Significance
Although infertility is a highly prevalent disease with a major genetic component, the underlying genetic causes are unknown in the vast majority of patients. The complexity of sperm and egg production has confounded most efforts to address the genetics of human infertility. We have developed an experimental strategy for identifying infertility-causing mutations that exist in human populations. This strategy exploits the expanding databases of human genetic variation to computationally identify potential infertility mutations, then mimic them in mice by CRISPR/Cas genome editing technology. The mice are then evaluated for whether the human mutation renders them infertile. This technology can be used to establish a permanent resource of validated disease-causing genetic variants in infertility and other diseases.
Keywords: CRISPR/Cas9, meiosis, spermatogenesis, cyclin, genome editing
Abstract
Infertility is a prevalent health issue, affecting ∼15% of couples of childbearing age. Nearly one-half of idiopathic infertility cases are thought to have a genetic basis, but the underlying causes are largely unknown. Traditional methods for studying inheritance, such as genome-wide association studies and linkage analyses, have been confounded by the genetic and phenotypic complexity of reproductive processes. Here we describe an association- and linkage-free approach to identify segregating infertility alleles, in which CRISPR/Cas9 genome editing is used to model putatively deleterious nonsynonymous SNPs (nsSNPs) in the mouse orthologs of fertility genes. Mice bearing “humanized” alleles of four essential meiosis genes, each predicted to be deleterious by most of the commonly used algorithms for analyzing functional SNP consequences, were examined for fertility and reproductive defects. Only a Cdk2 allele mimicking SNP rs3087335, which alters an inhibitory WEE1 protein kinase phosphorylation site, caused infertility and revealed a novel function in regulating spermatogonial stem cell maintenance. Our data indicate that segregating infertility alleles exist in human populations. Furthermore, whereas computational prediction of SNP effects is useful for identifying candidate causal mutations for diverse diseases, this study underscores the need for in vivo functional evaluation of physiological consequences. This approach can revolutionize personalized reproductive genetics by establishing a permanent reference of benign vs. infertile alleles.
Despite the high incidence of infertility, autosomal genetic causes of infertility attributable to gametogenesis defects are poorly characterized. In males, the most common known genetic causes of infertility are Y chromosome microdeletions, thought to be responsible for 6–18% of nonobstructive azoospermia (NOA) or severe oligozoospermia cases (1). In females, most of the known genetic causes are linked to syndromes that also affect the soma (e.g., Kallmann, Turner) or the neuroendocrine axis (2). In rare cases, families segregating infertility alleles have been mapped by linkage (3–6). Several candidate gene resequencing studies have implicated mutations or SNPs as being causative for azoospermia (7–13), but in the absence of genetic data, only a few reports (e.g., ref. 14) have made a compelling case. Recently, hemizygous deletions of the TEX11 gene, presumably catalyzed by unequal recombination between repetitive elements in the locus, have been linked to maturation arrest and infertility in azoospermic men (15).
Gene knockout and molecular genetic studies in mice have shown that germ cell development is genetically complex. A screen of the Mouse Genome Informatics database using MouseMine (www.mousemine.org) identifies 728 genes currently associated with infertility. Clearly, many more genes required for fertility remain to be identified. Furthermore, infertility is genetically heterogeneous; scores of distinct genes cause grossly identical phenotypes when mutated in mice (2, 16). This likely explains why genome-wide association studies (GWAS) have not been effective even in stratified cohorts, with only two reporting significant associations with NOA in Chinese populations (11, 17). Even if associations could be readily obtained, identification and validation of causative variants would remain problematic. Finally, the proportion of Mendelian infertilities that are caused by de novo mutations vs. segregating polymorphisms is unknown. Clearly, different approaches are needed to address the genetics of human infertility.
Here we describe a reverse genetics approach for identifying infertility alleles segregating in human populations that does not require linkage or association data; rather, it combines in silico prediction of deleterious allelic variants with functional validation in CRISPR/Cas9-edited “humanized” mouse models. We modeled four nonsynonymous human SNPs (nsSNPs) in genes that are essential for meiosis in mice. Each of these nsSNPs has been predicted to be deleterious to protein function by several widely used algorithms. Only one of the nsSNPs was found to cause infertility, highlighting the importance of experimentally evaluating computationally predicted disease SNPs.
Results
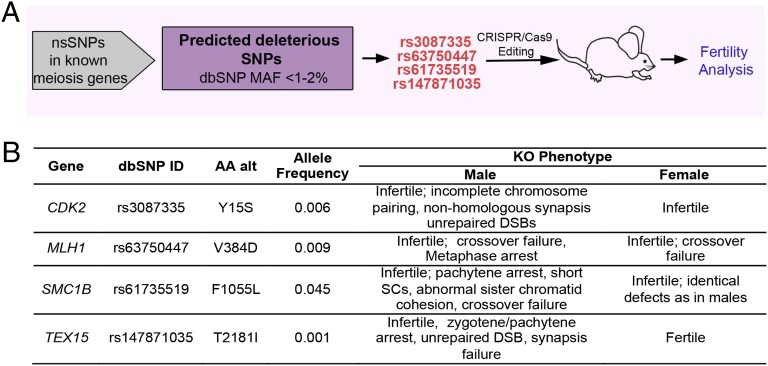
To address the question of whether human infertility can be caused by segregating Mendelian alleles, we applied a precision genome editing approach (Fig. 1A) to model variants of meiosis genes that when knocked out in mice cause the drastic infertility phenotype of NOA from meiotic arrest (also called “maturation arrest” in humans). To identify candidate infertility alleles in such genes, we screened the dbSNP database for potentially deleterious nsSNPs that met the following criteria: (i) they exist as minor (low frequency, preferably <1–2%) alleles in humans; (ii) they alter evolutionarily conserved amino acids; and (iii) the changes are predicted to be functionally deleterious by multiple algorithms.
Fig. 1.
Generating humanized mouse models to test potential human infertility SNPs. (A) Workflow for identifying and functionally testing potential infertility alleles, including the four (in red) studied here. (B) Candidate infertility SNPs modeled and tested in this study. MAF, minor allele frequency; AA alt, amino acid alteration; KO, knockout.
Meiosis gene nsSNPs were first ranked using two of the most commonly used computational tools for predicting the likelihood of a variant being deleterious: SIFT (18) and Polyphen-2 (19). Four of the highest-scoring SNPs identified by both algorithms, one each in the genes Cyclin-dependent kinase 2 (Cdk2), MutL homolog 1 (Mlh1), Structural maintenance of chromosomes protein 1B (Smc1b), and Testis-expressed gene 15 (Tex15) (Fig. 1B), were then evaluated with six additional tools that predict functional consequences of SNPs: Mutation Assessor, PANTHER, PhD-SNP, SNPs&GO, SNAP, and CADD (20–25). Overall, each of these four nsSNPs was predicted to be deleterious by at least five of the eight computational algorithms used (Table 1). These four genes have crucial roles in meiosis, affecting processes including recombination, homolog synapsis, DNA repair, and chromatid cohesion (Fig. 1B).
