucleus to the other in the context of serial endosymbiosis. Here I explore the magnitude and potential impact of nucleus-to-nucleus endosymbiotic gene transfer in the evolution of complex algae, and the extent to which such transfers compromise our ability to infer the deep structure of the eukaryotic tree of life. In addition to endosymbiotic gene transfer, horizontal gene transfer events occurring before, during, and after endosymbioses further confound our efforts to reconstruct the ancient mergers that forged multiple lines of photosynthetic microbial eukaryotes.
Keywords: eukaryotes, endosymbiosis, endosymbiotic gene transfer, genomics, plastids
A large and diverse body of evidence supports the notion that mitochondria and plastids (chloroplasts), the textbook membrane-bound organelles of eukaryotes, evolved from once free-living bacteria by endosymbiosis (1, 2). The plastids of plants and algae are demonstrably cyanobacterial in nature, and the host cell that took up the plastid progenitor perhaps 1.5 billion years ago was undoubtedly a fully fledged eukaryote (3–6). A common theme in the evolution of plastids is endosymbiotic gene transfer (EGT), the flow of genes from endosymbiont to host (7, 8). The establishment of dedicated machinery for importing the protein products of such genes is considered a critical step in the transition from endosymbiont to organelle (9), and a great deal is known about the process of plastid protein import (3, 10).
Yet we are still far from having a satisfactory explanation for the evolution of photosynthetic eukaryotes and the plastids they harbor. Like mitochondria, plastids are thought to have evolved from a prokaryotic endosymbiont on a single occasion, in this case in a common heterotrophic ancestor shared by red algae, glaucophyte algae, and green algae (and their land plant descendants); these so-called “primary” plastids are widely believed to have evolved vertically in these three core photosynthetic lines ever since (5, 11). Not everyone is so sure, however (e.g., refs. 12 and 13), and indeed we know that plastids have moved horizontally by endosymbiotic mergers involving unrelated eukaryotic donors and recipients (3, 14–16). Precisely how, and how many times, photosynthesis has spread among eukaryotes has been debated ever since the idea was first proposed in the 1970s (17, 18). Even with complete genome sequence data now available from a broad range of primary and secondary plastid-bearing algae, the evolutionary history of plastids across the tree of eukaryotic life is still unclear. Here I explore possible reasons why this might be, in light of EGT and the complex ways in which eukaryotic hosts and endosymbionts have amalgamated over the ∼1.5 billion years since plastids first evolved.
Evolving a Complex Alga
The broad strokes of primary plastid evolution can be inferred in a comparative genomic framework. Sequenced red, green, and glaucophyte plastid genomes are typically in the range of 100–200 kb-pairs in size and possess at most ∼250 genes (19–21), a small fraction of the ∼2,000–12,000 genes found in extant cyanobacteria (22). Most of the >1,000 proteins needed to support a functional plastid (23, 24) are encoded by nuclear genes; many (but not all) of these genes are of cyanobacterial ancestry, the product of EGT events that occurred during and after the integration of host and endosymbiont (7, 25, 26). Together with the migration of genes from endosymbiont to nucleus, dedicated translocation machineries known as TOC and TIC (translocon of the outer and inner chloroplast membrane) evolved from a mixture of host and endosymbiont-derived gene products to move essential proteins across the membranes of the nascent organelle (24, 27, 28). The general complexity and specific make-up of the TOC and TIC translocons speak to the single origin of the double-membrane primary plastids in red, green, and glaucophyte algae (2, 29, 30). Together these three groups comprise the Archaeplastida (31), one of a half-dozen proposed “supergroups” of eukaryotes, phylogenomic support for which has waxed and waned as sequence datasets have grown larger (32–35).
Attempts to quantify the “footprint’” of endosymbiosis in the nuclear genomes of primary plastid-bearing organisms have produced variable results. A 2002 study by Martin et al. (26) estimated that ∼18% of nuclear genes in the land plant Arabidopsis are of cyanobacterial ancestry (∼4,500 in total). Intriguingly, <50% of these genes encode proteins that were predicted to be plastid-localized: endosymbiotically derived gene products appear to contribute to a wide range of plastid-independent subcellular processes, including cell division, intracellular trafficking, gene expression, and metabolism (26). Similar analyses of the genomes of single-celled algae, including reds, greens, and glaucophytes, have produced much lower estimates of the cyanobacterial footprint, in the range of 6–12% (36–39), and with a higher proportion of the endosymbiont-derived proteins predicted to be plastid-targeted. There are both biological and methodological factors to consider when accounting for these differences (40). It is nevertheless clear that the biology of the primordial algal host cell was impacted by the endosymbiosis that gave rise to the plastid, and it is worth considering what the phylogenetic implications of this might be. In questioning the tree-based support for the sisterhood of red, green, and glaucophyte algae, Stiller (12) has argued that the influx of cyanobacterial DNA during the initial establishment of the plastid was sufficiently large (and cryptic) that it now compromises our ability to reliably reconstruct a host cell phylogeny based on nuclear genes. We will return to this general issue in due course.
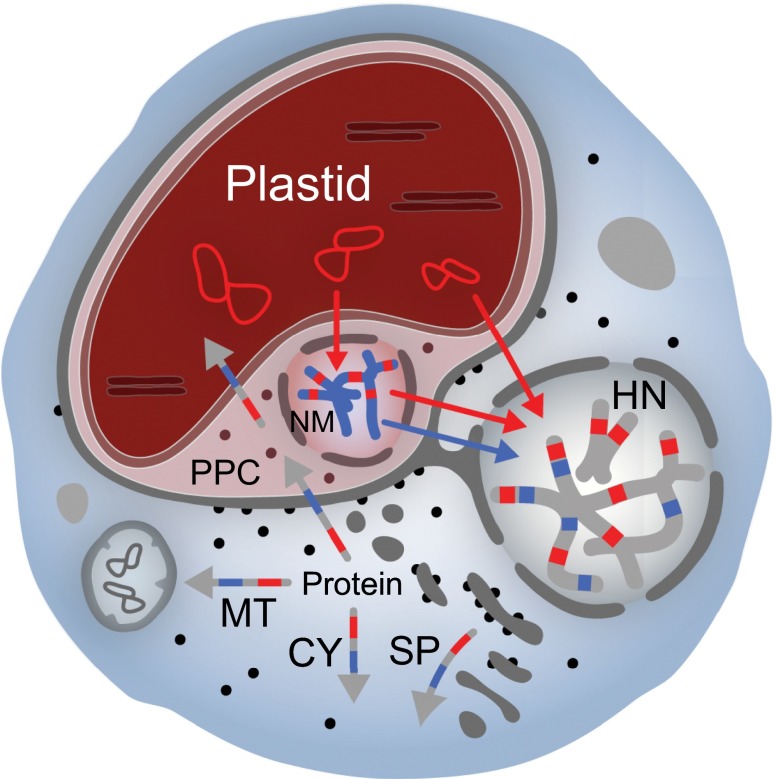
An important additional layer of organellar complexity is the process of secondary endosymbiosis, whereby a primary plastid-bearing alga is engulfed and assimilated by a heterotrophic host. A wealth of biochemical and comparative genomic data show that both red and green algal plastids have been captured in this manner. The “green” side of the equation is relatively straightforward. Two algal groups, the euglenids and chlorarachniophytes, harbor chlorophyll a+b-pigmented plastids, which are surrounded by three and four membranes, respectively (41–43). The chlorarachniophytes are of particular interest in that the nucleus of the green algal endosymbiont persists in a miniaturized form called a “nucleomorph” (44, 45). Nucleomorphs are considered the “smoking guns” of secondary endosymbiosis—most complex algae do not have them—although as we shall see their genomes are minuscule compared with the algal nuclear genomes from which they evolved; most of their genes have been lost or transferred to the host nucleus by EGT (45) (Fig. 1).
