Significance
Primary aldosteronism (PA) represents the most common adrenal disease and cause of secondary hypertension. However, little is known regarding adrenal cellular origins. Recently, subcapsular aldosterone-producing cell clusters (APCCs) were observed in normal adrenals. We hypothesize that APCCs are a contributor to PA. Here, we characterized the APCC transcriptome and show that CYP11B2 expression is increased compared with the rest of the adrenal cortex. We also show that many APCCs harbor known aldosterone-producing adenoma (APA)-related ion channels/pumps (ATPase, Na+/K+ transporting, α1-polypeptide and calcium channel, voltage-dependent, L-type, α1D-subunit) mutations that stimulate CYP11B2 expression and aldosterone production. Importantly, the mutation spectrum seen in APCCs differs from that reported for APA. These results provide molecular support for APCC as a precursor of PA.
Keywords: primary aldosteronism, aldosterone, adrenal, somatic mutations, aldosterone-producing cell cluster
Abstract
Primary aldosteronism (PA) represents the most common cause of secondary hypertension, but little is known regarding its adrenal cellular origins. Recently, aldosterone-producing cell clusters (APCCs) with high expression of aldosterone synthase (CYP11B2) were found in both normal and PA adrenal tissue. PA-causing aldosterone-producing adenomas (APAs) harbor mutations in genes encoding ion channels/pumps that alter intracellular calcium homeostasis and cause renin-independent aldosterone production through increased CYP11B2 expression. Herein, we hypothesized that APCCs have APA-related aldosterone-stimulating somatic gene mutations. APCCs were studied in 42 normal adrenals from kidney donors. To clarify APCC molecular characteristics, we used microarrays to compare the APCC transcriptome with conventional adrenocortical zones [zona glomerulosa (ZG), zona fasciculata, and zona reticularis]. The APCC transcriptome was most similar to ZG but with an enhanced capacity to produce aldosterone. To determine if APCCs harbored APA-related mutations, we performed targeted next generation sequencing of DNA from 23 APCCs and adjacent normal adrenal tissue isolated from both formalin-fixed, paraffin-embedded, and frozen tissues. Known aldosterone driver mutations were identified in 8 of 23 (35%) APCCs, including mutations in calcium channel, voltage-dependent, L-type, α1D-subunit (CACNA1D; 6 of 23 APCCs) and ATPase, Na+/K+ transporting, α1-polypeptide (ATP1A1; 2 of 23 APCCs), which were not observed in the adjacent normal adrenal tissue. Overall, we show three major findings: (i) APCCs are common in normal adrenals, (ii) APCCs harbor somatic mutations known to cause excess aldosterone production, and (iii) the mutation spectrum of aldosterone-driving mutations is different in APCCs from that seen in APA. These results provide molecular support for APCC as a precursor of PA.
Primary aldosteronism (PA) accounts for 8% of hypertension and is the most common adrenal disease (1–4). PA patients can be classified into those with aldosterone-producing adenomas (APAs), idiopathic hyperaldosteronism, or familial hyperaldosteronism (FH), which is further divided into FH types 1–3 (FHI–FHIII) (5). In 1992, FHI was shown to result from a gene fusion of cytochrome P450, family 11, subfamily B, polypeptide 2 (CYP11B2: aldosterone synthase) and cytochrome P450, family 11, subfamily B, polypeptide 1 (CYP11B1; cortisol synthase) that resulted in zona fasciculata (ZF) expression of CYP11B2 and excess aldosterone production (6). For almost two decades after the original report, no other genetic abnormalities were identified in the other forms of PA.
First reported in 2011, exome sequencing identified a series of germ-line and somatic mutations in genes that altered adrenal cell intracellular Ca2+ in PA. The most common mutations are somatic mutations of the gene encoding the potassium inwardly rectifying channel, subfamily J, member 5 (KCNJ5), which are found in at least 30% of APA (7–10). Germ-line KCNJ5 mutations were also identified as the cause of FHIII (7, 11). KCNJ5 mutations cause pathologic conductivity of Na+ ions, cell depolarization, and increased intracellular Ca2+, which results in CYP11B2 expression and aldosterone hypersecretion (7, 8, 11, 12). In addition to KCNJ5 mutations, ATPase, Na+/K+ transporting, α1-polypeptide (ATP1A1); ATPase, Ca2+-transporting, plasma membrane 3 (ATP2B3), and calcium channel, voltage-dependent, L-type, α1D-subunit (CACNA1D) mutations have been found in an additional ∼15% of APA (10, 13, 14). Like those in KCNJ5, mutations in ATP1A1, ATP2B3, and CACNA1D also increase adrenal cell intracellular Ca2+ and aldosterone production. Thus, in more than one-half of APA, the excess aldosterone production seems to relate to mutations in these four genes.
The pathophysiology of progression from normal adrenal to APA is not well-understood. However, the development of CYP11B2 antibodies enabled the identification of clusters of cells with increased expression of CYP11B2, which Nishimoto et al. (15) previously named aldosterone-producing cell clusters (APCCs). APCCs are CYP11B2-expressing nests of cells that are just below the adrenal capsule but protrude into cortisol-producing cells that are typically negative for CYP11B2 expression. These clusters or nests of cells, therefore, differ from the typical zonation seen in human and rodent adrenals [zona glomerulosa (ZG), ZF, and zona reticularis (ZR)]. Interestingly, APCCs are also frequently found in adrenal tissue adjacent to APA, despite the low circulating renin/angiotensin levels found in patients with APA, suggesting that APCC production of aldosterone is renin-independent (autonomous) (15–17). Although these studies suggest a role for APCCs in autonomous aldosterone production and potentially, PA, previous reports have been limited to immunohistochemical analysis.