Table 1.
Predicted functional consequences of human nsSNPs
| Algorithm | D Min | CDK2 | MLH1 | SMC1B | TEX15 |
| rs3087335 | rs63750447 | rs61735519 | rs147871035 | ||
| Cdk2Y15S | Mlh1V384D | Smc1bF1055L | Tex15T2181I | ||
| SIFT | ≤ 0.05 | D (0) | D (0) | D (0) | D (0) |
| PolyPhen 2 | >0.5 | D (1) | D (0.99) | D (0.99) | D (0.92) |
| Mutation Assessor | >0.65 | D (2.3) | D (3.2) | D (3.2) | D (1.8) |
| PANTHER | ≥0.5 | D (0.8) | D (0.5) | D (0.7) | T (0.4) |
| PhD-SNP | ≥0.5 | D (0.9) | D (0.9) | T (0.4) | D (0.7) |
| SNPs&GO | ≥0.5 | D (0.9) | D (0.9) | T (0.4) | D (0.7) |
| SNAP | ≥5 | D (5) | D (5) | T (2) | T (3) |
| CADD* | >15 | D (23) | D (19.5) | D (25.5) | D (24.3) |
The allele for each SNP (rs no.) modeled is indicated. D Min refers to the minimum score for deleterious classification; D, deleterious; T, tolerated.
CADD scores based on v1.1.
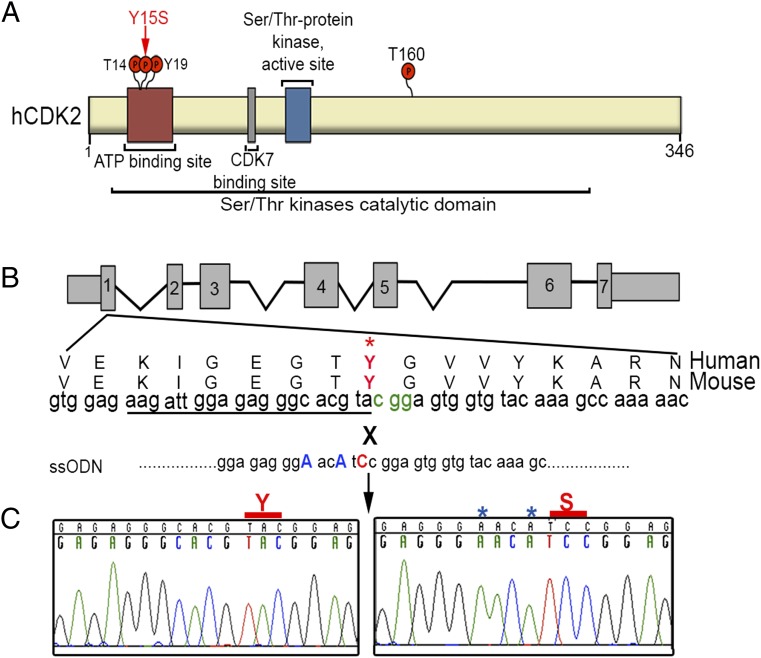
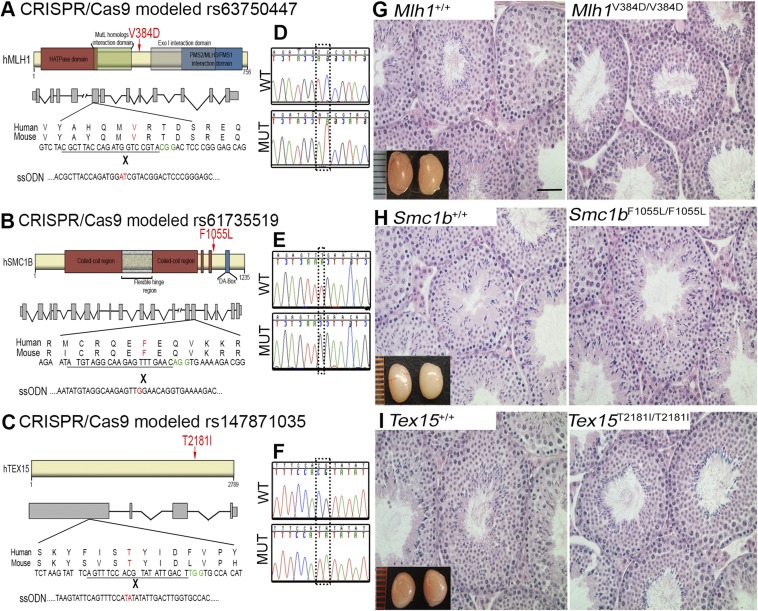
For each of the four SNPs, the orthologous amino acid changes were introduced into the mouse genes using CRISPR/Cas9-mediated genome editing in mouse embryos. Specifically, single-celled embryos were comicroinjected with Cas9 mRNA, sgRNA, and a synthetic single-stranded oligodeoxynucleotide (ssODN) containing nucleotide changes (Fig. 2B, Fig. S1 A–C, and Table S1) that, via homologous recombination stimulated by targeted Cas9 double-strand breaks (DSBs), introduced the desired amino acid change. We identified edited founder mice (Materials and Methods) carrying at least one desired humanized allele of Cdk2, Mlh1, Smc1b, and Tex15 at frequencies of 44.4%, 29.1%, 21.4%, and 56.2%, respectively (Table S2). In the experiments that generated the Cdk2 and Tex15 alleles used here, a Next-Gen sequencing strategy was used to confirm that there was no evidence of off-target editing at degenerate sgRNA recognition sites (26).
Fig. 2.
CRISPR/Cas9 modeling of CDK2 SNP rs3087335. (A) Domains in the hCDK2 protein and location of rs3087335 (red arrow). (B) CRISPR/Cas9 editing strategy for making the Y15S alteration. The Tyr15 site is conserved in humans and mice. PAM nucleotides are in green. Underscored nucleotides match the CRISPR gRNA. To avoid Cas9 recutting of an already-edited locus, two silent mutations (blue) were included into the ssODN recombination template, only part of which is shown (Table S1). (C) Sanger sequencing chromatograms of control and mutant mice showing correct CRISPR/Cas9 targeting.
Fig. S1.
Histological analysis of testes from mice homozygous for predicted deleterious human SNPs in Mlh1, Smc1b, and Tex15. (A–C) Schematics of human proteins with locations of the SNP-encoded amino acid changes (red arrows). Also shown are intron-exon structures, local sequence around the targeted SNPs, Cas9 PAM sites (green), sequences corresponding to gRNAs, and partial sequences of targeting ssODNs, including the edited nucleotides (red). (D–F) Sanger sequencing chromatograms of WT and mutant (MUT) animals confirming correct editing. (G–I) Testis histology of littermate pairs. Mice were 60 d old when killed. All are H&E-stained paraffin sections. (Insets) Whole testes (WT on left, mutant on right). (Scale bar: 100 μm.)
