Significance
For over two decades, inteins have been considered parasitic elements. However, recent examples of conditional protein splicing indicate that some inteins may have evolved a regulatory role in response to the environment. These cues result in fine-tuning of host protein function under conditions in which the proteins themselves are regulated. Here we report on the discovery that the Mycobacterium tuberculosis SufB intein when expressed in Escherichia coli possesses extraordinary sensitivity to oxidative and nitrosative stresses. These conditions are highly relevant to survival of mycobacterial pathogens. This work delivers a breakthrough in the understanding of regulation of protein splicing through cysteine chemistry, and proposes a new form of posttranslational control of the M. tuberculosis SufB protein.
Keywords: controllable protein splicing, posttranslational regulation, cysteine chemistry, [Fe-S] biogenesis, intein function
Abstract
Inteins are mobile genetic elements that self-splice at the protein level. Mycobacteria have inteins inserted into several important genes, including those corresponding to the iron-sulfur cluster assembly protein SufB. Curiously, the SufB inteins are found primarily in mycobacterial species that are potential human pathogens. Here we discovered an exceptional sensitivity of Mycobacterium tuberculosis SufB intein splicing to oxidative and nitrosative stresses when expressed in Escherichia coli. This effect results from predisposition of the intein’s catalytic cysteine residues to oxidative and nitrosative modifications. Experiments with a fluorescent reporter system revealed that reactive oxygen species and reactive nitrogen species inhibit SufB extein ligation by forcing either precursor accumulation or N-terminal cleavage. We propose that splicing inhibition is an immediate, posttranslational regulatory response that can be either reversible, by inducing precursor accumulation, or irreversible, by inducing N-terminal cleavage, which may potentially channel mycobacteria into dormancy under extreme oxidative and nitrosative stresses.
Inteins are mobile genetic elements that invade genes and are spliced out at the protein level (Fig. 1A) (1, 2). Mycobacterium tuberculosis has inteins in three important genes, dnaB, recA, and sufB, which encode a replicative helicase, recombinase, and iron-sulfur [Fe-S] cluster assembly protein, respectively. Of particular interest is that related mycobacteria have inteins in some of the same genes, but not always at the same location (3–5). Furthermore, inteins at different locations in the same gene tend to be divergent, suggesting independent intein invasion events and that the inteins have been maintained because they may benefit the cell (2). SufB is the most dramatic example of a mycobacterial protein with multiple intein insertion points, especially in mycobacterial species that are potential human pathogens (Fig. 1B) (6). Such examples of sporadic intein distribution have led to the hypothesis that the presence of the intein may confer some selective advantage through posttranslational regulation of the host protein (2, 5).
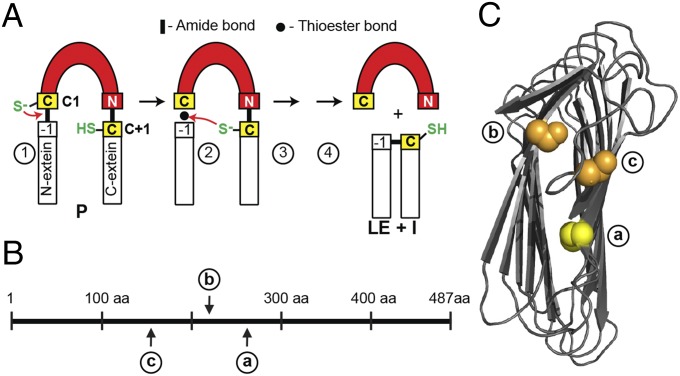
Fig. 1.
Splicing mechanism and intein insertions. (A) Simplified splicing mechanism using two catalytic cysteines (yellow). Initial nucleophilic attack (red arrow) in the precursor (P) by C1 at the N terminus of the intein (step 1). Second nucleophilic attack on the thioester by C+1 (step 2). Asparagine (N) cyclization to release intein (I) (step 3). Native peptide bond formation in ligated exteins (LE) (step 4). (B) Overview of SufB intein insertions. Three unique insertion points a, b, and c, with numbering based on M. tuberculosis SufB. (C) Simplified structure model of M. tuberculosis SufB exteins. Complete models of M. tuberculosis exteins are in Fig. S2. The C+1 at intein insertion site a is yellow, and S+1 at b and c are orange.
SufB functions as a subunit of the SufBCD complex to assemble [Fe-S] clusters (7, 8). The SUF system is required for metalloprotein biogenesis and plays an important role in both resisting iron limitation and in responding to oxidative and nitrosative stresses (9, 10), which are challenges that M. tuberculosis encounters in macrophages. Whereas many bacteria have multiple [Fe-S] cluster assembly pathways, mycobacteria use predominantly the SUF system (6, 11, 12), which is transcriptionally induced by oxidative and nitrosative stresses (13). Therefore, SufB is essential for survival and protein splicing is required for SufB function (14).
During protein splicing, the intein catalyzes a series of peptide bond rearrangements that release the intein from the two flanking host polypeptides called exteins, which are ligated to yield a functional protein (1). Protein splicing is usually initiated by a cysteine or serine residue at position 1 of the intein (Fig. 1A). This amino acid acts as a nucleophile that attacks the bond at the intein–N-terminal extein (N-extein) junction to form an ester or thioester bond (Fig. 1A, step 1). Next, the first residue of the C-terminal extein (C-extein), a cysteine, serine, or threonine residue, acts as the nucleophile that attacks the newly formed labile bond (Fig. 1A, step 2). After cyclization of the terminal asparagine, the free intein and ligated exteins are formed (Fig. 1A, steps 3 and 4). For the M. tuberculosis SufB intein, both nucleophiles are cysteines, designated C1 of the intein and C+1 of the C-extein.
The thiol side-chain of some cysteines can be highly reactive and undergo different modifications under oxidative and nitrosative stresses (15–17). Thiols can form disulfide bonds as well as sulfenates, sulfinates, and sulfonates under oxidizing conditions, and are susceptible to nitrosylation (16). Because oxidative and nitrosative stresses are relevant to M. tuberculosis biology (18), we wished to determine if splicing might be sensitive to these conditions. Here we show that, indeed, splicing of the SufB intein can be modulated by these stressors in Escherichia coli, and in different ways by reactive oxygen species (ROS) and reactive nitrogen species (RNS), and that the SufB intein is differentially sensitive among the three M. tuberculosis inteins. We further show that cysteine chemistry is involved in the effect. These findings suggest that the SufB intein could act as a sensor that regulates the SufB protein posttranslationally in response to ROS and RNS that are present in macrophages during infection.
Results
Diverse Intein Insertion Sites Are Proximal in SufB.
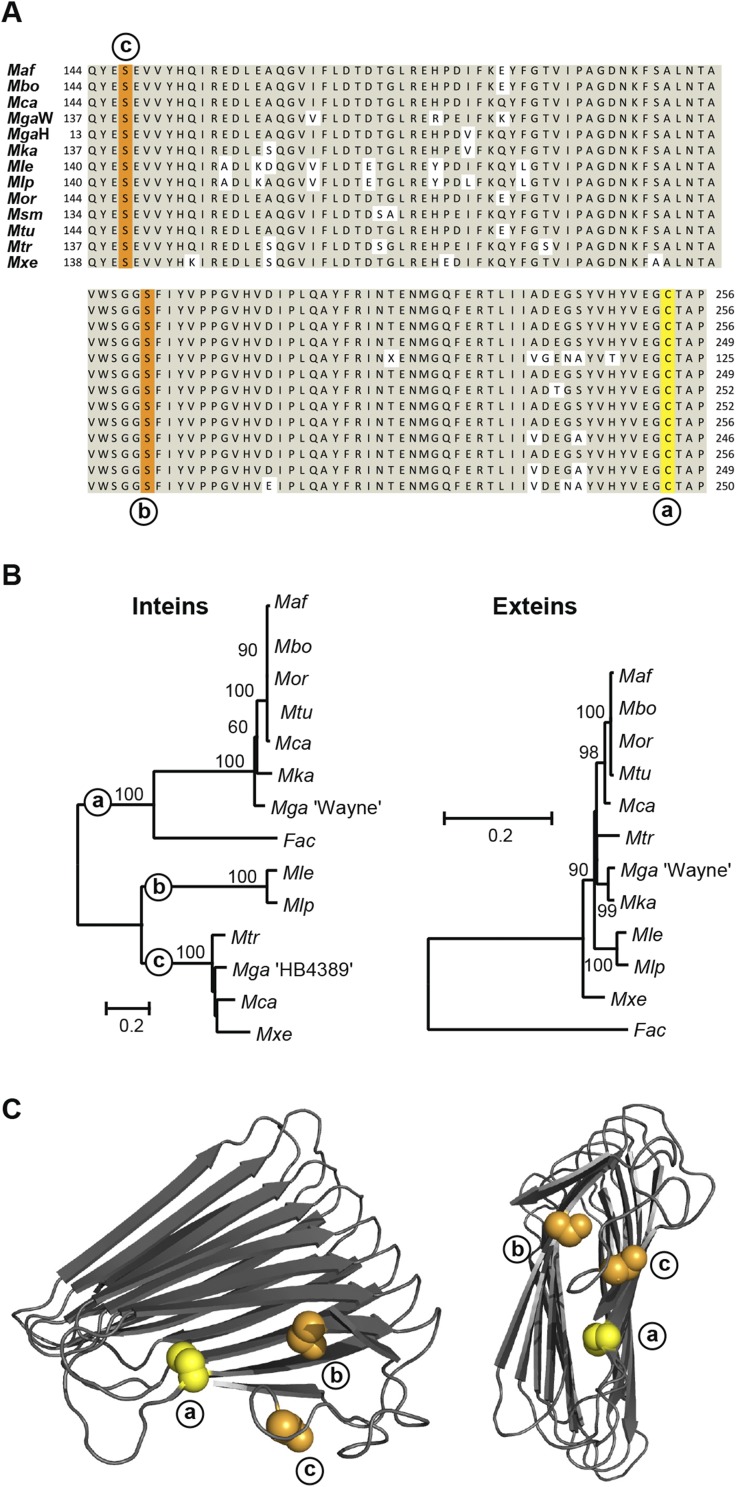
There are 3 unique insertion sites among 12 SufB intein-containing mycobacteria (Fig. 1B) (ref. 19 and NCBI database). These three sites, a, b, and c, separated by as many as 106 amino acids, suggest that three independent integration events occurred. The M. tuberculosis intein is at site a. Additionally, a SufB intein at site a was identified in a different domain of life, in the archaeon Ferroplasma acidarmanus. The site a inteins, including M. tuberculosis, have a cysteine at the +1 position, whereas the site b and c inteins both have serine (Fig. 1C and Fig. S1 A and C).