Fig. 1.
Genome and proteome mosaicism in complex algae. Diagram depicts a generic complex alga with a four-membrane secondary plastid and a nucleomorph (NM). When present, the nucleomorph resides within the periplastidial compartment (PPC), which corresponds to the cytosol of the engulfed primary algal cell. Sequenced nucleomorph genomes of cryptophyte and chlorarachniophyte algae possess at most ∼500 protein genes; most of the proteins needed to sustain the plastid and associated compartments are the products of genes residing in the host nucleus (HN). The host cell’s endomembrane system is involved in targeting nucleus-encoded proteins to the organelle. In some (but not all) complex algae, the outermost plastid membrane is physically continuous with the rough ER, allowing cotranslational insertion of organelle-targeted proteins. Blue and red arrows show endosymbiotic gene transfer events from the plastid to the primary and secondary host nuclei: their genomes are evolutionary mosaics. Multicolored arrows show possible destinations for nucleus-encoded proteins whose genes trace back to the plastid (red), nucleomorph (blue), and secondary host nucleus (gray). Evidence for all of these scenarios has come from comparative genomic investigations of several independently evolved complex algae (see text). Gene transfers involving mitochondrial DNA have been omitted for simplicity. CY, cytosol; MT, mitochondrion; SP, secretory pathway.
Red algal-derived complex plastids are found in a much wider range of algae and exhibit a broader spectrum of plastid-associated features. They are also more controversial. Some of these algae, such as the haptophytes, nucleomorph-bearing cryptophytes, and stramenopiles (e.g., diatoms and giant kelp) are well known and have well-studied plastids surrounded by four membranes (3, 46). Others, such as the coral reef-associated chromerids, have only come to light in the past decade (47, 48). Still others are secondarily nonphotosynthetic forms, such as the apicomplexan parasite Plasmodium; this organism harbors a red algal-derived plastid with a reduced genome (49), retained as the site of essential biochemical processes, such as fatty acid and iron-sulfur cluster biosynthesis (50).
One final group of organisms, the dinoflagellates, deserves special mention as the champions of plastid acquisition and exchange. Most photosynthetic species possess a peridinin-pigmented, red algal-type plastid surrounded by three membranes, which appears to be specifically related to the four-membrane plastids of apicomplexans and chromerids (48, 51). However, other dinoflagellates are more complicated in having replaced their “standard” peridinin plastid with an organelle taken from green algae by “serial secondary” endosymbiosis (52, 53) or from haptophytes (54) and diatoms (55) by “tertiary” endosymbiosis.
Cellular Mergers and Acquisitions—Who, How, and How Many?
What do the genomes of complex plastids look like and what do they tell us about the spread of photosynthesis? With the exception of the bizarre single-gene “minicircles” found in the peridinin plastids of dinoflagellates (e.g., refs. 56–58), which are essentially useless as phylogenetic markers, secondary plastid genomes are generally similar in structure, sequence, and coding capacity to the green and red primary plastid genomes from which they are derived (21). Simply put, primary plastids evolved and diversified into the red and green lines well before any of the secondarily derived organelles did. This of course is what allows us to label complex plastids as “red” or “green” in the first place, but it also provides the means with which to test more specific hypotheses about plastid gain. For example, phylogenomic evidence suggests that the green plastids of euglenids and chlorarachniophytes are derived from evolutionarily distinct green algal endosymbionts (43), a conclusion bolstered by the fact that the two hosts clearly belong to very different parts of the eukaryotic tree. Although the euglenozoans are “excavates,” the rhizarians (to which chlorarachniophytes belong) are related to stramenopiles and alveolates (33, 34, 59). Green algal secondary plastids evolved twice.
The chromalveolate hypothesis of Cavalier-Smith has dominated the field of plastid evolution for 15 years; it posits that all red complex plastids share a single secondary endosymbiotic origin (60). More specifically, there was a single engulfment of a red alga by a common heterotrophic ancestor shared by “chomists” and alveolates, a diverse collection of organisms sometimes referred to as CASH taxa: cryptophytes, alveolates (dinoflagellates, apicomplexans, and ciliates), stramenopiles, and haptophytes (61). A guiding principle behind the chromalveolate hypothesis is that evolving an organelle and everything that goes with it is difficult; the number of inferred endosymbiotic events should thus be minimized, even if it means that extensive plastid loss must also be invoked (e.g., in the plastid-lacking ciliates). The intricacies of the chromalveolate hypothesis have been discussed extensively elsewhere (e.g., refs. 14, 15, 62, and 63). What matters for the time being is that plastid genome sequences are generally consistent with a single origin scenario: multigene phylogenies show the plastid genomes of the CASH lineages to be monophyletic, albeit with variable statistical support (e.g., refs. 51, 57, 64, and 65).
An important development has been the realization that the plastid protein import machinery operating in all CASH taxa with four-membrane organelles is homologous. Where investigated, the standard TOC and TIC translocons mediate the import of nucleus-encoded proteins across the two innermost cyanobacterium-derived membranes of green and red complex plastids (see refs. 3, 29, 66, and 67 and references therein). Protein transport across the outermost host-derived plastid membrane in both red and green algal-derived complex plastids involves the host cell’s endomembrane system; the signal peptide secretion pathway has clearly been co-opted for this purpose multiple times independently (3). What about the remaining membrane? Maier and colleagues have shown that translocation across the second outermost membrane (originally the plasma membrane of the algal endosymbiont) in red-type four-membrane plastids is mediated by a multiprotein complex dubbed SELMA [symbiont-specific endoplasmic reticulum (ER)-associated degradation-like machinery (68–71)]. The evolution of SELMA appears to represent a singularity, an intricate complex cobbled together from a dozen or more proteins at least some of which originally functioned as part of the ER-associated degradation machinery in the red algal endosymbiont. SELMA is a robust character that supports a single origin of four-membrane CASH plastids (2).