Table S1.
Oligonucleotides used in the study
| Gene | Sequence (5′ to 3′) | |
| gRNA oligos annealed for plasmid cloning | Cdk2 | CACCAAGATTGGAGAGGGCACGTA |
| AAACTACGTGCCCTCTCCAATCT | ||
| Mlh1 | CACCCGCTTACCAGATGGTCCGTA | |
| AAACTACGGACCATCTGGTAAGCG | ||
| Smc1b | CACCGAATATGTAGGCAAGAGTTTGAAC | |
| AAACGTTCAAACTCTTGCCTACATATTC | ||
| Tex15 | CACCAGTTTCCACGTATATTGACT | |
| AAACAGTCAATATACGTGGAAACT | ||
| Homologous recombination ssODNs | Cdk2 | GAGCCGCCTCACTAGCGCTCCATGGAGAACTTCCAAAAGGTGGAGAAGATTGGAGAGGGAACATCCGGAGTGGTGTACAAAGCCAAAAACAAGTTGACGGGAGAAGTTGTGGCGCTTAAGAAGATCCG |
| Mlh1 | CACGACAGGGGTGGCTTCCTCATCCACTAGTGGAAGTGGCGACAAGGTCTACGCTTACCAGATGGATCGTACGGACTCCCGGGAGCAGAAGCTTGACGCCTTTCTGCAGCCTGTAAGCAGCCTTGGG | |
| Smc1b | TTCTTAGTTTTTGAGGCCAGCAGAAAGGAAGCCAGAATATGTAGGCAAGAGTTGGAACAGGTGAAAAGACGGAGGTACGATGCTTTCAGTCAATGTTTTGAACACATCTCAGTCTCAATTGATCAA | |
| Tex15 | AACTTACAGGCGTTAAAAGGCTTCTGAATAACTCTAAGTATTCAGTTTCCATATATATTGACTTGGTGCCACATACTGCATCTGTAAATTTTGGAAACACTGTGGCAGAATTAGAACATAACTACA | |
| Primers for making in vitro transcription templates by PCR | Cdk2 | TTAATACGACTCACTATAGCACCAAGATTGGAGAGGGCACGTA |
| Mlh1 | TTAATACGACTCACTATAGCACCCGCTTACCAGATGGTCCGTA | |
| Smc1b | TTAATACGACTCACTATAGCACCGAATATGTAGGCAAGAGTTTGAAC | |
| Tex15 | TTAATACGACTCACTATAGCACCAGTTTCCACGTATATTGACT | |
| Common reverse primer | AAAAGCACCGACTCGGTGCC |
Table S2.
Efficiency of CRISPR/Cas9-mediated targeting
| Gene | Concentration (Cas9/sgRNA/ ssODN), ng/μL | Two-cell stage embryos/zygotes surviving injection, n (%) | Recipients, n | Edited animals/ newborns, n (%) | HDR edited mice, n (%) | HDR heterozygotes/ HDR edited mice, n |
| Cdk2* | 50/50/100 | 69/86 (80.2) | 3 | 20/27 (74.1) | 10/27 (44.4) | 2/10 |
| Mlh1 | 100/50/100 | 95/135 (70.3) | 4 | 9/24 (37.5) | 7/24 (29.1) | 4/7 |
| Smc1b | 100/50/100 | 101/150 (67.3) | 4 | 5/14 (35.7) | 3/14 (21.4) | 2/3 |
| Tex15† | 50/50/100 | 35/40 (87.5) | 2 | 16/16 (100) | 9/16 (56.2) | 3/9 |
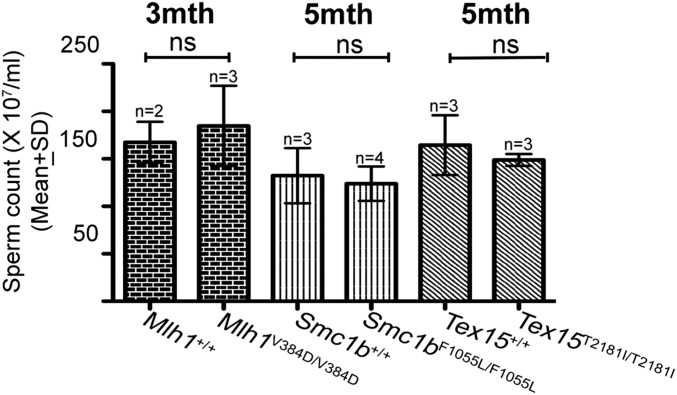
To determine whether the humanized alleles compromised reproduction, we bred each to homozygosity, tested fertility by mating, and performed a series of histological and cytological analyses. In males, we examined epididymal sperm number and morphology at progressive ages. In addition, we assessed testis size, weight, and histology. In the cases of adult males homozygous for the humanized alleles of rs63750447 (Mlh1V376D), rs61735519 (Smc1bF1055L), and rs147871035 (Tex152181I), histological analysis of testes of these mutants did not reveal any phenotypic defects; the seminiferous tubules exhibited normal spermatogenesis that was indistinguishable from that in wild type (WT) (Fig. S1). As predicted from the testes histology, the mutants exhibited no significant difference in epididymal sperm counts compared with WT, an assay that also reflects sperm motility (Fig. S2), and a homozygote of each was proven to be fertile in matings.
Fig. S2.
Sperm counts of Mlh1V384D/V384D, Smc1bF1055L/F1055L, and Tex15T2181I/T2181I mice. Graph represents cauda epididymal sperm counts (mean ± SD) from homozygous mutants for all three alleles and their respective littermate WT controls of indicated ages. The two-tailed unpaired t test revealed no significant differences (ns, not significant; P > 0.05). Two or more animals (as indicated) were analyzed for each genotype. mth, month.