Fig. S1.
Intein insertions in conserved exteins. (A) Sequence alignment of the mycobacterial SufB exteins, for Mycobacterium africanum (Maf; CCC26555), Mycobacterium bovis (Mbo; CDO42744), Mycobacterium canettii (Mca, two inteins; CCK59531), Mycobacterium gastri “Wayne” (MgaW; ETW23634), Mycobacterium gastri “HB4389” (MgaH; CAC37008), Mycobacterium kansasii (Mka; AGZ53558), Mycobacterium leprae (Mle; CAC30101), Mycobacterium lepromatosis (Mlp; WP_045842589), Mycobacterium orygis (Mor; EMT36244), Mycobacterium smegmatis (Msm; AIU08276), Mycobacterium triplex (Mtr; CDO88315), Mycobacterium tuberculosis (Mtu; AFN49373), and Mycobacterium xenopi (Mxe; EID15070). The alignment shows the conservation of extein sequences with +1 residues for the three insertion sites, yellow for cysteine, and orange for serines. Mca SufB has two inteins, at sites a and c within the same gene, whereas there are two different strains of Mga with inteins at these two different sites. (B) Phylogenetic trees of SufB extein and intein sequences. Inteins group based on insertion sites, whereas exteins are on a single branch, separate from the aquatic archaeal Ferroplasma acidarmanus (Fac; AGO60154) extein. MgaH exteins were excluded because of incomplete sequence availability. Bootstrap values provide statistical support. The bar represents the number of substitutions per site. (C) Simplified structure models of Mtu SufB exteins. Complete models of Mtu exteins are in Fig. S2. The C+1 at intein insertion site a is yellow, and S+1 at b and c are orange.
The mycobacterial phylogenetic extein tree reveals a high level of conservation (Fig. S1B). Conversely, the inteins show divergence with three branches, corresponding to the independent insertion sites, a, b, and c (Fig. S1B). Strikingly, the F. acidarmanus intein branches with the group a inteins, despite divergence of the extein sequences, suggesting interkingdom gene transfer of the intein.
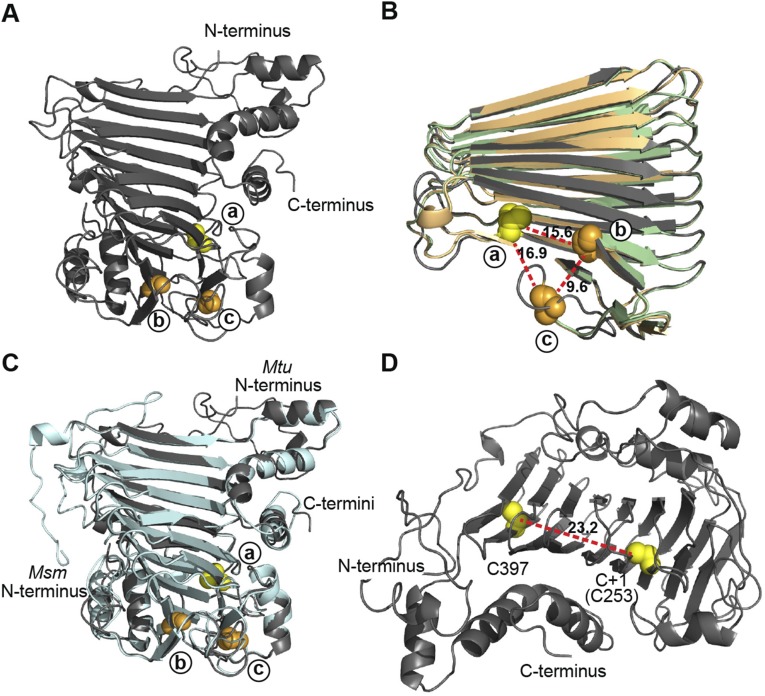
Structure models of mycobacterial SufB were generated using Phyre2 servers (20), showing a core right-handed parallel β-helix with helical N- and C-terminal domains and strong structural similarity (Fig. 1C and Fig. S2 A–C). Our models agree with the solved structure of SufD, which has been proposed to have arisen from a duplication event of SufB (8, 21). SufB functions as the scaffold for [Fe-S] cluster biogenesis, in complex with SufD and a SufC dimer (7, 8).
Fig. S2.
Structure modeling of SufB exteins. (A) Full structure model of M. tuberculosis SufB exteins. N- and C-termini are shown. (B) Overlay of SufB exteins representing the three distinct insertion sites. The SufB extein models of M. tuberculosis (dark gray; site a), M. leprae (light green; site b), and M. xenopi (light orange; site c) were aligned and simplified to the core β-helix. Alignment shows a high level of structural conservation and clustering of the insertion sites in 3D space. The distance between +1 residues was measured, based on the M. tuberculosis extein model: site a to site b 15.6 Å; site b to site c 9.6 Å; site c to site a 16.9 Å. (C) Overlay of full SufB extein models from intein-containing M. tuberculosis (dark gray) and inteinless M. smegmatis (light cyan). Alignment shows a high level of structural conservation and the conservation of the +1 residues at the three insertion sites. (D) Distance between cysteines in the SufB exteins. The distance between the two exclusive cysteines (C397 and C+1[C253]) in the M. tuberculosis SufB exteins was measured to be 23.2 Å. This distance suggests that C397 and C+1 are not cooperatively involved in [Fe-S] cluster coordination. All displayed models were made using Phyre2 servers. PyMOL was used to manipulate models and generate distances. Cysteines are indicated as yellow spheres and, with the exception of C397 in D, represent the C+1 of the site a insertions. Serines are shown as orange spheres and represent the S+1 of either the site b or site c insertions as indicated.
The three intein insertion sites were mapped onto a structure model of M. tuberculosis SufB exteins within the core β-helix (Fig. 1C and Fig. S1C). Despite the distance between the intein locations in primary sequence, they are close in 3D space, within 17 Å of each other (Fig. S2B). Their proximity hints at a functional importance of the inteins. It is also noteworthy that cysteines are often residues involved in coordination of [Fe-S] clusters, and that there are only two cysteines in the M. tuberculosis SufB (Fig. S2D). One of these two cysteines is Cys-253, which corresponds to the C+1 extein residue of the SufB precursor that is critical for splicing. This observation suggests that C+1 (Cys-253) is a pivotal residue, participating in both intein splicing and SufB function.
Splicing of the SufB Intein Is Hypersensitive to Oxidation.
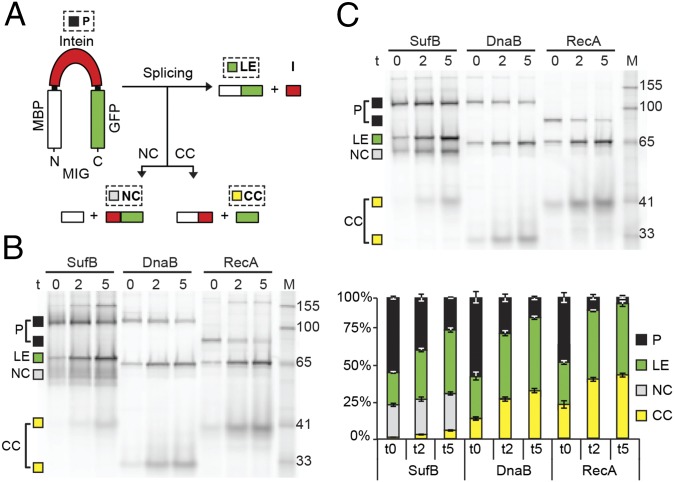
To conduct functional comparisons among the three M. tuberculosis inteins, we developed a fluorescence reporter system that would allow us to observe splicing in E. coli cell lysates. The sufB intein, as well as the two other M. tuberculosis intein, dnaB and recA, were cloned into a reporter construct encoding maltose binding protein (MBP) and green fluorescent protein (GFP) flanking the intein. These MIG (MBP-intein-GFP) fusions (Fig. 2A) contain short native exteins (8–10 amino acids) (Table S1) to preserve effects of extein residues at the splice junctions (22–24). Protein bands of the GFP-containing MIG precursors and splice products were visualized in unboiled extracts by GFP fluorescence in SDS-polyacrylamide gels (Fig. 2B). All three constructs had accumulated an unspliced precursor (P) during induction at time 0, the point of inhibition of protein synthesis, after which the precursor was auto-processed.
Fig. 2.
