Abstract
Chloroplasts and mitochondria are subcellular bioenergetic organelles with their own genomes and genetic systems. DNA replication and transmission to daughter organelles produces cytoplasmic inheritance of characters associated with primary events in photosynthesis and respiration. The prokaryotic ancestors of chloroplasts and mitochondria were endosymbionts whose genes became copied to the genomes of their cellular hosts. These copies gave rise to nuclear chromosomal genes that encode cytosolic proteins and precursor proteins that are synthesized in the cytosol for import into the organelle into which the endosymbiont evolved. What accounts for the retention of genes for the complete synthesis within chloroplasts and mitochondria of a tiny minority of their protein subunits? One hypothesis is that expression of genes for protein subunits of energy-transducing enzymes must respond to physical environmental change by means of a direct and unconditional regulatory control—control exerted by change in the redox state of the corresponding gene product. This hypothesis proposes that, to preserve function, an entire redox regulatory system has to be retained within its original membrane-bound compartment. Colocation of gene and gene product for redox regulation of gene expression (CoRR) is a hypothesis in agreement with the results of a variety of experiments designed to test it and which seem to have no other satisfactory explanation. Here, I review evidence relating to CoRR and discuss its development, conclusions, and implications. This overview also identifies predictions concerning the results of experiments that may yet prove the hypothesis to be incorrect.
Keywords: chloroplast, mitochondrion, photosynthesis, oxidative phosphorylation, CoRR hypothesis
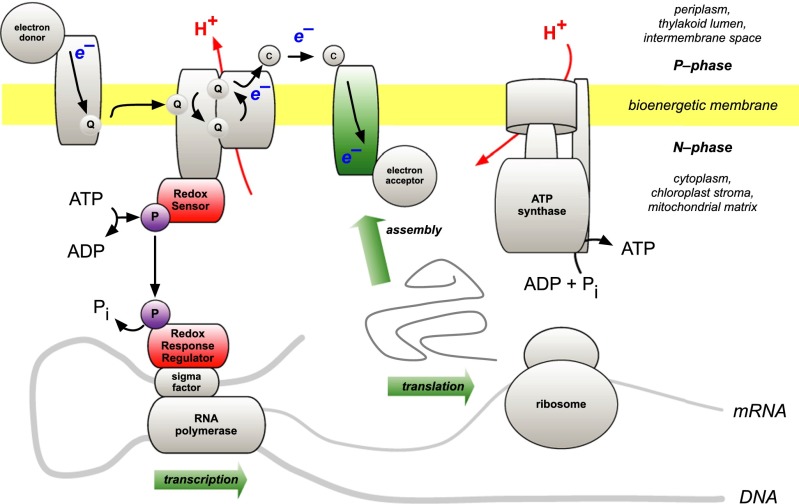
Regulatory control of gene expression in response to changes in redox state of gene products is proposed as the selective advantage of genome retention by chloroplasts and mitochondria. Changes in the physical environment are then able to induce compensatory effects on the composition of photosynthetic and respiratory electron transport chains. Fig. 1 outlines a redox feedback control of transcription by means of a characteristically prokaryotic two-component regulatory system. The short and direct signal transduction pathway contains two proteins: a sensor kinase and a response regulator. Phosphorylation of the sensor kinase responds to redox state, and its phosphorylated form transfers the phosphate group to a response regulator.
Fig. 1.
Two-component redox regulatory control of transcription. The bioenergetic membrane is a diagrammatic composite of photosynthetic and respiratory membranes that couple electron transport with ATP synthesis/hydrolysis by means of a transmembrane gradient of hydrogen ion (H+) concentration and electrical potential difference between the outer, positive aqueous phase (P-phase) and the inner, negative aqueous phase (N-phase). A redox sensor responds, by autophosphorylation, to a change in the redox state of an electron carrier. Phosphoryl transfer to a specific response regulator then initiates or inhibits a DNA-dependent RNA polymerase through the action of a regulatory sigma factor that is specific to a particular promoter. Transcription, translation, and assembly of an electron carrier then serve to regulate electron transfer at a specific point in the chain, optimizing it in response to a change in environmental conditions. Adapted from ref. 1.
Genes within chloroplasts and mitochondria derive from subsets of endosymbiont genes whose products regulate their own gene transcription. These genes remain, along with their protein products, within a single membrane-bound compartment. This hypothesis was put forward explicitly for both chloroplasts and mitochondria (1), and separately for chloroplasts alone (2). The term “CoRR” (also “CORR”) was introduced (3, 4): It stands for “colocation (of gene and gene product) for redox regulation of gene expression.”
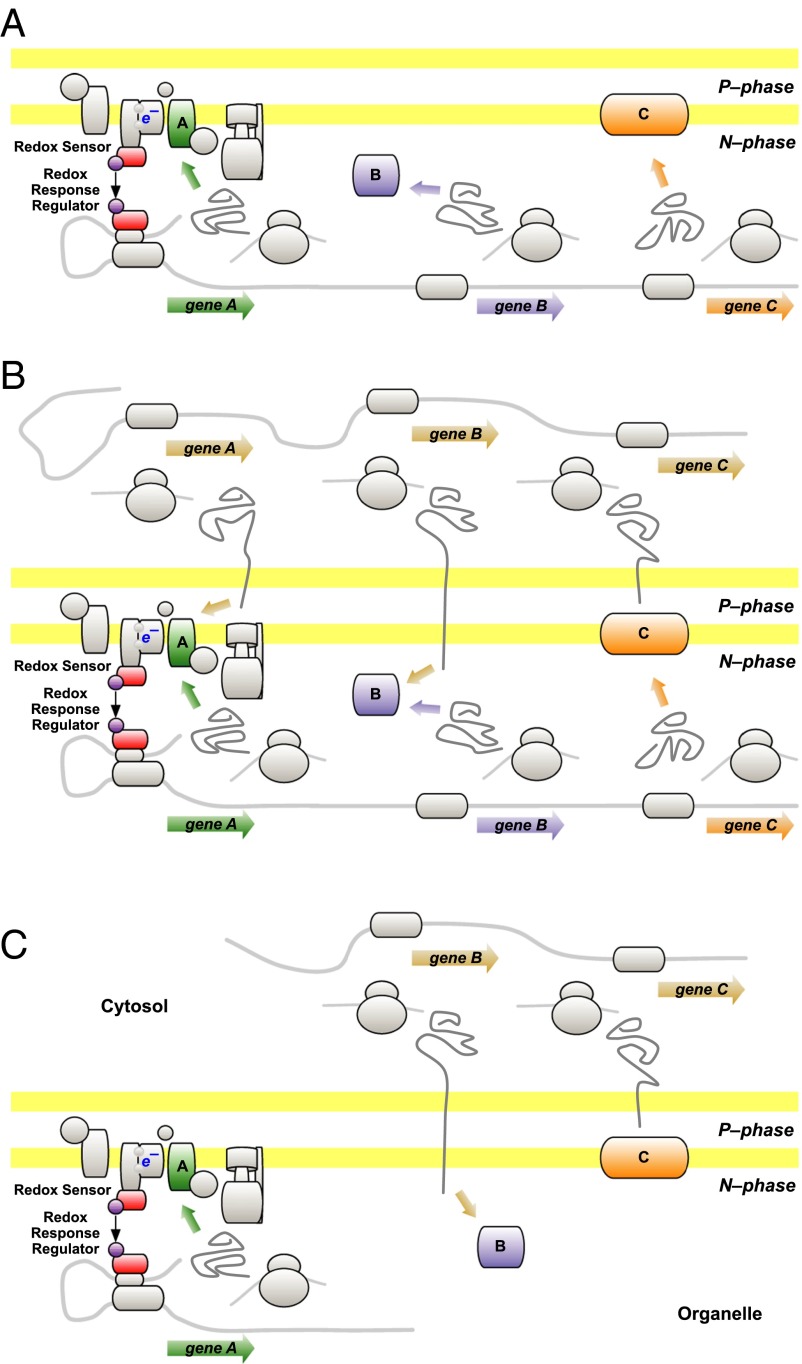
Fig. 2A depicts a simplified redox two-component regulatory system in a prokaryotic endosymbiont. The endosymbiont has three token genes, A, B and C, of which only A is subject to redox control. Fig. 2B illustrates the two possible pathways of synthesis of each of the three token proteins, A, B, and C. Synthesis may begin with transcription of genes in the endosymbiont or of gene copies acquired by the host. CoRR proposes that gene location by itself has no structural or functional consequence for the mature form of any protein whereas natural selection nevertheless operates to determine which of the two copies is retained. Selection favors continuity of redox regulation of gene A, and this regulation is sufficient to render the host’s unregulated copy redundant. In contrast, there is a selective advantage to location of genes B and C in the genome of the host (5), and thus it is the endosymbiont copies of B and C that become redundant and are lost. The resulting, complementary distribution of the three genes is depicted in Fig. 2C.