Herein, we hypothesized that APCCs arise from ZG cells as a result of somatic mutations that result in renin-independent aldosterone production. If the hypothesis was true, then APCCs could consist of cells with APA-related somatic mutations. To test this hypothesis, we pursued microarray analysis to determine if the ZG and APCCs have similar transcriptomes and next generation sequencing (NGS) to determine if APCCs have APA-related mutations. We show that many APCCs harbor known APA-related ion channels/pumps (ATP1A1 and CACNA1D) mutations that stimulate CYP11B2 expression and aldosterone production.
Results
Transcriptome Comparison Between APCC, ZG, ZF, and ZR.
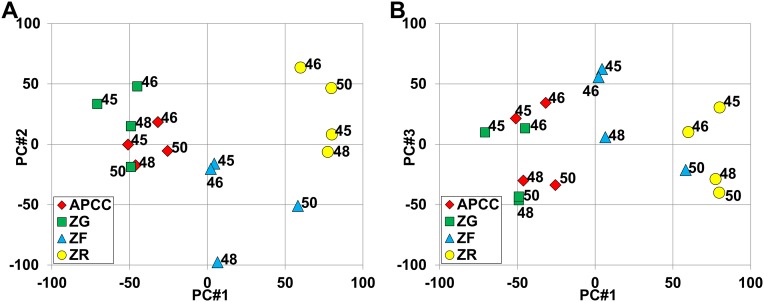
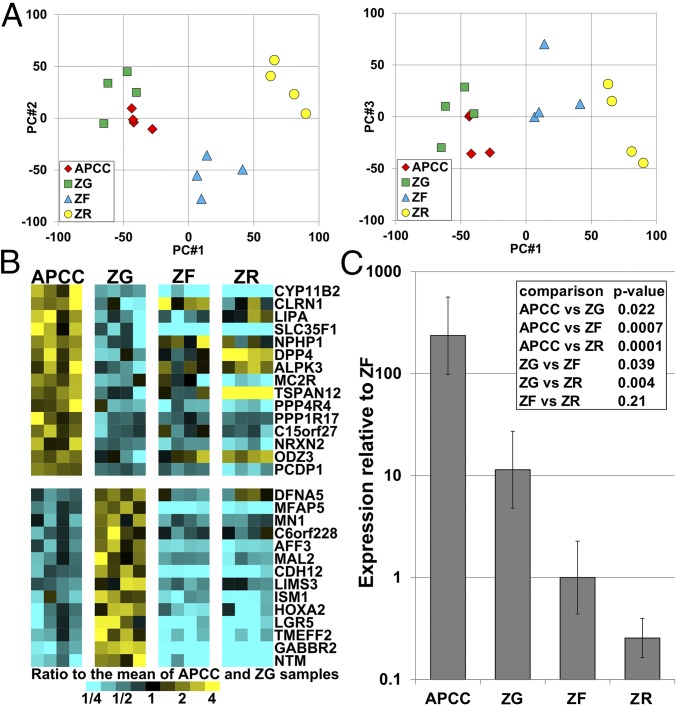
To analyze the transcriptome of APCCs, we compared RNA microarray data between APCC, ZG, ZF, and ZR. Laser capture microdissection (LCM) was used to acquire enriched populations of cells from each zone using frozen sections from four adrenal glands (marked M in Table 1). Because of limited source material, pools of APCC RNA were prepared from each adrenal gland. From these samples, RNA was isolated, amplified, labeled with biotin, and used for microarray analysis. To visualize overall transcriptome differences, we calculated principal components using all 54,675 probe sets, with the first three components shown in Fig. S1. The principal component analysis showed that the third component held considerable main effects caused by subjects (individual adrenal glands), and therefore, we elected to model subject effects as well as tissue effects in our subsequent statistical analysis of each probe set. Fig. 1A shows similar plots after estimated subject effects were removed for each probe set and indicates large differences between ZG, ZF, and ZR, with ZR being the most separated. APCCs were highly similar to ZG, with lower similarity to ZF samples.
Table 1.