SufB intein is sensitive to oxidation. (A) Reporter construct MIG. Precursor (P), ligated exteins (LE), and off-pathway N- and C-terminal cleavage products (NC and CC) were visualized using in-gel fluorescence. (B) Diffuse bands and N-terminal cleavage are pronounced in SufB. Splicing was monitored over time (t: 0, 2, and 5 h) for SufB, DnaB, and RecA MIG. There are migration anomalies unrelated to oxidative effects. First, LE bands migrate slightly differently, because of distinct extein residues flanking the three inteins. Second, CC migrates spuriously because of distinct extein residues and folding anomalies of GFP without boiling (22). Third, RecA P runs lower than SufB or DnaB P due to a minimized RecA intein (37). Marker (M) bands are in kilodaltons. (C) SufB species are reversibly modified. Samples from B were rerun with TCEP. Bands were quantitated using ImageQuant. The diminished band intensity at t0 is likely because of reduced GFP chromophore formation. Sizes of splicing products are listed in Table S2. Data are representative of ≥three independent experiments.
Table S1.
Bacterial strains and plasmids
| Strain/plasmid | Relevant features; comments | Source |
| E. coli strains | ||
| DH5α | F– endAI recAl hsdRl7 (rK− mK−) deoR supE44 thi-J gyrA96 relA | Gibco-BRL |
| MG1655 (DE3) | F− (λDE3) ilvG rfb50 rph1 | James Imlay, University of Illinois, Urbana, IL |
| BL21 (DE3) | F– ompT hsdSB(rB–mB–) gal dcm rne131 (DE3) | Life Technologies |
| Plasmids | ||
| pACYCDuet-1 | Expression vector, T7 promoter, CamR | Novagen |
| pACYCDuet-1 FL SufB endo− | Mtu full-length sufb gene cloned into EcoRI/HindIII sites, with endonuclease mutations | |
| pACYC MIG-SufB | Mtu SufB intein flanked by short extein sequences (N-extein: ADEGSYVHYVEG; C-extein: CTAPIYKSDSLH) cloned into ClaI/SphI sites between MBP and GFP coding sequences in pACYCDuet-1 backbone, endonuclease mutations | This study |
| pACYC MIG-SufB C+1S | Site-directed mutagenesis of the pACYC MIG-SufB plasmid to mutate C+1 residue to S, endonuclease mutations | This study |
| pACYC MIG-RecA K-1P | Mtu minimized RecA intein flanked by short extein sequences (N-extein: RTRVKVVKNK; C-extein: CSPPFKQAEF) cloned into ClaI/SphI sites between MBP and GFP coding sequences in pACYCDuet-1 backbone, site directed mutagenesis carried out to mutate K-1 to P which inhibits splicing | This study |
| pACYC MIG-DnaB G-1F | Mtu DnaB intein flanked by short extein sequences (N-extein: PMLADLRESG; C-extein: SLEQDAD) cloned into ClaI/SphI sites between MBP and GFP coding sequences in pACYCDuet-1 backbone, site directed mutagenesis carried out to mutate G-1 to F which delays splicing | This study |
| pET30b | Cloning Vector | Novagen |
| pET30b/WT SufB Intein 4C/S | Four cysteines in endonuclease domain mutated to serine | This study |
| pET30b/C+1S SufB Intein 4C/S | Four cysteines in endonuclease domain mutated to serine | This study |
Table S2.
Sizes of splicing products
| Construct | P (kDa) | LE (kDA) | NC (kDa) | CC (kDa) |
| MIG SufB | 114.9 | 74.8 | 68.6 | 28.3 |
| MIG DnaB | 120.6 | 74.1 | 74.3 | 27.8 |
| MIG RecA | 93.6 | 74.6 | 47.1 | 28.1 |
| Full-length SufB | 96.0 | 56.0 | 66.0 | N/A |
All three intein precursors generated ligated exteins (LE) and a small amount of off-pathway C-terminal cleavage product (CC). Additionally, WT MIG SufB processing specifically resulted in accumulation of off-pathway N-terminal cleavage product (NC) (Fig. 2 A and B). Curiously, the WT MIG SufB samples showed diffuse precursor and N-terminal cleavage bands, as well as faint bands above the precursor. Treatment with the reducing agent Tris (2-carboxyethyl) phosphine (TCEP) sharpened the bands and removed the high molecular-weight products (Fig. 2C), suggesting that the band artifacts were because of oxidative modifications of the cysteines present in the precursor and the cleavage product. The well-resolved TCEP-treated bands facilitated quantitative analysis of intein splicing. The stack plot, where the GFP-containing products (P, LE, NC, and CC) total 100%, shows precursor disappearance, accumulation of ligated exteins, and more C-terminal cleavage product with time for all three inteins (Fig. 2C). However, processing of the SufB precursor exclusively resulted in N-terminal cleavage.
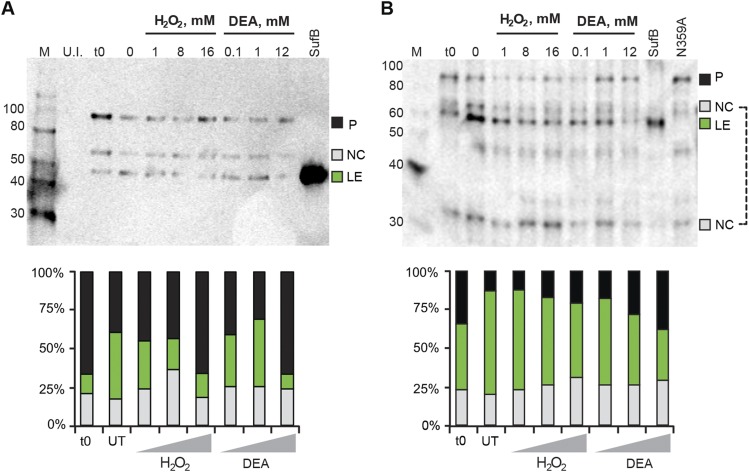
To ensure that the N-terminal cleavage observed with WT MIG SufB precursor is not an artifact of splicing from nonnative exteins, full-length SufB precursor, with the native SufB exteins, was analyzed using extein antibodies. Again, we observed formation of ligated exteins and N-terminal cleavage product, indicating that N-terminal cleavage is an innate property of the SufB intein (Fig. S3). The sharpening of the precursor and cleavage product bands by TCEP (Fig. 2 B and C), and the SufB intein-specific development of N-terminal cleavage with different extein combinations suggest cysteine-based oxidative reactions of the SufB precursor.
Fig. S3.
SufB intein with native exteins undergoes N-terminal cleavage. (A) C-extein antibody blot. Full-length, His-tagged SufB was overexpressed and treated with H2O2 and DEA, similar to Fig. 3 C and D. The lysates were separated by 12% SDS/PAGE and transferred to Immuno-Blot PVDF (Bio-Rad). An anti–C-extein antibody was used to probe splicing products at 1:5,000 dilution. The blot shows N-terminal cleavage, which increases with 1 mM and 8 mM H2O2 and all concentrations of DEA treatment. Again, similar to Fig. 3 C and D, the highest concentrations of H2O2 and DEA cause precursor accumulation. The SufB lane is purified inteinless M. smegmatis SufB protein as size control for ligated exteins. Products detected are: precursor (P; 96 kDa), N-terminal cleavage product (NC, intein+C-extein; 66 kDa) and ligated exteins (LE; 56 kDa). (B) SufB (ligated extein) antibody blot. The same lysates from panel A were separated and probed using a SufB antibody at 1:5,000 dilution. N-terminal cleavage is seen at all concentrations of H2O2 and DEA, and as in A, precursor accumulation occurs at the highest concentrations for both treatments. The 66-kDa NC band was used for N-terminal cleavage quantitation. An additional N-terminal cleavage product, the N-extein (NC; 30 kDa), is seen with this antibody. Western blots were scanned on a Typhoon imager and quantified using ImageQuant. Polyclonal antibodies were produced in rabbits against purified SufB protein of M. smegmatis or a synthetic C-extein peptide CVEPIAKELPMEYALE by Covance. Because of differential reactivity with polyclonal antibodies, ratios of different species in the Western blots are arbitrary, unlike fluorescence quantitation.
ROS and RNS Inhibit SufB Intein Splicing.
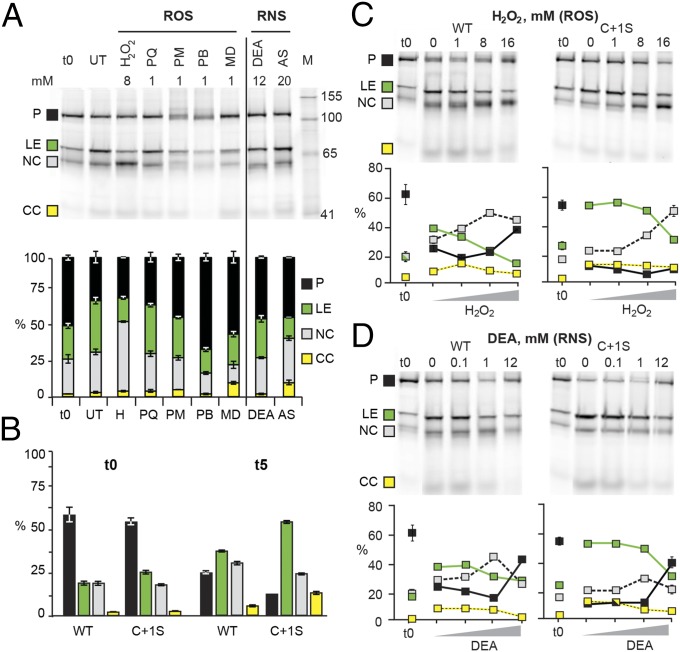
ROS and RNS are the most common agents responsible for intracellular modification of activated cysteines. To test if SufB splicing is sensitive to ROS and RNS, the WT MIG SufB precursor was allowed to process for 5 h in cells inhibited for protein synthesis and subjected to reagents that induce intracellular accumulation of these species (Fig. 3A). The t0 lane in Fig. 3A represents splicing products at the time of translation inhibition, whereas products in the untreated lane are made during incubation without treatments. The different ROS and RNS reagents generally inhibited protein splicing, reducing the level of ligated exteins from 37% for the untreated sample by more than one-half, to as low as 17% for the ROS reagent hydrogen peroxide (H2O2), and 15% for the RNS reagent Angeli’s salt (AS). The other ROS reagents, paraquat (PQ), plumbagin (PB), phenazine methosulfate (PM), and menadione (MD), as well as the other RNS reagent DEA NONOate (DEA) inhibited extein ligation to intermediate levels. Interestingly, apart from uniform inhibition of splicing and minimal effects on C-terminal cleavage, these reagents had different effects, where some induced N-terminal cleavage (H2O2), others caused precursor accumulation (PQ, PM, PB, MD, and DEA) and some stimulated both N-terminal cleavage and precursor accumulation (AS). The PB- and MD-treated samples showed less N-terminal cleavage than the t0, which suggests that some N-terminal cleavage occurs upon cell lysis. More generally, the data reveal that ROS and RNS interfere with SufB intein splicing by inhibiting ligated extein formation and causing accumulation of SufB precursor or N-terminal cleavage product.