However, there are limits to what plastids and their genomes can tell us. The monophyly of red algal complex plastids in CASH lineages is consistent with a single origin in the common ancestor of all of the organisms that contain them, but it does not prove it. Similarly, although the presence of the same highly distinct protein import apparatus in CASH taxa (excluding the three-membrane organelles of dinoflagellates) speaks strongly to the single origin of red algal-type complex plastids, there are other possible explanations for how the organelles—and SELMA—have come to reside in such diverse algal lineages. Based on complementary lines of inquiry, it now seems very likely that at least some of the red algal-derived plastids traditionally thought to be of secondary origin are in fact serial acquisitions involving tertiary and perhaps even quaternary endosymbiotic events. Algal nuclear genome sequences have provided the key to begin unlocking some of the mysteries; at present they raise as many questions as they answer.
The past decade has seen support and enthusiasm for the chromalveolate hypothesis gradually diminished, in large part because of the results of global eukaryote phylogenomic investigations. Recent analyses of data matrices containing >100 concatenated nucleus-encoded proteins do not support the hypothesis that the CASH taxa are a monophyletic assemblage. Most notably, the cryptophytes and haptophytes, once thought to be specifically related (e.g., refs. 32 and 72), do not appear to be each other’s closest relatives in an analysis of 258 nuclear genes (59). Instead, the haptophytes show strong ties to the stramenopiles, alveolates, and rhizarians, the latter group being a large and mostly nonphotosynthetic lineage (the green complex plastid-containing chlorarachniophytes being a notable exception). The cryptophytes branch as sister to the primary plastid-bearing red, green, and glaucophyte algae (59). It would be prudent to consider the jury still out over many aspects of the supergroup level relationships among eukaryotes, including the position of the root. Nevertheless, it has become increasingly difficult to rationalize a single, ancient red algal plastid acquisition in a common ancestor shared by such an apparently large fraction of eukaryotic diversity.
Complementary evidence against the chromalveolate hypothesis has come from a critical assessment of the strength of phylogenetic signals contained in the nuclear and organellar genomes of CASH taxa. Baurain et al. (61) showed that the plastid genomes of these groups appear to have diverged from each other more recently than their mitochondrial and nuclear genomes have. In essence, these analyses show that the evolutionary histories of the plastid and nucleocytoplasmic compartments of the CASH lineages are incongruent with one another, a red flag for the existence of cryptic plastid spread (73).
Stiller et al. (74) have recently gone an interesting step further in proposing an explicit scenario for how red complex plastids have evolved: from red algae to cryptophytes by secondary endosymbiosis, from nucleomorph-bearing cryptophytes to a subgroup of the stramenopiles by tertiary endosymbiosis (more specifically, to an ancestor of modern-day photosynthetic ochrophytes), and from stramenopiles to haptophytes by quaternary transfer. Their novel statistical approaches add empirical meat to earlier, largely speculative, proposals for the serial transfer of plastids (e.g., refs. 15 and 63). They also speak to the importance of a well-known but often neglected aspect of algal evolution: nucleus-to-nucleus gene transfer.
The Fate of Endosymbiotically Derived Nuclei and Their Genes
The evolution of a complex alga entails two rounds of EGT, first the movement of genes from the cyanobacterium-turned-plastid to the primary algal nucleus, and second from the primary nucleus to the secondary host nucleus (Fig. 1). In most complex algae the second wave of EGT has gone to completion: all essential genes for the maintenance of the plastid now reside in the secondary nucleus, and their protein products are imported to the organelle in a stepwise fashion as described above. Sequenced nucleomorph genomes from cryptophytes and chlorarachniophytes are all <1-Mb pairs in size and possess at most ∼500 genes and ∼30 genes for plastid-targeted proteins (44, 75–79). Consequently, the cyanobacterial footprint in the host nuclear genomes of complex algae should be roughly similar to that found in primary plastid-bearing algae, assuming their plastid proteomes are similarly sized (i.e., comprised of 1,000 or more proteins; see below). Less attention has been paid to the fate of eukaryotic genes transferred from one nucleus to the other.
Stiller et al. (74) probe the genomic footprint of serial endosymbiosis in an unorthodox fashion. They capitalize on the strong correlation that exists between (i) the overall similarity between a given query genome and a target group of genomes (inferred by top BLAST hits) and (ii) the size of the sequence database for that target; basically, the larger the number of target genes and genomes, the more likely it is that top matches between query and target will be obtained by chance (80). This correlation was used to identify “outlier” relationships between CASH taxa: that is, relationships between specific (and apparently distantly related) algal groups that rise above statistical background. It is the identification of these outliers that lead Stiller et al. (74) to posit specific serial endosymbiotic interactions (and EGT events) between cryptophytes, plastid-bearing stramenopiles (ochrophytes), and haptophytes. The sequence of events is polarized with cryptophytes as the source of the original red algal secondary plastid mainly because of the presence of a nucleomorph in this group. Interestingly, when the authors exclude genes involved in photosynthesis from their analyses, a significant EGT signal can still be detected between cryptophytes and plastid-bearing stramenopiles, but not between cryptophytes and plastid-lacking stramenopiles, which we would expect to see if endosymbiosis predated the divergence of photosynthetic and nonphotosynthetic stramenopile lineages.
These analyses are far outside the box of standard phylogenomic procedures. As such, they beg numerous questions. First and foremost, what is the precise nature of the data that is giving rise to the signal? What do we see when we look at the genes underlying the statistically significant outlier relationships between specific CASH taxa? It is too early to say, but at face value one important message seems to be that serial endosymbioses involve the transfer and retention of genes of eukaryotic ancestry from donors to recipients. Left undetected, such transfers have the potential to greatly confound phylogenomic analyses. So how big is this eukaryotic footprint, how important is it, and how good are we at detecting it?
Detailed insight into the fate of endosymbiotically derived genes in complex algae has come from investigation of the nuclear genomes of two independently evolved nucleomorph-bearing algae, the chlorarachniophyte Bigelowiella natans and the cryptophyte Guillardia theta (81). From a set of ∼6,200 and 7,500 broadly distributed proteins in B. natans and G. theta, respectively, 353 (5.7%) and 508 (6.8%) were found to be of clear algal endosymbiont origin (81). One curious observation is that genes of apparent green and red algal ancestry were found in both genomes. (Recall that B. natans has a green algal-derived plastid and nucleomorph, whereas G. theta has a plastid and nucleomorph of red algal origin.) This red-green mosaicism mirrors that seen in an earlier study (82) and in analyses of other complex algae, such as diatoms (83, 84) and Chromera (85, 86). The magnitude and significance of this red-green footprint is as yet unclear, but it seems reasonable to assume that a good chunk of it is the result of methodological artifacts and taxonomic biases in the datasets presently available for analysis (see below and refs. 2, 84, and 87 for discussion).
