Significance
Hsp70 chaperones are key components of the cellular proteostasis network. The ATP-dependent interaction of Hsp70 with its substrates prevents aggregation and promotes their correct folding and maturation. Here we investigate the impact of Hsp70 binding on the conformation of a client substrate using nuclear magnetic resonance spectroscopy. Our experiments provide a model for Hsp70 action in which the substrate can adopt substantial amounts of secondary structure even in the globally unfolded Hsp70-bound state. Moreover, the substrate conformation does not evolve as a function of the Hsp70 nucleotide state, demonstrating that the large structural changes in Hsp70 resulting from ATP binding and hydrolysis do not perform conformational work on the bound substrate or lead to changes in client protein conformation.
Keywords: Hsp70, protein folding, NMR, molecular chaperones, CEST
Abstract
The 70 kDa heat shock protein (Hsp70) chaperone system is ubiquitous, highly conserved, and involved in a myriad of diverse cellular processes. Its function relies on nucleotide-dependent interactions with client proteins, yet the structural features of folding-competent substrates in their Hsp70-bound state remain poorly understood. Here we use NMR spectroscopy to study the human telomere repeat binding factor 1 (hTRF1) in complex with Escherichia coli Hsp70 (DnaK). In the complex, hTRF1 is globally unfolded with up to 40% helical secondary structure in regions distal to the binding site. Very similar conformational ensembles are observed for hTRF1 bound to ATP-, ADP- and nucleotide-free DnaK. The patterns in substrate helicity mirror those found in the unfolded state in the absence of denaturants except near the site of chaperone binding, demonstrating that DnaK-bound hTRF1 retains its intrinsic structural preferences. To our knowledge, our study presents the first atomic resolution structural characterization of a client protein bound to each of the three nucleotide states of DnaK and establishes that the large structural changes in DnaK and the associated energy that accompanies ATP binding and hydrolysis do not affect the overall conformation of the bound substrate protein.
The Hsp70 chaperone system forms a central hub in the cellular proteostasis network, performing a large number of functions, all of which are predicated upon its relatively simple ATP-dependent interaction with a client protein (1, 2). Hsp70 is a 70-kDa protein containing both nucleotide (NBD) and substrate (SBD) binding domains connected by a conserved linker (2). The best-studied Hsp70 is Escherichia coli DnaK, which recognizes an approximate seven-residue protein segment rich in large aliphatic hydrophobic residues flanked by positively charged amino acids (3). Client substrates enter the Hsp70 functional cycle by binding the ATP form of the chaperone, which has lower substrate affinity but faster binding and release rates compared with the ADP state (2). The crystal structure of ATP-DnaK (4, 5) shows that the two domains of Hsp70 are docked on each other, with the helical lid flanking the DnaK binding cleft in an open state. Subsequent ATP hydrolysis, stimulated by both Hsp40 and the bound substrate, locks the substrate in the binding pocket (2) and disengages the two domains of DnaK from each other, leading to major structural differences between the ATP- (4, 5) and ADP-bound (6) forms of the chaperone. Finally, ADP release, facilitated by nucleotide exchange factors (7), and rebinding of ATP free the bound substrate and reset the chaperone cycle.
Although the structural transitions of DnaK and the role of allostery throughout the chaperone reaction cycle are becoming increasingly well understood (7), far less information is available on the substrate conformation in the DnaK-bound state. Our current view is derived primarily from studies on the isolated SBD of DnaK in conjunction with peptide substrates incapable of independent folding (8). High-resolution structures of DnaK SBD in complex with the seven-residue peptide, NRLLLTG, show that the peptide binds the DnaK binding cleft in an extended conformation and is stabilized by hydrophobic contacts and backbone hydrogen bonds (9, 10). Larger fragments of apomyoglobin bound to the DnaK SBD were found to be predominantly unfolded but adopted small amounts of native and nonnative helical structure (11, 12). In addition, low-resolution studies on protein substrates have suggested that the substrate is globally unfolded (13, 14) and expanded in the DnaK-bound conformation (15). Although these studies have provided important insights into DnaK-substrate binding, a more comprehensive structural characterization of this interaction is crucial for a detailed understanding of DnaK function. Herein we present an atomic resolution-based structural analysis of a small, folding-competent protein domain in nucleotide-free (NF) and ATP- and ADP-DnaK bound states. Our results establish that in the DnaK bound form the substrate is globally unfolded, while retaining the structural propensities encoded by its primary sequence in regions outside of the chaperone binding site.
Results
The hTRF1 Protein Binds to DnaK in a Canonical Manner.
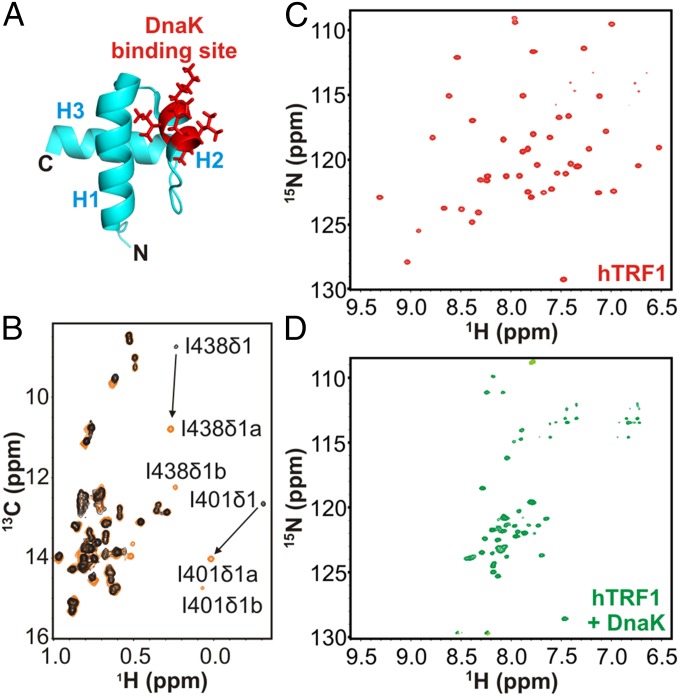
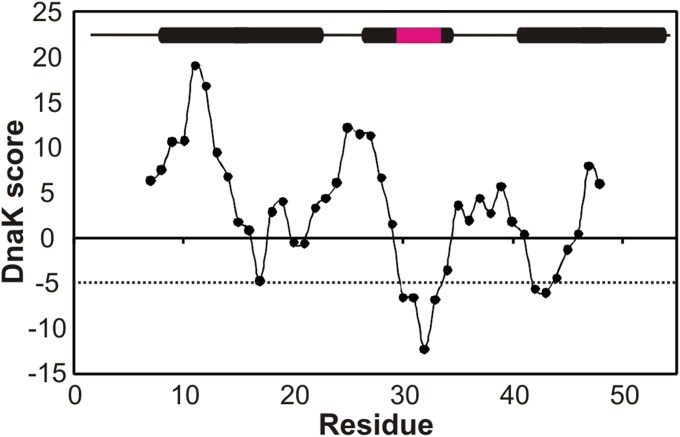
To probe DnaK–client interactions structurally we used the DNA binding domain of human telomere repeat binding factor 1 (hTRF1) (16) as a substrate. hTRF1 is a three-helix bundle belonging to the homeodomain protein family (17) (Fig. 1A) and is predicted (3) to have a DnaK binding site in helix 2 involving residues Lys-28–His-32 (Fig. 1A and Fig. S1). Moreover, hTRF1 is marginally stable with a folding free energy difference of −2.8 kcal/mol (35 °C) (17). This implies that the population of the unfolded state is ∼1% at 35 °C, and in this state the DnaK binding site is potentially surface-exposed and therefore available for binding. hTRF1 is thus an ideal folding-competent substrate for studying interactions with DnaK.
Fig. 1.
hTRF1 binding to DnaK results in a globally unfolded conformation. (A) Structure of the three-helix bundle hTRF1 [PDB ID code 1BA5 (16)] with the predicted DnaK binding site highlighted in red. (B) Ile δ1region of a 1H-13C HMQC spectrum of ILVM-13CH3/2H ADP-DnaK free (black) and bound (orange) to 12C/2H hTRF1 (35 °C). Changes in chemical shifts of a number of Ile residues proximal to the DnaK binding groove upon addition of hTRF1 are indicated by arrows. Duplicate peaks are observed for a number of methyl groups and denoted by a and b.1H-15N TROSY HSQC spectra of 15N-labeled hTRF1 in the absence (C) and presence (D) of 1.6-fold excess unlabeled ADP-DnaK.