Fig. 3.
ROS and RNS modulate SufB splicing. (A) MIG SufB is sensitive to ROS and RNS. ROS (left of line) and RNS (right of line) treatments of MIG SufB at 5 h (t5). The stack plot shows the percentage of GFP-containing species. ROS compounds: H2O2 (H), paraquat (PQ), phenazine methosulfate (PM), plumbagin (PB), and menadione (MD); RNS compounds: DEA NONOate (DEA) and Angeli’s salt (AS). (B) C+1S MIG SufB mutant splices more readily than WT. The bar graph is derived from 0- and 5-h untreated samples (0 mM) of WT and C+1S MIG SufB from C. (C and D) H2O2 and DEA modulate splicing in a concentration-dependent manner. The gels are representative images of treatments of WT and C+1S MIG SufB, quantitated in scatter plots. Results with full-length exteins are similar (Fig. S3).
We surmise that the cysteines, C1 and C+1, which catalyze the first two steps of intein splicing (Fig. 1A), are responsible for the ROS and RNS effects. This suspicion is based on these cysteines having increased nucleophilicity, making them prone to modifications. We therefore made a mutant with the C+1 mutated to serine, as this amino acid can act as a nucleophile and facilitate splicing (1), yet has a hydroxyl group instead of a sulfhydryl. Comparison of WT MIG SufB with C+1S MIG SufB allows dissection of multiple aspects of C1 and C+1 chemistry during protein splicing and ROS/RNS treatment (Fig. 3 B–D). Upon induction, WT and C+1S MIG SufB accumulated 62% and 55% precursor (t0), whereas within 5 h of incubation (t5), 24% of precursor remained for WT (∼60% decrease) and 11% for C+1S (∼80% decrease), revealing the dynamic range of the experiment. Increased processing of untreated C+1S MIG SufB was also reflected in more ligated exteins totaling 55% at t5, compared with 38% for WT, suggesting that either C+1S accelerates splicing (Fig. 1A, step 2) or that C+1 modification reduces extein ligation. Simultaneously, there was less N-terminal cleavage product for C+1S MIG SufB, with 22% at t5 compared with 30% for WT. Taken together, these results indicate a stronger propensity of WT MIG SufB to promote N-terminal cleavage than C+1S, suggesting a role for the C+1 residue in N-terminal cleavage.
H2O2 and DEA, which respectively cause N-terminal cleavage product and precursor accumulation, were selected to further study the consequences of ROS and RNS on splicing. These compounds have concentration-dependent effects on M. tuberculosis, which exhibits a high degree of resistance to these reagents (13). Precursor processing of WT and C+1S MIG SufB was allowed to proceed for 5 h in the presence of 1 mM, 8 mM, and 16 mM H2O2 and 0.1 mM, 1 mM, and 12 mM DEA (Fig. 3 C and D). After treatment with H2O2 (Fig. 3C) or DEA (Fig. 3D), splicing of both WT and C+1S MIG SufB was inhibited in a concentration-dependent manner, as reflected by a decrease in formation of ligated exteins, with a sharper decrease for H2O2. C+1S MIG SufB required higher concentrations of both H2O2 and DEA for inhibition of splicing, reinforcing the idea that C+1 is involved in oxidative inhibition of splicing. In addition, at the lower concentrations of H2O2 and DEA, inhibition of WT MIG SufB splicing was accompanied by induction of N-terminal cleavage, whereas at the highest concentration of both reagents, splicing was inhibited and precursor accumulation was triggered. For C+1S MIG SufB, H2O2 and DEA had similar effects of inhibiting splicing, but at high concentrations, only DEA induced precursor accumulation, whereas H2O2 resulted in N-terminal cleavage. These data show that in WT MIG SufB, precursor accumulated after both high-level ROS and RNS treatment, whereas C+1S only demonstrated precursor accumulation with DEA treatment. This observation not only suggests that splicing inhibition can occur by different mechanisms after oxidative and nitrosative stresses, but also that the extein C+1 residue is directly involved in H2O2-induced precursor accumulation.
Identification of Modified Cysteines Under Oxidative Stress.
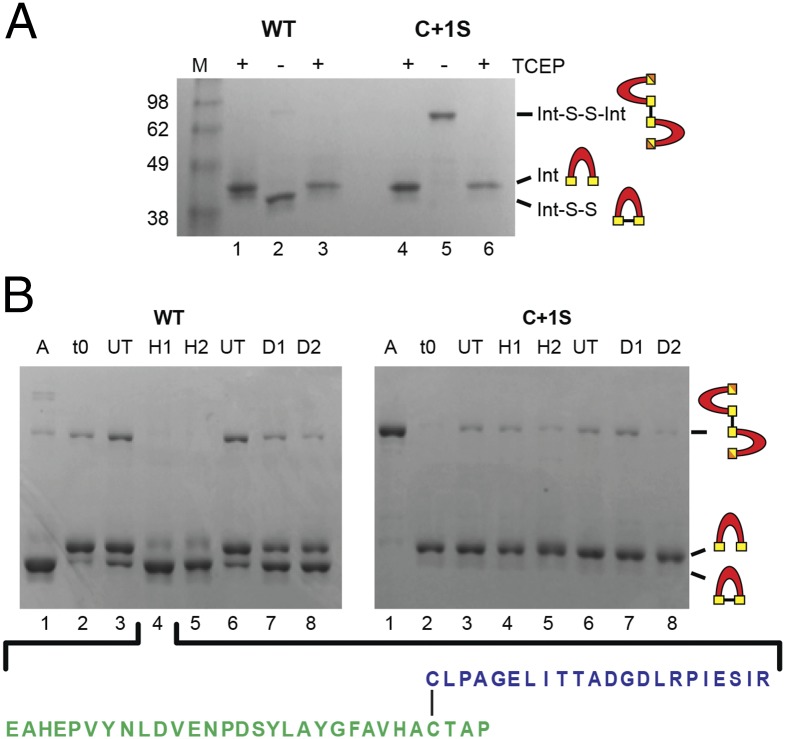
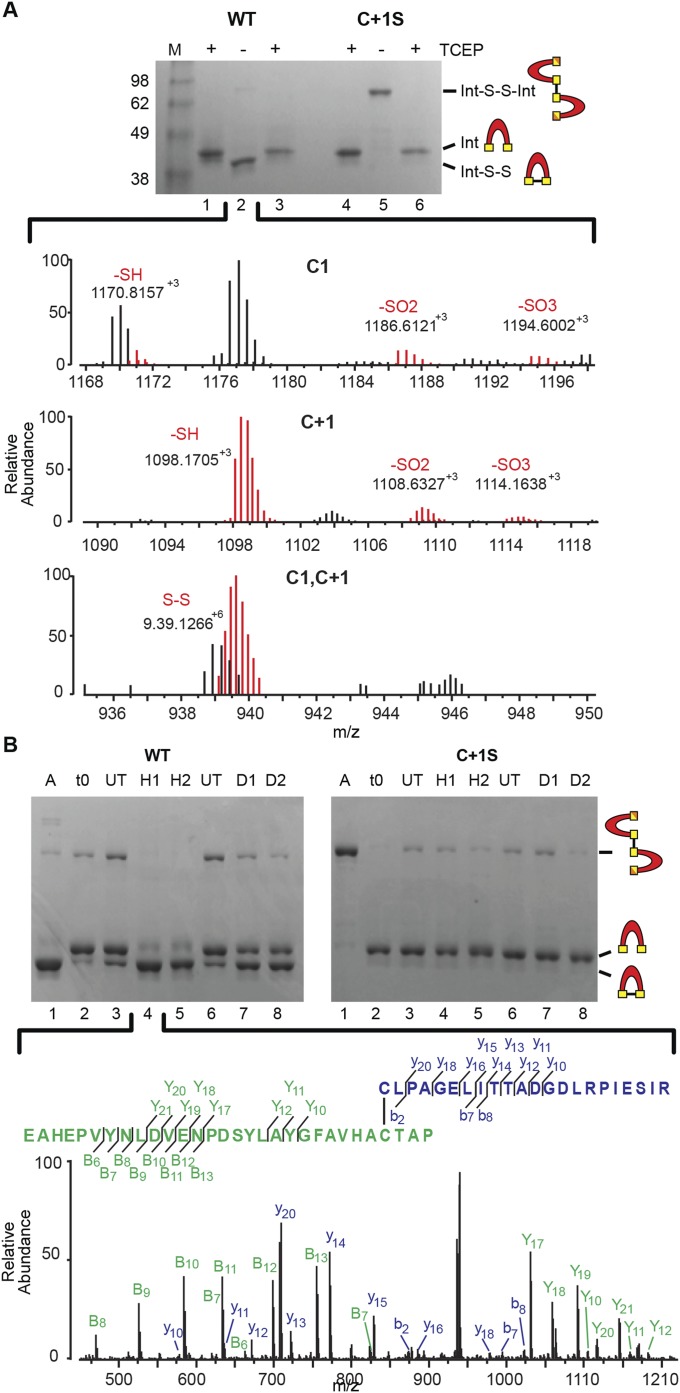
To identify cysteine modifications responsible for splicing modulation in WT and the C+1S mutant, SufB intein purification constructs (WT SufBi-PC and C+1S SufBi-PC) were overexpressed. Both inteins have short native exteins and mutations that abolish splicing (N359A), endonuclease toxicity, and remove noncatalytic cysteines. WT and C+1S SufBi-PC samples were supplemented with TCEP during lysis and purification, to protect cysteines from oxidation and allow for characterization of cysteine modification in vitro. Interestingly, upon TCEP removal, both the WT and C+1S mutant precursors changed mobility on a polyacrylamide gel (Fig. 4A, lanes 2 and 5). WT showed a shift from ∼41 kDa to ∼38 kDa and also a faint band was apparent at ∼90 kDa. In contrast, the C+1S mutant had a strong band at ∼90 kDa and no band at ∼38 kDa. These mobility shifts could be reversed by TCEP treatment before sample loading (Fig. 4A, lanes 3 and 6), suggestive of cysteine oxidation under aerobic conditions and formation of disulfide bonds. We propose that WT precursor formed a mixture of intramolecular and intermolecular disulfide bonds, where the intramolecular disulfide dominated, consistent with the proximity of the C1 and C+1 residues in 3D space. Accordingly, the C+1S mutant formed only intermolecular disulfide bonds. Ion-trap mass spectrometry of these samples indicated that under aerobic conditions, C1 and C+1 of the WT exist in disulfide bonded form, alongside thiol, sulfinic (dioxidized, -SO2) and sulfonic (trioxidized, -SO3) forms (Fig. S4A, red traces). These data support the in vivo indication of the hypersensitivity of the SufB intein to oxidation.