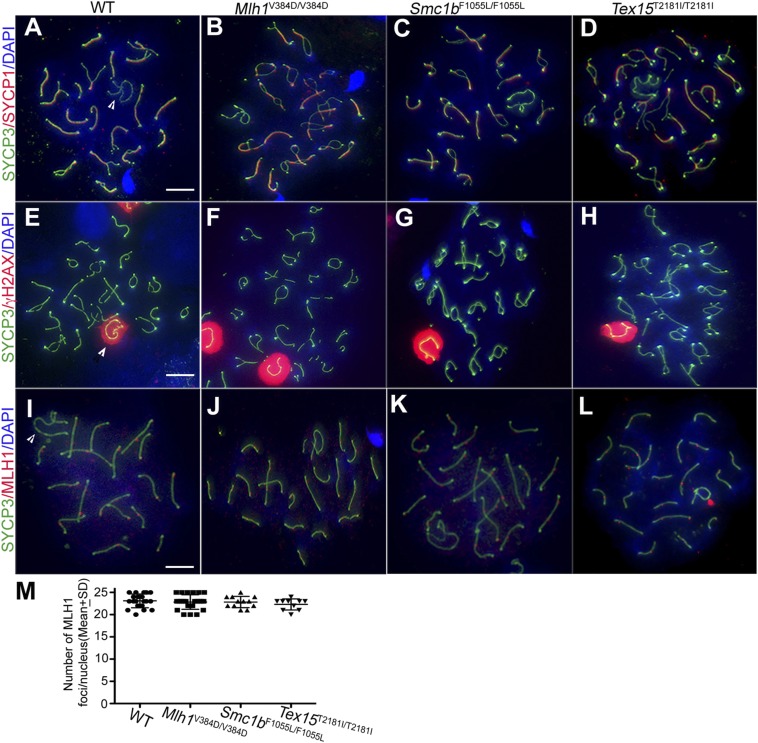
Null alleles of these genes, which encode proteins involved in meiotic chromosome cohesion, crossing-over, and repair of meiotic DSBs, respectively, cause meiotic arrest with characteristic defects in meiotic prophase I chromosomes (27–29). To determine whether there was evidence of any such defects in these mice, we immunolabeled surface-spread spermatocyte nuclei for proteins diagnostic of key meiotic events. For each humanized allele, patterns of γH2AX (present at meiotic DSBs and on asynapsed chromosomes), SYCP1 (chromosome synapsis marker), SYCP3 (a synaptonemal complex axial element protein), and MLH1 foci (which mark sites of chiasmata) revealed no differences between WT and mutants (Fig. S3 A–L), including in the number of MLH1 foci (Fig. S3M). In addition, whereas mouse knockouts of Smc1b and Mlh1 cause female infertility from meiotic failure, the female homozygotes for each humanized allele were fertile and normally fecund (Fig. S4A). These data indicate that rs61735519 (SMC1B), rs63750447 (MLH1), and rs147871035 (TEX15) do not impact fertility, despite the high algorithm prediction scores.
Fig. S3.
Immunocytologic analysis of spermatocytes homozygous for predicted deleterious human SNPs in Mlh1, Smc1b, and Tex15. Surface-spread nuclei were immunolabeled with a synaptonemal complex lateral element marker, SYCP3, in combination with markers for fully synapsed regions (SYCP1), unrepaired DSBs and silenced chromatin (γH2AX), or MLH1 (chiasmata). In A, E, and I, XY pairs are denoted by arrowheads. (A–H) Late pachytene/diplotene nuclei. (I–L) Pachytene nuclei. (M) Quantification of MLH1 foci on meiotic chromosomes of WT, Mlh1V384D/V384D, Smc1bF1055L/F1055L, and Tex15T2181I/T2181I pachytene spermatocytes. At least 10 total spreads from two or more animals for each genotype were scored. There was no significant difference in focus number in any of the mutants compared with WT. Data are presented as mean ± SD from two or more animals. (Scale bars: 10 μm.)
Fig. S4.
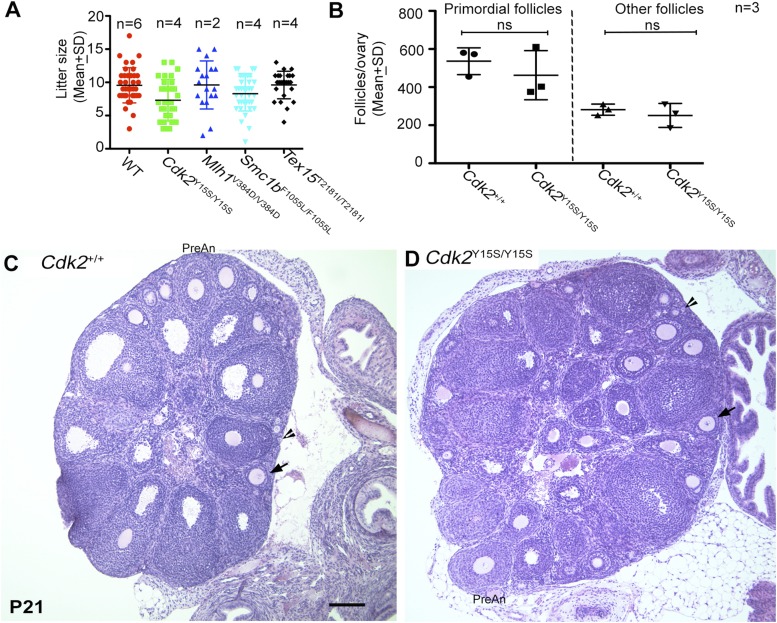
Female mice with humanized SNP alleles are normally fertile. (A) Litter sizes of humanized models. The WT data are pooled from littermates combined from the different mutant alleles. (B) Quantification of ovarian follicle types (± SD). “Other follicles” consist of primary, secondary, preantral, and antral follicles. (C and D) The Cdk2Y15S allele does not impact ovarian histology. Shown are H&E-stained paraffin sections of 3-wk-old mouse ovaries. Arrowheads indicate a primordial follicle, and the arrow indicates a secondary follicle. A preantral follicle from each section is also labeled. ns, not significant; wk, week. (Scale bar: 100 μm.)