Fig. S1.
hTRF1 is predicted to bind DnaK. The sequence-dependent DnaK score for hTRF1 derived from a computational DnaK prediction algorithm (3). Regions of the primary sequence with scores less than −5 (dotted line) are predicted to bind DnaK. The secondary structure of hTRF1 is denoted on the plot and the predicted DnaK binding site is highlighted in magenta.
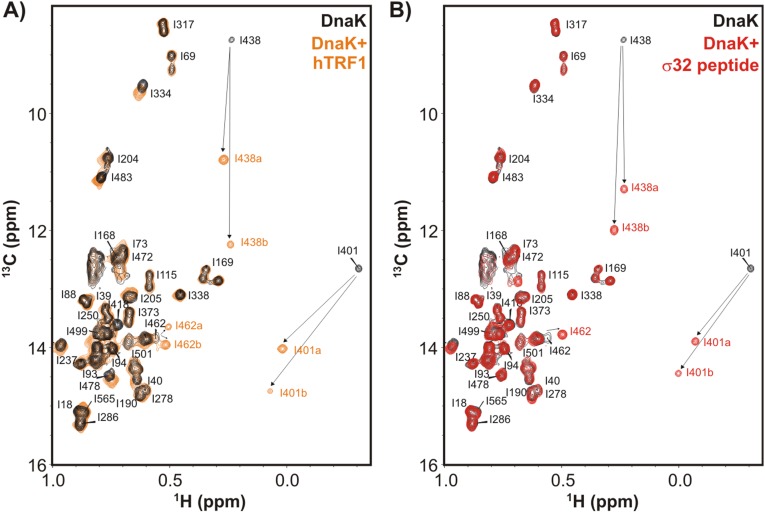
Fig. 1B (black) shows the Ile δ1 methyl region from a 1H-13C heteronuclear multiple quantum coherence (HMQC) spectrum of highly deuterated, I,L,V,M methyl-labeled (ILVM-13CH3/2H) DnaK in the ADP state. Upon addition of threefold molar excess of 12C/2H hTRF1 (orange), resonances belonging to Ile-401 and Ile-438, near the DnaK binding cleft, change significantly in chemical shift (Fig. 1B, arrows), showing that hTRF1 binds DnaK at the canonical binding site. The changes in the ADP-DnaK spectrum, observed at the binding pocket upon interaction with hTRF1 (Fig. 1B and Fig. S2), are very similar to those seen when DnaK binds a peptide from the E. coli heat shock transcription factor σ32 (corresponding to residues Gln-132–Gln-144) (18). Moreover, the resonances from hTRF1-bound DnaK are in slow exchange with the free form on the NMR chemical shift timescale, consistent with the slow binding and release of substrates from ADP-DnaK (2). Taken together, these results confirm that hTRF1 binds DnaK in a manner similar to what has been identified in previous studies.
Fig. S2.
hTRF1 binds DnaK similarly to the well-studied canonical substrate σ32 peptide. (A and B) Ile δ1 region of a 1H-13C HMQC spectrum of ILVM-13CH3/2H ADP-DnaK in the absence (black) and presence (orange) of 2H/12C hTRF1 (A) or without (black) and with (red)1H/12C σ32 peptide (B), 35 °C. The residues of DnaK near the substrate binding site move as indicated by arrows upon addition of substrate. Duplicate peaks are observed for a number of methyl groups, as denoted by a and b in the figure. Assignments of ADP-DnaK are those from a previously published report (18).
Binding of hTRF1 to DnaK Results in a Globally Unfolded Structural Ensemble.
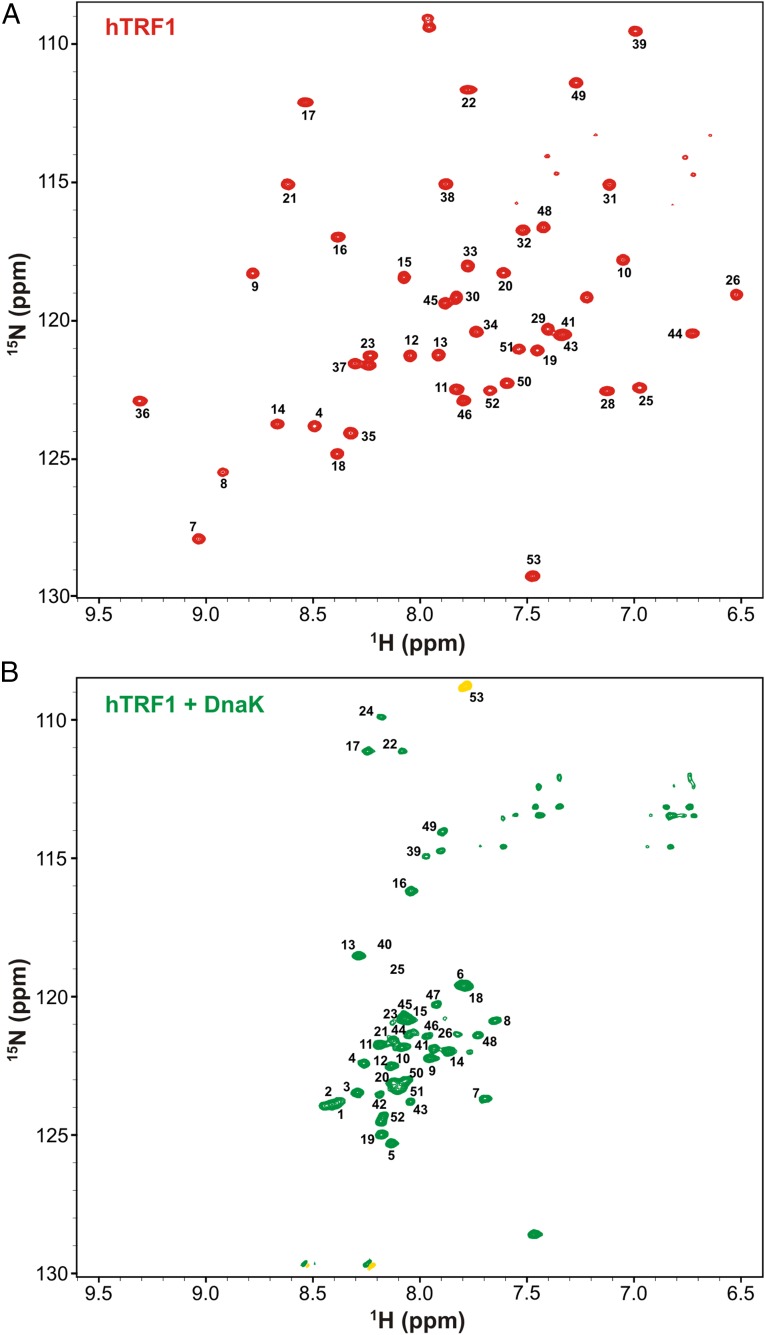
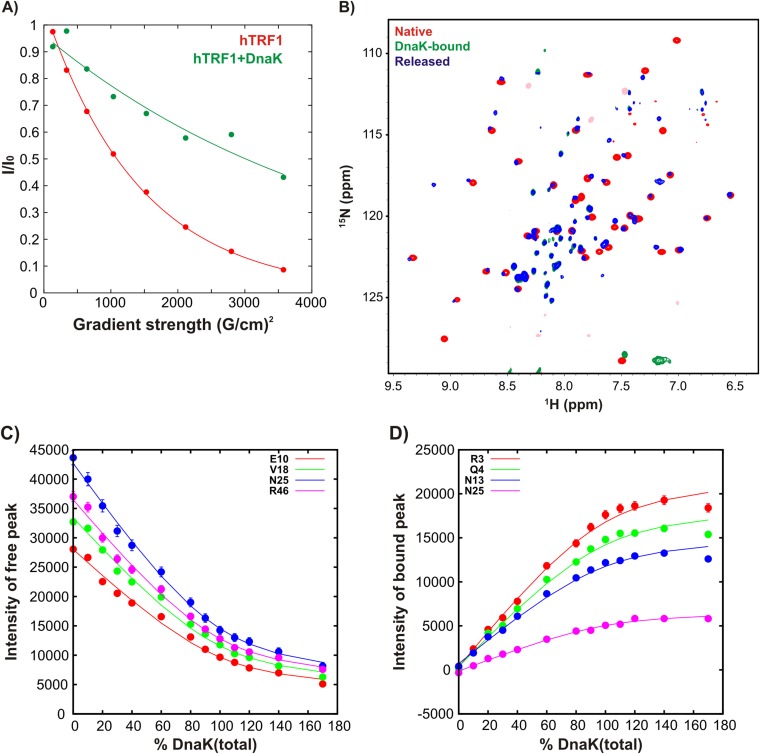
The 1H-15N transverse relaxation-optimized spectroscopy heteronuclear single quantum coherence (TROSY HSQC) (19) spectrum of hTRF1 (Fig. 1C and Fig. S3, Top) is well dispersed in both 1H and 15N chemical shift dimensions, as expected for a well-folded protein. When a 1.6-fold molar excess of 2H/14N ADP-DnaK is added, however, there is a dramatic shift to a poorly dispersed spectrum (Fig. 1D and Fig. S3, Bottom) typically seen for globally unfolded proteins lacking tertiary structure. The translational diffusion coefficients of hTRF1 in the absence and presence of ADP-DnaK differ by a factor of 3.2 ± 0.3 (Fig. S4A), establishing that hTRF1 is indeed DnaK-bound. In a subsequent experiment in which we added 14N-labeled hTRF1 to a sample of 15N-hTRF1 fully bound to ADP-DnaK, resonances corresponding to the native state of hTRF1 can clearly be observed in the 1H-15N TROSY HSQC spectrum (Fig. S4B). This indicates that unlabeled hTRF1 displaces the labeled DnaK-bound substrate, which then rapidly folds to the native conformation, thus demonstrating the reversibility of the binding reaction. DnaK-hTRF1 binding isotherms can be fit well to a 1:1 binding model (SI Discussion and Fig. S4 C and D) in which hTRF1 first unfolds and then binds one molecule of DnaK with an affinity of 1.4 ± 0.2 μM. The globally unfolded conformation of DnaK-bound hTRF1 observed in our experiments is consistent with previous low-resolution circular dichroism (13, 14) and single-molecule fluorescence data (15) showing that DnaK-bound clients are expanded and have a diminished helical content compared with the native substrate. In addition, the 1:1 stoichiometry of the DnaK–hTRF1 complex is in line with expectations from the small size of hTRF1 and the presence of only one strong DnaK binding site along its primary sequence (Fig. S1).