Setting aside for a moment the issue of red versus green, what is interesting about the algal proteins in B. natans and G. theta is the diversity of their subcellular locations. Only ∼20% of these proteins were predicted to be plastid-targeted in each organism and only 3% appear to be targeted to the periplastidial compartment (PPC), that is, the remnant algal cytosolic compartment where the nucleomorph resides (Fig. 1). Proteins of algal ancestry were also predicted to be targeted to the host ER/Golgi apparatus and to the mitochondrion (81). The bulk of these algal proteins—76% and 66% in B. natans and G. theta, respectively—appear to be cytosolic. The secondary endosymbionts of both organisms clearly contributed to biochemical pathways and processes taking place in the host. Conversely, host-derived genes appear to play a significant role in the maintenance of plastid- and nucleomorph-associated processes in B. natans and G. theta, and duplication of both host and endosymbiont-derived nuclear genes has resulted in the production of recently diverged paralogs that appear to be targeted to different compartments in the cell (e.g., the PPC and host cytosol) (81). These in silico predictions need to be looked at much more closely, but the emerging picture is one of mix-and-match biochemistry (Fig. 1).
A similar picture can be drawn from a recent study of EGT in two tertiary plastid-bearing dinoflagellate lineages, the kareniaceans, which have a haptophyte-derived plastid, and the so-called “dinotoms,” dinoflagellates with diatom plastids (88). A merit of the transcriptome-based datasets used in this analysis is that the tertiary acquisitions are relatively recent and the donor lineages (haptophytes and diatoms) are well defined, in principle making it easier to detect EGT against the dinoflagellate nuclear genomic background. Burki et al. (88) identified 90 and 9 examples of EGT-derived genes in the host nuclear genomes of kareniaceans and dinotoms, respectively. As in B. natans and G. theta, the predicted functions of at least some of these EGT proteins extend beyond the plastid. Nevertheless, Burki et al. (88) deemed the level of EGT to be low: 4.7% of analyzable proteins in the kareniaceans (90 of 1,923 trees), ∼1% in dinotoms. The dinotoms are interesting in that the diatom “endosymbiont” (it is not clear whether “endosymbiont” or “organelle” is more appropriate) possesses an apparently unreduced nucleus and mitochondrion (55), which may partly explain the low levels of observed EGT.
The analyses of Burki et al. (88) underscore the complexity of signals that invariably emerge from a careful gene-by-gene phylogenomic analysis that includes manual inspection. Most analyzable proteins yield phylogenies sufficiently poorly resolved that no meaningful conclusions can be drawn, and there is a background of phylogenetic noise, a variable number of trees in which the proteins of interest branch robustly with homologs from a wide range of prokaryotes and eukaryotes. It is usually not clear where this “noise” ends and meaningful phylogenetic signal (in terms of the proportion of total trees) begins. Considering these and other factors, Burki et al. speculate that the impact of EGT on the nuclear genomes of complex algae has been more modest than often stated, “…in the range of a few hundred genes or less” (88).
This may be so, but we still need to account for the large size of the organellar proteomes in complex algae and their antecedents in primary plastid-bearing algae. Curtis et al. predicted no fewer than ∼1,000 and 2,400 nucleus-encoded, PPC-/nucleomorph-targeted proteins in B. natans and G. theta, respectively, and ∼700 plastid-targeted proteins each (81). In a companion study, a proteomics approach was used to identify ∼300 candidate plastid-PPC-nucleomorph proteins in B. natans (89). A thorough investigation of two diatoms predicted 900–1,000 nucleus-encoded, plastid-targeted proteins, comprising no less than ∼8% of the genes in the Thalassiosira pseudonana and Phaeodactylum tricornutum genomes (90). These numbers are similar to the ∼1,000 predicted plastid proteins in the green alga Chlamydomonas reinhardtii (∼7% of the total gene number), ∼900 of which are experimentally verified (see ref. 23 and references therein). All things considered, even accounting for proteins encoded in the plastid (43, 91) and nucleomorph genomes (44, 75), there are nowhere near enough host nucleus-encoded algal proteins to service the plastid and PPC-nucleomorph compartments of B. natans and G. theta (and as noted above, only 20–25% of the identified algal proteins actually are predicted to be targeted). So from which nuclear genome, host or endosymbiont, do the bulk of the missing genes ultimately derive?
Secondary host nuclear genes have clearly been recruited to function in the endosymbiont-derived compartments in both G. theta and B. natans, for example, via gene duplication and differential targeting of paralogs (81). It may be that much, or even most, of the organellar proteomes of these and other complex algae are made up of host-derived proteins. However, the “repurposing” of endosymbiont-derived proteins has also clearly taken place, and there is no obvious reason to assume that during the course of host–endosymbiont integration, the plastid and PPC-nucleomorph proteomes (which were already large at the time of secondary endosymbiosis) were heavily modified by proteins of host origin but that the host cell was somehow not similarly impacted in reverse. After all, the evidence for extensive nucleus-to-nucleus gene transfer in the evolution of complex algae is beyond refute—nucleomorphs have either disappeared entirely or become frozen in a highly reduced state (81)—and mechanistically the DNA transfer process must surely be random with respect to the biochemical functions of the migrating genes. Either the cryptophyte and chlorarachniophyte host cells have donated many more proteins to their endosymbionts than vice versa, or the algal footprint in the nuclear genomes of these two lineages (and presumably other complex algae) has been vastly underestimated. The truth is probably somewhere in between, and the implications are significant.
Gene Transfer and Genome Mosaicism: Causes and Consequences
… we probably need to keep our expectations more relaxed when it comes to the phylogenetic behavior of genes that eukaryotes acquired from plastids and mitochondria. If we do that, endosymbiotic theory explains a lot as it is.
Zimorski et al. (2)
The presence of red and green genes in independently evolved complex algae is vexing. It has been explained as a consequence of phagotrophy in the case of green plastid-bearing mixotrophic chlorarachniophytes (82) and it has prompted proposals for the existence of a cryptic green algal endosymbiont in an ancestor of red plastid-bearing algae (e.g., refs. 83 and 92). The reality is that there are lots of possible explanations, both biological and experimental (84, 85, 87, 93, 94), and when strict a priori criteria are applied, such as requiring that algal genes be present in two or more primary plastid-bearing groups to be considered algal (e.g., ref. 84), thousands of candidate EGTs quickly becomes hundreds (87).
Nevertheless, as I have argued above, if we keep tabs on the number of nucleus-encoded proteins required to sustain primary and complex plastids, the algal footprint seems insufficient. The number of genes that we can unambiguously track (by careful phylogenomic analysis) from cyanobacteria to the nuclear genomes of red and green algae and on to complex algae is small, but this does not mean that these are the only genes that were inherited from reds and greens. The cellular mergers we are trying to dissect are ancient, and it is important to consider how much the nuclear genomes of present-day donor and recipient lineages actually reflect those at the time of serial endosymbiosis. If the situation is even remotely like that in prokaryotes, where mosaic genomes are the norm (see refs. 2 and 95 and references therein), then the answer may be “not very much.” If on top of EGT we add horizontal gene transfer (HGT) before, during, and after serial endosymbiotic events, the challenge of quantifying the endosymbiotic footprint in complex algae becomes even more difficult.