Fig. 2.
(A) The CoRR hypothesis outlined—ancestral prokaryote. Genes A, B, and C are transcribed and translated to give proteins A, B, and C. Protein A is a membrane-intrinsic component of the prokaryote’s bioenergetic (energy-transducing) membrane; its redox state regulates its own synthesis by the action of a two-component regulatory system on transcription of gene A. (B) The CoRR hypothesis outlined—endosymbiont. Genes A, B, and C are copied from the endosymbiont to the genome of the host cell. They are transcribed and translated, on host ribosomes, to give precursor proteins that are exported from the host into the endosymbiont. Each of proteins A, B, and C then has two possible sites of synthesis. Natural selection determines which of these sites is maintained. (C) The CoRR hypothesis outlined—bioenergetic organelle. Genes B and C are lost from their original location whereas a continued requirement for regulation of gene A by the redox state of protein A maintains colocation of gene A with gene product. The endosymbiont has become a bioenergetic organelle. Adapted from ref. 1.
The CoRR hypothesis originated from considering protein phosphorylation in photosynthesis (6), an example of redox regulation by reversible, posttranslational, covalent modification of preexisting proteins. Absorbed excitation energy becomes distributed optimally between chloroplast photosystems I and II as a result of activation of a protein kinase. The kinase is activated by the reduced form of an electron carrier, plastoquinone, whose redox state is determined by the difference between the rates of electron transport through photosystem I and photosystem II (7). This purely posttranslational redox regulatory control enables chloroplast photosynthesis to adjust to changes in light quality that favor one photosystem over the other. Reversible, redox-dependent protein phosphorylation distributes absorbed excitation energy optimally between the two photosystems, thereby correcting any imbalance that would otherwise be detrimental to overall quantum yield. Cyanobacteria lack the key chlorophyll-binding protein kinase substrate found in chloroplasts while nevertheless exhibiting redox control of excitation energy distribution between photosystem I and photosystem II (8). Attempts to identify a cyanobacterial substrate for redox-controlled protein phosphorylation resulted in the unexpected discovery of elements of a cyanobacterial two-component regulatory system (9).
Two-component systems are widespread in prokaryotes, where they exert specific control, usually over transcription, in response to any one of a wide variety of environmental signals (10). A two-component regulatory system reporting on plastoquinone redox state was proposed for cyanobacteria (6, 9), and the terms “redox sensor” and “redox response regulator” were introduced (1, 6, 11). Two-component redox regulation of transcription is known to control the mode of primary energy metabolism in anoxygenic photosynthetic bacteria (12) and to adjust the stoichiometry of photosystem I and photosystem II in oxygenic cyanobacteria (13). The cyanobacterial ancestor of chloroplasts is likely to have carried such a system into the chloroplasts as a transcriptional counterpart to the posttranslational control that balances light energy conversion. A conserved, ancestral redox two-component system might then explain photosystem stoichiometry adjustment (14, 15). Genes for the major reaction center apoproteins of both photosystem I and photosystem II are located in chloroplast DNA (16, 17). The general inference is that “the requirement for regulation of gene expression by redox potential may in principle explain the evolutionary maintenance, in eukaryotic cells, of the cytoplasmic genomes of chloroplasts and mitochondria” (1).
Experimental Evidence Bearing Directly on CoRR
Evidence 1: Selection of Proteins for Synthesis Within Isolated Chloroplasts and Mitochondria.
Protein synthesis by isolated chloroplasts can be studied using incorporation of [35S]methionine as a radioactive label (18). Notable products include the large subunit of Rubisco (19) and the chloroplast psbA gene product or D1 protein of the reaction center of photosystem II (20). Synthesis of D1 is light-dependent (20, 21) and required for resynthesis after breakdown in a photosystem II repair cycle (22, 23).
Allen et al. (24) incubated isolated chloroplasts from pea (Pisum sativum) in the presence of [35S]methionine in the light and dark, both with and without site-specific inhibitors of photosynthetic electron transport. Synthesis of a number of polypeptides was seen from autoradiography of protein gels to be light-dependent and unaffected by the presence of the inhibitor DBMIB, which blocks oxidation of the plastoquinone pool, but inhibited in the presence of DCMU, which blocks plastoquinone reduction. Additional experiments included redox reagents in the incubation medium in darkness, from the oxidizing agent ferricyanide to reducing agents such as ascorbate, DTT, and dithionite. Synthesis of chloroplast proteins with a broad range of molecular masses was observed when DTT was present. The presence of ascorbate or dithionite also supported synthesis, with marked qualitative differences between the subsets of polypeptides produced.
Isolated pea leaf mitochondria gave comparable results (24)—the pattern of [35S]methionine incorporation depended on the redox reagent included in the incubation, and effects of respiratory chain donors and inhibitors suggested that the redox state of respiratory electron carriers determined patterns of protein synthesis. Duroquinol, an electron donor to the quinone pool, gave a distinctive pattern of in vitro protein synthesis in both chloroplasts and mitochondria. Further experiments with isolated mitochondria used additional site-specific electron transport inhibitors and supported the conclusion that a site of redox control of mitochondrial protein synthesis lay at, or close to, respiratory complex II (25).
There are a number of possible reasons why patterns of protein synthesis “in organello” seem to be specific to isolated sections of the electron transport chains and responsive to added redox reagents (26). Further experiments will be required to distinguish between them. Given these caveats, Allen et al. (24) concluded that “our results are consistent with the hypothesis that the primary function of organelle genomes is the encoding in situ of proteins whose synthesis is thereby able to respond rapidly to changes in redox potential.”
Evidence 2: Redox Regulatory Control of Chloroplast DNA Transcription.
Pfannschmidt et al. (27, 28) provided evidence for redox regulatory control of transcription of chloroplast genes in mustard (Sinapis alba) by measurement of both mRNA quantity and rate of mRNA accumulation in run-on transcription. Changes in chloroplast transcription followed changes in wavelength of incident light that selected partially for either photosystem I or photosystem II. Corresponding effects were also observed after addition to isolated chloroplasts, under constant illumination, of site-specific inhibitors of photosynthetic electron transport. It was concluded that changes in quantity and rate of synthesis of specific mRNAs followed from perturbation of the redox state of a component of the electron transport chain.
Three genes of particular interest were psbA, encoding the D1 reaction center apoprotein of photosystem II, and psaAB, encoding two corresponding reaction center apoproteins of photosystem I. When experimental conditions were changed to make photosystem I rate-limiting for photosynthesis, psaAB was induced whereas psbA was repressed. Conversely, if photosystem II was made rate-limiting, then psaAB was repressed and psbA was induced. These responses are functionally intelligible as initial steps taken by the chloroplast itself to rectify imbalance in the stoichiometry of photosystem I to photosystem II and signify a rebalancing of the two photosystems by redox control of chloroplast DNA transcription (27, 28). Although posttranslational redox regulation of light-harvesting structure and function balances the two photosystems and so readjusts the redox state of plastoquinone (7), a transcriptional mechanism operates to achieve the same effect.