Fig. S3.
hTRF1 binding to DnaK results in a globally unfolded conformation. 1H-15N HSQC spectra of hTRF1 in the absence (A) and presence (B) of ADP-DnaK as shown in Fig. 1 C and D, but with assignments indicated next to the peaks. The peak in yellow (Bottom) is of opposite phase to the other correlations in the spectrum and is aliased.
Fig. S4.
hTRF1 binds reversibly to DnaK. (A) Normalized intensity as measured by integration over all peaks in the amide region of 1D 15N edited 1H spectra of1H/15N hTRF1 recorded in the absence (red) and presence (green) of 2H/14N ADP-DnaK (35 °C) as a function of squared gradient strength. The curves scale as I/I0 = exp(−dG2) where d is proportional to the translational diffusion coefficient of the protein in the sample. (B) 1H-15N TROSY HSQC spectrum (blue) of a sample made by addition of 670 μM 1H/14N hTRF1 to 215 μM 2H/15N-hTRF1 that is fully bound to ADP-DnaK (360 μM) (pH 6, 35 °C), which results in the appearance of resonances belonging to native 2H/15N hTRF1. The spectra of native hTRF1 (red) and hTRF1 fully bound to ADP-DnaK (green) are also shown for comparison. (C and D) Binding curves following the intensity of free (C) and DnaK-bound (D) hTRF1 peaks as a function of total DnaK concentration. Solid lines are fits of the data to an apparent two-state model, as described in SI Materials and Methods.
DnaK-Bound hTRF1 Contains Substantial Residual Secondary Structure.
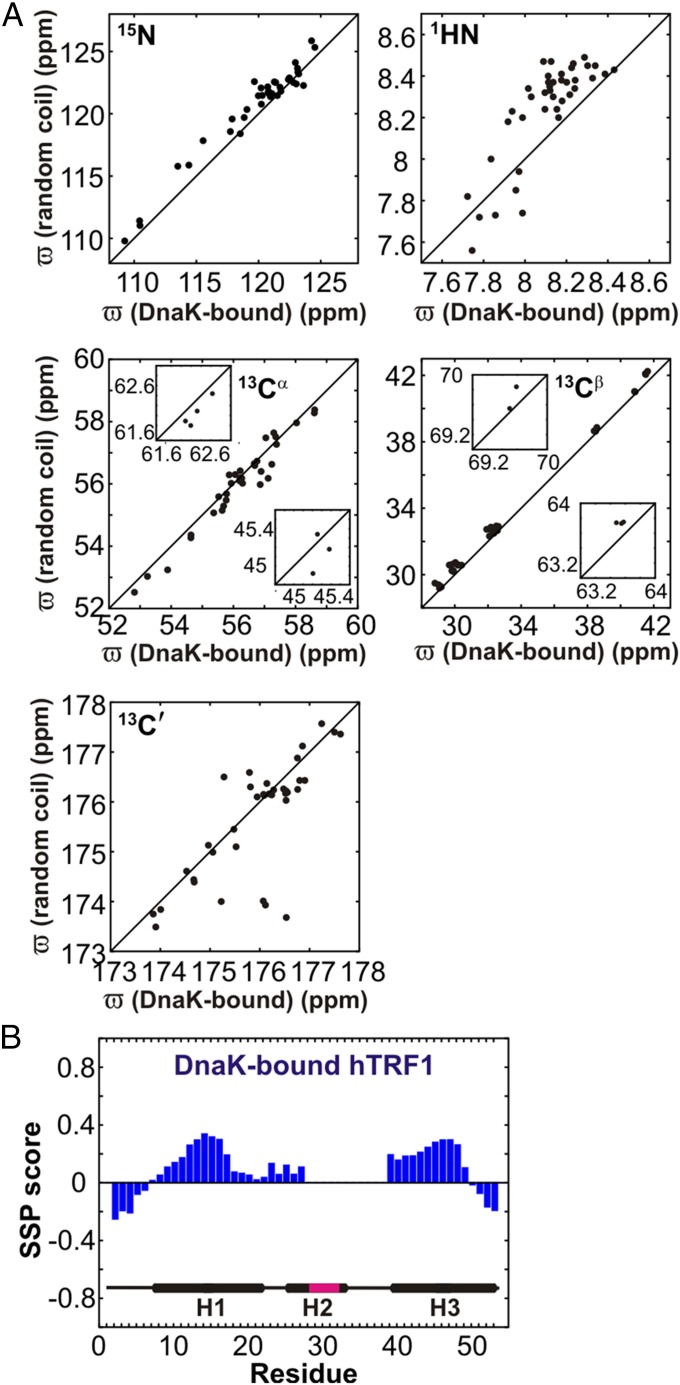
The strong correlations between measured chemical shifts of hTRF1 in the ADP-DnaK–bound form and those of a random coil (20) (Fig. 2A) confirm that this substrate exists in a globally unfolded conformation when bound to DnaK. However, on average,15N, 1HN, and 13Cβ chemical shifts from DnaK-bound hTRF1 are smaller than random coil values whereas the corresponding 13Cα and 13C′ shifts are larger, consistent with residual helical structure in the DnaK-bound conformation. To estimate the secondary structure content in the DnaK-bound hTRF1 ensemble, we computed the secondary structural propensity (SSP) score (21) using chemical shifts of the bound state. The SSP score ranges from −1 for a fully formed β-strand to +1 for a stable α-helix. The sequence-specific SSP score for hTRF1 bound to ADP-DnaK (Fig. 2B) shows the presence of substantial amounts of helicity (up to ∼40%). Strikingly, the helical content is maximum in regions of the protein that form helices 1 and 3 in the native state and that are far from the DnaK binding site. The presence of considerable residual helicity in the bound hTRF1 substrate suggests that, in general, a client protein may begin to sample conformational space and form local secondary structure even when associated with the DnaK chaperone.
Fig. 2.