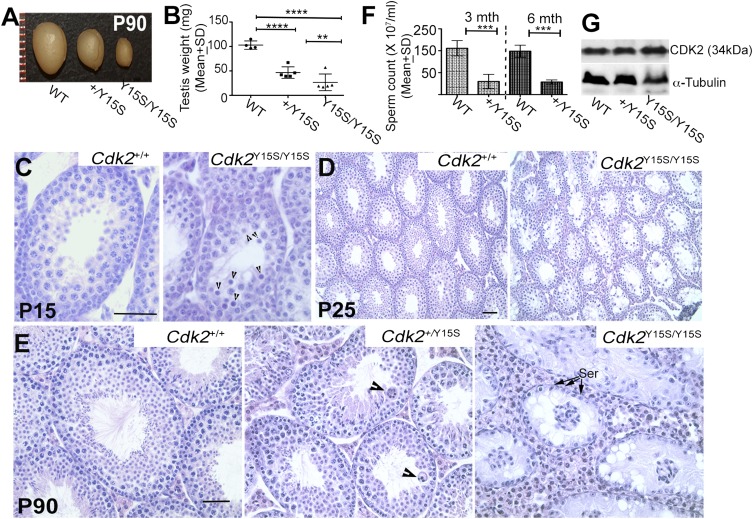
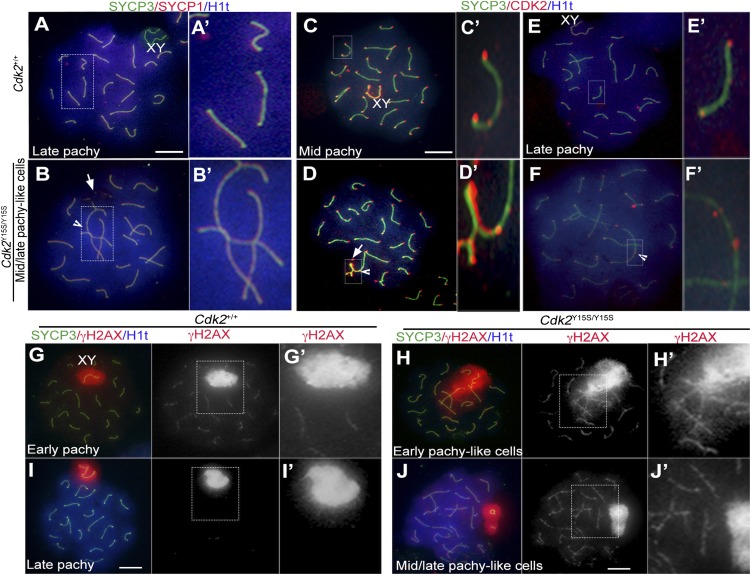
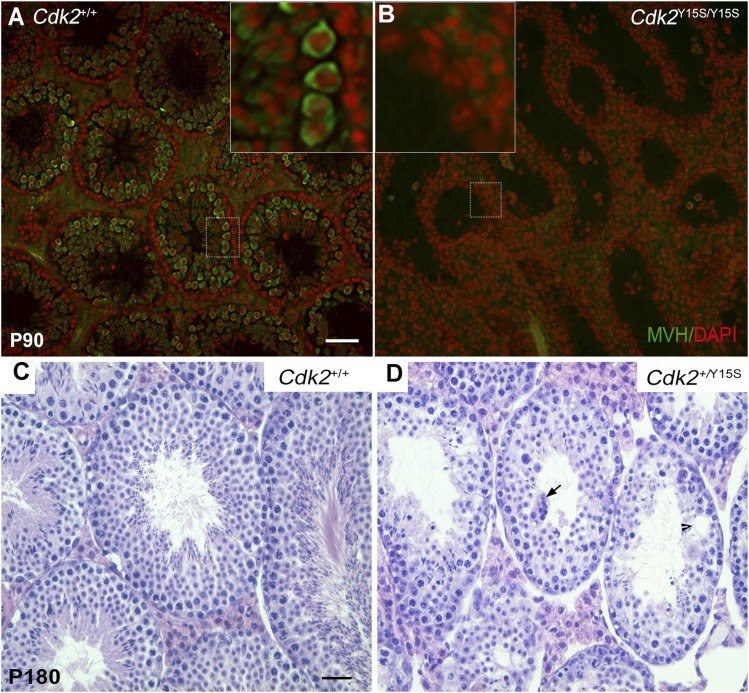
In contrast, male mice homozygous for the CDK2-Y15S alteration, mimicking human rs3087335, severely disrupted spermatogenesis. Cdk2Y15S/Y15S males (n = 13) had markedly smaller testes than WT siblings (n = 12), weighing ∼80% less (Fig. 3 A and B). These mutants were devoid of sperm in their epididymides, and failed to impregnate WT female partners despite plugging them. Histological examination of Cdk2Y15S/Y15S testes revealed an arrest of meiotic progression at the pachytene spermatocyte stage; no metaphase spermatocytes or postmeiotic spermatids were observed (Fig. 3 C–E), consistent with the phenotype of Cdk2 null mice (30, 31). Furthermore, spermatocyte meiotic prophase I chromosomes exhibited abnormalities common in null mutants (32), including telomere fusions, XY body defects, and unrepaired DSBs, all of which prevent mutant spermatocytes from progressing past the midpachytene stage (Fig. 4). There were three differences from the knockout, however. First, unlike null females, Cdk2Y15S/Y15S females were fertile, delivering an average of approximately seven pups per litter (Fig. S4A). The mutant ovaries had grossly normal histology and numbers of follicles (both primordial pool and developing follicles) that were not significantly different from that in WT at postnatal day (P) 21 (Fig. S4 B and C). Second, whereas Cdk2−/− seminiferous tubules retained spermatogonia that continued to initiate waves of spermatogenesis until at least P120 (31), Cdk2Y15S/Y15S seminiferous tubules became devoid of germ cells, marked by mouse vasa homolog (MvH), by P90; they exhibited a Sertoli cell-only phenotype (Fig. 3E and Fig. S5B). As discussed later, this is indicative of spermatogonial stem cell maintenance or a proliferation defect. We also noticed pyknotic cells (presumably spermatocytes) appearing as early as P15, when germ cells in the first spermatogenic wave progress as far as the pachytene spermatocyte stage (Fig. 3C).
Fig. 3.
The CDK2Y15S alteration encoded by rs3087335 causes male infertility. (A) Testes (90 d old) from littermates of indicated genotypes. (B) Testes weights of P90 heterozygous and homozygous mice were 54.6% and 81.5% lower, respectively, than those of WT. (C–E) H&E-stained seminiferous tubule cross-sections of 15-, 25-, and 90-d-old testes. Arrowheads indicate degenerating cells. (F) Sperm counts of 3- and 6-mo-old WT and heterozygous males. (G) Western blot analysis of CDK2 in spleen. Genotypes are indicated, as are approximate molecular weights of the detected species with anti-CDK2 antibody. Spleens were used instead of testes, because differences in cellularity of mutant testes would compromise interpretation of relative protein amounts. ****P < 0.0002; ***P < 0.0001; **P < 0.001. Data are presented as mean ± SD of at least three animals. Ser, Sertoli cells. (Scale bars: 100 μm).
Fig. 4.
Meiotic defects in Cdk2Y15S/Y15S spermatocytes. (A–J) Immunolabeling of surface-spread spermatocyte nuclei. Genotypes, antibodies used, and pachytene substages are indicated. Histone H1t appears in mid-pachynema. Note abnormal nonsynaptic fusions (arrowheads in B and D) and abnormal XY body (arrows in B and D). Meiotic DSBs (indicated by γH2AX foci) in WT spermatocytes (G) are repaired by late pachynema (I); however, mutant chromosomes retain γH2AX (which marks both unrepaired DSBs and areas of asynapsed chromosomes) and these spermatocytes do not progress beyond a mid/late pachynema-like stage (J and J′). Boxed areas are enlarged to highlight specific abnormalities (A′–J′). XY, XY body (also known as the “sex body”); arrow, abnormal XY body; arrowhead, telomeric fusion; pachy, pachynema. (Scale bars: 10 μm.)