Individual adrenal (DAN) sample information and aldosterone-producing cell cluster (APCC) score
| DAN no. | Ethnicity | Sex | Age (yr) | APCC score | |
| Mean | SE | ||||
| 2 | Caucasian | Male | 17 | 2.00 | 0.63 |
| 3 | Caucasian | Male | 17 | 3.00 | 0.00 |
| 7 | Caucasian | Female | 24 | 1.20 | 0.20 |
| 8 P | AA | Female | 54 | 4.00 | 0.00 |
| 9 | Caucasian | Male | 51 | 3.20 | 0.20 |
| 10 | Caucasian | Male | 59 | 1.80 | 0.20 |
| 11 P | Caucasian | Female | 45 | 4.60 | 0.24 |
| 12 | AA | Female | 33 | 2.00 | 0.63 |
| 13 | AA | Male | 40 | 1.60 | 0.24 |
| 14 | Caucasian | Female | 45 | 3.20 | 0.20 |
| 15 P | Caucasian | Female | 49 | 3.80 | 0.37 |
| 16 | Caucasian | Female | 49 | 3.00 | 0.00 |
| 17 P | AA | Male | 44 | 1.60 | 0.40 |
| 18 P | Caucasian | Female | 65 | 1.60 | 0.24 |
| 20 P | AA | N.A. | N.A. | N.A. | N.A. |
| 21 P | Caucasian | Female | 35 | 3.80 | 0.20 |
| 22 P | AA | Male | 49 | 1.80 | 0.20 |
| 23 | AA | Male | 49 | 0.00 | 0.00 |
| 24 | Hispanic | Male | 30 | 2.60 | 0.68 |
| 25 | Caucasian | Female | 52 | 1.20 | 0.37 |
| 26 | AA | Male | 58 | 3.60 | 0.40 |
| 27 | N.A. | N.A. | N.A. | 0.00 | 0.00 |
| 28 | Caucasian | Female | 43 | 1.40 | 0.24 |
| 29 | AA | Male | 59 | 1.40 | 0.40 |
| 30 | Caucasian | Male | 30 | 0.60 | 0.40 |
| 31 | AA | Male | 29 | 0.40 | 0.24 |
| 32 | Caucasian | Male | 17 | 0.80 | 0.20 |
| 33 | AA | Male | 43 | 0.00 | 0.00 |
| 34 | Caucasian | Male | 23 | 0.60 | 0.24 |
| 35 P | AA | Male | 38 | 3.00 | 0.00 |
| 36 | Caucasian | Male | 16 | 1.00 | 0.00 |
| 37 P | AA | Female | 43 | 3.20 | 0.20 |
| 40 P* | N.A. | N.A. | N.A. | 2.20 | 0.49 |
| 41 | Caucasian | Male | 31 | 1.60 | 0.51 |
| 43 | Caucasian | Female | 18 | 2.20 | 0.20 |
| 44 | Caucasian | Female | 54 | 1.60 | 0.24 |
| 45 M, F | Caucasian | Female | 37 | 0.80 | 0.20 |
| 46 M | Caucasian | Female | 63 | 2.80 | 0.37 |
| 47 | Caucasian | Male | 46 | 2.20 | 0.20 |
| 48 M | AA | Male | 39 | 2.00 | 0.00 |
| 49 | AA | Male | 43 | 2.00 | 0.00 |
| 50 M, F | AA | Male | 29 | N.A. | N.A. |
| 52 | Hispanic | Male | 28 | 0.60 | 0.40 |
| 53 | Hispanic | Male | 28 | 1.70 | 0.30 |
AA, African American; F, analyzed by NGS using frozen sections; M, analyzed by microarray; N.A., not available; P, analyzed by NGS using paraffin sections.
DAN40 yielded low library quality and was excluded from additional NGS analysis.
Fig. S1.
Principal component analysis of the microarray. Plots of the first three principal components [PC #1–3 (#1 vs. #2 for A and #1 vs. #3 for B)] using log2-transformed data for all probe sets. Subject numbers (DAN sample numbers) are labeled to show that PC3 indicates important subject effects (individual adrenals), with DAN samples 45 and 46 having greater PC3 than DAN samples 48 and 50 within every tissue.
Fig. 1.
APCC transcriptome comparison with adrenal ZG, ZF, and ZR. (A) Principal component analyses using microarray analysis after estimated subject effects were removed for four adrenal cell populations. Log2-transformed values are used for the graphs. PC, principal component. (B) Heat map of genes with a mean expression variation of greater than threefold between APCC and ZG (P < 0.01). Only probe sets annotated as representing a known gene are shown, and the probe set for each gene shown is the one with largest APCC vs. ZG fold change. (C) qPCR analysis of CYP11B2 in four adrenocortical tissues (APCC/ZG/ZF/ZR) from four subjects. P values are from two-way ANOVA models with terms for subjects and tissues. Error bars are SEMs.
Array data were analyzed using two-way ANOVA models with each probe set tested for differences between every pair of tissues. Probe sets with significantly different expression under different selection criteria are given in Table S1. Pairwise comparisons showed many probe sets differentially expressed, except for the APCC vs. ZG. At a significance of P < 0.01 and threefold change, only 39 probe sets were differentially expressed between APCC and ZG, and approximately one-half are expected to be false positives by permutation testing (Table S1). Strikingly, as shown in Fig. 1B, CYP11B2 was significantly higher in APCC compared with ZG (5.9-fold, P = 0.0008). Importantly, quantitative real-time PCR (qPCR) analysis confirmed increased CYP11B2 expression in APCC [mean (SE range) = 20.7 (12.0–28.7) fold] compared with ZG [1.0 (0.6–1.4), P = 0.02] (Fig. 1C). This is likely because of the increased dynamic range for qPCR expression analysis compared with microarray. The 39 probe sets that were elevated in APCCs represented 29 distinct annotated genes, the data for which are shown in Fig. 1B. We note that, for transcripts lower in APCC than ZG, expression was also lower in ZF than ZG. However, for transcripts that had elevated expression in APCC vs. ZG, most were also higher than either ZF or ZR. This also shows that the contamination of APCCs with ZF cells is likely low. Overall, our microarray and qPCR analyses expanded our understanding of the APCC transcriptome, including its increased capacity to produce aldosterone.
Table S1.