DnaK-bound hTRF1 contains substantial residual secondary structure. (A) Correlations between predicted 15N, 1HN, 13Cα, 13Cβ, and 13C′ random coil chemical shifts of hTRF1 and measured chemical shifts of hTRF1 bound to ADP-DnaK. Predicted values are obtained from Tamiola and Mulder (20). (B) SSP (21) score of DnaK-bound hTRF1. The secondary structure of native hTRF1 is also indicated on the plot and the predicted DnaK binding site is shown in magenta. Correlations from hTRF1 residues near the DnaK binding site were not observable in 1H-15N HSQC spectra.
DnaK Binding Preserves the Structural Propensities of hTRF1.
Next, we wished to establish whether binding of hTRF1 to DnaK is directly responsible for secondary structure formation or whether the presence of this level of structure is a property inherent to the unfolded free form of the substrate. To address this properly chemical shifts of bound hTRF1 must be compared with those from an unfolded ensemble in the absence of denaturants and under identical experimental conditions. However, the population of the unfolded state is sparse and cannot be directly observed in the NMR spectrum of native hTRF1. To overcome this limitation, we exploited chemical exchange saturation transfer (CEST) NMR methodology (22, 23), by which it is possible to detect sparsely populated (0.5% or higher) and transiently formed protein conformers (so-called invisible or excited states) exchanging with the native (visible or ground) state at rates ranging from ∼20–400 s−1. In this experiment a very weak rf field is applied during a mixing period when the magnetization of interest is longitudinal that can modulate the ground state peak intensities (I) relative to the case where the weak field is absent (I0). When the field is positioned close to an invisible excited state resonance the resultant perturbation is transferred to the corresponding peak derived from the visible, ground state via chemical exchange, leading to a decrease in its intensity (I/I0 < 1). Similarly, application of the field at a frequency proximal to the ground state peak leads to its disappearance owing to a saturation effect (I/I0 ∼ 0). A large set of spectra is typically recorded whereby the position of the weak perturbing field is varied from one spectrum to the next and a plot of the relative intensity of each ground state peak as a function of the position of the rf field gives a series of CEST profiles, each consisting of a pair of dips for a two-state conformational exchange process. The main dip is located at the position of the chemical shift of the resonance belonging to the major state conformer and a smaller dip at the position of the frequency of the corresponding excited state.
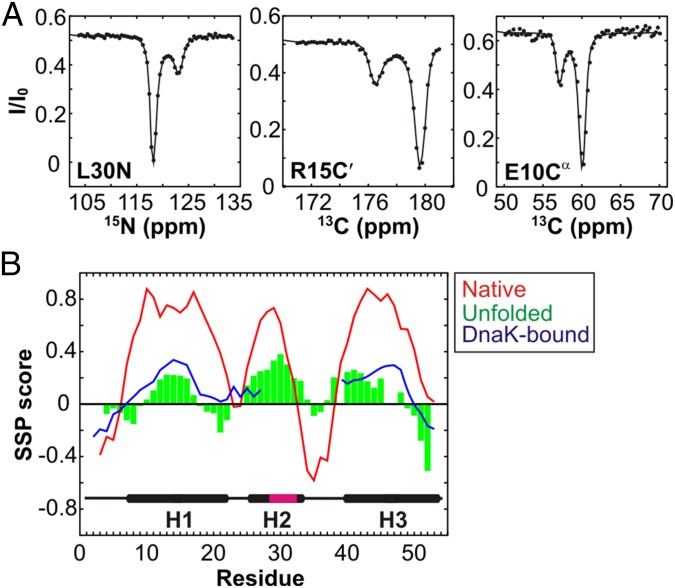
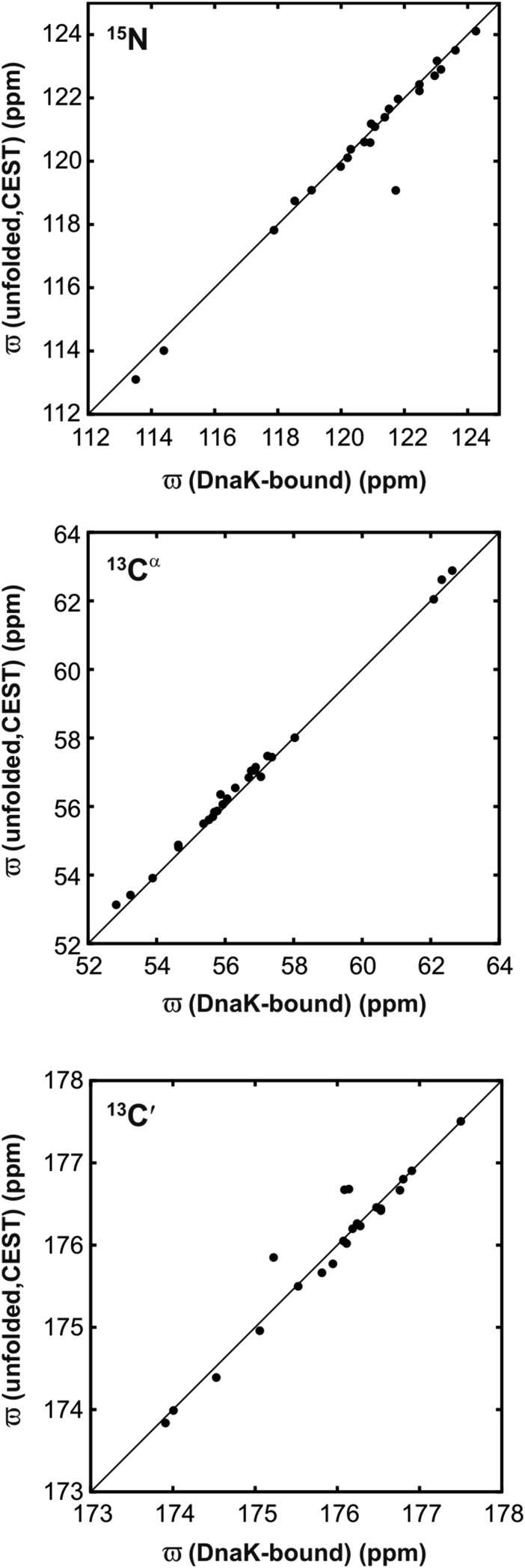
15N, 13Cα, and 13C′ CEST profiles of hTRF1 were acquired in the absence of DnaK at 35 °C (Fig. 3A and Fig. S5) and these show a pair of dips, one large and a second small, that is characteristic of two-state chemical exchange. The small dips are at frequency positions that correspond to an unfolded polypeptide and all of the profiles are well fit together to a protein folding reaction with an unfolded state population of 4.22 ± 0.03% and a lifetime of 2.89 ± 0.04 ms that is consistent with expectations from previous work on hTRF1 folding (17). The 15N, 13Cα, and 13C′ chemical shifts of unfolded hTRF1 obtained from CEST profiles match very well with values for DnaK-bound hTRF1 (Fig. S6). As expected, therefore, the sequence-specific pattern of SSP scores for the unfolded state of hTRF1 calculated from CEST-derived chemical shifts is similar to the SSP scores for DnaK-bound hTRF1 (compare green and blue curves, Fig. 3B). Specifically, the helical content of unfolded hTRF1 in regions corresponding to helices 1 and 3 in the native state is comparable to the helicity in the DnaK-bound chain. This strongly suggests that interaction of DnaK with the client substrate does not alter the subtle structural tendencies of the polymer chain outside the binding region.
Fig. 3.
DnaK binding does not disrupt hTRF1 secondary structure propensities outside the binding region. (A) 15N, 13C′, and 13Cα CEST profiles probing the folding transition of hTRF1 (35 °C). Each profile graphs the peak intensity (I) for a spin probe of a given residue as a function of the position of a small-amplitude rf field, normalized to the intensity in the absence of the field (I0). Solid lines are fits of the data to a two-state model of conformational exchange (23). The positions of the large and small dips directly correspond to the chemical shifts of the appropriate nucleus (L30N, R15C′, or E10Cα) in the folded and unfolded states, respectively. (B) SSP scores for the unfolded state (green) calculated from CEST-derived chemical shifts for 15N, 13Cα, and 13C′ nuclei. The red and blue lines are the SSP scores for the native and ADP-DnaK bound states of hTRF1, respectively. Cross-peaks for amide correlations derived from hTRF1 residues in and immediately surrounding the binding site could not be observed in the DnaK-bound state. Also shown is the secondary structure of native hTRF1 with the DnaK-predicted binding site in magenta.