The rate of chloroplast run-on transcription changed rapidly after the change in wavelength of light, with a 25–30% decrease in psaAB rate, for example, being detected 7.5 min after switching from light favoring photosystem II to light favoring photosystem I (28). These kinetics suggest that the transcriptional and posttranslational responses of the photosynthetic apparatus, although arising from the same redox signal, are initiated independently (29). Comparable results were obtained using pea (P. sativum) (30). In all cases, transcriptional regulation specific to psbA did not remain after chloroplasts isolation, as subsequently observed also for Arabidopsis thaliana (31). This result suggests that photosystem stoichiometry is sufficiently maintained by redox control of synthesis of photosystem I alone, as envisaged for cyanobacteria (32).
Nine different chloroplast genes were investigated by Pfannschmidt et al. (27), with rbcL transcription being found to be affected in the same way, and in the same direction, as psbA, although with a smaller amplitude of redox response. RbcL is the large subunit of the Rubisco, the enzyme catalyzing the primary carboxylation step of the Benson–Calvin cycle. Also of interest are chloroplast genes whose transcription seemed not to be under plastoquinone redox regulatory control relative to the reference, which was, in all cases, rrn16. In the experiment of Pfannschmidt et al. (27), apparently redox-independent chloroplast genes were as follows: petA (cytochrome f); atpA (ATP synthase subunit A); ribosomal subunits rpl2 and rps16; trnG (glycine tRNA); and rpoB (RNA polymerase subunit).
The results of Pfannschmidt and coworkers (27, 28, 30, 33) are in agreement with the predictions of the 1993 hypothesis (1) for the function of genomes in chloroplasts, and, by inference, in mitochondria. Newer techniques, including RNA-seq, can be expected to allow more quantitative analysis whereas redox titration of the kind applied to chloroplast protein kinase activity (34) should permit precise identification of the site(s) of transcriptional redox control.
The functional significance of chloroplasts keeping genomes is thus conservation of an essential mechanism for adjustment of the stoichiometry of photosystems I and II. The chloroplast genome permits the photosynthetic apparatus to continue to compensate for an imbalance and inefficiency otherwise produced by changes in spectral composition (wavelength) of light. The results of Pfannschmidt et al. (27, 28) on chloroplast transcription support the CoRR hypothesis in a direct way, and it is difficult to envisage an alternative explanation. Pfannschmidt et al. (28) conclude that “...such rapid and direct regulatory coupling may depend upon the genes concerned being present in the same intracellular compartment as the electron-transport chain that regulates their expression, and upon the persistence there of prokaryotically derived, redox signal-transduction pathways to provide the means of control.”
Evidence 3: Persistence of Bacterial Redox Signaling Components in Chloroplasts.
Chloroplast sensor kinase (CSK) is a protein imported into chloroplasts (31). In A. thaliana, CSK is the product of the nuclear gene At1g67840, which has homologs in representatives of all major groups of photosynthetic eukaryotes (35, 36). The amino acid sequence of CSK contains motifs characteristic of a bacterial sensor histidine kinase (31, 35, 37).
Location of CSK in the chloroplast was demonstrated using two independent methods: by fluorescence microscopy of tobacco leaf epidermis after its transformation with a CSK-GFP gene construct, transiently expressed; and by uptake and import into isolated pea chloroplasts of [35S]methionine-labeled precursor protein, a product of coupled transcription and translation of cDNA in vitro, in a wheat germ cell-free system (31).
CSK acquires a radioactive label from [γ-32P]ATP in an autophosphorylation assay, and phosphorylation seems to be redox-dependent (31). The phosphorylated histidine characteristic of bacterial sensor kinases is present in CSK of eukaryotic red, green, and brown algae (35, 37), but not in land plants, where the histidine is replaced by glutamate.
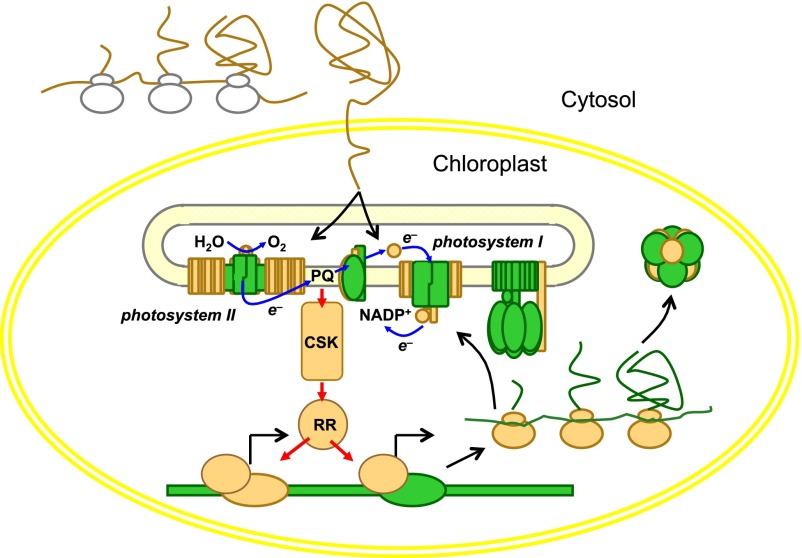
In Arabidopsis, inactivation of At1g67840 by T-DNA insertion results in a change in the response of photosystem I reaction center gene psaA transcription to a switch between light selective for photosystem I and photosystem II. In the experiments of Pfannschmidt et al. (27, 28) with mustard seedlings, the switch from photosystem II to photosystem I resulted in a decrease in psaA transcription, an effect contributing to a decrease in the stoichiometry of photosystem I to photosystem II. In Arabidopsis, the same response was observed in the WT whereas inactivation of CSK in two independent T-DNA–insertion lines was accompanied, paradoxically, by an increase in psaA transcription (31). This aberrant behavior of CSK knock-out mutants implicates CSK in redox regulatory control of chloroplast transcription. Despite the overlap in the kinetics of posttranslational and transcriptional responses to altered wavelength of light (29), the two signal transduction pathways are not known to contain any component downstream of the plastoquinone pool itself (36, 38). The proposed role of CSK in adjusting photosynthetic reaction center gene transcription is outlined in Fig. 3.
Fig. 3.
Two-component redox regulation of chloroplast transcription. Chloroplast sensor kinase (CSK) selectively switches on and off chloroplast genes in response to perturbations in the photosynthetic electron transport chain (depicted as electron flow from H2O to NADP+) within the thylakoid membrane. CSK is a redox sensor and reports on electron flow through plastoquinone (PQ). A response regulator (RR) mediates CSK’s control over transcription of genes for reaction center apoproteins of photosystem I and photosystem II, giving autoregulatory adjustment of photosystem stoichiometry. Chloroplast genes and gene products are shown in green. The nuclearly encoded components, imported into the chloroplast, are shown in light brown. Adapted from ref. 35.
In chloroplasts of red algae, a cognate two-component response regulator is likely to interact with CSK (35, 37). In contrast, CSK in the land plant Arabidopsis has at least two other interaction partners that mediate redox control of transcription. These interaction partners are the eukaryotically derived plastid transcription kinase and a typically prokaryotic RNA polymerase sigma factor (37, 39). A possible mechanism for chloroplast transcriptional control is that CSK responds to an oxidized plastoquinone pool by becoming active as a protein kinase that catalyzes phosphorylation of RNA polymerase sigma factor 1 (σ1) (37, 39). Promoter specificity and repression of photosystem I transcription is then determined by the phosphate group compensating for positive charges of basic residues adjacent to the σ1 phosphorylation site and resulting in reversible formation of a compact protein secondary structure known to be present as a permanent feature of bacterial sigma factors. Activation of photosystem I transcription is then predicted to result from the action of a phospho-sigma factor phosphatase (39).