Pairwise comparisons between APCC, ZG, ZF, and ZR
| Comparison | Total (FDR) | |||||
| P < 0.01 | P < 0.001 | P < 0.01, FC > 1.3 | P < 0.01, FC > 2 | P < 0.01, FC > 3 | P < 0.001, FC > 2 | |
| P value from F test, people | 2,084 | 412 | ||||
| P value from F test, tissues | 4,088 | 1,213 | ||||
| APCC vs. ZG | 600 (0.80) | 53 (0.86) | 506 (0.78) | 131 (0.64) | 39 (0.47) | 23 (0.43) |
| APCC vs. ZF | 1,872 (0.26) | 430 (0.11) | 1,682 (0.24) | 767 (0.11) | 326 (0.06) | 278 (0.04) |
| APCC vs. ZR | 3,643 (0.13) | 1,177 (0.04) | 3,449 (0.11) | 1,867 (0.04) | 905 (0.02) | 867 (0.01) |
| ZG vs. ZF | 3,128 (0.16) | 770 (0.06) | 2,855 (0.14) | 1,158 (0.07) | 527 (0.04) | 457 (0.02) |
| ZG vs. ZR | 4,066 (0.12) | 1,447 (0.03) | 3,874 (0.10) | 2,281 (0.04) | 1,099 (0.02) | 1,077 (0.01) |
| ZF vs. ZR | 3,525 (0.14) | 715 (0.07) | 3,185 (0.13) | 973 (0.09) | 376 (0.05) | 339 (0.03) |
Two-way ANOVA models with terms for four tissues and four subjects were fit to each of 54,675 probe sets using log-transformed data. The number of probe sets qualifying for several selection criteria are given. The first two rows of numbers give the numbers of probe sets having P < 0.01 and P < 0.001 for the overall F tests that ask if there are differences between any of the subjects or tissues; naively, 546.75 and 54.675 would be expected by chance alone, respectively, and therefore, the people effects were important to include in the model. The last six rows show results for all pairwise comparisons of four groups of tissues. We also ask for a minimum fold change (FC) in either direction, where FC was computed as the antilogarithm of the difference between the means of the log-transformed values. False discovery rates (FDRs) are given in parentheses and were computed by analyzing 1,000 additional datasets that were like the actual data, except that the tissue labels within each person were randomly permuted (we excluded permuted datasets that gave the same groupings as the actual data). The FDR given is simply the average of the number of qualifying probe sets in the permuted datasets divided by the actual number that we obtained. This analysis shows that the data easily delivered many significant differences between all of the pairs of tissues, except for APCC vs. ZG, where even fairly stringent selection criteria do not give very small FDRs.
Age, Sex, and Race Associations with Adrenal APCCs.
Using CYP11B2 immunohistochemistry, we evaluated APCCs in 40 adrenals from 16 women and 24 men for association with age, sex, and ethnicity (two samples without available race, sex, and ethnicity data were excluded) (Table 1). APCC scoring was based on APCC size normalized to the adrenal surface length on each section (Fig. 2 A and B shows examples of small and large APCCs, respectively). Scores from five independent observers were averaged, with good agreement between individual observer scores and the average APCC score (correlations ranging from r = 0.86 to r = 0.91). APCC scores were greater in women (mean ± SD = 2.53 ± 1.18) than men (1.63 ± 1.00, P = 0.014, two-sample t test). There was no difference in APCC scores between 23 Caucasians (2.09 ± 1.14, 10 males), 14 African Americans (1.90 ± 1.24, 3 males), and 3 Hispanics (1.63 ± 1.00, 3 males; P = 0.77, one-way ANOVA). The disparity in APCC scores between males and females necessitated additional analysis. Multivariable models including both ethnicity and sex showed no observable difference between Caucasians and African Americans (P = 0.72). The overall correlation between APCC score and age was nonsignificant (r = 0.28, P = 0.08); however, in view of the sex difference, we fit multivariable models. The age by sex interaction was negligible (P = 0.92) (Fig. 2C), where the two lines are nearly parallel. In models with just age and sex, sex remained significant (P = 0.036), but the significance of age decreased (score increasing 0.014 per year, P = 0.23).
Fig. 2.
APCC score (frequency and size) during aging. (A) CYP11B2 immunohistochemistry from a normal adrenal (DAN22) showing an example of small APCCs (blue arrows). (B) CYP11B2 immunohistochemistry for DAN11 with examples of large APCCs (red arrows). (C) Scatter plot of average APCC score (from five observers) vs. age and sex of patient.