Fig. 4.
Oxidative modifications of the SufB intein. (A) Intra- and intermolecular disulfide bond formation. Purified WT and C+1S SufBi-PC were run with TCEP (+, lanes 1 and 4). Upon TCEP removal, WT forms intramolecular (Int-S-S) and intermolecular (Int-S-S-Int) disulfide bonds (–, lane 2), as visualized by band shifts. The C+1S precursor forms only the intermolecular disulfide bond (–, lane 5). TCEP treatment reversed disulfide bond formation to yield reduced intein (+, lane 3 and 6). LC-MS confirmation presented in Fig. S4A. (B) ROS and RNS promote disulfide bond formation only in WT precursor. A, Aerobic; t0, protein capped with iodoacetamide immediately after the removal of TCEP; UT, untreated; H1, 10×-fold [H2O2] over protein; H2, 100×-fold [H2O2]; D1, 10×-fold [DEA]; D2, 100×-fold [DEA]. The C1−C+1 intramolecular disulfide-bonded peptide (lane 4) was further validated by LC-MS/MS (Fig. S4B). The peptide coverage is shown below and in Fig. S4B.
Fig. S4.
Oxidative modifications of the SufB intein confirmed by mass spectrometry. (A) Intra- and intermolecular disulfide bond formation. WT precursor without TCEP (lane 2) was trypsin-digested and analyzed by LC-MS. The Top and Middle show the various oxidative states of C1 and C+1, respectively. The Bottom shows the disulfide bonded C1-C+1 peptide, which is colored red to distinguish from other overlapping peaks. Gels are described in Fig. 4A. (B) ROS and RNS promote disulfide bond formation only in WT precursor. The C1−C+1 intramolecular disulfide-bonded peptide identified in H2O2 -treated WT SufBi-PC (H1) was further validated by LC-MS/MS (lane 4). The peptide coverage is shown in the Inset. The spectra represent the peaks generated by fragmentation of the C1−C+1 disulfide-bonded peptide. The “y” and “b” ions correspond to C1 peptide fragments, and the “Y” and “B” ions correspond to C+1 peptide fragments. Gels are described in Fig. 4B. Disulfide bond formation between C1 and C+1 was detected with both treatments, although other NO-based modifications were not detected, likely due to the labile nature of these modifications (41–43).
Because of the extreme sensitivity of the SufB intein to oxidation, WT and C+1S proteins from SufBi-PC were purified in the presence of TCEP and then placed in an anaerobic chamber. H2O2 and DEA treatments were carried out anaerobically after TCEP removal. Similar to molecular oxygen, treatment of the WT precursor with H2O2 or DEA resulted primarily in intramolecular disulfide bond formation (Fig. 4B, Left, lanes 4, 5, 7, and 8). Interestingly, the C+1S precursor formed minimal intermolecular disulfides upon treating with H2O2 and DEA (Fig. 4B, Right), suggesting that either C1 is differentially reactive or accessible relative to C+1, or that C+1 is the primary oxidation target.
Liquid chromatography-mass spectrometry (LC-MS) was carried out on H2O2- and DEA-treated WT SufBi-PC (Fig. S4A). Disulfide bond formation between C1 and C+1 was detected with both treatments. To further confirm disulfide bond formation between C1 and C+1 of H2O2- and DEA-treated WT precursor, tandem LC-MS/MS was performed on the peak corresponding to the disulfide-bonded peptide (Fig. 4B, lane 4). Good coverage of the peptide sequence further validated the formation of an intramolecular disulfide bond for both reagents (Fig. 4B, below gel, and Fig. S4B). Together, these findings indicate that the catalytic cysteines of the SufB intein are sensitive to modifications induced by oxidative and nitrosative stresses.
Discussion
Three independent intein invasion events in mycobacterial sufB (Fig. 1 B and C and Fig. S1), together with intein retention by streamlined pathogenic genomes (25), provoke the question of the importance of inteins in regulating SufB function and impacting mycobacterial fate. The SufB intein has an exceptional predisposition to oxidation and nitrosylation among the three M. tuberculosis inteins (Figs. 2–4) (15). Protein sensitivity to oxidation is primarily mediated by reactive sulfur in cysteine residues (26), which often act as redox switches (17). Most cysteines are not reactive because they are buried or are not activated by their microenvironment, such as proximal charged residues that can lower their pKa (27). Protein splicing by canonical inteins uses nucleophilic sulfhydryl groups of catalytic cysteines, C1 and C+1 (Fig. 1A), which have low pKas to enhance their nucleophilic properties (28–30). Both C1 and C+1 participate in oxidative reactions modulating SufB precursor processing. The presence of these reactive cysteines also holds promise for drug development, as redox-active cysteines were shown to be potently inhibited by auranofin, leading to death of M. tuberculosis (31).
Whereas precursor accumulation can be achieved by either C1-C+1 disulfide bond formation or C1 modification, which inhibits the C1 residue from forming a thioester bond (Figs. 1A, step 1, and 5, Left), N-terminal cleavage can happen in one of three ways. First, a C1-derived labile thioester bond can be cleaved by an external nucleophile. Second, C+1 forms a second thioester bond that can be cleaved by an external nucleophile. Third, when C+1 is oxidatively modified, the C1-derived labile thioester is unable to proceed down the splicing pathway, and can be cleaved by an external nucleophile (Figs. 1A, step 2, and 5, Center). C-terminal cleavage occurs when the intein’s terminal asparagine cyclizes before the N-extein is connected to the C-extein (Figs. 1A, step 3, and 5, Right). C-terminal cleavage was not considered in the analysis for SufB because it is not dramatically affected by ROS or RNS, and is also observed for all three mycobacterial inteins.
Fig. 5.
Pathways for intein processing under H2O2 and NO stress. SufB splicing outcomes include precursor accumulation, N-terminal and C-terminal cleavage. C1 and C+1, yellow rectangles; where intermediates or products can be formed by both WT and C+1S, the +1 residue is shown as a yellow/orange rectangle. (Left) Precursor accumulation occurs by C1−C+1 disulfide bonding or C1 modification (star), both of which inhibit the first step of splicing. (Center) N-terminal cleavage occurs when an external nucleophile (Nu:) attacks the thioester bond formed by C1, or by C+1, or with oxidative modification of C+1 arresting intein splicing after step 1 (Fig. 1A), allowing Nu: to cleave the C1 thioester bond. (Right) C-terminal cleavage occurs when the terminal asparagine of the intein cyclizes before the N-extein is connected to the C-extein. The ability of H2O2 or DEA to accelerate any of these reactions is indicated.
Precursor accumulation occurs after exposure to both high ROS and RNS concentrations, although these species do not react identically, as revealed by the behavior of C+1S MIG SufB. H2O2-dependent precursor accumulation occurs exclusively through formation of a disulfide bond between C1 and C+1, as only WT, and not C+1S MIG SufB, accumulated precursor (Figs. 3C and 5, Left). In contrast, the NO-donor DEA induced precursor accumulation in both the WT and C+1S MIG SufB (Fig. 3D), which implies that NO can directly affect the C1 residue. Although, gel mobility of DEA-treated WT SufBi-PC suggests that NO-dependent precursor accumulation also occurs mostly through C1-C+1 disulfide bonding (Fig. 4), the C+1 mutant indicates that ROS and RNS can act differently.