Chloroplast sensor kinase is a redox sensor and a prediction of the CoRR hypothesis depicted in Fig. 2. In view of the experimental progress with chloroplasts, what can be said concerning redox control of gene expression in mitochondria?
Independent Evidence and Observations Bearing on CoRR
Redox Regulatory Control of Mitochondrial DNA Transcription.
Wilson et al. (40) estimated a redox midpoint potential of +270 mV, n = 1, for incorporation of radiolabeled UTP into RNA by purified potato mitochondria. Corresponding effects of electron donors and site-specific inhibitors of photosynthetic electron transport were also observed in chloroplasts (41). These results supported the conclusion (40, 42) that the redox state of the Rieske iron–sulfur center governs transcription of mitochondrial and chloroplast DNA, with an oxidized center resulting in RNA synthesis and a reduced center inhibiting it. The Rieske iron–sulfur center is a component of cytochrome b–c1 and b6–f complexes, the sites of oxidation of the ubiquinone and plastoquinone pools in the proton-motive Q-cycle (43–45). Acting as a transcriptional redox sensor, the Rieske protein will respond to imbalance in electron flow into, and out of, the Q-cycle. Because Wilson et al. (40) report that an oxidized Rieske Fe-S promotes general mitochondrial transcription, it might be predicted that a major component of the induced RNA synthesis is mRNA for an electron donor, most obviously respiratory complex I, core subunits of which are encoded in mitochondrial DNA.
From experiments with shoots of Arabidopsis seedlings. Zubo et al. (46) report effects of electron transport chain inhibitors on transcription of a range of genes in mitochondrial DNA. Inhibition of electron transport through respiratory complex IV by KCN was accompanied by increases in transcription of nad genes, which encode subunits of respiratory complex I. The respiratory complex III inhibitor, antimycin A, increased nad transcription in the experiment of Zubo et al. (46), as did inhibition of the alternative oxidase. Although Zubo et al. investigated redox effects on transcription of specific genes, the site(s) of redox control are less easily resolved than in the experiments of Wilson et al. (40) on total mitochondrial RNA synthesis. It would be useful to combine aspects of these two approaches to resolve redox control of mitochondrial transcription, and revealing to make exact measurement of quantities of all mitochondria mRNAs as they respond in simultaneous potentiometric redox titration. The midpoint(s) and direction of the responses would be expected to reveal the number of sites at which redox control is exerted and the potential functional significance of each.
Protein phosphorylation is likely to be involved in redox signal transduction in both mitochondria and chloroplasts. Redox titration of chloroplast thylakoid protein phosphorylation indicates a wide range of phosphoproteins, with most becoming phosphorylated when the plastoquinone pool is reduced (34). “Reverse titrators” at 46 and 63 kDa are phosphorylated with the same midpoint, but when plastoquinone is oxidized. These components could be involved in chloroplast redox sensing whereas the results together indicate a single point of redox control with a midpoint of +38 mV, n = 1. This result cannot reflect control by the Rieske iron–sulfur center, as concluded for potato mitochondria by Wilson et al. (40). For both mitochondria and chloroplasts, resolution and identification of the site(s) of redox control of transcription and of protein phosphorylation are promising areas for future research.
Alternatives to Two-Component Transcriptional Control.
RNA polymerases in animal and plant mitochondria have properties in common with RNA polymerase of bacteriophage viruses (47). A bacterial-type RNA polymerase is one of two polymerases found in chloroplasts of higher plants (48). Its major subunits are encoded in chloroplast DNA whereas those of its phage-type counterpart are imported as the products of nuclear genes (49). Mitochondria of many protists have bacterial-type RNA polymerases (50), and it will be useful to know whether they interact with response regulators that are themselves under the control of redox sensor kinases.
Nucleic acid topology is suggested as a basis of redox control of transcription by RNA helicase whereas it is the helicase gene itself, crhR, that seems to be under the control of transcription in the cyanobacterium Synechocystis PCC 6803 (51).
Mitochondria from mammalian fibroblasts contain a specific DNA topoisomerase, topoisomerase I. Sobek et al. (52) report increased transcription of respiratory chain mitochondrial genes when DNA topoisomerase I is either deficient or depleted. It is concluded that topoisomerase I inhibits mitochondrial DNA transcription and that release of transcriptional inhibition is accompanied by increased respiratory rate and superoxide production (52). DNA topoisomerase is also implicated in redox control of transcription of mitochondrial DNA in plant tissue (53). Redox regulation of topoisomerase I is predicted to play a role in mitochondrial homeostasis (54). It is possible to envisage transcriptional regulation by DNA and RNA topology accompanying the acquisition of a phage-type RNA polymerase in specific eukaryotic lineages, this control mechanism eventually superseding the bacterial mode of redox control of transcription present in the endosymbiotic ancestor of all mitochondria.
Gene expression in the context of the CoRR hypothesis has received less direct experimental investigation for mitochondria than for chloroplasts. Nevertheless a range of circumstantial evidence is consistent with respiratory electron transport exerting redox control over mitochondrial gene expression and requiring mitochondrial DNA (55–57).
Mitosomes.
The CoRR hypothesis proposes that DNA in chloroplasts and mitochondria confers autonomous gene regulation of their bioenergetic systems. A clear prediction is that organelles that lose their bioenergetic function will also eventually lose their genomes. This prediction is borne out with the identification of mitosomes as specialist mitochondria whose primary function is no longer energy transduction. Relict mitochondria of unicellular parasites are evidence for CoRR because restriction or loss of aerobic oxidative phosphorylation is accompanied by reduction or loss of mitochondrial DNA (58). Hydrogenosomes are found for example in Trichomonas vaginalis; anaerobic mitochondria are found in Blastocystis sp., mitosomes in Giardia sp., Entamoeba histolytica, and Cryptosporidium parvum–causative agents of water-borne human gastrointestinal disease (59). Mitosomes do not carry out oxidative phosphorylation and contain no respiratory chain components. No mitosome genome has been detected, either directly (60) or from genomic inference (61–63). The retained function of mitosomes seems primarily to be production of iron–sulfur clusters (64).
What is Forbidden? How to Disprove CoRR
To count as science, a hypothesis must be falsifiable, and a single forbidden observation can be sufficient to disprove an otherwise successful theory (65). In principle, the CoRR hypothesis could have been disproved by the redox independence of chloroplast transcription and protein synthesis. However, functionally intelligible redox control of chloroplast photosynthesis and genome function was discovered from experiments designed to test CoRR, and the results of these experiments must count as evidence in its favor. I suggest that attention can usefully be given to predictions that may yet demonstrate that the CoRR hypothesis is false. Some of these predictions are now discussed.
Primary Electron Transport Components Imported into Organelles as Precursors.
A single example would be a nuclear gene for an imported precursor protein that functions in primary vectorial electron transport and/or proton translocation: for example photosynthetic reaction center core subunits and respiratory chain subunits. Rubisco activity, too, has direct consequences for the redox state of photosynthetic electron carriers (66).
Vectorial Electron Transport Without a Genome.
Mitosomes exemplify clear association of oxidative phosphorylation with mitochondrial DNA. There seems so far to be no example of a chemiosmotically functional subcellular compartment that does not carry genes for some of its own components. One example would suffice to disprove CoRR.
Genomes Retained Without Electron Transport.
There are DNA-containing and nonphotosynthetic plastids of parasitic plants (67) and apicoplasts (68). In the absence of photosynthesis, it is possible that secondary functions, such as heme synthesis and iron–sulfur cluster assembly, have become primary, that these processes require redox regulation at the level of gene expression, and that chloroplast genomes cannot therefore be replaced. It may also be that additional obstacles to gene relocation apply in these cases. One suggestion is that transfer of plastid RNA to the nucleus is forbidden (69). Tetrapyrrole synthesis requires glutamyl-tRNA and mitochondrial protein synthesis depends on N-formyl-methionine-tRNA in the plastid (69). It should also be noted that some species, notably of Chlamydomonas, successfully import tRNAs to the chloroplast.