Mutation Analyses.
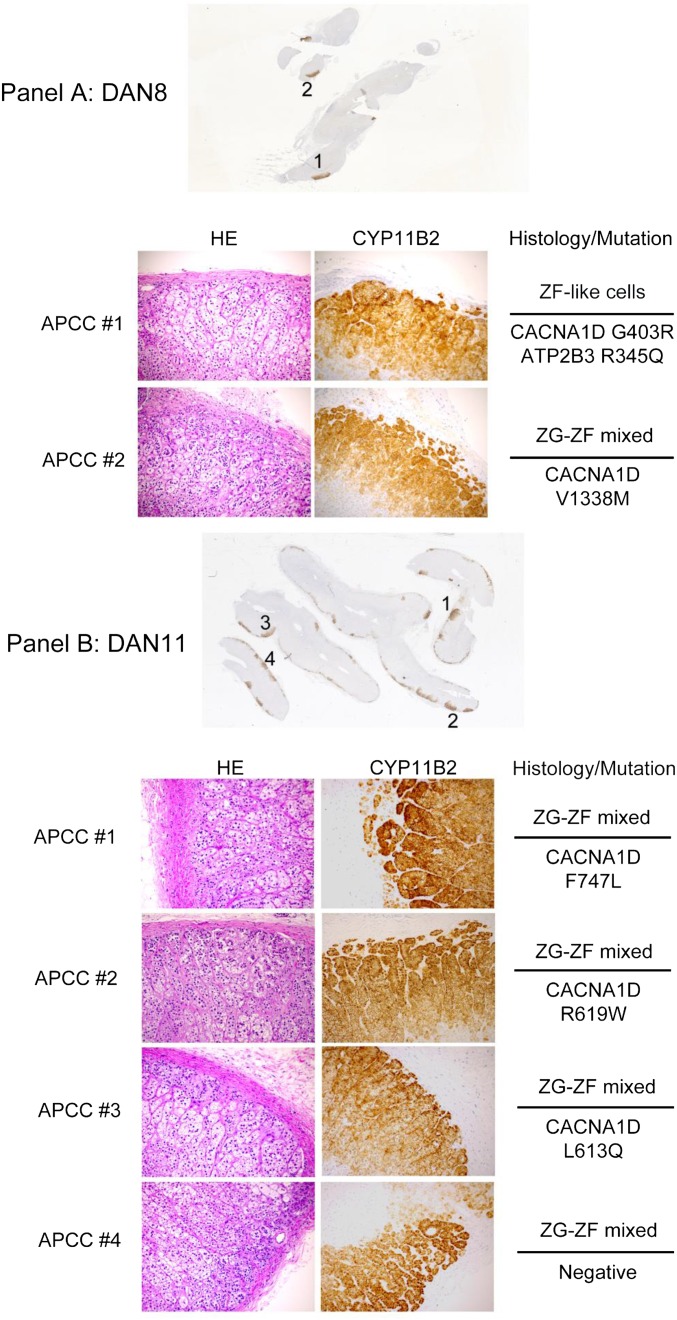
To assess whether APCCs harbored somatic mutations seen in APA, we developed a custom Ion Torrent AmpliSeq Panel (APA_v1) with 310 multiplexed amplicons targeting the entire coding sequences of genes with described somatic mutations in APA (ATP1A1, ATP2B3, CACNA1D, and KCNJ5) as well as genes shown to harbor germ-line or somatic variants associated with adrenal hyperplasia [phosphodiesterase 11A (PDE11A), phosphodiesterase 8B (PDE8B), and protein kinase, cAMP-dependent, regulatory, type Iα (PRKAR1A)] (7, 10, 13, 18–26). NGS was performed on APCCs and paired control adrenal tissue (adjacent ZF and/or medulla), both of which were isolated from formalin-fixed, paraffin-embedded (FFPE) adrenal samples (marked P in Table 1). APCCs were identified by CYP11B2 immunostaining and subsequently, isolated by macrodissection on intervening unstained FFPE sections (Fig. 3). In total, we macrodissected 22 APCCs and paired normal adrenal tissue from 11 cases (Table 2). These adrenals lacked overt pathology by histologic evaluation, and although two adrenals had small micronodules, these lacked CYP11B2 and did not contain APCCs used for assessment.
Fig. 3.
Identification of APCCs in FFPE tissues for targeted NGS (only mutated samples are shown). Six consecutive 5-µm FFPE sections were cut from blocks containing histologically benign adrenal glands. APCCs were identified after CYP11B2 immunohistochemistry of the first and last sections. Careful macrodissection was performed on intervening sections to isolate APCCs or adjacent normal adrenal tissue. Boxed areas indicate the APCC or normal tissue regions that were isolated. For each case with an identified somatic mutation (Table 2), APCCs and normal adrenal tissue subjected to sequencing are indicated. (Scale bar: 1 cm.)
Table 2.
Sequencing statistics for APCC NGS
| Sample | Mapped reads | Mean depth | Uniformity (%) | On target (%) | Variants |
| FFPE samples | |||||
| DAN8 | |||||
| N | 925,991 | 186 | 44 | 12 | 190 |
| 1 | 1,404,546 | 910 | 60 | 23 | 223 |
| 2 | 1,806,327 | 1,248 | 62 | 24 | 166 |
| DAN11 | |||||
| N | 1,370,920 | 1,860 | 72 | 43 | 106 |
| 1 | 1,575,078 | 1,752 | 69 | 36 | 104 |
| 2 | 1,486,679 | 1,108 | 69 | 26 | 133 |
| 3 | 1,097,462 | 986 | 62 | 30 | 97 |
| 4 | 1,247,491 | 1,343 | 67 | 35 | 91 |
| DAN15 | |||||
| N | 1,273,712 | 1,680 | 62 | 42 | 113 |
| 1 | 1,109,118 | 893 | 61 | 27 | 145 |
| 2 | 1,098,406 | 599 | 61 | 20 | 166 |
| DAN17 | |||||
| N | 825,723 | 84 | 24 | 14 | 222 |
| 1 | 593,981 | 66 | 54 | 8 | 160 |
| DAN18 | |||||
| N | 637,288 | 273 | 70 | 16 | 127 |
| 1 | 563,252 | 40 | 44 | 8 | 173 |
| DAN20 | |||||
| N | 695,115 | 230 | 64 | 13 | 115 |
| 1 | 1,983,659 | 2,816 | 62 | 44 | 103 |
| 2 | 1,535,371 | 1,601 | 63 | 34 | 128 |
| 3 | 1,356,271 | 1,015 | 65 | 26 | 119 |
| DAN21 | |||||
| N | 873,390 | 344 | 60 | 15 | 129 |
| 1 | 206,339 | 48 | 9 | 39 | 140 |
| 2 | 796,601 | 41 | 45 | 6 | 131 |
| 3 | 884,877 | 49 | 46 | 6 | 158 |
| DAN22 | |||||
| N | 929,125 | 232 | 67 | 10 | 125 |
| 1 | 1,508,814 | 1,772 | 61 | 38 | 127 |
| DAN35 | |||||
| N | 2,119,384 | 2,585 | 63 | 39 | 116 |
| 1 | 1,350,485 | 1,330 | 62 | 33 | 124 |
| 2 | 2,369,804 | 781 | 56 | 15 | 236 |
| DAN37 | |||||
| N | 1,025,881 | 973 | 69 | 32 | 108 |
| 1 | 689,170 | 84 | 54 | 8 | 153 |
| 2 | 891,467 | 156 | 51 | 10 | 181 |
| LCM samples | |||||
| DAN45 | |||||
| N | 315,051 | 934 | 75 | 98 | 55 |
| 1 | 85,826 | 240 | 71 | 96 | 65 |
| DAN50 | |||||
| N | 51,232 | 157 | 86 | 99 | 46 |
| 1 | 52,588 | 161 | 86 | 99 | 40 |
Sequencing statistics are provided for each FFPE and LCM isolated APCC (numbered) and matched normal sample (N) from donor adrenal samples (DAN) subjected to NGS. The number of called variants before any filtering are shown. Mapped reads, number of mapped reads per sample; Mean depth, average read coverage depth over targeted bases; On target, percentage of reads on target; Uniformity, uniformity of mapped reads; Variants, number of variants.