Fig. S5.
The Cdk2Y15S allele causes spermatogonial loss in homozygotes and age-related testis pathology in heterozygotes. (A and B) Immunolabeling of testis sections from P90 mice with the cytoplasmic germ cell marker MvH (green). Cdk2Y15S/Y15S seminiferous tubules are devoid of MvH-positive spermatogonia. Nuclei were stained with DAPI (red). (C and D) H&E-stained cross-sections of seminiferous tubules from Cdk2+/+ and Cdk2+/Y15S mutants. The arrow points to an abnormal multinucleate cell structure. Note the reduced elongating spermatids in the lumen of Cdk2+/Y15S tubules. The arrowhead indicates an abnormal vacuole. (Scale bar: 100 μm.)
The third distinction between the Cdk2Y15S and null alleles appeared in heterozygous males. At P90, Cdk2Y15S/+ males exhibited smaller testes of approximately one-half the weight of WT testes (Fig. 3 A and B), abnormal seminiferous tubule histology characterized by large multinucleate cells and decreased luminal elongated spermatids (Fig. 3E), and a ∼3.5-fold reduction in epididymal sperm count compared with WT littermates (Fig. 3F). The seminiferous tubules became more disorganized by P180 (Fig. S5 C and D). Mice heterozygous for a Cdk2 null allele do not exhibit meiotic chromosome abnormalities or infertility (32), and the Cdk2Y15S allele did not affect CDK2 levels (Fig. 3G). Thus, Cdk2Y15S appears to be a semidominant, gain-of-function allele.
Discussion
In this paper, we introduce an association- and linkage-free approach to studying the genetics of human infertility that can be applicable to other diseases, implemented either in a purely reverse genetic paradigm as described here or as a validation tool for candidate SNPs implicated by genetic mapping. The speed and efficacy with which CRISPR/Cas9 can be used to generate precisely engineered mouse models enables powerful and physiologically relevant functional assessment of human genetic variants. This is especially important in cases of infertility, for which accurate in vitro systems are lacking (33).
A surprising outcome of the present study was the poor accuracy of computational tools designed to predict functional consequences of SNPs. Whereas all eight algorithms correctly predicted that the SNP in CDK2 would disrupt gene function, all classified the MLH1 SNP as being functionally damaging, and most also incorrectly predicted the same for the SNPs in SMC1B and TEX15. SNAP, a neural network-based tool (24), was the most accurate (75% correctly predicted) for the small sample size tested here. Nonetheless, it remains possible that the SNPs affect protein function in ways that do not grossly impact germ cell function. It also should be cautioned that the SNPs may be deleterious in humans but not in mice. Also noteworthy is the fact that the CDK2 SNP was the only SNP tested in which the affected amino acid resided in an annotated functional domain of the protein, and in this case, the amino acid itself was known to be a target of WEE1 kinase (Fig. 2A and Fig. S1 A–C). We surmise that manual scrutiny of nsSNPs will complement algorithm predictions.
Our finding that CDK2Y15S disrupts spermatogenesis is remarkable in two respects. First, it provides evidence that semidominant infertility alleles can segregate in human populations. This particular allele exists at a low frequency, having been identified in only 2 of 688 alleles sampled (dbSNP build 142). Because it affects only male mice (with full penetrance in homozygotes and with a partial, age-related germ cell depletion phenotype in heterozygotes), transmission may occur only through women. If men heterozygous for CDK2Y15S are fertile (perhaps only at earlier ages), then homozygous males would occur at a rate of ∼1 in 236,672. The allele frequencies in different demographic populations remain unclear, however. In mice, a fully penetrant, dominant male-specific sterile allele of the meiotic recombination gene Dmc1 has provided evidence that dominant infertility mutations can arise in genes that have a recessive character as null alleles, and that such mutations can be transmitted through the unaffected sex (34). Finally, these cases indicate the difficulty in predicting the mode of inheritance and associated phenotypes purely by computational means.
Second, the CDK2Y15S allele reveals a novel role for the function of CDK2 in vivo. Histological and cytological analyses indicated that, along with the null-like meiotic arrest of Cdk2Y15S/Y15S spermatocytes during the first wave of spermatogenesis, homozygotes became germ cell-depleted, unlike the null alleles. Given that phosphorylation of Tyr15 by WEE1 kinase inhibits CDK2 activity and thus cell cycle progression in mitotic cells (35–37), we speculate that mutation of the inhibitory phosphorylation site in this allele disturbs the proper cell cycle control of spermatogonial stem cells, affecting the regulation of self-renewal and differentiation of stem cells. It will be of interest to determine whether Cdk2Y15S allele triggers inappropriate proliferation or differentiation of spermatogonial stem cells at the expense of proliferation or renewal of the stem cell population. Consistent with this, a previous study found that when both of the CDK2 inhibitory phosphorylation sites at threonine 14 and tyrosine 15 were changed to alanine and phenylalanine, respectively, heterozygous male mice were rendered subfertile or sterile, and that somatic cells bearing these mutations entered S phase improperly (38). We suspect that in Cdk2 null mice, control of the spermatogonial cell cycle or meiotic entry is accomplished by another cyclin, such as Cdk1 (39). The absence of a self-renewing oogonial stem cell population may explain the fertility of CdkY15S homozygous females. We caution that our alleles were generated in a different genetic background (FvB × C57BL/6) than that of mice bearing null alleles (mixes of 129 and C57BL/6, as far as can be judged from the literature) (30–32).
The complexity of male germ cell differentiation, coupled with limited information on the biochemistry of germ cell-specific genes, complicates the task of identifying true segregating infertility alleles in humans. Our results demonstrate that whereas computational predictions of SNP effects are useful for identifying candidate causal mutations for diverse diseases, including those mapped by GWAS, functional evaluation in vivo is essential to define true physiological consequences. It is likely that computational tools can be improved with increased knowledge of protein structure and function, coupled with a larger body of training data generated by methods described in this report; however, a present shortcoming for identifying human disease SNPs in mice is that this is limited to nsSNPs that alter conserved amino acids, and that addressing noncoding SNPs will be more difficult. Nevertheless, the efficiency of this approach can revolutionize reproductive genetics by establishing a permanent reference of deleterious vs. benign alleles, which will help clarify the molecular basis of idiopathic infertilities in individual patients. Furthermore, because this approach can be applied to any genetic disease, it should be a powerful tool in the era of personalized genomic medicine.
Materials and Methods
Computational Identification of Potential Infertility SNPs.
The Ensembl Genome Browser (40) was used to extract lists of nsSNPs in several meiosis genes that were predicted to be deleterious based on precomputed scores from the SIFT (v5.0.2) (18) and PolyPhen-2 (v2.2.2) (19) algorithms. The SNPs selected were limited to those with frequency information from multiple sources (e.g., 1000 Genomes Project, HapMap Project) or multiple individuals. The highest-scoring SNPs were screened with the other six algorithms listed in Table 1.