N-terminal cleavage is a major side reaction for the SufB intein, unlike the DnaB and RecA inteins (Fig. 2C). WT MIG SufB displayed higher levels of N-terminal cleavage than C+1S in E. coli (Fig. 3B), indicating the role of C+1 in this phenomenon. This effect was exaggerated by ROS and RNS treatment (Figs. 3 C and D and 5, Center). Importantly, H2O2 can directly stimulate N-terminal cleavage by acting as the external nucleophile to cleave thioester bonds in the precursor, as evident for the C+1S mutant, and explaining the enhanced sensitivity of the SufB intein to H2O2 over DEA (Fig. 3 C and D).
Relative reactivity of C1 and C+1 toward H2O2 presents an interesting conundrum. On one hand, the highly nucleophilic C1 of the C+1S precursor can form an intermolecular disulfide bond in the presence of molecular oxygen (Fig. 4A); on the other hand, it was much less able to form the intermolecular disulfide bond anaerobically in the presence of a strong, albeit larger, oxidant H2O2 (Fig. 4B). In accord with these data, H2O2-dependent precursor accumulation in the MIG splicing reporter occurred exclusively in the WT, and not in the C+1S mutant. We propose that steric inaccessibility or differential reactivity of C1 accounts for such distinct responses. This finding suggests that formation of the intramolecular disulfide bond by WT after H2O2 treatment (Fig. 5C, Left) occurs through initial oxidation of C+1. Thus, C+1 may act as the sentinel that senses the oxidation signals. Interestingly, the other two intein insertion points have serine as the +1 residue, which implies that a mechanism focused on C+1 modification is specific for inteins in insertion point a, such as in M. tuberculosis (Fig. 1C).
Considering that oxidative modifications favor the C+1 residue, we propose that C+1 functions as a pivot during conditions of extreme oxidative and nitrosative stresses to control SufB precursor processing, and ultimately SUF function. Intriguingly, the C+1 residue is one of two cysteine residues of the SufB protein available to coordinate the [Fe-S] cluster (Fig. S2D). We therefore hypothesize that C+1 has a direct role in [Fe-S] cluster assembly in SufB. In the presence of the intein, this [Fe-S] cluster pocket would be disrupted, rendering SufB nonfunctional (Fig. 6). Upon intein splicing, the [Fe-S] cluster binding pocket would be restored, allowing [Fe-S] cluster coordination. Thus, the C+1 residue might act as a toggle, directly participating in both modulation of the stress response of the SufB intein by ROS and RNS, and [Fe-S] cluster assembly by the SufB protein, as a posttranslational regulator of SufB function.
Fig. 6.
Model of SufB precursor processing under ROS/RNS stresses. We propose that the C+1 residue, crucial for splicing, also coordinates the [Fe-S] cluster of the active SufB protein. Under ROS/RNS stresses, the C+1 of the SufB precursor can either disulfide bond, resulting in precursor accumulation (PA), or become modified (star), leading to N-terminal cleavage (NC). See Discussion for detailed explanation.
In organisms with multiple [Fe-S] cluster biogenesis systems, the SUF operon is usually devoted to [Fe-S] cluster assembly under conditions of oxidative stress or iron limitation (32, 33). However, in mycobacteria the SUF pathway is the primary [Fe-S] cluster assembly system, functioning during both normal and stress conditions (6, 11, 12). Therefore, the suf operon in M. tuberculosis is both constitutively transcribed and induced by oxidative and nitrosative stress (13). Although this work does not consider the impact of the complex interactions of the SufB protein with other SUF components, we show that superimposed upon transcriptional control, splicing regulation might act as an immediate switch of SufB function.
Our data suggest that ROS and RNS interference with SufB splicing is a dynamic process, where the concentration of these species affects the outcome of precursor processing (Fig. 3 C and D). We propose that these dynamics depend on whether the catalytic cysteine residues participate in splicing before they become modified through oxidative reactions. We also suggest that oxidative modifications inhibit the splicing pathway and diminish activity of the [Fe-S] assembly machinery. This would provide instantaneous opposition to stimulation of transcription of the suf operon by ROS and RNS in M. tuberculosis (13). Suppression of SUF function by splicing inhibition at high ROS and RNS concentrations could occur because it may not be efficient to repair [Fe-S] clusters under such extreme conditions. Therefore, SufB intein splicing could be an adaptive response to provide posttranslational down-regulation of SufB function, similar to that proposed for the redox-sensitive Pyrococcus abyssi MoaA intein (34). The responsiveness of M. tuberculosis to ROS and RNS is a significant factor in pathogen survival (18, 35). Whereas precursor accumulation is reversible and may act as a rheostat, N-terminal cleavage is not (Fig. 6). It is unclear if the N-terminal cleavage products might have an independent functional role or signal the transition of M. tuberculosis to a latent, persistent stage.
Methods
M. tuberculosis SufB Extein Modeling.
The M. tuberculosis SufB extein sequence was submitted to Phyre2 servers (www.sbg.bio.ic.ac.uk/∼phyre2) (20) for modeling using the intensive mode. The SufB extein model returned a high confidence level with 418 residues (86%) modeled at >90% accuracy. The model was manipulated in PyMOL (v1.7.2) and the conserved insertion site +1 residues highlighted. Extended information on modeling can be found in SI Methods.
Bacterial Strains and Plasmid Construction.
Bacterial strains, growth conditions, plasmid methodology, restriction enzymes, and oligonucleotides are found in SI Methods and Tables S1 and S3, following several protocols previously described in ref. 36.
Table S3.
Oligonucleotides used in this study
| Oligo ID | Sequence (5′ to 3′) | Application |
| MB73 | gcatgcgaattcggatgacactcaccccagaggcc | Amplify full-length sufb gene with EcoRI end |
| IDT3158 | gtcatgcaagctttcatccgaccgcgccctccatctg | Amplify full-length sufb gene with HindIII end |
| IDT3513 | gggatcgatatgagcaaaggagaagaacttttcactgg | Amplify sfGFP with ClaI end |
| IDT3514 | cccgcggccgcggtaccttatttgtagagctcatccatgccatgtg | Amplify sfGFP with NotI end |
| IDT3507 | actacctggcggcgggtcacgcgtg | Mtu SufB E143A mutation to disrupt endonuclease activity |
| IDT3508 | cctatgtgaatggagccggcaacgtcacgcg | Mtu SufB D242A mutation to disrupt endonuclease activity |
| IDT3588 | ggggcatgcagatgagggctcttacgtgcactacgtagag | SufB intein amplification for MIG-SufB with SphI end |
| IDT3589 | cccatcgatgtgcaatgaatccgatttgtagatcggtgcggtgc | SufB intein amplification for MIG-SufB with ClaI end |
| IDT3732 | gccgtgcacaacagcaccgcaccga | C+1S mutagenesis of MIG-SufB |
| IDT3733 | tcggtgcggtgctgttgtgcacggc | C+1S mutagenesis of MIG-SufB |
| IDT3539 | ggggcatgcacgcacccgggtcaaggtcgtcaagaacaagtgcctcgcagagggcactcggatcttc | RecA minimized intein amplification for MIG-RecA with SphI end |
| IDT3540 | cacgggccgcgcatgaagcgttccgtccttggcagcagccac | RecA minimized intein amplification for MIG-RecA to remove internal SphI site |
| IDT3541 | gccaaggacggaacgcttcatgcgcggcccgtggtgtcctgg | RecA minimized intein amplification for MIG-RecA to remove internal SphI site |
| IDT3542 | cccatcgatgaactcggcctgcttgaatggtggcgaacagttgtgcacgacaaccccttcggcgac | RecA minimized intein amplification for MIG-RecA with ClaI end |
| IDT3773 | caaggtcgtcaagaacccgtgcctcgcagagggc | Mutagenesis of RecA K-1 to P |
| IDT3774 | gccctctgcgaggcacgggttcttgacgaccttg | Mutagenesis of RecA K-1 to P |
| IDT3590 | ggggcatgcaccgatgctggccgacctcagggaatcgggc | DnaB intein amplification for MIG-DnaB with SphI end |
| IDT3591 | cccatcgatgtcggcatcttgttccagcgaattgtgcaaac | DnaB intein amplification for MIG-DnaB with ClaI end |
| IDT3758 | gacctcagggaatcgttctgcctgaccgcgtc | Mutagenesis of DnaB intein G-1 to F |
| IDT3759 | gacgcggtcaggcagaacgattccctgaggtc | Mutagenesis of DnaB intein G-1 to F |
| IDT4036 | catatgcatcatcatcatcatcatgaaaacctgtatttccagggggtagagggctgcctgcccgcc | Amplification of SufB intein from MIG with NdeI site, His-tag, and TEV protease site |
| IDT4037 | aagctttcacggtgcggtgcatgcgtgcacggcgaacccgtaggcgaggtag | Amplification of SufB intein from MIG with HindIII site, introduces terminal N to A |
| IDT4017 | atgtgcgccaagcgagcaagtcgctgtac | Mutagenesis of C142S |
| IDT4018 | gtacagcgacttgctcgcttggcgcacat | Mutagenesis of C142S |
| IDT4019 | aggcccgcgacagcggcgactac | Mutagenesis of C319S |
| IDT4020 | gtagtcgccgctgtcgcgggcct | Mutagenesis of C319S |
| IDT4179 | tggcggcgggtcacgcgagtctcaccaatggcagtgagtcgctgatcttc | Multisite mutagenesis of C147S and C152S |
| IDT4180 | gaagatcagcgactcactgccattggtgagactcgcgtgacccgccgcca | Multisite mutagenesis of C147S and C152S |
Splicing Assays and ROS/RNS Treatments.