Fig. S5.
Native hTRF1 exchanges on the millisecond timescale with a sparsely populated unfolded state.15N, 13C′, and 13Cα CEST profiles of native hTRF1 acquired at 35 °C.
Fig. S6.
DnaK-bound and unfolded hTRF1 have very similar conformations. 15N, 13Cα, and 13C′ chemical shifts of the unfolded state obtained from CEST measurements correlate very well with chemical shifts of DnaK-bound hTRF1.
The Structural Ensemble of Bound DnaK Does Not Change During the DnaK Cycle.
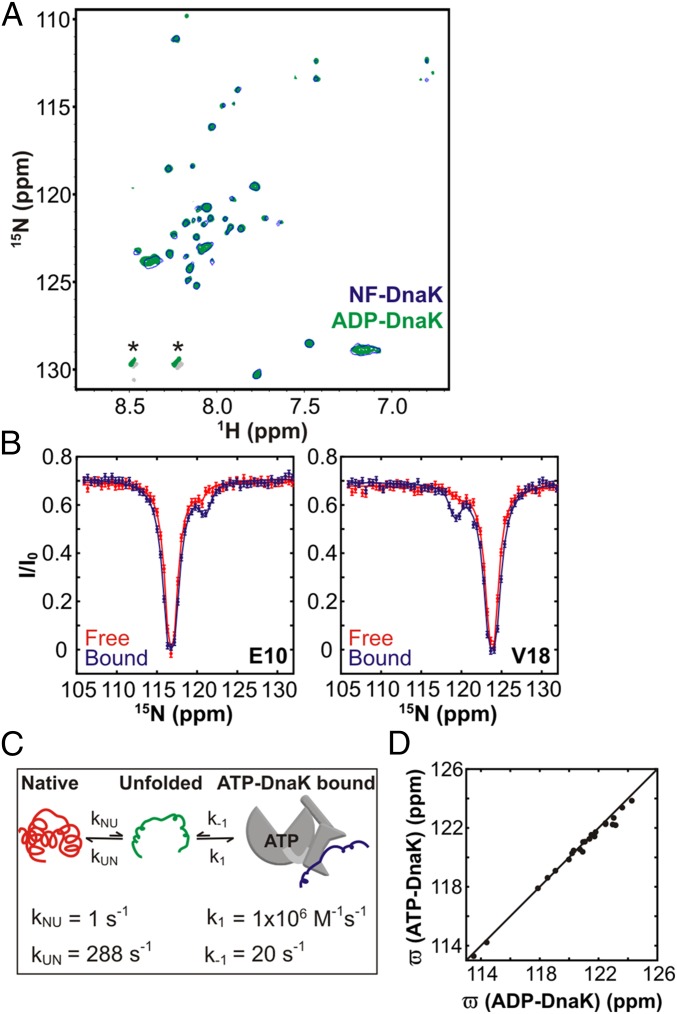
During the course of its function DnaK cycles through ADP-bound, ATP-bound, and NF states (2). In both NF and ADP-bound conformations the NBD and SBD of DnaK are separated from each other, and the helical lid is closed, with small kon,koff values and high affinities for substrate (2, 6). In contrast, in the low-affinity, fast kon,koff ATP-DnaK state the two domains are docked onto each other and the helical lid is open (2, 4, 5). The energy of ATP binding and/or hydrolysis as well as the large allosteric changes occurring in DnaK upon ATP binding have been hypothesized to remodel the bound substrate conformation (24). To investigate the impact of the DnaK nucleotide state on the substrate, we acquired a 1H-15N TROSY HSQC spectrum of hTRF1 bound to NF-DnaK and compared it to the corresponding spectrum of the ADP-DnaK bound state. The spectra are identical (Fig. 4A), consistent with very similar hTRF1 conformations in both NF and ADP states.
Fig. 4.
hTRF1 conformation is independent of DnaK nucleotide state. (A) 1H-15N TROSY HSQC (19) spectra of 2H/15N hTRF1 bound to either NF- (blue, single contour) or ADP-DnaK (green). Peaks indicated with an asterisk arise from buffer components. (B) CEST profiles of free (red) and ATP-DnaKT199A–bound hTRF1 (blue), 25 °C. Solid lines are fits to a two-state (red) or a three-state (blue) model as shown in C. (C) Three-state model for binding of hTRF1 to ATP-DnaKT199A, wherein the native substrate unfolds before interaction with DnaK. Kinetic parameters derived from fitting CEST data (B, blue) to this three-state model are indicated. (D) Correlation between the 15N chemical shifts of hTRF1 bound to ADP- and ATP-DnaKT199A. The latter values are derived from CEST profiles like the ones in B.
Next, experiments were carried out to probe the conformational states of hTRF1 bound to ATP-DnaK. Because DnaK hydrolyzes ATP, these experiments were performed with the hydrolysis-deficient mutant T199A DnaK (25) (hereafter referred to as DnaKT199A). In addition, an ATP-regeneration system was used to ensure that DnaKT199A remained in the ATP-bound state (Materials and Methods). A high-quality spectrum of the ATP complex, (hTRF1/ATP-DnaKT199A) is, however, difficult to obtain. The lower-affinity of ATP-DnaK for substrate relative to the ADP and NF states (2) necessitates the use of high DnaKT199A concentrations to obtain a sufficient fraction of bound substrate for NMR studies and the concomitant increase in solution viscosity significantly degrades spectral quality. Use of only fractionally bound samples, in turn, led to spectra for which only a small percentage of the expected peaks in the hTRF1 bound state could be observed. We thus used CEST experiments to obtain hTRF1 chemical shifts in the ATP-DnaKT199A–bound state, exploiting the relatively more rapid rates of exchange of substrate between free and bound forms in the ATP vs. ADP states of DnaK. These experiments have the advantage that only relatively small amounts of DnaKT199A-bound hTRF1 are required, with the shifts of the bound conformer obtained indirectly from peak intensities of the unbound native state of the substrate, in spectra that are of high quality. It is possible to “tune” the experimental conditions to reduce interference from exchange of unbound hTRF1 between native and unfolded states by using a temperature at which the unfolded state population is very small (25 °C was used in the present analysis where the unfolded state is populated at 0.3%, relative to 4.2% at 35 °C). Moreover, the experiment is insensitive to any residual hTRF1 bound to ADP-DnaK, because the exchange rate between free and ADP-bound hTRF1 is slower than that required to observe minor state dips in CEST (23).
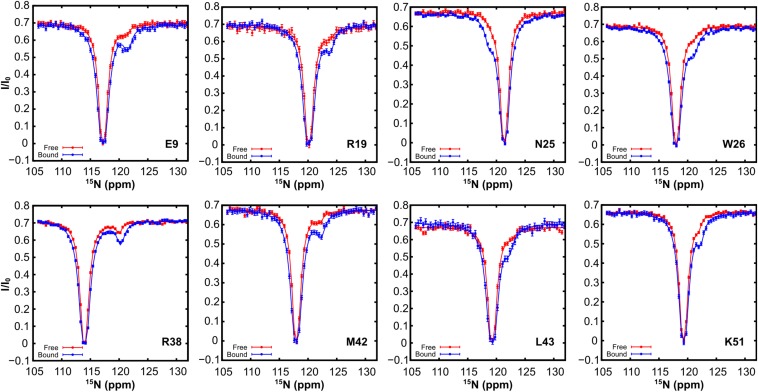
Upon addition of ATP-DnaKT199A to a sample of 15N hTRF1 the size of the minor dips significantly increased, clearly demonstrating the presence of the ATP-DnaKT199A–bound form (Fig. 4B and Fig. S7). CEST profiles acquired at 2B1 fields were fitted to a three-state model of exchange involving the interconversion of native, unfolded, and bound hTRF1 states (, Fig. 4C). The obtained values of kon = (1.1 ± 0.2) × 106 M−1⋅s−1, koff = 20.4 ± 0.2 s−1, and Kd = 18 ± 3 µM are consistent with rates and affinities reported in the literature for peptide and protein binding to ATP-DnaK (2). Under our experimental conditions (500 µM hTRF1 and 600 µM ATP-DnaKT199A) the population of ATP-DnaKT199A–bound hTRF1 is calculated to be 9 ± 2%. Notably, the 15N chemical shifts of the ATP-DnaKT199A–bound state of hTRF1, generated from fits of the CEST data, correlate very well with those of the ADP-DnaK–bound form (Fig. 4D). Because chemical shifts are powerful probes of structure in proteins (26, 27), the absence of shift changes implies that cycling of the DnaK chaperone between ATP, ADP, and NF states does not perform conformational work on the bound substrate, so that the substrate conformation does not evolve as a function of the DnaK nucleotide state.