Production of CRISPR/Cas9-Edited Mice.
sgRNAs (Table S2) were designed and produced as described previously (26). The ssODN source was “Ultramer” oligos (Integrated DNA Technologies). The plasmid pST1374-NLS-flag-linker-Cas9 (44758; Addgene) was used for generating Cas9 mRNA after linearization with AgeI. In vitro transcription and capping were performed using the mMESSAGE mMACHINE T7 Transcription Kit (AM1345; Life Technologies). Embryo microinjections into zygotes [F1 hybrids between strains FVB/NJ and B6(Cg)-Tyrc-2J/J] were performed as described previously (26) using the reagent concentrations listed in Table S1. Edited founders were identified either by Next-Gen sequencing or by subcloning followed by Sanger sequencing as described previously (26). Genotyping was performed by sequencing or by restriction enzyme analysis of PCR products produced from DNA isolated from ear punch biopsy specimens. Primer sequences and their respective amplicon sizes for genotyping for each mutant mouse line are listed in Table S3. Annealing temperature was 64 °C for Cdk2, 60 °C for Mlh1, 62 °C for Smc1b, and 60 °C for Tex15.
Table S3.
Genotyping primers
| Gene | Direction | Sequence (5′ to 3′) | Product, bp | |
| Cdk2 | F | TGCAAGAAACTTTCCCCTGG | 470 | |
| R | GCTCTCCTTGCGTTCCATC | |||
| Mlh1 | F | GCTGCGTCTGAAGTCTCCAT | 422 | |
| R | CACTGCTGTTCTGTTTTGGGA | |||
| Smc1b | F | CACTGGCTTTGCACCTATGT | 440 | |
| R | ACACTGTCAGGATTTGCTAACA | |||
| Tex15 | F | CATTGCCTTTGGAAACTGGT | 308 | |
| R | CCAAAATCTTTCTGAGGGACA | |||
Mice and Reproductive Phenotypes.
The alleles generated in this study and their abbreviations (in parentheses) were as follows: Cdk2em1Jcs (Cdk2Y15S), Mlh1em1Jcs (Mlh1V384D), Smc1bem1Jcs (Smc1bF1055L), and Tex15em1Jcs (Tex15T2181I). For female fertility tests, 8-wk-old animals were housed with age-matched FVB/NJ males. For the Cdk2Y15S allele, there was perfect genotype–phenotype correlation; all 13 homozygous males (P25–P180) and 8 heterozygous males (P90 and P180) examined, but none of the 12 WT littermates, had small gonads.
Histology and Immunohistochemistry.
Testes and ovaries were fixed for 24 h at 23 °C in Bouin’s solution, paraffin-embedded, sectioned at 5 μm, and then stained with H&E. Ovarian follicle quantification was done by counting the every fifth section, as described previously (41). For immunofluorescence, testes were fixed in 4% paraformaldehyde for ∼24 h, paraffin-embedded, sectioned at 7 μm, deparaffinized, and then boiled for 30 min in 10 mM citric acid (pH 6.0) for antigen retrieval. Sections were blocked in PBS containing 5% goat serum for 30 min at 23 °C, followed by incubation with primary antibody (rabbit anti-MVH; 1:500, ab13840; Abcam) for 12 h at 4 °C and detection by the secondary antibody (goat anti-rabbit IgG 488; 1:1,000, A11008; Molecular Probes).
Immunocytology of Meiotic Chromosomes.
Preparation of surface-spread spermatocyte chromosomes and immunolabeling was done as described previously (42, 43). Primary antibodies were rabbit anti-CDK2 (1:50, ab7954-1; Abcam), rabbit anti-SYCP3 (1:500, ab15093; Abcam), mouse anti-SYCP3 (1:500, ab97672; Abcam), rabbit anti-SYCP1 (1:500, ab15087; Abcam), mouse anti-γH2AX (1:500, 05-636; Millipore), guinea pig anti-H1t (1:1,000, a gift from M. A. Handel, The Jackson Laboratory, Bar Harbor, ME), and mouse anti-MLH1 (1:50, 550838; BD Pharmingen). Secondary antibodies were goat anti rabbit-IgG 488 (1:1,000, A11008; Molecular Probes), goat ant-rabbit IgG 594 (1:1,000, A11012; Molecular Probes), goat anti-guinea pig IgG 647 (1:1,000, A21450; Molecular Probes), and goat anti-mouse IgG 594 (1:1,000, A11005; Molecular Probes). Nuclei were counterstained with DAPI, and slides were mounted with ProLong Gold antifade reagent (P36930; Molecular Probes).
Western Blot Analysis.
Protein was extracted from spleens using RIPA buffer (9806S; Cell Signaling Technology) supplemented with protease inhibitors (cOmplete, Mini, 11836153001; Roche). Extracts were separated by SDS/PAGE (12% acrylamide), followed by electrotransfer to a PVDF membrane (Immobilon-P membrane, IPVH00010; EMD Millipore), and then blocked in 5% nonfat milk. Membranes were probed with rabbit anti-CDK2 (1:200, ab7954-1; Abcam) for 2 h at room temperature and then washed with Tris-buffered saline with 0.1% Tween-20, followed by a 1-h incubation in anti-rabbit IgG HRP-linked secondary antibody (7074S; Cell Signaling Technology). Signal was detected using Luminata Classico Western HRP substrate (WBLUC0100; EMD Millipore).
Imaging.
Surface-spread preparations were imaged with an Olympus BX51 microscope with a 60×/0.90 NA infinity/0.11–0.23 objective, and Magnafire 2.0 software (Olympus). Histological sections were imaged with the same microscope using a 20× (20×/0.05; NA infinity/0.17) objective. Cropping, color, and contrast adjustments were made with Adobe Photoshop CC 2014, using identical background adjustments for all images.
Sperm Counts.
Cauda epididymides (3–6 mo old) were minced in 2 mL of PBS and then incubated for 15 min at 37 °C to allow spermatozoa to swim out. The sperm were diluted 1:5 in PBS, incubated for 1 min at 60 °C, and then counted with a hematocytometer.
Acknowledgments
We thank Rob Munroe, Christian Abratte, and Ewelina Bolcun for assistance with CRISPR/Cas9 microinjections and CRISPR design. This work was supported by National Institutes of Health Grant GM45415 (to J.C.S.) and Contract CO29155 from the New York State Stem Cell Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1506974112/-/DCSupplemental.