Epistasis Leading to Improved Fitness.
Relocation of organellar genes to the nucleus has been achieved experimentally (66, 70). An accompanying gain of fitness would indicate selective value to relocation and would falsify CoRR.
Selective Barriers to Gene Transfer.
DNA transfer from organelle to nucleus has given rise to nuclear genes that originated in the organelle and is a frequent occurrence (71–73). Specific sequence motifs correlating with gene transfer could be sought. However, mechanisms currently envisaged center on chemical and mechanical DNA damage, with no role for specificity of nucleotide sequence (74–76).
The Limited Transfer Window Hypothesis.
If copying genes to the nucleus depends on organelle lysis, then it is likely to be lethal to a cell with a single organelle. Gene transfer to the nucleus may therefore be restricted to organisms containing multiple organelles per cell. This “limited transfer window hypothesis” is in agreement with observed frequencies of nuclear copies of mitochondrial and plastid genes in protists (69, 77, 78) and with the frequency of plastid copies of mitochondrial genes (79). Retention by the Plasmodium plastid of two structural genes, clpC and sufB (68), could be cited as a counter example and falsification of CoRR. However, nonphotosynthetic plastids are derived from photosynthetic plastids, and so genes in nonphotosynthetic plastids “might not reflect a need to have them expressed in the plastid, but rather an inability to get them out.” (69).
Selective Barriers to Import of Precursor Proteins—Testing Unim-portability.
Hydrophobicity has been suggested as a barrier to reimport of membrane proteins whose organellar genes move to the nucleus (80). It could be informative to devise worst-case import requirements—a stringent test to see whether it is truly impossible for isolated organelles to import any precursor for, for example, polyphenylalanine.
Endosymbiosis in Progress—Forbidden Gene Relocations in Paulinella chromatophora.
The amoeba Paulinella chromatophora carries an endosymbiotic cyanobacterium, or “chromatophore” (81). More than 30 genes found in cyanobacteria have moved to the Paulinella cell nucleus, functionally replacing endosymbiont copies (82). Endosymbiont-derived products of nuclear genes are include three apopoteins of electron carriers of photosystem I (83). Evidence so far indicates that relocated, functional genes encode only secondary components of the photosynthetic electron transport chain, and never reaction center proteins or cytochrome b6. The CoRR hypothesis will be disproved if Paulinella or an analogous endosymbiosis (84, 85) can be shown to depend on primary bioenergetics operating solely with imported protein products of relocated genes.
Transcription of a Redox-Regulated Subset of Cyanobacterial Genes.
Redox regulation of cyanobacterial transcription is predicted by CoRR to operate on genes homologous with those in chloroplasts (86). If there is no correspondence, then CoRR, without modification, will be false.
Redox Titration of Transcription.
Redox titration of transcription would be a stringent test of the CoRR hypothesis. The example of Wilson et al. (40) was on total mitochondrial RNA. Identification of redox midpoint potentials for transcription of specific genes is arguably long overdue. Chloroplast phosphoprotein phosphatases have been shown to be strictly redox-independent using potentiometric redox titration (87). This technique could be applied to transcription. Rigorous demonstration of strict redox independence of nonribosomal protein gene transcription would disprove CoRR.
Consequences and Implications of CoRR
The Energetics of Genome Complexity.
Lane and Martin (88) ask why prokaryotes have not evolved to match the size and complexity of eukaryotes. They propose that the answer lies in intracellular compartmentalization of energy transduction: that is, in mitochondria, defining features of eukaryotic cells (89). Lane and Martin propose that the demand for energy for gene expression, coupled with the simple constraint of surface area-to-volume, sets a limit to the size of prokaryotic cells (88). With the advent of mitochondria, each mitochondrion being as constrained in size as a bacterium, the eukaryotic cell was able to multiply mitochondria and greatly increase its size and number of genes. In support of this proposal, Lane and Martin summarize data on the rate of respiration, cell size, and genome size in a range of bacteria and eukaryotes (88). Their broad conclusion is that a bacterium scaled to the volume, shape, and genome size of a representative protozoan would have around a 200,000-fold decrease in power available per gene. Genomes large enough to support the complexity of eukaryotic cells are therefore metabolically and energetically impossible without multiple copies of dedicated internal organelles housing energy-transducing membranes that synthesize ATP for gene expression, particularly protein synthesis. Lane and Martin support the proposal that mitochondrial DNA exists to allow redox regulation of gene expression, while noting that DNA replication and synthesis of respiratory chain components can then be optimized by individual mitochondria to match their specific intracellular environmental conditions (88). A prerequisite for multicellularity is not just mitochondria, but colocation of the mitochondrial genome with the respiratory chain that it encodes.
Convergent Evolution for Ribosomal Protein Gene Content in Plastid and Mitochondrial Genomes.
The CoRR hypothesis, as originally formulated (1) and subsequently developed (4), makes no prediction concerning redox control of organelle-located genetic system genes. Indeed, chloroplast ribosomal protein gene expression was found to be redox-independent in the experiments of Pfannschmidt et al. (27, 28), in contrast to the marked redox dependency of photosynthetic reaction center gene transcription.
From genome sequence comparisons, Maier et al. (90) report conservation of genes for the same conserved subset of ribosomal proteins in both mitochondria and chloroplasts (Fig. 4). Because chloroplasts and mitochondria had independent origins, this observation indicates convergent evolution, with natural selection operating on gene location. By comparison with Escherichia coli, Maier et al. observe that the conserved proteins are implicated in 30S and 50S ribosomal subunit assembly and in initial binding of rRNA (90). Maier et al. propose that the presence of these genes in organelle DNA enhances their function in ribosome synthesis whereas the requirement for organelle ribosomes in the first place is sufficiently explained by CoRR.
Fig. 4.

Convergent evolution of gene content in mitochondria and chloroplasts. The ancestors of both organelles were prokaryotes with genomes encoding around 5,000 genes. During the course of endosymbiosis, genes are transferred from each organelle to the hosts’ nuclear genome, and the corresponding gene products are imported back to the organelles. The initial genome size of around 5,000 genes decreased to 3–67 genes in mitochondria and 23–200 genes in chloroplasts. The color coding within compartments in the lower part of the figure illustrates the convergent evolution of genes retained in the two bioenergetic organelles: genes for components of oxidative phosphorylation, photosynthesis, and proteins of 50S and 30S ribosomal subunits. Organellar-encoded genes are colored brown for mitochondria and green for plastids. TIC/TOC, protein translocator of the inner/outer chloroplast membrane; TIM/TOM, protein translocator of the inner/outer mitochondrial membrane. Schemes for oxidative phosphorylation and photosynthesis are adapted from ref. 4. Reproduced from ref. 90.
Chloroplast and Mitochondrial Genome Content.
In photosynthetic eukaryotes, chloroplast DNA encodes between 80 and 200 proteins, much less than 1% of proteins in the cell, and much less than 10% of the proteins in the chloroplast, to which a higher plant’s nuclear–cytosolic system successfully supplies around 3,000 proteins for import (91). A common subset of less than 50 chloroplast genes encodes proteins of only five functional classes: photosystem I, photosystem II, secondary electron transport and ATP synthesis, ribosomal subunits and RNA polymerase, and the Rubisco large subunit. All mitochondria that contain a respiratory chain also contain a genome. Mitochondrial DNA typically encodes between 3 and 63 proteins (92); in metazoan, the number is 13 (93). Among the smaller mitochondrial genomes are those of Plasmodium spp., Toxoplasma gondii, and the basal dinoflagellate Oxyrrhis marina. These mitochondrial genomes encode three polypeptides: apocytochrome b and two subunits, I and III, of cytochrome oxidase. Although the ranges of gene content in bioenergetic organelles indicate additional factors that must be considered in specific lineages, the universal retention of genes for central components of chemiosmotic energy transduction is consistent with the CoRR hypothesis. CoRR could also account for selection and convergent evolution that retains, for example, respiratory complex II subunit genes in mitochondria of only very distantly related eukaryotes (94).