Fig. S7.
hTRF1 bound to ATP-DnaK is globally unfolded. 15N CEST profiles for hTRF1 in the absence (red) and presence (blue) of ATP-DnaKT199A, 25 °C. An increase in the size of the minor dip establishes DnaK binding. Solid lines are fits of the data to a two-state (red) or three-state (blue) model of exchange depicted in Fig. 4C. Note that the minor state dips obtained in the absence of DnaKT199A are much smaller than in Fig. 3 and Fig. S5 because of the lower temperature used, which significantly decreases the population of the unfolded state, from 4.2% (35 °C) to 0.3% (25 °C).
Discussion
This work provides key insights into the impact of Hsp70 binding on the conformational ensemble of a client protein substrate through the chaperone cycle. A model for Hsp70 action emerges whereby the chaperone binds its hTRF1 substrate in a globally unfolded conformation while allowing it to explore secondary structural propensities inherent to the polypeptide chain. In the case of hTRF1, the client protein cycles through ATP-, ADP-, and NF DnaK-bound states without changing structure, so that the DnaK functional cycle, which involves cochaperones DnaJ and GrpE, is essentially a molecular timer that sets the duration for which the substrate is associated with DnaK.
Central to the study has been the use of recently developed CEST experiments for studies of sparsely populated, transiently formed protein conformational states. Using these experiments, we were able to measure the backbone heteronuclear chemical shifts of the unfolded ensemble of free hTRF1 as well as of hTRF1 in the ATP-DnaK–bound conformation. The populations of both of these hTRF1 states were below the threshold where workable “visible” spectra could be obtained, severely limiting the utility of conventional NMR approaches in our studies. In contrast, by reading out “invisible state” chemical shifts from well-resolved cross-peaks of the native hTRF1 conformer quantitative insights into structure could be readily obtained.
Our experiments suggest a scheme for chaperone-assisted de novo folding (Fig. 5) in which the protein chain progressively acquires structure in a manner that depends on the nature of the interactions with each element of the chaperone assembly awaiting the nascent chain. In this picture a newly synthesized polypeptide is kept unfolded after synthesis on the ribosome by trigger factor (28). Subsequently, it is transferred to DnaK, where it acquires secondary structure as dictated by the intrinsic properties of the substrate itself, and then finally released into the cytosol, where it can either independently, or through interaction with GroEL/ES (1, 29), fold into its native state. Because the DnaK-released client protein is an important starting conformation for folding in vivo, our data further imply that the folding process may begin from a state with significant secondary structure.
Fig. 5.
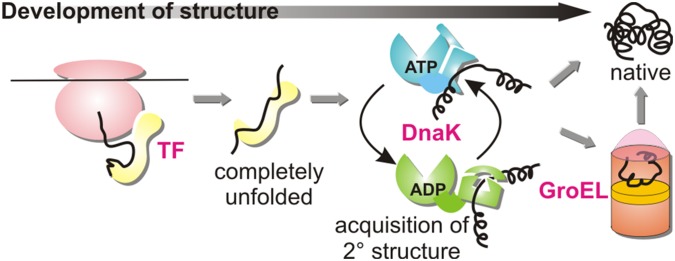
Tandem acquisition of structure during de novo protein folding. Model of chaperone-assisted de novo folding, suggested by data on hTRF1. In this scheme a newly synthesized polypeptide is kept unfolded after synthesis on the ribosome by trigger factor (TF). Next, it is transferred to DnaK, where it acquires secondary structure as dictated by the intrinsic properties of the substrate itself, and then finally released into the cytosol where it can either independently, or through interaction with GroEL/ES, fold into its native structure.
The impact of Hsp70 binding on the structure of hTRF1 provides some insight into how this chaperone can perform its diverse set of functions. For example, Hsp70 plays important roles in the translocation, cotranslational folding, and disaggregation of protein substrates. Hsp70-bound client proteins are predominantly unstructured while traversing the membrane (30) or upon transfer from trigger factor and before folding to the native state (28, 31) or during the handover to ClpB (32). However, Hsp90 recognizes native but locally unfolded substrates bound to Hsp70 (33). Our data suggest that these disparate requirements for Hsp70 action on its substrates can be achieved by the judicious positioning of Hsp70 binding sites on the substrate coupled with the intrinsic conformational tendencies of the client protein. In the case of Hsp70-bound hTRF1, for instance, the Hsp70 binding site is located in the middle of the second helix in the three-helix bundle. This results in a bound client protein ensemble lacking global tertiary structure but with significant amounts of local secondary structure in the terminal helices H1 and H3. A binding site closer to the N or C terminus of a substrate would be expected to be less perturbing and result in local unfolding, while maintaining native structure in other regions of the protein.
Our data cannot distinguish whether Hsp70 binds the unfolded state of hTRF1 via a holdase type of mechanism (8, 24) or interacts with the native state and unfolds it upon binding (unfoldase) (8, 34). However, the similar conformations of hTRF1 bound to NF-, ADP-, and ATP-DnaKT199A rules out a power stroke model for Hsp70 function, whereby one of the steps in the Hsp70 ATPase cycle results in substrate unfolding, and clarifies that conformational changes occurring in Hsp70 during the ATPase cycle do not affect the conformation of the bound client protein.
Materials and Methods
Protein Overexpression and Purification.
The purification of wild-type DnaK and DnaKT199A [ATP hydrolysis-deficient mutant (25)] were as described previously, with several modifications (35). Wild-type and T199A DnaK(1–638) were expressed in E. coli BL21(DE3) cells from a pCA528 plasmid, a gift from B. Bukau, Universität Heidelberg, Germany, carrying an N-terminal polyhistidine SUMO-fusion tag. The cells were grown at 37 °C in M9 D2O media supplemented with 14NH4Cl and [2H,12C]glucose as the sole nitrogen and carbon sources, respectively. Methyl labeling of samples (i.e., U-[2H], Met-ε-[13CH3], Ileδ1-[13CH3], Leu,Val-[13CH3, 12CD3]-) followed the procedure of Tugarinov et al. (36). Cells were grown to OD600 ∼ 0.8, at which point expression was induced by addition of 1.0 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and allowed to proceed overnight at 25 °C. Following expression, bacteria were harvested and the proteins were purified on nickel nitrilotriacetic acid (Ni-NTA) resin (GE Healthcare) under denaturing conditions (6 M GuHCl) to ensure complete removal of bound nucleotides. DnaK was refolded on the Ni-NTA column by gradually reducing the concentration of denaturing agent in the wash buffer (from 6 M to 0 M GuHCl), and the purification tag was removed via cleavage using Ulp1 protease. The cleaved protein was further purified on DEAE (GE Healthcare) and HiLoad 16/60 Superdex 200 pg gel filtration columns (GE Healthcare), equilibrated with 50 mM Hepes, 50 mM KCl (pH 7.4), 2 mM tris(2-carboxyethyl)phosphine (TCEP), and 0.03% NaN3.
The human TRF1 protein (hTRF1) was expressed in E. coli BL21(DE3) CodonPlus cells containing a pET29b (kanR) plasmid encoding the hTRF1 377–430 fragment. The cells were grown at 37 °C in M9 D2O media supplemented with 15NH4Cl or 14NH4Cl and [2H,12C]glucose or [2H,13C]glucose as the sole nitrogen and carbon sources, respectively. Cells were grown to OD600 ∼ 0.8, after which expression was induced by addition of 1.0 mM IPTG and allowed to proceed overnight at 25 °C. Following expression, bacteria were harvested and the proteins were purified under denaturing conditions on Ni-NTA resin (GE Healthcare), followed by on-column refolding by the gradual removal of denaturant through a linear gradient extending from 6 M GuHCl to 0 M GuHCl in the buffer. The N-terminal hexaHis tag was cleaved using TEV protease followed by an additional Ni-NTA purification step. Cleaved protein was concentrated on an Amicon Ultra-15 3K molecular weight cutoff filter (Millipore) and further purified on a HiLoad 16/60 Superdex 75 pg gel filtration column (GE Healthcare), equilibrated with 50 mM Hepes, 250 mM KCl (pH 7.4), 2 mM TCEP, and 0.03% NaN3. The purity of all proteins was confirmed by SDS/PAGE.