References
- 1.Foresta C, Moro E, Ferlin A. Y chromosome microdeletions and alterations of spermatogenesis. Endocr Rev. 2001;22(2):226–239. doi: 10.1210/edrv.22.2.0425. [DOI] [PubMed] [Google Scholar]
- 2.Matzuk MM, Lamb DJ. The biology of infertility: Research advances and clinical challenges. Nat Med. 2008;14(11):1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dieterich K, et al. Homozygous mutation of AURKC yields large-headed polyploid spermatozoa and causes male infertility. Nat Genet. 2007;39(5):661–665. doi: 10.1038/ng2027. [DOI] [PubMed] [Google Scholar]
- 4.Dam AH, et al. Homozygous mutation in SPATA16 is associated with male infertility in human globozoospermia. Am J Hum Genet. 2007;81(4):813–820. doi: 10.1086/521314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieterich K, et al. The Aurora Kinase C c.144delC mutation causes meiosis I arrest in men and is frequent in the North African population. Hum Mol Genet. 2009;18(7):1301–1309. doi: 10.1093/hmg/ddp029. [DOI] [PubMed] [Google Scholar]
- 6.Brioude F, et al. Two families with normosmic congenital hypogonadotropic hypogonadism and biallelic mutations in KISS1R (KISS1 receptor): Clinical evaluation and molecular characterization of a novel mutation. PLoS One. 2013;8(1):e53896. doi: 10.1371/journal.pone.0053896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mandon-Pépin B, et al. Infertilité humaine: des gènes de méiose comme candidats potentiels [Human infertility: Meiotic genes as potential candidates] Gynecol Obstet Fertil. 2002;30(10):817–821. doi: 10.1016/s1297-9589(02)00444-7. French. [DOI] [PubMed] [Google Scholar]
- 8.Stouffs K, Vandermaelen D, Tournaye H, Liebaers I, Lissens W. Mutation analysis of three genes in patients with maturation arrest of spermatogenesis and couples with recurrent miscarriages. Reprod Biomed Online. 2011;22(1):65–71. doi: 10.1016/j.rbmo.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Sato H, et al. Polymorphic alleles of the human MEI1 gene are associated with human azoospermia by meiotic arrest. J Hum Genet. 2006;51(6):533–540. doi: 10.1007/s10038-006-0394-5. [DOI] [PubMed] [Google Scholar]
- 10.Sakugawa N, et al. LMTK2 and PARP-2 gene polymorphism and azoospermia secondary to meiotic arrest. J Assist Reprod Genet. 2009;26(9-10):545–552. doi: 10.1007/s10815-009-9347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyamoto T, et al. Single-nucleotide polymorphisms in HORMAD1 may be a risk factor for azoospermia caused by meiotic arrest in Japanese patients. Asian J Androl. 2012;14(4):580–583. doi: 10.1038/aja.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzmin A, et al. Identification of potentially damaging amino acid substitutions leading to human male infertility. Biol Reprod. 2009;81(2):319–326. doi: 10.1095/biolreprod.109.076000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, et al. Excess of rare variants in genes that are key epigenetic regulators of spermatogenesis in the patients with non-obstructive azoospermia. Sci Rep. 2015;5:8785. doi: 10.1038/srep08785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes AM, et al. Human spermatogenic failure purges deleterious mutation load from the autosomes and both sex chromosomes, including the gene DMRT1. PLoS Genet. 2013;9(3):e1003349. doi: 10.1371/journal.pgen.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatsenko AN, et al. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med. 2015;372(22):2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matzuk MM, Lamb DJ. Genetic dissection of mammalian fertility pathways. Nat Cell Biol. 2002;4(Suppl):s41–s49. doi: 10.1038/ncb-nm-fertilityS41. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, et al. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat Commun. 2014;5:3857. doi: 10.1038/ncomms4857. [DOI] [PubMed] [Google Scholar]
- 18.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 19.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas PD, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31(1):334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R. Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum Mutat. 2009;30(8):1237–1244. doi: 10.1002/humu.21047. [DOI] [PubMed] [Google Scholar]
- 23.Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22(22):2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- 24.Bromberg Y, Rost B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007;35(11):3823–3835. doi: 10.1093/nar/gkm238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh P, Schimenti JC, Bolcun-Filas E. A mouse geneticist’s practical guide to CRISPR applications. Genetics. 2015;199(1):1–15. doi: 10.1534/genetics.114.169771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revenkova E, Eijpe M, Heyting C, Gross B, Jessberger R. Novel meiosis-specific isoform of mammalian SMC1. Mol Cell Biol. 2001;21(20):6984–6998. doi: 10.1128/MCB.21.20.6984-6998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edelmann W, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85(7):1125–1134. doi: 10.1016/s0092-8674(00)81312-4. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol. 2008;180(4):673–679. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13(20):1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Ortega S, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35(1):25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- 32.Viera A, et al. CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J Cell Sci. 2009;122(Pt 12):2149–2159. doi: 10.1242/jcs.046706. [DOI] [PubMed] [Google Scholar]
- 33.Handel MA, Eppig JJ, Schimenti JC. Applying “gold standards” to in vitro-derived germ cells. Cell. 2014;157(6):1257–1261. doi: 10.1016/j.cell.2014.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bannister LA, et al. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5(5):e105. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu Y, Rosenblatt J, Morgan DO. Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J. 1992;11(11):3995–4005. doi: 10.1002/j.1460-2075.1992.tb05493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker LL, Atherton-Fessler S, Piwnica-Worms H. p107wee1 is a dual-specificity kinase that phosphorylates p34cdc2 on tyrosine 15. Proc Natl Acad Sci USA. 1992;89(7):2917–2921. doi: 10.1073/pnas.89.7.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parker LL, Piwnica-Worms H. Inactivation of the p34cdc2-cyclin B complex by the human WEE1 tyrosine kinase. Science. 1992;257(5078):1955–1957. doi: 10.1126/science.1384126. [DOI] [PubMed] [Google Scholar]
- 38.Zhao H, Chen X, Gurian-West M, Roberts JM. Loss of cyclin-dependent kinase 2 (CDK2) inhibitory phosphorylation in a CDK2AF knock-in mouse causes misregulation of DNA replication and centrosome duplication. Mol Cell Biol. 2012;32(8):1421–1432. doi: 10.1128/MCB.06721-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravnik SE, Wolgemuth DJ. Regulation of meiosis during mammalian spermatogenesis: The A-type cyclins and their associated cyclin-dependent kinases are differentially expressed in the germ-cell lineage. Dev Biol. 1999;207(2):408–418. doi: 10.1006/dbio.1998.9156. [DOI] [PubMed] [Google Scholar]
- 40.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42(Database issue):D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers M, Britt KL, Wreford NG, Ebling FJ, Kerr JB. Methods for quantifying follicular numbers within the mouse ovary. Reproduction. 2004;127(5):569–580. doi: 10.1530/rep.1.00095. [DOI] [PubMed] [Google Scholar]
- 42.Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40(3):184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- 43.Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5(1):66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]