NGS of barcoded libraries prepared with the APA_v1 Panel was performed on a single Ion Proton P1 Chip (Life Technologies), generating an average of 1,155,842 mapped reads with an average of 849 times coverage depth over targeted bases per sample (Table 2). Average uniformity and on target reads (58% and 23%, respectively) were lower than observed with typical AmpliSeq libraries, consistent with the use of FFPE-derived DNA amplified with additional cycles needed for the low-input DNA amount. We have previously confirmed that point mutations observed with 20 ng FFPE DNA can also be defined using as little as 600 pg FFPE DNA and additional amplification cycles using the AmpliSeq technology. Of 22 APCCs and 11 normal adrenal samples, only 1 [APCC from donor adrenal normal 40 (DAN40)] had a low-quality library that precluded assessment of variants; this case was excluded from all subsequent analyses.
Across 31 informative samples (APCCs and paired normal adrenal DNA of the FFPE cohort), we identified a total of 4,510 called sequence variants (3,058 in APCCs and 1,452 in paired normal tissue) (Table 2), of which 11 variants (i) passed rigorous filtering criteria and visual inspection and (ii) were exclusively present in APCCs and not normal adrenal tissues (Table 3). Of these 11 somatic mutations, 7 (64%) occurred at 1 of 31 residues previously reported as a somatic mutation in APA (7, 10, 13, 18–22, 24–26) (deletions with unique start and stop sites but spanning the same residues in ATP2B3 were considered as separate mutations) (Table 3, asterisk and Table S2). In addition to the above seven mutations previously reported as somatic in APA, we identified 4 well-supported somatic variants of unknown function in 21 APCCs (Fig. 3 and Table 3). We performed logistic regression analysis of mutation status of the samples with age, race, sex, and average APCC score. There were no significant associations, but a trend was seen for higher APCC score in samples with detected mutations (P = 0.087).
Table 3.
Somatic nonsynonymous mutations identified in APCCs
| Age/sex/race | Sample | Cohort | Type | Gene | Reference allele | Variant allele | Amino acid change | FAO | FDP | Variant allele frequency (FAO/FDP; %) | Variant allele frequency in matched normal |
| 54/F/C | APCC 8–1* | FFPE* | APCC* | CACNA1D* | G* | C* | p.G403R* | 540* | 1,530* | 35* | 2* |
| 54/F/C | APCC 8–1 | FFPE | APCC | ATP2B3 | G | A | p.R345Q | 97 | 453 | 21 | 0 |
| 54/F/C | APCC 8–2* | FFPE* | APCC* | CACNA1D* | G* | A* | p.V1338M* | 28* | 91* | 31* | N.A.*† |
| 45/F/C | APCC 11–1* | FFPE* | APCC* | CACNA1D* | T* | C* | p.F747L* | 308* | 1,762* | 17* | 0* |
| 45/F/C | APCC 11–2 | FFPE | APCC | CACNA1D | C | T | p.R619W | 293 | 1,993 | 15 | 0 |
| 45/F/C | APCC 11–3 | FFPE | APCC | CACNA1D | T | A | p.L613Q | 440 | 1,934 | 23 | 0 |
| 49/F/C | APCC 15–1* | FFPE* | APCC* | ATP1A1* | C* | G* | p.L104V* | 165* | 1,416* | 12* | 0* |
| N.A. | APCC 20–1* | FFPE* | APCC* | ATP1A1* | T* | G* | p.V332G* | 235* | 1,986* | 12* | 1* |
| 38/M/AA | APCC 35–1* | FFPE* | APCC* | CACNA1D* | T* | G* | p.F747V* | 382* | 1,999* | 19* | 0* |
| 38/M/AA | APCC 35–2* | FFPE* | APCC* | CACNA1D* | G* | C* | p.G403R* | 112* | 770* | 15* | 0* |
| 43/F/AA | APCC 37–1 | FFPE | APCC | ATP1A1 | A | T | p.M734L | 13 | 84 | 15 | 2 |
| 29/M/AA | APCC 50* | LCM* | APCC* | CACNA1D* | C* | G* | p.F747L* | 41* | 123* | 33* | 0* |
| 65/F/C | APCC 18–N | FFPE | Normal | PDE11A§ | G | A | p.R307X | 48 | 90 | 53 | N.A.‡ |
All high-confidence somatic nonsynonymous variants (Materials and Methods) identified in APCC are shown. The gene, reference and variant alleles, amino acid change, and read level information are shown. The variant allele frequency in the matched normal tissue is shown for comparison. AA, African American; C, Caucasian; F, female; FAO, flow-corrected variant allele-containing read; FDP, flow-corrected read depth; M, male; N.A., not available.
Variants affecting residues previously reported as somatically altered in APA.
No coverage in the paired normal tissue.
Variant occurred in paired normal tissue (no coverage in the paired APCC).
Truncating mutations in PDE11A have been reported to predispose to a variety of adrenal neoplasms (25).
Table S2.