To determine the splicing activity of the MIG constructs, an overnight culture was diluted 1:100 into fresh LB medium and grown at 37 °C with aeration to midlog phase (OD600 ∼0.5). Induction was with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h at 30 °C with aeration. Protein synthesis was stopped with spectinomycin (100 µg/mL) and cultures were incubated on ice for 10 min. A 1.5-mL aliquot of induced culture was immediately pelleted at 13,000 rpm for 1 min and frozen at −80 °C as a time 0 (t0) control. Aliquots of 1.5 mL of induced cells were incubated at 30 °C without shaking, with or without treatment, for the specified time, and samples pelleted and frozen as above. Pellets were thawed on ice, 400 µL of lysis buffer [50 mM Tris pH 8.0, 10% (vol/vol) glycerol] was added and lysis was performed using a tip sonicator. Lysates were left untreated or treated with 20 mM TCEP on ice for 10 min before gel loading. Classic dye [10% (wt/vol) SDS, 10% (vol/vol) glycerol, 12.5% (vol/vol) stacking buffer [10% (wt/vol) SDS, Tris pH 6.8], 0.1% Bromophenol blue] without β-mercaptoethanol (to prevent induced N-terminal cleavage) was added post-TCEP treatment. Samples were not boiled, to preserve the folding and fluorescence of GFP. Proteins were separated on a 12% SDS/PAGE gel (Jules Precast) and GFP-containing bands were visualized using a Typhoon 9400 scanner (GE Healthcare), with excitation and emission wavelengths of 488 nm and 526 nm, respectively. Aliquots of 1.5 mL of induced cells were treated at 30 °C without shaking for 2 h or 5 h. For ROS compounds, catalase was added to scavenge unreacted species. Cell pellets were frozen at −80 °C, lysed, separated by 12% SDS/PAGE, and analyzed as above.
SufB Intein Expression and Purification.
WT and C+1S SufBi-PC were grown as above. Protein was induced with 0.5 mM IPTG for 18 h at 16 °C with aeration, pelleted at 4,000 rpm for 10 min at 4 °C, and frozen at −80 °C. Pellets were lysed with a tip sonicator in lysis buffer (20 mM Mops pH 7.0, 200 mM NaCl, 25 mM imidazole, 5% glycerol, 5 mM TCEP). The His-tagged proteins were purified using nickel-chelating chromatography at 4 °C. Buffer components were as follows: Binding buffer is identical to the above lysis buffer, wash buffer is 20 mM Mops pH 7.0, 200 mM NaCl, 40 mM imidazole, 5% glycerol, 5 mM TCEP, and elution buffer is 20 mM Mops pH 7.0, 200 mM NaCl, 250 mM imidazole, 5% glycerol, 5 mM TCEP. The 1-mL HisTrap HP columns (GE Healthcare Life Sciences) containing Ni Sepharose High Performance were used following manufacturer instructions. Ni-NTA-purified proteins were separated on a 12% SDS/PAGE gel and visualized by staining with Coomassie brilliant blue.
Analysis of Disulfide Bond Formation.
SufB intein disulfide bonding was observed by in-gel analysis carried out under anaerobic conditions (Type B Vinyl Anaerobic Chamber, Coy) after ROS or RNS treatment. Further details, including confirmation of disulfide bonding by mass spectrometry, are described in SI Methods.
SI Methods
Computational Analysis and Expanded Modeling.
Sequences for the intein-containing mycobacterial SufB proteins and one intein-containing archeal SufB were accessed from the National Center for Biotechnology Information (NCBI; www.ncbi.nlm.nih.gov). Alignments of the intein and extein sequences were performed by ClustalW2 (https://www.ebi.ac.uk/Tools/msa/clustalw2/) (38). Jalview (www.jalview.org) (39) was used to analyze the aligned extein sequences and color the +1 residues. Trees were built using the neighbor-joining method in MEGA5.2.1 (40). The neighbor-joining trees were evaluated for statistical support by bootstrapping (1,000 replicates).
SufB extein sequences for Mycobacterium leprae, M. tuberculosis, Mycobacterium xenopi and SufB protein of inteinless Mycobacterium smegmatis were submitted to Phyre2 servers (www.sbg.bio.ic.ac.uk/∼phyre2) (20) for modeling using the intensive mode. Levels of confidence for the SufB proteins were high, with the M. leprae SufB extein model at a confidence of 420 residues (87%), the M. smegmatis SufB protein model at a confidence of 414 residues (87%), the M. tuberculosis SufB extein model at a confidence of 418 residues (86%), and the M. xenopi SufB extein model at a confidence of 423 residues (88%), all modeled at >90% accuracy. The models were aligned in PyMOL (v1.7.2) and the conserved insertion site +1 residues highlighted. Distances of the +1 residues relative to each other, as well as the distance between the two cysteines present in the M. tuberculosis extein model, were estimated using PyMOL.
Bacterial Strains and Growth Conditions.
All strains and plasmids used in this study are listed in Table S1. Escherichia coli DH5α and BL21(DE3) were grown in Luria Broth (LB) with aeration at 250 rpm. Where appropriate, the medium contained kanamycin (50 µg/mL) or chloramphenicol (25 µg/mL). Electroporation was performed with a Gene Pulser apparatus (Bio-Rad). Transformants were recovered for 1 h at 37 °C in SOC medium (0.5% yeast extract, 2% tryptone, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose), plated on LB agar with appropriate antibiotic and incubated overnight at 37 °C.
Plasmid Methodology, Enzymes, and Oligonucleotides.
Plasmid DNA was isolated and purified using a QIAprep Spin Miniprep Kit (Qiagen). The digests and PCR fragments were visualized by electrophoresis in 1% agarose gels using EZ-Vision DNA Dye (Amresco). DNA fragments were purified from agarose gels using the Zymoclean Gel DNA Recovery Kit (Zymo Research). Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (NEB), and manufacturer protocols were followed. A list of oligonucleotides, synthesized by Integrated DNA Technologies (IDT), is located in Table S3.
Construction of Plasmids.
A fluorescent splicing reporter MIG, based on MBP, the intein (I) and superfolder GFP, was constructed, with a His-tag at the N terminus of MBP, in pACYCDuet-1 (Novagen). The recA (C1, C+1), dnaB (C1, S+1), and sufB (C1, C+1) intein genes, with short flanking sequences coding for 8–10 native N- and C-extein residues (Table S1), were cloned into the ClaI and SphI sites of pACYCDuet-1, between the MBP and superfolder GFP coding sequences. The intein fragments were generated by PCR from M. tuberculosis genomic DNA with primer pairs IDT3590/IDT3591 (dnaB), IDT3539/IDT3540 and IDT3541/IDT3542 (recA), and IDT3588/IDT3589 (sufB). The RecA intein was cloned in the form of ΔI-SM, a splicing proficient mutant with deleted endonuclease domain (37). The SufB intein has E143A and D242A mutations (IDT3507 and IDT3508, respectively) in its endonuclease domain, to inhibit its endonuclease activity. The C+1S MIG SufB mutant was generated using QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent) using primer pair IDT3732/IDT3733. The DnaB and RecA MIG constructs have mutations in the upstream exteins in the −1 position, G399F (IDT3758/IDT3759) and K251P (IDT3773/IDT3774), respectively, to slow down protein splicing.
The full-length (FL) sufB gene from M. tuberculosis was amplified with primer pairs MB73/IDT3158, and cloned into the EcoRI and HindIII sites of pACYCDuet-1. The endonuclease of the SufB intein was inactivated with the E143A and D242A mutations, as above.
The SufB intein purification construct (SufBi-PC) was cloned into pET30b. The intein fragment was generated by PCR from the WT MIG SufB plasmid using primer pair IDT4036/IDT4037. The forward primer introduced an NdeI site, His-tag, and tobacco etch virus (TEV) protease site, whereas the reverse primer introduced a terminal N359A mutation, to prevent splicing by blocking C-terminal cleavage, and a HindIII site. N-extein residues VEG and C-extein residues CTAP were included in the following configuration: His6-TEV-VEG-SufB intein-CTAP. The PCR product was ligated into a TOPO vector using standard TA cloning. The insert was excised using NdeI and HindIII and ligated into pET30b. The C+1S SufBi-PC was generated from the WT SufBi-PC construct using the same mutagenic primers as the C+1S MIG SufB construct (IDT3732/IDT3733). The constructs have the two mutations in the catalytic center of the endonuclease domain (E143A, D242A). Finally, C147S, C152S, C174S, and C319S mutations were made in the endonuclease domain (4C/S) to prevent possible interference by disulfide bonding, using primers IDT4017, IDT4019, and IDT4179 and the QuikChange Multi Site-Directed Mutagenesis Kit (Agilent).
Anaerobic Protein Treatments for In-Gel Analysis of Disulfide Bond Formation.
To identify oxidative modifications of cysteines of WT and C+1S SufBi-PC, the proteins were purified over Ni-NTA columns in the presence of TCEP. The protein samples were further processed under anaerobic conditions (Type B Vinyl Anaerobic Chamber, Coy), and TCEP was removed using Zeba spin desalting columns (Thermo Scientific). An aliquot containing 20 µM protein was immediately capped with 20 mM iodoacetamide (IAM) at t0. The rest of the samples were treated with 200 µM and 2 mM H2O2 and DEA for 15 min. Catalase was added to H2O2-treated samples to remove unreacted H2O2. After 15 min, IAM was added to all of the samples for 1 h at 25 °C in the dark to cap unreacted cysteines. A portion of each sample was separated on 12% SDS/PAGE gel.