What is the evidence that HGT leads to algal genome mosaicism? Prokaryotic contributions to the nuclear genomes of diatoms, dinoflagellates, glaucophytes, and red algae have been documented (e.g., refs. 37 and 96–98). How substantial these contributions are is debatable; methodologies and datasets used to detect HGT vary, as do interpretations of the data. At any rate, evidence suggests that algal genomes can be remarkably dynamic over short evolutionary timescales. For example, two strains of the planktonic green alga Micromonas, whose small subunit ribosomal RNA genes are ∼97% identical, share only 90% of their genes (99). In an analysis of genome sequences from more than a dozen strains of the haptophyte Emiliania huxleyi, Read et al. found that almost 25% of genes present in the reference genome were not found in at least three other strains (100). These authors went as far as to invoke the concept of the “pan-genome,” now broadly accepted for prokaryotes as the sum-total of genes present in a species, only a small subset of which is actually present in a given strain (101).
There is (and should be) debate over the frequency and detection of HGT in eukaryotes. However, the evidence that prokaryote-to-eukaryote and eukaryote-to-eukaryote transfer happens is now overwhelming and it is not limited to single-celled organisms (see refs. 102 and 103 and references therein). HGT is known to have played a role in lineage-specific adaptations via transfer of metabolic enzymes (104) and it can also impact classic housekeeping markers, such as tubulin (105). We should also not discount viruses as agents of HGT in eukaryotes as they are in prokaryotes. For example, the cryptophyte G. theta and the haptophyte E. huxleyi were recently shown to possess genes derived from nucleocytoplasmic large DNA viruses (megaviruses). The G. theta genome possesses a gene encoding a megavirus-type D5 primase/helicase, which appears to have replaced the standard archaeal-eukaryotic DNA primase gene (106). An analysis by Yutin et al. (107) showed that megaviruses, such as the recently described pandoravirus (108), have acquired numerous genes from their amoeba hosts, including transcription and translation factors. It is not unreasonable to think that they could mediate the spread of such genes within and between distinct eukaryotic lineages.
It is also not unreasonable to ask how much EGT and HGT contamination exists in the large protein supermatrices used to construct global eukaryote phylogenies. Recent work from Katz and Grant (109) serves as a cautionary tale. Upon close inspection, these authors found that ∼10% of 1,554 genes present in most or all of the major eukaryotic lineages showed evidence of cyanobacterial EGT in photosynthetic taxa (109). Removal of these genes changed the relative branching order of red, green, and glaucophyte algae. Methods for the systematic detection of problem genes exist (e.g., refs. 110 and 111) but concatenated datasets are becoming increasingly unwieldy, and the possible reasons for incongruence between the data partitions within them are numerous and not limited simply to nonvertical inheritance. We do the best we can to rid our data matrices of obvious EGTs and HGTs, usually on a taxon-by-taxon basis, acknowledging that factors influencing sequence evolution can change dramatically when a gene moves from one genome to another. However, we are only as good at detecting incongruence as our methods allow us to be, and we are invariably dependent on an imperfect phylogenomic framework when deciding whether a given protein should be retained or excluded. Prokaryote-to-eukaryote HGTs are the easiest to detect and are arguably more likely to impart useful novelty to their recipients. Eukaryote-to-eukaryote transfers should be easier in that the donors and recipients are more closely related and have more compatible gene-expression systems; they will also be harder to detect. These factors apply equally well to genes acquired by HGT and EGT.
All this could mean that the spread of complex plastids by serial endosymbiosis is even more convoluted than even the most complex models propose. Or it could mean that it is simpler. It will be interesting to see how the outlier data of Stiller et al. (74) stack up against traditional gene-by-gene phylogenomic results, including the “rhodoplex hypothesis” of Petersen et al. (112). These are hypotheses that can be tested by seeing whether there are indeed significant patterns of gene exchange between candidate plastid donors and recipients. We still need to account for the presence of the conserved multiprotein translocon SELMA in CASH taxa (71). Is it simply nucleus-to-nucleus EGT? There is precedent for this if we consider genes encoding the core components of the TIC and TOC translocons, widely considered markers for the uniqueness of primary plastid evolution. Although TIC-TOC evolved only once, the genes for TIC-TOC components have passed from endosymbiont to host on multiple occasions; that is, in the green algal secondary plastids of chlorarachniophytes (and presumably euglenids as well) (67) and at least once into the red complex plastid line (3). The precise history of the TIC-TOC and SELMA translocons in red complex plastids remains to be determined, but it would be a mistake to dismiss nucleus-to-nucleus EGT as an explanation for their dispersal. Zimorski et al. (2) wisely caution against expecting too much from individual gene trees. If we relax our expectations, endosymbiotic theory does indeed explain a lot. However, it still has some explaining to do.
Are All of the Players at the Table?
With the now substantial number of organellar and nuclear genome sequences available for study, it is tempting to assume that we have enough data; we just need to figure out what it all means. In that regard we are regularly proven wrong, however, as underscored by exciting discoveries from the realm of uncultured phototrophs. For example, the rappemonads are a recently named group of marine and freshwater plastid-bearing eukaryotes whose plastid ribosomal RNA sequences ally them with haptophytes and cryptophytes (113). Essentially nothing is known about their biology and it will be important to determine the nature of the host cell in which these plastids reside. Janouškovec et al. recently showed that prokaryotic environmental sequence datasets in fact contain thousands of plastid-derived sequences mislabeled as “novel bacteria” (114). Analysis of these “new” sequences revealed the existence of a rich diversity of organisms related to apicomplexans, far beyond Chromera and Vitrella (47, 48).
Research over the past decade has also revealed the presence of heterotrophic protist lineages with phylogenetic affinities to secondary plastid-harboring groups. These groups include the telonemids, centrohelids, katablepharids, and Picomonas (32, 115–118). As protist taxonomic sampling continues to expand, plastid-bearing lineages such as the cryptophytes have begun to look increasingly like isolated photosynthetic tips on a tree comprised mainly of heterotrophic branches. This contrasts the situation described above for apicomplexans, where the phylogenetic neighborhood is steadily being filled in with photosynthetic members. The bottom line is that the pool of unexplored eukaryotic biodiversity is deep, and it is both sobering and exhilarating to realize that we have only begun to dip beneath its surface. Rapidly advancing genomic and metagenomic technologies will yield sequence data from these depths sooner rather than later, and will no doubt force us to adjust our framework for interpreting the diversification of photosynthetic life on Earth. In some ways, the field of eukaryotic comparative genomics is in the same place the field of prokaryotic genomics was 20 years ago, grappling with the relative importance of vertical versus horizontal inheritance, and knowing that we have only glimpsed the true diversity of organisms that exist and must be accounted for.