NMR Spectroscopy.
All NMR experiments were carried out on 11.7 T (500 MHz), 14.1 T (600 MHz), or 18.8 T (800 MHz) Varian INOVA spectrometers. The 600-MHz spectrometer was equipped with a cryogenically cooled probe. The sample temperature was measured using a thermocouple connected to a digital thermometer inserted into an NMR tube containing D2O. NMRPipe (37) and Sparky (38) were used to process and visualize NMR spectra, respectively.
CEST.
15N, 13Cα, and 13C′ CEST spectra of hTRF1 were acquired using previously published pulse sequences (23, 39, 40). CEST experiments at 35 °C were performed to obtain chemical shifts of the unfolded state of the protein. Here a 1.0 mM 1H/13C/15N sample of hTRF1 in 50 mM Hepes (pH 6.8), 50 mM KCl, 1 mM TCEP, 0.03% NaN3, 90% H2O/10% D2O buffer was used. Exchange times (TCEST) and B1 field strengths of 250 ms, 18 Hz, and 35 Hz were used for 15N CEST, 175 ms and 25 Hz for 13Cα CEST, and 200 ms and 20 Hz for 13C′ CEST. The B1 field was varied in frequency between 103.8 ppm and 133.5 ppm (74 planes with 18-Hz spacing and 38 planes spaced at 35-Hz intervals), 50.1 ppm and 70.1 ppm (77 planes), and 171.0 ppm and 180.1 ppm (52 planes) for 15N, 13Cα, and 13C′, respectively, and a 2D spectrum was acquired for each frequency value of the B1 field.
A second set of 15N CEST experiments was performed both without and with ATP-DnaKT199A at 25 °C to obtain 15N chemical shifts of hTRF1 in the ATP-DnaKT199A bound form. 2H/15N hTRF1 samples (0.5 mM) in 50 mM MES (pH 6.0), 50 mM KCl, 5 mM MgCl2, 5 mM ATP, 0.1 mM EDTA, 1 mM TCEP, 0.03% NaN3, and 90% H2O/10% D2O supplemented with an ATP regeneration system (41) and 5 μM GrpE were used, either without or with 0.6 mM ATP-DnaKT199A. Experiments were recorded with TCEST = 350 ms and a B1 field frequency range from 105.9 to 131.8 ppm using a pair of B1 field strengths, 27.6 Hz (62 planes) and 53.8 Hz (44 planes), for the sample containing ATP-DnaK and 27.6 Hz for the sample without ATP-DnaKT199A. Reference spectra were also acquired in each case without the TCEST period. Peak intensities were measured by fitting the lineshape of the respective peaks using FuDA (pound.med.utoronto.ca/software) and CEST profiles were obtained as the ratio of the peak intensity with and without the TCEST period as a function of the frequency of the B1 field (23). CEST profiles of hTRF1 recorded in the absence of DnaK were fit to a two-state model of chemical exchange, , using the custom program Chemex (https://github.com/gbouvignies/chemex), which numerically propagates the Bloch–McConnell equations (42), as described previously (23). For 15N CEST profiles of hTRF1 recorded with ATP-DnaKT199A (25 °C) a model of three-site chemical exchange was used, as described in the text. In this case fits were performed in two stages. First, profiles recorded on a sample without ATP-DnaKT199A at 25 °C were fit to a two-state model to extract the population of the unfolded state (pU), and the rate of exchange (kex,UN = kU,N + kN,U) along with chemical shift differences between native and unfolded states (ΔϖUN). The value of pU was then used to calculate the equilibrium constant for unfolding, . Subsequently, TRF1 profiles recorded in the presence of ATP-DnaKT199A were fit to a three-state model, , where B is the DnaKT199A bound form of TRF1 by fixing KNU, kex,NU, and ΔϖNU to the values obtained from the two-state fits of hTRF1 in the absence of ATP-DnaKT199A and parameters kon, koff , and ΔϖUB were allowed to vary freely.
SI Materials and Methods
Resonance Assignments.
Backbone resonance assignments of hTRF1 were carried out on a 0.8 mM 1H/15N/13C sample in 50 mM sodium acetate (pH 5.7), 50 mM NaCl, 1 mM EDTA, 1 mM NaN3, and 90% H2O/10% (vol/vol) D2O buffer using 2D 1H-15N HSQC and 3D HNCACB, CBCA(CO)NH, and HNCO (43, 44) experiments recorded at 500 MHz. Assignments of DnaK-bound hTRF1 were obtained from 2D 1H-15N HSQC and 3D HNCA, HN(CA)CO, HN(CO)CA, HNCACB, and HNCO spectra recorded at 600 MHz using a 0.5 mM 2H/13C/15N hTRF1 sample with 1.4 mM ADP-DnaK. The sample was dissolved in 50 mM MES (pH 6.0), 50 mM KCl, 5 mM MgCl2, 5 mM ADP, 0.1 mM EDTA, 1 mM TCEP, 0.03% NaN3, and 90% H2O/10% D2O buffer.
Ile δ1 methyl group assignments of ADP-DnaK bound to hTRF1 were confirmed by recording a NOESY dataset on a sample of 0.8 mM ILVM-13CH3/2H DnaK, 1.8 mM 12C/2H hTRF1 in 50 mM Hepes (pH 8),100% D2O buffer containing 50 mM KCl, 5 mM MgCl2, 5 mM ADP, 1 mM EDTA, 0.03% NaN3, and 1 mM TCEP. In this experiment (200-ms mixing time) correlations of the form (13Cj-NOE-13Ck-1Hk) were recorded. Assignments of the complex were facilitated by previously published Ile methyl resonance assignments of ADP-DnaK (18), as well as the NMR structure of ADP-DnaK (6).
Translational Diffusion.
Translational diffusion coefficients of hTRF1 either free or fully bound to DnaK were obtained from measurements on 1.0 mM samples of 1H/15N hTRF1 without or with 1.5 mM 2H/14N ADP-DnaK, respectively, in 50 mM Hepes (pH 6.8), 25 mM KCl, 5 mM MgCl2, 5 mM ADP, 1 mM TCEP, 0.03% NaN3, and 93% H2O/7% D2O buffer. Pulsed field gradient diffusion experiments were run in 1D mode with a 15N-edited longitudinal encode decode pulse sequence (45). A diffusion delay of 200 ms was used in conjunction with encode/decode gradient strengths ranging from 11–60 G/cm. The amide region of each 1D spectrum was collectively integrated using NMRPipe to quantify intensity as a function of gradient strength and the intensity (I) vs. squared gradient strength (G2) profile was fit to a single exponential of the form I = I0 exp(−dG2), to extract a decay rate (d) that is proportional to the translational diffusion coefficient of the protein in the sample.
SI Discussion
Data Acquisition for ADP-DnaK Binding Titrations.
The affinity of ADP-DnaK for hTRF1 was determined by an NMR-detected titration experiment. Because peaks of free and DnaK-bound hTRF1 are in slow exchange on the NMR chemical shift timescale, peak intensities were used to determine the fraction of free and bound hTRF1. The titration was carried out using a constant concentration (400 μM) of hTRF1 and varying the concentration of DnaK from 0 to 680 μM (1.7-fold molar excess). 1H-15N TROSY HSQC spectra (800-MHz field strength) were acquired at each DnaK concentration and intensities of free and bound resonances, which are proportional to the concentration of the respective molecular species, were extracted by fitting the lineshape globally across all concentrations using FuDA (pound.med.utoronto.ca/software.html).
Fitting Data to a Three-State Model of Binding.