Previously reported APA somatic mutations used to assess APCC mutation significance
| Protein symbol | CACNA1D | ATP2B3 | ATP1A1 | KCNJ5 |
| Reference sequence | NP_001122311 | NP_068768 | NP_000692 | NP_000881 |
| No. of amino acid residues | 2137 | 1173 | 1023 | 419 |
| Previously reported mutations | V259D | L424_V425del | G99R | W126R |
| Previously reported mutations | G403R | L425_V426del | F100_104del | E145G/Q/K |
| Previously reported mutations | S652L | V426_V427del | L104R | G151R |
| Previously reported mutations | L655P | V332G | I157del | |
| Previously reported mutations | Y741C | del960_963S | T158A | |
| Previously reported mutations | F747V/L | L168R | ||
| Previously reported mutations | I750M/F | |||
| Previously reported mutations | V979E | |||
| Previously reported mutations | L981N | |||
| Previously reported mutations | R990H | |||
| Previously reported mutations | A998I/V | |||
| Previously reported mutations | V1151F | |||
| Previously reported mutations | I1152N | |||
| Previously reported mutations | P1336R | |||
| Previously reported mutations | V1338M | |||
| Previously reported mutations | M1354I | |||
| Previously reported mutations | V1373M |
Of interest, one adrenal (DAN8) had two APCCs (8–1 and 8–2) with unique, somatic, previously reported aldosterone-driving mutations. APCC 8–1 had a well-supported CACNA1D G403R somatic variant, whereas APCC 8–2 had a well-supported CACNA1D V1338M variant (Table 3) (10, 13, 22). Further supporting their somatic nature, neither variant was detected at significant frequency in the other DAN8 APCC or adjacent normal tissue (no coverage of V1338M was present in the matched normal tissue).
In addition to the variants passing stringent filtering as just described, two additional previously reported APA variants and a deleterious mutation in PDE11A were observed in APCCs at variant allele frequencies between 5% and 10% in APCCs (PDE11A Y137X in APCC 15–2 and CACNA1D R990H in APCCs 20–1 and 20–3). These variants may represent somatic events in samples with low-purity APCC content given the challenges of macrodissection; however, they did not pass stringent filtering and were not considered further.
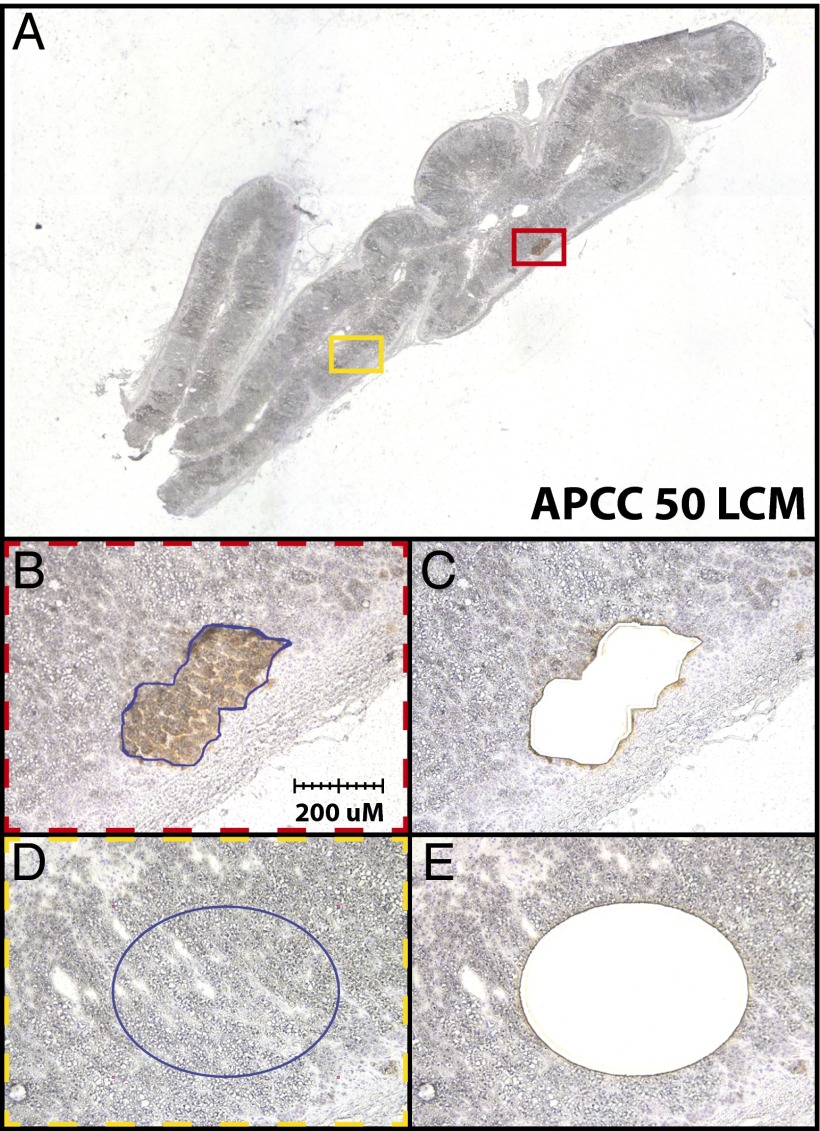
To further examine APCCs for known APA somatic mutations, we used the same NGS strategy on two cases of paired APCCs and normal tissue (ZF) isolated by LCM from fresh frozen adrenal tissue (DAN45 and -50) (F in Table 1). Using the APA_v1 Panel and sequencing on a single-ion Torrent 318 Chip on the Ion Personal Genome Machine Sequencer, we generated an average of 126,174 mapped reads and an average coverage depth of 373 times over targeted bases per sample (Table 2). Average uniformity and % on target reads (98% and 80%, respectively) were improved compared with the FFPE-derived DNA described above, likely because of isolation from non-FFPE tissue (Table 2). Across four samples, we identified a total of 206 called sequence variants (105 in APCCs and 101 in paired normal tissue) (Table 1), of which only a single variant passed all filtering criteria, which was not present in the paired normal adrenal tissue. In the APCC isolated from DAN50, we identified an F747L mutation in CACNA1D, which was not present in the paired normal adrenal DNA (Fig. 4 and Table 3). This sequence variant has previously been reported in APA as a somatic mutation resulting in Ca2+ influx (10, 13). The high frequency for this mutation in the APCC (an estimated 66% of cells) is consistent with it being a clonal event.