Redox Chemistry and Gene Regulation in Cyanobacteria and Chloroplasts, Eubacteria, and Mitochondria
Prokaryotes exhibit diversity in their sources of energy, electrons, carbon, and nitrogen. Free-living prokaryotes are often able to switch between disparate modes of primary metabolism: for example, between phototrophy and chemotrophy, between autotrophy and heterotrophy, and between aerobic and anaerobic lifestyles.
At the base of primary energy metabolism is redox (reduction–oxidation) chemistry. For sustained metabolism—to be alive—all living things require an external source of free energy and use an ultimate electron donor (source) and acceptor (sink) that are not, together, already at thermodynamic equilibrium (95). An apparent exception is photosynthesis, where light energy moves electrons in the opposite direction to that in which they would otherwise pass, using light to set in motion chains of electron transfer.
Some individual bacterial species are able to switch between these different modes of primary metabolism. The purple nonsulfur photosynthetic bacteria, for example, are phototrophic under illumination and anaerobic conditions, but chemotrophic in the dark or in the presence of oxygen. Redox control was envisaged to account for this metabolic versatility (96). A redox genetic switch selects between expression of different sets of genes (97). Cyanobacteria are mostly obligate photolithoautotrophs, and even here a redox genetic switch selects between sets of photosynthesis genes to compensate for changes in light quality (wavelength) and quantity (intensity) (13, 98, 99).
Given the evolutionary origin of chloroplasts and mitochondria from free-living bacteria (100–102), it would be unremarkable for redox control of gene expression to have been retained during the initial transition to endosymbiosis. In the case of chloroplasts, light quality and quantity must have continued to change so it is to be expected that redox control persisted as the endosymbiosis became permanent. In the case of mitochondria, too, physiological and metabolic demand would continue to change during the transition, through endosymbiosis, to subcellular organelle, particularly because mitochondria are known to exhibit diversity and versatility (103) comparable with that of their evolutionary precursor (89).
Genetic adaptation to varying redox conditions in photosynthesis and respiration may have been continuous and essential for subcellular, bioenergetic organelles, no less than for their endosymbiotic ancestors. The vital requirement for continued operation of redox regulatory control over gene expression is proposed as the primary reason for the retention of chloroplast and mitochondrial DNA. The redox chemistry of biological energy transduction is then the primary factor determining which genes this DNA contains.
Acknowledgments
I thank William Martin, Nick Lane, Carol Allen, Sujith Puthiyaveetil, Iskander Ibrahim, and Wilson de Paula for discussion, and Uwe Maier for granting permission to include Fig. 4. Support from the Leverhulme Trust as Research Grant F07476AQ is gratefully acknowledged.
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. P.J.K. is a guest editor invited by the Editorial Board.
References
- 1.Allen JF. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol. 1993;165(4):609–631. doi: 10.1006/jtbi.1993.1210. [DOI] [PubMed] [Google Scholar]
- 2.Allen JF. Redox control of gene expression and the function of chloroplast genomes: An hypothesis. Photosynth Res. 1993;36(2):95–102. doi: 10.1007/BF00016274. [DOI] [PubMed] [Google Scholar]
- 3.Allen JF. Why chloroplasts and mitochondria contain genomes. Comp Funct Genomics. 2003;4(1):31–36. doi: 10.1002/cfg.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen JF. The function of genomes in bioenergetic organelles. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):19–37, discussion 37–38. doi: 10.1098/rstb.2002.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen JF, Raven JA. Free-radical-induced mutation vs redox regulation: Costs and benefits of genes in organelles. J Mol Evol. 1996;42(5):482–492. doi: 10.1007/BF02352278. [DOI] [PubMed] [Google Scholar]
- 6.Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098(3):275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- 7.Allen JF, Bennett J, Steinback KE, Arntzen CJ. Chloroplast protein phosphorylation couples plastoquinone redox state to distribution of excitation energy between photosystems. Nature. 1981;291(5810):25–29. [Google Scholar]
- 8.Allen JF, Sanders CE, Holmes NG. Correlation of membrane protein phosphorylation with excitation energy distribution in the cyanobacterium Synechococcus 6301. FEBS Lett. 1985;193(2):271–275. [Google Scholar]
- 9.Harrison MA, Keen JN, Findlay JBC, Allen JF. Modification of a glnB-like gene product by photosynthetic electron transport in the cyanobacterium Synechococcus 6301. FEBS Lett. 1990;264(1):25–28. doi: 10.1016/0014-5793(90)80755-8. [DOI] [PubMed] [Google Scholar]
- 10.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 11.Allen JF. Redox control of transcription: Sensors, response regulators, activators and repressors. FEBS Lett. 1993;332(3):203–207. doi: 10.1016/0014-5793(93)80631-4. [DOI] [PubMed] [Google Scholar]
- 12.Elsen S, Swem LR, Swem DL, Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68(2):263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Sherman LA. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. Strain PCC 6803. J Bacteriol. 2000;182(15):4268–4277. doi: 10.1128/jb.182.15.4268-4277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chow WS, Melis A, Anderson JM. Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA. 1990;87(19):7502–7506. doi: 10.1073/pnas.87.19.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujita Y, Murakami A. Regulation of electron transport composition in cyanobacterial photosynthetic system: Stoichiometry among photosystem I and photosystem II complexes and their light-harvesting antennae and cytochrome-b6 cytochrome-f complex. Plant Cell Physiol. 1987;28(8):1547–1553. [Google Scholar]
- 16.Ohyama K, et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322(6079):572–574. [Google Scholar]
- 17.Shinozaki K, et al. The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 1986;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellis RJ. Protein synthesis by isolated chloroplasts. Biochim Biophys Acta. 1977;463(2):185–215. [Google Scholar]
- 19.Blair GE, Ellis RJ. Protein synthesis in chloroplasts. I. Light-driven synthesis of the large subunit of fraction I protein by isolated pea chloroplasts. Biochim Biophys Acta. 1973;319(2):223–234. doi: 10.1016/0005-2787(73)90013-0. [DOI] [PubMed] [Google Scholar]
- 20.Eaglesham ARJ, Ellis RJ. Protein synthesis in chloroplasts. 2. Light-driven synthesis of membrane proteins by isolated pea chloroplasts. Biochim Biophys Acta. 1974;335(3):396–407. doi: 10.1016/0005-2787(73)90013-0. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman-Falk H, Mattoo AK, Marder JB, Edelman M, Ellis RJ. General occurrence and structural similarity of the rapidly synthesized, 32,000-dalton protein of the chloroplast membrane. J Biol Chem. 1982;257(8):4583–4587. [PubMed] [Google Scholar]
- 22.Ohad I, Kyle DJ, Arntzen CJ. Membrane protein damage and repair: Removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J Cell Biol. 1984;99(2):481–485. doi: 10.1083/jcb.99.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puthiyaveetil S, et al. Compartmentalization of the protein repair machinery in photosynthetic membranes. Proc Natl Acad Sci USA. 2014;111(44):15839–15844. doi: 10.1073/pnas.1413739111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Allen CA, Håkansson G, Allen JF. Redox conditions specify the proteins synthesized by isolated chloroplasts and mitochondria. Redox Rep. 1995;1(2):119–123. doi: 10.1080/13510002.1995.11746969. [DOI] [PubMed] [Google Scholar]