Peak intensities as a function of DnaK concentration were first fit to the three-state binding model used in the analysis of CEST data recorded on ATP-DnaK, . Here, N, U, and B are the native, unfolded, and DnaK-bound states of hTRF1. The equilibrium constant for folding (KUN) and the binding dissociation constant (Kd) are defined as and , where [N], [U], and [B] are the equilibrium concentrations of the native, unfolded, and DnaK-bound forms of hTRF1, respectively. Using the definitions of the equilibrium constants, as well as the two mass balance equations and , it can be shown that
| [S1] |
where
and
| [S2] |
where
Peak intensities reporting on [N] and [B] were identified and globally fit to either Eq. S1 or Eq. S2. In the fits, KUN was held constant at the value obtained from CEST experiments (KUN = 22.7), whereas Kd was allowed to vary as a global parameter. In addition, two local parameters were included in each fit, corresponding to the proportionality constant between concentration and intensity, and the plateau value of the intensity at a large DnaK concentration. Free and bound resonances, totaling 35 peaks, were fit globally to this model to obtain an estimate of 1.4 μM for the dissociation constant.
Fitting Data to a Pseudo-Two-State Model.
We next considered the pseudo-two-state model as an alternate way to fit the titration data, where the apparent dissociation constant is given by . It can be shown that is related to Kd as . In this model, the equilibrium concentrations [N] and [B] are given by
| [S3] |
where
and
| [S4] |
Intensities from 35 free and bound peaks were globally fit to Eq. S3 or Eq. S4 using as a global parameter and the same residue-specific local parameters as above. Fits provide an estimate of as 32 μM, and using KUN = 22.7, Kd can be determined to be 1.4 μM, which is identical to what is obtained using fits to the three-state model.
Estimating Errors in Kd Using a Bootstrap Method.
Errors in the dissociation constant for binding of hTRF1 to ADP-DnaK were estimated by bootstrapping (46) the fits to the pseudo-two-state model. Thirty-five residues were chosen randomly with replacement from the list of free and bound residues and their intensity vs. [DnaK] fit to Eq. S3 or Eq. S4 as described above. This procedure was repeated 700 times and the SD of the Kd values obtained from the bootstrapping analysis was taken as the error in the dissociation equilibrium constant. In this manner Kd for the binding of hTRF1 to ADP-DnaK was 1.4 ± 0.2 μM.
Acknowledgments
We thank Lila Gierasch for helpful discussions and Bernd Bukau for DnaK plasmids. This work was supported by grants from the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada. L.E.K holds a Canada Research Chair in Biochemistry.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: NMR chemical shifts have been deposited in the BioMagResBank, www.bmrb.wisc.edu [accession nos. 26594 (hTRF1) and 26595 (hTRF1 bound to ADP-DnaK)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508504112/-/DCSupplemental.
References
- 1.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 2.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62(6):670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16(7):1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48(6):863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 5.Qi R, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20(7):900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ER. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106(21):8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci. 2013;38(10):507–514. doi: 10.1016/j.tibs.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol. 2015;427(7):1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu X, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272(5268):1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevens SY, Cai S, Pellecchia M, Zuiderweg ER. The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG. Protein Sci. 2003;12(11):2588–2596. doi: 10.1110/ps.03269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Kurt N, Rajagopalan S, Cavagnero S. Secondary structure mapping of DnaK-bound protein fragments: Chain helicity and local helix unwinding at the binding site. Biochemistry. 2006;45(40):12325–12333. doi: 10.1021/bi0612263. [DOI] [PubMed] [Google Scholar]
- 12.Kurt N, Cavagnero S. Nonnative helical motif in a chaperone-bound protein fragment. Biophys J. 2008;94(7):L48–L50. doi: 10.1529/biophysj.107.127647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palleros DR, Shi L, Reid KL, Fink AL. hsp70-protein complexes. Complex stability and conformation of bound substrate protein. J Biol Chem. 1994;269(18):13107–13114. [PubMed] [Google Scholar]
- 14.Sekhar A, Santiago M, Lam HN, Lee JH, Cavagnero S. Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci. 2012;21(7):1042–1055. doi: 10.1002/pro.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellner R, et al. Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc Natl Acad Sci USA. 2014;111(37):13355–13360. doi: 10.1073/pnas.1407086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishikawa T, Nagadoi A, Yoshimura S, Aimoto S, Nishimura Y. Solution structure of the DNA-binding domain of human telomeric protein, hTRF1. Structure. 1998;6(8):1057–1065. doi: 10.1016/s0969-2126(98)00106-3. [DOI] [PubMed] [Google Scholar]
- 17.Gianni S, et al. Unifying features in protein-folding mechanisms. Proc Natl Acad Sci USA. 2003;100(23):13286–13291. doi: 10.1073/pnas.1835776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell. 2012;151(6):1296–1307. doi: 10.1016/j.cell.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamiola K, Mulder FA. Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans. 2012;40(5):1014–1020. doi: 10.1042/BST20120171. [DOI] [PubMed] [Google Scholar]
- 21.Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 2006;15(12):2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fawzi NL, Ying J, Ghirlando R, Torchia DA, Clore GM. Atomic-resolution dynamics on the surface of amyloid-β protofibrils probed by solution NMR. Nature. 2011;480(7376):268–272. doi: 10.1038/nature10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vallurupalli P, Bouvignies G, Kay LE. Studying “invisible” excited protein states in slow exchange with a major state conformation. J Am Chem Soc. 2012;134(19):8148–8161. doi: 10.1021/ja3001419. [DOI] [PubMed] [Google Scholar]
- 24.Goloubinoff P, De Los Rios P. The mechanism of Hsp70 chaperones: (Entropic) pulling the models together. Trends Biochem Sci. 2007;32(8):372–380. doi: 10.1016/j.tibs.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 25.McCarty JS, Walker GC. DnaK as a thermometer: Threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88(21):9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wishart DS. Interpreting protein chemical shift data. Prog Nucl Magn Reson Spectrosc. 2011;58(1–2):62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Spera S, Bax A. Empirical correlation between protein backbone conformation and C. alpha. and C. beta. 13C nuclear magnetic resonance chemical shifts. J Am Chem Soc. 1991;113(14):5490–5492. [Google Scholar]
- 28.Saio T, Guan X, Rossi P, Economou A, Kalodimos CG. Structural basis for protein antiaggregation activity of the trigger factor chaperone. Science. 2014;344(6184):1250494. doi: 10.1126/science.1250494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigler PB, et al. Structure and function in GroEL-mediated protein folding. Annu Rev Biochem. 1998;67(1):581–608. doi: 10.1146/annurev.biochem.67.1.581. [DOI] [PubMed] [Google Scholar]
- 30.Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: Machineries and mechanisms. Cell. 2009;138(4):628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agashe VR, et al. Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell. 2004;117(2):199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 32.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119(5):653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 33.Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell. 2014;157(7):1685–1697. doi: 10.1016/j.cell.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma SK, De los Rios P, Christen P, Lustig A, Goloubinoff P. The kinetic parameters and energy cost of the Hsp70 chaperone as a polypeptide unfoldase. Nat Chem Biol. 2010;6(12):914–920. doi: 10.1038/nchembio.455. [DOI] [PubMed] [Google Scholar]
- 35.Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286(3):915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 36.Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1(2):749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Goddard T, Kneller D. Sparky—NMR assignment and integration software. Univ of California; San Francisco: 2006. [Google Scholar]
- 39.Long D, Sekhar A, Kay LE. Triple resonance-based ¹³C(α) and ¹³C(β) CEST experiments for studies of ms timescale dynamics in proteins. J Biomol NMR. 2014;60(4):203–208. doi: 10.1007/s10858-014-9868-5. [DOI] [PubMed] [Google Scholar]
- 40.Vallurupalli P, Kay LE. Probing slow chemical exchange at carbonyl sites in proteins by chemical exchange saturation transfer NMR spectroscopy. Angew Chem Int Ed Engl. 2013;52(15):4156–4159. doi: 10.1002/anie.201209118. [DOI] [PubMed] [Google Scholar]
- 41.Karagöz GE, et al. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci USA. 2011;108(2):580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McConnell HM. Reaction rates by nuclear magnetic resonance. J Chem Phys. 1958;28(3):430–431. [Google Scholar]
- 43.Cavanagh J, Fairbrother WJ, Palmer AG, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic; New York: 1995. [Google Scholar]
- 44.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nucl Magn Reson Spectrosc. 1999;34(2):93–158. [Google Scholar]
- 45.Altieri AS, Hinton DP, Byrd RA. Association of biomolecular systems via pulsed field gradient NMR self-diffusion measurements. J Am Chem Soc. 1995;117(28):7566–7567. [Google Scholar]
- 46.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C. Cambridge Univ Press; Cambridge, UK: 1996. [Google Scholar]