Acknowledgments
Thanks to Sven Gould, Daniel Moog, Ugo Cenci, Bruce Curtis, and Ford Doolittle for helpful discussion and comments; two anonymous reviewers for comments on an earlier version of the manuscript; and apologies to those whose primary research could not be cited because of space constraints. Research on endosymbiosis in the J.M.A. laboratory is supported by the Natural Sciences and Engineering Research Council of Canada, The Canadian Institutes of Health Research, and the Centre for Comparative Genomics and Evolutionary Bioinformatics at Dalhousie University. J.M.A. acknowledges long-term support from the Canadian Institute for Advanced Research.
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. P.J.K. is a guest editor invited by the Editorial Board.
References
- 1.Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 2.Zimorski V, Ku C, Martin WF, Gould SB. Endosymbiotic theory for organelle origins. Curr Opin Microbiol. 2014;22:38–48. doi: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 3.Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 4.Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108(33):13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reyes-Prieto A, Weber AP, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- 6.Yoon HS, Hackett JD, Ciniglia C, Pinto G, Bhattacharya D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol Biol Evol. 2004;21(5):809–818. doi: 10.1093/molbev/msh075. [DOI] [PubMed] [Google Scholar]
- 7.Martin W, Brinkmann H, Savonna C, Cerff R. Evidence for a chimeric nature of nuclear genomes: Eubacterial origin of eukaryotic glyceraldehyde-3-phosphate dehydrogenase genes. Proc Natl Acad Sci USA. 1993;90(18):8692–8696. doi: 10.1073/pnas.90.18.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 9.Cavalier-Smith T, Lee JJ. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J Protozool. 1985;32(3):376–379. [Google Scholar]
- 10.Jarvis P, Soll J. Toc, Tic, and chloroplast protein import. Biochim Biophys Acta. 2001;1541(1-2):64–79. doi: 10.1016/s0167-4889(01)00147-1. [DOI] [PubMed] [Google Scholar]
- 11.Palmer JD. The symbiotic birth and spread of plastids: How many times and whodunnit? J Phycol. 2003;39(1):4–11. [Google Scholar]
- 12.Stiller JW. Plastid endosymbiosis, genome evolution and the origin of green plants. Trends Plant Sci. 2007;12(9):391–396. doi: 10.1016/j.tplants.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Nozaki H, et al. Phylogenetic positions of Glaucophyta, green plants (Archaeplastida) and Haptophyta (Chromalveolata) as deduced from slowly evolving nuclear genes. Mol Phylogenet Evol. 2009;53(3):872–880. doi: 10.1016/j.ympev.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19(2):R81–R88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- 15.Bodył A, Stiller JW, Mackiewicz P. Chromalveolate plastids: Direct descent or multiple endosymbioses? Trends Ecol Evol. 2009;24(3):119–121, author reply 121–122. doi: 10.1016/j.tree.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor FJR. Implications and extensions of the serial endosymbiosis theory of the origin of eukaryotes. Taxon. 1974;23(2-3):229–258. [Google Scholar]
- 18.Gibbs SP. The chloroplasts of Euglena may have evolved from symbiotic green algae. Can J Bot. 1978;56(22):2883–2889. [Google Scholar]
- 19.Bélanger AS, et al. Distinctive architecture of the chloroplast genome in the chlorophycean green alga Stigeoclonium helveticum. Mol Genet Genomics. 2006;276(5):464–477. doi: 10.1007/s00438-006-0156-2. [DOI] [PubMed] [Google Scholar]
- 20.Janouškovec J, et al. Evolution of red algal plastid genomes: Ancient architectures, introns, horizontal gene transfer, and taxonomic utility of plastid markers. PLoS ONE. 2013;8(3):e59001. doi: 10.1371/journal.pone.0059001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E, Archibald JM. Diversity and evolution of plastids and their genomes. In: Aronsson H, Sandelius AS, editors. The Chloroplast-Interactions with the Environment, Plant Cell Monographs. Springer; Berlin: 2009. pp. 1–39. [Google Scholar]
- 22.Dagan T, et al. Genomes of Stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol Evol. 2013;5(1):31–44. doi: 10.1093/gbe/evs117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terashima M, Specht M, Hippler M. The chloroplast proteome: A survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Curr Genet. 2011;57(3):151–168. doi: 10.1007/s00294-011-0339-1. [DOI] [PubMed] [Google Scholar]
- 24.Shi LX, Theg SM. The chloroplast protein import system: From algae to trees. Biochim Biophys Acta. 2013;1833(2):314–331. doi: 10.1016/j.bbamcr.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Martin W, et al. Gene transfer to the nucleus and the evolution of chloroplasts. Nature. 1998;393(6681):162–165. doi: 10.1038/30234. [DOI] [PubMed] [Google Scholar]
- 26.Martin W, et al. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA. 2002;99(19):12246–12251. doi: 10.1073/pnas.182432999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14(12):787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 28.McFadden GI. Plastids and protein targeting. J Eukaryot Microbiol. 1999;46(4):339–346. doi: 10.1111/j.1550-7408.1999.tb04613.x. [DOI] [PubMed] [Google Scholar]
- 29.McFadden GI, van Dooren GG. Evolution: Red algal genome affirms a common origin of all plastids. Curr Biol. 2004;14(13):R514–R516. doi: 10.1016/j.cub.2004.06.041. [DOI] [PubMed] [Google Scholar]
- 30.Reumann S, Inoue K, Keegstra K. Evolution of the general protein import pathway of plastids (review) Mol Membr Biol. 2005;22(1-2):73–86. doi: 10.1080/09687860500041916. [DOI] [PubMed] [Google Scholar]
- 31.Adl SM, et al. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52(5):399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 32.Burki F, et al. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, Telonemia and Centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1(1):231–238. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampl V, et al. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci USA. 2009;106(10):3859–3864. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parfrey LW, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59(5):518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodríguez-Ezpeleta N, et al. Monophyly of primary photosynthetic eukaryotes: Green plants, red algae, and glaucophytes. Curr Biol. 2005;15(14):1325–1330. doi: 10.1016/j.cub.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 36.Moustafa A, Bhattacharya D. PhyloSort: A user-friendly phylogenetic sorting tool and its application to estimating the cyanobacterial contribution to the nuclear genome of Chlamydomonas. BMC Evol Biol. 2008;8:6. doi: 10.1186/1471-2148-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price DC, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335(6070):843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- 38.Reyes-Prieto A, Hackett JD, Soares MB, Bonaldo MF, Bhattacharya D. Cyanobacterial contribution to algal nuclear genomes is primarily limited to plastid functions. Curr Biol. 2006;16(23):2320–2325. doi: 10.1016/j.cub.2006.09.063. [DOI] [PubMed] [Google Scholar]
- 39.Sato N, Ishikawa M, Fujiwara M, Sonoike K. Mass identification of chloroplast proteins of endosymbiont origin by phylogenetic profiling based on organism-optimized homologous protein groups. Genome Inform. 2005;16(2):56–68. [PubMed] [Google Scholar]