- 25.Escobar Galvis ML, Allen JF, Hâkansson G. Protein synthesis by isolated pea mitochondria is dependent on the activity of respiratory complex II. Curr Genet. 1998;33(5):320–329. doi: 10.1007/s002940050343. [DOI] [PubMed] [Google Scholar]
- 26.Allen JF, Alexciev K, Håkansson G. Photosynthesis: Regulation by redox signalling. Curr Biol. 1995;5(8):869–872. doi: 10.1016/s0960-9822(95)00176-x. [DOI] [PubMed] [Google Scholar]
- 27.Pfannschmidt T, Nilsson A, Tullberg A, Link G, Allen JF. Direct transcriptional control of the chloroplast genes psbA and psaAB adjusts photosynthesis to light energy distribution in plants. IUBMB Life. 1999;48(3):271–276. doi: 10.1080/713803507. [DOI] [PubMed] [Google Scholar]
- 28.Pfannschmidt T, Nilsson A, Allen JF. Photosynthetic control of chloroplast gene expression. Nature. 1999;397(6720):625–628. [Google Scholar]
- 29.Allen JF, Pfannschmidt T. Balancing the two photosystems: Photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 2000;355(1402):1351–1359. doi: 10.1098/rstb.2000.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tullberg A, Alexciev K, Pfannschmidt T, Allen JF. Photosynthetic electron flow regulates transcription of the psaB gene in pea (Pisum sativum L.) chloroplasts through the redox state of the plastoquinone pool. Plant Cell Physiol. 2000;41(9):1045–1054. doi: 10.1093/pcp/pcd031. [DOI] [PubMed] [Google Scholar]
- 31.Puthiyaveetil S, et al. The ancestral symbiont sensor kinase CSK links photosynthesis with gene expression in chloroplasts. Proc Natl Acad Sci USA. 2008;105(29):10061–10066. doi: 10.1073/pnas.0803928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aizawa K, Fujita Y. Regulation of synthesis of PSI in the cyanophytes Synechocystis PCC6714 and Plectonema boryanum during the acclimation of the photosystem stoichiometry to the light quality. Plant Cell Physiol. 1997;38(3):319–326. [Google Scholar]
- 33.Pfannschmidt T. Chloroplast redox signals: How photosynthesis controls its own genes. Trends Plant Sci. 2003;8(1):33–41. doi: 10.1016/s1360-1385(02)00005-5. [DOI] [PubMed] [Google Scholar]
- 34.Silverstein T, Cheng LL, Allen JF. Redox titration of multiple protein phosphorylations in pea chloroplast thylakoids. Biochim Biophys Acta. 1993;1183(1):215–220. [Google Scholar]
- 35.Puthiyaveetil S, Allen JF. Chloroplast two-component systems: Evolution of the link between photosynthesis and gene expression. Proc Biol Sci. 2009;276(1665):2133–2145. doi: 10.1098/rspb.2008.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puthiyaveetil S, Ibrahim IM, Allen JF. Oxidation-reduction signalling components in regulatory pathways of state transitions and photosystem stoichiometry adjustment in chloroplasts. Plant Cell Environ. 2012;35(2):347–359. doi: 10.1111/j.1365-3040.2011.02349.x. [DOI] [PubMed] [Google Scholar]
- 37.Puthiyaveetil S, et al. Transcriptional control of photosynthesis genes: The evolutionarily conserved regulatory mechanism in plastid genome function. Genome Biol Evol. 2010;2:888–896. doi: 10.1093/gbe/evq073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen JF, Santabarbara S, Allen CA, Puthiyaveetil S. Discrete redox signaling pathways regulate photosynthetic light-harvesting and chloroplast gene transcription. PLoS ONE. 2011;6(10):e26372. doi: 10.1371/journal.pone.0026372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puthiyaveetil S, Ibrahim IM, Allen JF. Evolutionary rewiring: a modified prokaryotic gene-regulatory pathway in chloroplasts. Philos Trans R Soc Lond B Biol Sci. 2013;368(1622):20120260. doi: 10.1098/rstb.2012.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson SB, Davidson GS, Thomson LM, Pearson CK. Redox control of RNA synthesis in potato mitochondria. Eur J Biochem. 1996;242(1):81–85. doi: 10.1111/j.1432-1033.1996.0081r.x. [DOI] [PubMed] [Google Scholar]
- 41.Pearson CK, Wilson SB, Schaffer R, Ross AW. NAD turnover and utilisation of metabolites for RNA synthesis in a reaction sensing the redox state of the cytochrome b6f complex in isolated chloroplasts. Eur J Biochem. 1993;218(2):397–404. doi: 10.1111/j.1432-1033.1993.tb18389.x. [DOI] [PubMed] [Google Scholar]
- 42.Pearson CK, Wilson SB. NAD turnover in plant mitochondria. Biochem Soc Trans. 1997;25(4):S652–S652. doi: 10.1042/bst025s652. [DOI] [PubMed] [Google Scholar]
- 43.Allen JF. Cytochrome b6f: Structure for signalling and vectorial metabolism. Trends Plant Sci. 2004;9(3):130–137. doi: 10.1016/j.tplants.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Cramer WA, Hasan SS, Yamashita E. The Q cycle of cytochrome bc complexes: A structure perspective. Biochim Biophys Acta. 2011;1807(7):788–802. doi: 10.1016/j.bbabio.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hasan SS, Yamashita E, Baniulis D, Cramer WA. Quinone-dependent proton transfer pathways in the photosynthetic cytochrome b6f complex. Proc Natl Acad Sci USA. 2013;110(11):4297–4302. doi: 10.1073/pnas.1222248110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zubo YO, et al. Inhibition of the electron transport strongly affects transcription and transcript levels in Arabidopsis mitochondria. Mitochondrion. 2014;19(Pt B):222–230. doi: 10.1016/j.mito.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 47.Ringel R, et al. Structure of human mitochondrial RNA polymerase. Nature. 2011;478(7368):269–273. doi: 10.1038/nature10435. [DOI] [PubMed] [Google Scholar]
- 48.Shiina T, Tsunoyama Y, Nakahira Y, Khan MS. Plastid RNA polymerases, promoters, and transcription regulators in higher plants. Int Rev Cytol. 2005;244:1–68. doi: 10.1016/S0074-7696(05)44001-2. [DOI] [PubMed] [Google Scholar]
- 49.Gray MW, Lang BF. Transcription in chloroplasts and mitochondria: A tale of two polymerases. Trends Microbiol. 1998;6(1):1–3. doi: 10.1016/S0966-842X(97)01182-7. [DOI] [PubMed] [Google Scholar]
- 50.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5(2):418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kujat SL, Owttrim GW. Redox-regulated RNA helicase expression. Plant Physiol. 2000;124(2):703–714. doi: 10.1104/pp.124.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sobek S, et al. Negative regulation of mitochondrial transcription by mitochondrial topoisomerase I. Nucleic Acids Res. 2013;41(21):9848–9857. doi: 10.1093/nar/gkt768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Konstantinov Y, Tarasenko VI, Rogozin IB. Redox modulation of the activity of DNA topoisomerase I from carrot (Daucus carota) mitochondria. Dokl Biochem Biophys. 2001;377(2):82–84. doi: 10.1023/a:1011523522080. [DOI] [PubMed] [Google Scholar]
- 54.Sobek S, Boege F. DNA topoisomerases in mtDNA maintenance and ageing. Exp Gerontol. 2014;56:135–141. doi: 10.1016/j.exger.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 55.Allen CA, van der Giezen M, Allen JF. Origin, function and transmission of mitochondria. In: Martin W, Müller M, editors. Origins of Mitochondria and Hydrogenosomes. Springer; Berlin: 2007. pp. 39–56. [Google Scholar]
- 56.Allen JF, Puthiyaveetil S, Ström J, Allen CA. Energy transduction anchors genes in organelles. BioEssays. 2005;27(4):426–435. doi: 10.1002/bies.20194. [DOI] [PubMed] [Google Scholar]
- 57.de Paula WBM, Allen JF, van der Giezen M. Mitochondria, hydrogenosomes and mitosomes in relation to the CoRR hypothesis for genome function and evolution. In: Bullerwell CE, editor. Organelle Genetics: Evolution of Organelle Genomes and Gene Expression. Springer; Berlin: 2011. pp. 105–119. [Google Scholar]
- 58.van der Giezen M, Tovar J. Degenerate mitochondria. EMBO Rep. 2005;6(6):525–530. doi: 10.1038/sj.embor.7400440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Giezen M. Hydrogenosomes and mitosomes: Conservation and evolution of functions. J Eukaryot Microbiol. 2009;56(3):221–231. doi: 10.1111/j.1550-7408.2009.00407.x. [DOI] [PubMed] [Google Scholar]