Mass Spectrometry Analysis.
Selected samples were trypsin-digested for 18 h at 37 °C under anaerobic conditions. Sequencing-grade modified trypsin (Promega) was added at a protease-to-protein ratio of 1:20 (wt/wt). To ensure that cysteines were fully available, 1 M urea was added to denature the protein. IAM was immediately added, and samples were trypsin-digested as above. Samples were then purified over Pierce C-18 spin columns (Thermo Scientific) and eluted with 70% LC-MS grade acetonitrile. Mass spectrometry analysis of the peptides was carried out on an LTQ Orbitrap Velos (Thermo Scientific) instrument. The peaks corresponding to the peptides of interest (C1 and C+1 peptides) were isolated and tandem mass spectrometry was carried out for further validation. MS-Bridge and MS-Product programs (University of California, San Francisco) were used to analyze the data.
Acknowledgments
We thank Seemanti Ramanath for construction of the FL-SufB endonuclease mutant; Mike Pugliese for construction of various SufB intein mutants; Daniele Fabris and Matteo Scalabrin for help with the mass spectrometry experiments; Olga Novikova for assistance with SufB intein distribution analysis; Seth Pearson for useful discussions and the SufBi-PC construct; Kathleen McDonough for valuable exchanges; James Imlay for useful discussions and training; and Rebecca McCarthy for manuscript preparation. Work in our laboratory is supported by National Institutes of Health Grants GM39422, GM44844, and T32AI055429.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1512777112/-/DCSupplemental.
References
- 1.Mills KV, Johnson MA, Perler FB. Protein splicing: How inteins escape from precursor proteins. J Biol Chem. 2014;289(21):14498–14505. doi: 10.1074/jbc.R113.540310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novikova O, Topilina N, Belfort M. Enigmatic distribution, evolution, and function of inteins. J Biol Chem. 2014;289(21):14490–14497. doi: 10.1074/jbc.R114.548255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander P, et al. Inteins in mycobacterial GyrA are a taxonomic character. Microbiology. 1998;144(Pt 2):589–591. doi: 10.1099/00221287-144-2-589. [DOI] [PubMed] [Google Scholar]
- 4.Saves I, et al. Specificities and functions of the recA and pps1 intein genes of Mycobacterium tuberculosis and application for diagnosis of tuberculosis. J Clin Microbiol. 2002;40(3):943–950. doi: 10.1128/JCM.40.3.943-950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis EO, Thangaraj HS, Brooks PC, Colston MJ. Evidence of selection for protein introns in the recAs of pathogenic mycobacteria. EMBO J. 1994;13(3):699–703. doi: 10.1002/j.1460-2075.1994.tb06309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huet G, Daffé M, Saves I. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: Evidence for its implication in the pathogen’s survival. J Bacteriol. 2005;187(17):6137–6146. doi: 10.1128/JB.187.17.6137-6146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry. 2009;48(44):10644–10653. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd ES, Thomas KM, Dai Y, Boyd JM, Outten FW. Interplay between oxygen and Fe-S cluster biogenesis: Insights from the Suf pathway. Biochemistry. 2014;53(37):5834–5847. doi: 10.1021/bi500488r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imlay JA. Iron-sulphur clusters and the problem with oxygen. Mol Microbiol. 2006;59(4):1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 10.Sharma AK, Naithani R, Kumar V, Sandhu SS. Iron regulation in tuberculosis research: Promise and challenges. Curr Med Chem. 2011;18(11):1723–1731. doi: 10.2174/092986711795471301. [DOI] [PubMed] [Google Scholar]
- 11.Singh A, et al. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc Natl Acad Sci USA. 2007;104(28):11562–11567. doi: 10.1073/pnas.0700490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rybniker J, et al. The cysteine desulfurase IscS of Mycobacterium tuberculosis is involved in iron-sulfur cluster biogenesis and oxidative stress defence. Biochem J. 2014;459(3):467–478. doi: 10.1042/BJ20130732. [DOI] [PubMed] [Google Scholar]
- 13.Voskuil MI, Bartek IL, Visconti K, Schoolnik GK. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol. 2011;2:105. doi: 10.3389/fmicb.2011.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huet G, Castaing JP, Fournier D, Daffé M, Saves I. Protein splicing of SufB is crucial for the functionality of the Mycobacterium tuberculosis SUF machinery. J Bacteriol. 2006;188(9):3412–3414. doi: 10.1128/JB.188.9.3412-3414.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttmann RP. Redox regulation of cysteine-dependent enzymes. J Anim Sci. 2010;88(4):1297–1306. doi: 10.2527/jas.2009-2381. [DOI] [PubMed] [Google Scholar]
- 16.Lo Conte M, Carroll KS. The chemistry of thiol oxidation and detection. In: Jakob U, Reichmann D, editors. Oxidative Stress and Redox Regulation. Springer; The Netherlands: 2013. pp. 1–42. [Google Scholar]
- 17.Groitl B, Jakob U. Thiol-based redox switches. Biochim Biophys Acta. 2014;1844(8):1335–1343. doi: 10.1016/j.bbapap.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell Microbiol. 2009;11(8):1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perler FB. InBase: The Intein Database. Nucleic Acids Res. 2002;30(8):383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 21.Wada K, et al. Molecular dynamism of Fe-S cluster biosynthesis implicated by the structure of the SufC(2)-SufD(2) complex. J Mol Biol. 2009;387(1):245–258. doi: 10.1016/j.jmb.2009.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amitai G, Callahan BP, Stanger MJ, Belfort G, Belfort M. Modulation of intein activity by its neighboring extein substrates. Proc Natl Acad Sci USA. 2009;106(27):11005–11010. doi: 10.1073/pnas.0904366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shemella PT, et al. Electronic structure of neighboring extein residue modulates intein C-terminal cleavage activity. Biophys J. 2011;100(9):2217–2225. doi: 10.1016/j.bpj.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwai H, Züger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580(7):1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 25.Moran NA. Microbial minimalism: Genome reduction in bacterial pathogens. Cell. 2002;108(5):583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Dutton RJ, Beckwith J, Boyd D. Membrane topology and mutational analysis of Mycobacterium tuberculosis VKOR, a protein involved in disulfide bond formation and a homologue of human vitamin K epoxide reductase. Antioxid Redox Signal. 2011;14(8):1413–1420. doi: 10.1089/ars.2010.3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witt AC, et al. Cysteine pKa depression by a protonated glutamic acid in human DJ-1. Biochemistry. 2008;47(28):7430–7440. doi: 10.1021/bi800282d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du Z, et al. Highly conserved histidine plays a dual catalytic role in protein splicing: A pKa shift mechanism. J Am Chem Soc. 2009;131(32):11581–11589. doi: 10.1021/ja904318w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pereira B, et al. Spontaneous proton transfer to a conserved intein residue determines on-pathway protein splicing. J Mol Biol. 2011;406(3):430–442. doi: 10.1016/j.jmb.2010.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Z, Zheng Y, Patterson M, Liu Y, Wang C. pK(a) coupling at the intein active site: Implications for the coordination mechanism of protein splicing with a conserved aspartate. J Am Chem Soc. 2011;133(26):10275–10282. doi: 10.1021/ja203209f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbut MB, et al. Auranofin exerts broad-spectrum bactericidal activities by targeting thiol-redox homeostasis. Proc Natl Acad Sci USA. 2015;112(14):4453–4458. doi: 10.1073/pnas.1504022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Outten FW, Djaman O, Storz G. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol Microbiol. 2004;52(3):861–872. doi: 10.1111/j.1365-2958.2004.04025.x. [DOI] [PubMed] [Google Scholar]
- 33.Jang S, Imlay JA. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol Microbiol. 2010;78(6):1448–1467. doi: 10.1111/j.1365-2958.2010.07418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Callahan BP, Topilina NI, Stanger MJ, Van Roey P, Belfort M. Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications. Nat Struct Mol Biol. 2011;18(5):630–633. doi: 10.1038/nsmb.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garbe TR, Hibler NS, Deretic V. Response of Mycobacterium tuberculosis to reactive oxygen and nitrogen intermediates. Mol Med. 1996;2(1):134–142. [PMC free article] [PubMed] [Google Scholar]
- 36.Topilina NI, Novikova O, Stanger M, Banavali NK, Belfort M. Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing. Nucleic Acids Res 2015 doi: 10.1093/nar/gkv612. , 10.1093/nar/gkv612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hiraga K, Derbyshire V, Dansereau JT, Van Roey P, Belfort M. Minimization and stabilization of the Mycobacterium tuberculosis recA intein. J Mol Biol. 2005;354(4):916–926. doi: 10.1016/j.jmb.2005.09.088. [DOI] [PubMed] [Google Scholar]
- 38.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 39.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torta F, Elviri L, Bachi A. Direct and indirect detection methods for the analysis of S-nitrosylated peptides and proteins. Methods Enzymol. 2010;473:265–280. doi: 10.1016/S0076-6879(10)73014-7. [DOI] [PubMed] [Google Scholar]
- 42.Qin Y, Dey A, Daaka Y. Protein S-nitrosylation measurement. Methods Enzymol. 2013;522:409–425. doi: 10.1016/B978-0-12-407865-9.00019-4. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y-JJ, Ching W-CC, Lin Y-PP, Chen Y-JJ. Methods for detection and characterization of protein S-nitrosylation. Methods. 2013;62(2):138–150. doi: 10.1016/j.ymeth.2013.04.016. [DOI] [PubMed] [Google Scholar]