- 40.Archibald JM. Algal genomics: Exploring the imprint of endosymbiosis. Curr Biol. 2006;16(24):R1033–R1035. doi: 10.1016/j.cub.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Hallick RB, et al. Complete sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 1993;21(15):3537–3544. doi: 10.1093/nar/21.15.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hibberd DJ, Norris RE. Cytology and ultrastructure of Chlorarachnion reptans (Chlorarachniophyta divisio nova, Chlorarachniophyceae classis nova) J Phycol. 1984;20(2):310–330. [Google Scholar]
- 43.Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: Evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24(1):54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- 44.Gilson PR, et al. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus. Proc Natl Acad Sci USA. 2006;103(25):9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore CE, Archibald JM. Nucleomorph genomes. Annu Rev Genet. 2009;43:251–264. doi: 10.1146/annurev-genet-102108-134809. [DOI] [PubMed] [Google Scholar]
- 46.Not F, et al. Diversity and ecology of eukaryotic marine phytoplankton. In: Piganeau G, editor. Genomic Insights into the Biology of Algae. Academic; Waltham, MA: 2012. pp. 1–53. [Google Scholar]
- 47.Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451(7181):959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 48.Oborník M, Lukeš J. Cell biology of chromerids: Autotrophic relatives to apicomplexan parasites. Int Rev Cell Mol Biol. 2013;306:333–369. doi: 10.1016/B978-0-12-407694-5.00008-0. [DOI] [PubMed] [Google Scholar]
- 49.Waller RF, McFadden GI. The apicoplast: A review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7(1):57–79. [PubMed] [Google Scholar]
- 50.Seeber F, Soldati-Favre D. Metabolic pathways in the apicoplast of apicomplexa. Int Rev Cell Mol Biol. 2010;281:161–228. doi: 10.1016/S1937-6448(10)81005-6. [DOI] [PubMed] [Google Scholar]
- 51.Janouškovec J, Horák A, Oborník M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci USA. 2010;107(24):10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto T, et al. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist. 2011;162(2):268–276. doi: 10.1016/j.protis.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Minge MA, et al. A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol Biol. 2010;10:191. doi: 10.1186/1471-2148-10-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tengs T, et al. Phylogenetic analyses indicate that the 19’Hexanoyloxy-fucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin. Mol Biol Evol. 2000;17(5):718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- 55.Dodge JD. A dinoflagellate with both a mesocaryotic and a eucaryotic nucleus. I. Fine structure of the nuclei. Protoplasma. 1971;73(2):145–157. doi: 10.1007/BF01275591. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, Green BR, Cavalier-Smith T. Single gene circles in dinoflagellate chloroplast genomes. Nature. 1999;400(6740):155–159. doi: 10.1038/22099. [DOI] [PubMed] [Google Scholar]
- 57.Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist. 2004;155(1):65–78. doi: 10.1078/1434461000165. [DOI] [PubMed] [Google Scholar]
- 58.Barbrook AC, Santucci N, Plenderleith LJ, Hiller RG, Howe CJ. Comparative analysis of dinoflagellate chloroplast genomes reveals rRNA and tRNA genes. BMC Genomics. 2006;7:297. doi: 10.1186/1471-2164-7-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: Phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279(1736):2246–2254. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46(4):347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 61.Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 2010;27(7):1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- 62.Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- 63.Sanchez-Puerta MV, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol. 2008;44(5):1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- 64.Iida K, Takishita K, Ohshima K, Inagaki Y. Assessing the monophyly of chlorophyll-c containing plastids by multi-gene phylogenies under the unlinked model conditions. Mol Phylogenet Evol. 2007;45(1):227–238. doi: 10.1016/j.ympev.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Yoon HS, Hackett JD, Pinto G, Bhattacharya D. The single, ancient origin of chromist plastids. Proc Natl Acad Sci USA. 2002;99(24):15507–15512. doi: 10.1073/pnas.242379899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bullmann L, et al. Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. J Biol Chem. 2010;285(9):6848–6856. doi: 10.1074/jbc.M109.074807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hirakawa Y, Burki F, Keeling PJ. Genome-based reconstruction of the protein import machinery in the secondary plastid of a chlorarachniophyte alga. Eukaryot Cell. 2012;11(3):324–333. doi: 10.1128/EC.05264-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Felsner G, et al. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol Evol. 2011;3:140–150. doi: 10.1093/gbe/evq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hempel F, Bullmann L, Lau J, Zauner S, Maier UG. ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Mol Biol Evol. 2009;26(8):1781–1790. doi: 10.1093/molbev/msp079. [DOI] [PubMed] [Google Scholar]
- 70.Sommer MS, et al. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24(4):918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- 71.Stork S, et al. Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Eukaryot Cell. 2012;11(12):1472–1481. doi: 10.1128/EC.00183-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17(10):887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- 73.Larkum AW, Lockhart PJ, Howe CJ. Shopping for plastids. Trends Plant Sci. 2007;12(5):189–195. doi: 10.1016/j.tplants.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Stiller JW, et al. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat Commun. 2014;5:5764. doi: 10.1038/ncomms6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Douglas S, et al. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410(6832):1091–1096. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- 76.Lane CE, et al. Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc Natl Acad Sci USA. 2007;104(50):19908–19913. doi: 10.1073/pnas.0707419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tanifuji G, et al. Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol Evol. 2011;3:44–54. doi: 10.1093/gbe/evq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moore CE, Curtis B, Mills T, Tanifuji G, Archibald JM. Nucleomorph genome sequence of the cryptophyte alga Chroomonas mesostigmatica CCMP1168 reveals lineage-specific gene loss and genome complexity. Genome Biol Evol. 2012;4(11):1162–1175. doi: 10.1093/gbe/evs090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanifuji G, et al. Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: Convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics. 2014;15:374. doi: 10.1186/1471-2164-15-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stiller JW, Huang J, Ding Q, Tian J, Goodwillie C. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics. 2009;10:484. doi: 10.1186/1471-2164-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Curtis BA, et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature. 2012;492(7427):59–65. doi: 10.1038/nature11681. [DOI] [PubMed] [Google Scholar]