- 60.León-Avila G, Tovar J. Mitosomes of Entamoeba histolytica are abundant mitochondrion-related remnant organelles that lack a detectable organellar genome. Microbiology. 2004;150(Pt 5):1245–1250. doi: 10.1099/mic.0.26923-0. [DOI] [PubMed] [Google Scholar]
- 61.Clark CG, et al. Structure and content of the Entamoeba histolytica genome. Adv Parasitol. 2007;65:51–190. doi: 10.1016/S0065-308X(07)65002-7. [DOI] [PubMed] [Google Scholar]
- 62.Loftus B, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433(7028):865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 63.Morrison HG, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317(5846):1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 64.Lill R, Mühlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci. 2005;30(3):133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 65.Popper KR. The Logic of Scientific Discovery. Routledge & Kegan Paul; London: 1959. [Google Scholar]
- 66.Kanevski I, Maliga P. Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc Natl Acad Sci USA. 1994;91(5):1969–1973. doi: 10.1073/pnas.91.5.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bungard RA. Photosynthetic evolution in parasitic plants: Insight from the chloroplast genome. BioEssays. 2004;26(3):235–247. doi: 10.1002/bies.10405. [DOI] [PubMed] [Google Scholar]
- 68.Wilson RJM, et al. Parasite plastids: Maintenance and functions. Philos Trans R Soc Lond B Biol Sci. 2003;358(1429):155–162, discussion 162–164. doi: 10.1098/rstb.2002.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barbrook AC, Howe CJ, Purton S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006;11(2):101–108. doi: 10.1016/j.tplants.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 70.Bietenhader M, et al. Experimental relocation of the mitochondrial ATP9 gene to the nucleus reveals forces underlying mitochondrial genome evolution. PLoS Genet. 2012;8(8):e1002876. doi: 10.1371/journal.pgen.1002876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin W. Gene transfer from organelles to the nucleus: Frequent and in big chunks. Proc Natl Acad Sci USA. 2003;100(15):8612–8614. doi: 10.1073/pnas.1633606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richly E, Leister D. NUPTs in sequenced eukaryotes and their genomic organization in relation to NUMTs. Mol Biol Evol. 2004;21(10):1972–1980. doi: 10.1093/molbev/msh210. [DOI] [PubMed] [Google Scholar]
- 73.Richly E, Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004;21(6):1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- 74.Bock R, Timmis JN. Reconstructing evolution: Gene transfer from plastids to the nucleus. BioEssays. 2008;30(6):556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- 75.Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5(2):123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- 76.Timmis JN, Wang D. Endosymbiotic evolution: The totalitarian nucleus is foiled again. Curr Biol. 2013;23(1):R30–R32. doi: 10.1016/j.cub.2012.11.038. [DOI] [PubMed] [Google Scholar]
- 77.Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philos Trans R Soc Lond B Biol Sci. 2010;365(1541):785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smith DR, Crosby K, Lee RW. Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol Evol. 2011;3:365–371. doi: 10.1093/gbe/evr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith DR. Extending the limited transfer window hypothesis to inter-organelle DNA migration. Genome Biol Evol. 2011;3:743–748. doi: 10.1093/gbe/evr068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.von Heijne G. Why mitochondria need a genome. FEBS Lett. 1986;198(1):1–4. doi: 10.1016/0014-5793(86)81172-3. [DOI] [PubMed] [Google Scholar]
- 81.Marin B, Nowack ECM, Melkonian M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist. 2005;156(4):425–432. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 82.Nowack ECM. Paulinella chromatophora: Rethinking the transition from endosymbiont to organelle. Acta Soc Bot Pol. 2014;83(4):387–397. [Google Scholar]
- 83.Nowack ECM, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci USA. 2012;109(14):5340–5345. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakayama T, et al. Complete genome of a nonphotosynthetic cyanobacterium in a diatom reveals recent adaptations to an intracellular lifestyle. Proc Natl Acad Sci USA. 2014;111(31):11407–11412. doi: 10.1073/pnas.1405222111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thompson AW, et al. Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science. 2012;337(6101):1546–1550. doi: 10.1126/science.1222700. [DOI] [PubMed] [Google Scholar]
- 86.Hihara Y, Sonoike K, Kanehisa M, Ikeuchi M. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2003;185(5):1719–1725. doi: 10.1128/JB.185.5.1719-1725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Silverstein T, Cheng L, Allen JF. Chloroplast thylakoid protein phosphatase reactions are redox-independent and kinetically heterogeneous. FEBS Lett. 1993;334(1):101–105. doi: 10.1016/0014-5793(93)81690-2. [DOI] [PubMed] [Google Scholar]
- 88.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467(7318):929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 89.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392(6671):37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 90.Maier UG, et al. Massively convergent evolution for ribosomal protein gene content in plastid and mitochondrial genomes. Genome Biol Evol. 2013;5(12):2318–2329. doi: 10.1093/gbe/evt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allen JF, de Paula WBM, Puthiyaveetil S, Nield J. A structural phylogenetic map for chloroplast photosynthesis. Trends Plant Sci. 2011;16(12):645–655. doi: 10.1016/j.tplants.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 92.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283(5407):1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 93.Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 94.Burger G, Lang BF, Reith M, Gray MW. Genes encoding the same three subunits of respiratory complex II are present in the mitochondrial DNA of two phylogenetically distant eukaryotes. Proc Natl Acad Sci USA. 1996;93(6):2328–2332. doi: 10.1073/pnas.93.6.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lane N, Martin WF, Raven JA, Allen JF. Energy, genes and evolution: introduction to an evolutionary synthesis. Philos Trans R Soc Lond B Biol Sci. 2013;368(1622):20120253. doi: 10.1098/rstb.2012.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cohen-Bazire G, Sistrom WR, Stanier RY. Kinetic studies of pigment synthesis by non-sulfur purple bacteria. J Cell Physiol. 1957;49(1):25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 97.Cheng Z, et al. Activity of the tetrapyrrole regulator CrtJ is controlled by oxidation of a redox active cysteine located in the DNA binding domain. Mol Microbiol. 2012;85(4):734–746. doi: 10.1111/j.1365-2958.2012.08135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujita Y, Murakami A, Ohki K. Regulation of photosystem composition in the cyanobacterial photosynthetic system: The regulation occurs in response to the redox state of the electron pool located between the 2 photosystems. Plant Cell Physiol. 1987;28(2):283–292. [Google Scholar]
- 99.Melis A, Mullineaux CW, Allen JF. Acclimation of the photosynthetic apparatus to photosystem I or photosystem II light: Evidence from quantum yield measurements and fluorescence spectroscopy of cyanobacterial cells. Z Naturforsch C. 1989;44(1-2):109–118. [Google Scholar]
- 100.Archibald JM. One Plus One Equals One. Symbiosis and the Evolution of Complex Life. Oxford Univ Press; Oxford: 2014. [Google Scholar]
- 101.Margulis L. Symbiosis in Cell Evolution. W. H. Freeman; New York: 1981. [Google Scholar]
- 102.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14(3):255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 103.Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76(2):444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]