- 82.Archibald JM, Rogers MB, Toop M, Ishida K, Keeling PJ. Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc Natl Acad Sci USA. 2003;100(13):7678–7683. doi: 10.1073/pnas.1230951100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moustafa A, et al. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324(5935):1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- 84.Deschamps P, Moreira D. Reevaluating the green contribution to diatom genomes. Genome Biol Evol. 2012;4(7):683–688. doi: 10.1093/gbe/evs053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burki F, et al. Re-evaluating the green versus red signal in eukaryotes with secondary plastid of red algal origin. Genome Biol Evol. 2012;4(6):626–635. doi: 10.1093/gbe/evs049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Woehle C, Dagan T, Martin WF, Gould SB. Red and problematic green phylogenetic signals among thousands of nuclear genes from the photosynthetic and apicomplexa-related Chromera velia. Genome Biol Evol. 2011;3:1220–1230. doi: 10.1093/gbe/evr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moreira D, Deschamps P. What was the real contribution of endosymbionts to the eukaryotic nucleus? Insights from photosynthetic eukaryotes. Cold Spring Harb Perspect Biol. 2014;6(7):a016014. doi: 10.1101/cshperspect.a016014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burki F, et al. Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryot Cell. 2014;13(2):246–255. doi: 10.1128/EC.00299-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hopkins JF, et al. Proteomics reveals plastid- and periplastid-targeted proteins in the chlorarachniophyte alga Bigelowiella natans. Genome Biol Evol. 2012;4(12):1391–1406. doi: 10.1093/gbe/evs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gruber A, Rocap G, Kroth PG, Armbrust EV, Mock T. Plastid proteome prediction for diatoms and other algae with secondary plastids of the red lineage. Plant J. 2015;81(3):519–528. doi: 10.1111/tpj.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Douglas SE, Penny SL. The plastid genome of the cryptophyte alga, Guillardia theta: Complete sequence and conserved synteny groups confirm its common ancestry with red algae. J Mol Evol. 1999;48(2):236–244. doi: 10.1007/pl00006462. [DOI] [PubMed] [Google Scholar]
- 92.Frommolt R, et al. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol Biol Evol. 2008;25(12):2653–2667. doi: 10.1093/molbev/msn206. [DOI] [PubMed] [Google Scholar]
- 93.Dagan T, Martin W. Microbiology. Seeing green and red in diatom genomes. Science. 2009;324(5935):1651–1652. doi: 10.1126/science.1175765. [DOI] [PubMed] [Google Scholar]
- 94.Elias M, Archibald JM. Sizing up the genomic footprint of endosymbiosis. BioEssays. 2009;31(12):1273–1279. doi: 10.1002/bies.200900117. [DOI] [PubMed] [Google Scholar]
- 95.Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3(2):180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456(7219):239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- 97.Schönknecht G, et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science. 2013;339(6124):1207–1210. doi: 10.1126/science.1231707. [DOI] [PubMed] [Google Scholar]
- 98.Shoguchi E, et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 2013;23(15):1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- 99.Worden AZ, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324(5924):268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- 100.Read BA, et al. Emiliania huxleyi Annotation Consortium Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature. 2013;499(7457):209–213. doi: 10.1038/nature12221. [DOI] [PubMed] [Google Scholar]
- 101.Mira A, Martín-Cuadrado AB, D’Auria G, Rodríguez-Valera F. The bacterial pan-genome: A new paradigm in microbiology. Int Microbiol. 2010;13(2):45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- 102.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9(8):605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 103.Hirt RP, Alsmark C, Embley TM. Lateral gene transfers and the origins of the eukaryote proteome: A view from microbial parasites. Curr Opin Microbiol. 2015;23:155–162. doi: 10.1016/j.mib.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Keeling PJ. Functional and ecological impacts of horizontal gene transfer in eukaryotes. Curr Opin Genet Dev. 2009;19(6):613–619. doi: 10.1016/j.gde.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 105.Simpson AG, Perley TA, Lara E. Lateral transfer of the gene for a widely used marker, alpha-tubulin, indicated by a multi-protein study of the phylogenetic position of Andalucia (Excavata) Mol Phylogenet Evol. 2008;47(1):366–377. doi: 10.1016/j.ympev.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 106.Filée J. Multiple occurrences of giant virus core genes acquired by eukaryotic genomes: The visible part of the iceberg? Virology. 2014;466-467:53–59. doi: 10.1016/j.virol.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 107.Yutin N, Wolf YI, Koonin EV. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466-467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Philippe N, et al. Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science. 2013;341(6143):281–286. doi: 10.1126/science.1239181. [DOI] [PubMed] [Google Scholar]
- 109.Katz LA, Grant JR. Taxon-rich phylogenomic analyses resolve the eukaryotic tree of life and reveal the power of subsampling by sites. Syst Biol December 23, 2014 doi: 10.1093/sysbio/syu126. , 10.1093/sysbio/syu126. [DOI] [PubMed] [Google Scholar]
- 110.de Vienne DM, Ollier S, Aguileta G. Phylo-MCOA: A fast and efficient method to detect outlier genes and species in phylogenomics using multiple co-inertia analysis. Mol Biol Evol. 2012;29(6):1587–1598. doi: 10.1093/molbev/msr317. [DOI] [PubMed] [Google Scholar]
- 111.Leigh JW, Susko E, Baumgartner M, Roger AJ. Testing congruence in phylogenomic analysis. Syst Biol. 2008;57(1):104–115. doi: 10.1080/10635150801910436. [DOI] [PubMed] [Google Scholar]
- 112.Petersen J, et al. Chromera velia, endosymbioses and the rhodoplex hypothesis—Plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages) Genome Biol Evol. 2014;6(3):666–684. doi: 10.1093/gbe/evu043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim E, et al. Newly identified and diverse plastid-bearing branch on the eukaryotic tree of life. Proc Natl Acad Sci USA. 2011;108(4):1496–1500. doi: 10.1073/pnas.1013337108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Janouškovec J, Horák A, Barott KL, Rohwer FL, Keeling PJ. Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr Biol. 2012;22(13):R518–R519. doi: 10.1016/j.cub.2012.04.047. [DOI] [PubMed] [Google Scholar]
- 115.Cavalier-Smith T, von der Heyden S. Molecular phylogeny, scale evolution and taxonomy of centrohelid heliozoa. Mol Phylogenet Evol. 2007;44(3):1186–1203. doi: 10.1016/j.ympev.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 116.Okamoto N, Chantangsi C, Horák A, Leander BS, Keeling PJ. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS ONE. 2009;4(9):e7080. doi: 10.1371/journal.pone.0007080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Seenivasan R, Sausen N, Medlin LK, Melkonian M. Picomonas judraskeda gen. et sp. nov.: The first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘picobiliphytes’. PLoS ONE. 2013;8(3):e59565. doi: 10.1371/journal.pone.0059565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shalchian-Tabrizi K, et al. Telonemia, a new protist phylum with affinity to chromist lineages. Proc Biol Sci. 2006;273(1595):1833–1842. doi: 10.1098/rspb.2006.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]