Fig. 4.
LCM of APCC for targeted NGS. APCCs from DAN45 and -50 were identified by CYP11B2 immunostaining on frozen tissue sections for NGS. (A) Low-magnification view of DAN50 with the regions captured for APCC 50 (red box) and paired normal adrenal tissue (yellow box) shown. (B–E) Photomicrographs showing (B and D) pre- and (C and E) post-LCM images that confirm isolation of desired cell populations. A somatic CACNA1D F747L mutation was identified exclusively in the APCC component (Table 2).
Given that 4,752 residues were targeted by the APA_v1 Panel in four genes previously reported as somatically mutated in APA, the fact that 8 of 12 observed APCC somatic mutations across the FFPE and LCM cohorts occurred at 31 residues previously reported as somatically altered in APA is consistent with significant overrepresentation at these residues (P = 1.6E-15, binomial test). Taken together, our sequencing of APCCs from FFPE and LCM tissues shows that APCCs can harbor acquired mutations in genes affecting aldosterone production as seen in APA.
There have been several reports that find cellular histologic correlations with aldosterone-stimulating mutations (9, 13, 17, 27). Therefore, we examined the cell histology of APCC samples that were captured for sequence analysis (Fig. S2). The majority of samples (76%) had a histology that included a mixture of ZG and ZF cell types (Table S3). Indeed, many of the APCCs exhibited a zonation pattern, with ZG-like cells closest to the capsule and ZF-like cells away from the capsule. However, as opposed to normal zonation, the ZF-like cells associated with the APCC retained CYP11B2 protein expression. Only five APCCs had a single-histologic cell makeup (either ZG- or ZF-like). Genetically, the ZF-like APCC phenotype carried mutations in both CACNA1D and ATP2B3 (DAN8#1). The two APCCs with a ZG-like histology carried mutations in ATP1A1 (DAN15#1 and DAN20#1), and two were negative for the mutations examined with our panel (DAN18#1 and DAN21#3). All other APCCs had a histology that included both ZG- and ZF-like cell types.
Fig. S2.
(Continued)
Table S3.
Cell morphology and mutation status of sequenced APCCs
| DAN no. and APCC no. | Histology | Mutation | ||
| ZG–ZF mixed | ZG-like | ZF-like | ||
| DAN8 | ||||
| 1 | + | CACNA1D (G403R),* ATP2B3 (R345Q) | ||
| 2 | + | CACNA1D (V1338M)* | ||
| DAN11 | ||||
| 1 | + | CACNA1D (F747L)* | ||
| 2 | + | CACNA1D (R619W) | ||
| 3 | + | CACNA1D (L613Q) | ||
| 4 | + | Negative | ||
| DAN15 | ||||
| 1 | + | ATP1A1 (L104V)* | ||
| 2 | + | Negative | ||
| DAN17 | ||||
| 1 | + | Negative | ||
| DAN18 | ||||
| 1 | + | Negative | ||
| DAN20 | ||||
| 1 | + | ATP1A1 (V332G)* | ||
| 2 | + | Negative | ||
| 3 | + | Negative | ||
| DAN21 | ||||
| 1 | + | Negative | ||
| 2 | + | Negative | ||
| 3 | + | Negative | ||
| DAN22 | ||||
| 1 | + | Negative | ||
| DAN35 | ||||
| 1 | + | CACNA1D (F747V)* | ||
| 2 | + | CACNA1D (G403R)* | ||
| DAN37 | ||||
| 1 | + | ATP1A1 (M734L) | ||
| 2 | + | Negative | ||
Discussion
Recently developed antibodies to CYP11B2 have been used to identify APCCs in human adrenal glands. However, other than increased CYP11B2 expression, little is known regarding these cell clusters. In this study, we expanded our understanding of APCCs using histopathology, expression profiling, and targeted DNA sequencing. Our findings indicate that APCCs have a transcriptome phenotype that is similar to the adjacent ZG but also, that many APCCs harbor gene mutations previously shown to cause autonomous aldosterone production.
CYP11B2 immunohistochemistry of our human adrenal biorepository provides the first sizable analysis, to our knowledge, of APCCs in normal adrenals and the study of APCC association with age and sex. We observed a clear sex difference in APCC score, with higher APCC scores in adrenals from women. Generally, there is no sex difference in prevalence of APA (28, 29). Nonetheless, it has been reported that APA from women harbors KCNJ5 mutations with significantly higher rates (72%) than that from males (28%, P < 0.001) (22).
Transcriptome analysis of APCC and the three cortical zones indicated that APCC is highly similar to ZG compared with ZF or ZR. Of note, in addition to CYP11B2 (which we confirmed by qPCR), differentially expressed transcripts between APCC and ZG may have a role of APCC function. Interestingly, we did not see a decrease in DAB2 expression, which had previously been reported by Boulkroun et al. (30). One possible explanation for this discrepancy is our differences in the definition of APCC. Boulkroun et al. (30) had three categories for CYP11B2-expressing cell clusters that included foci, megafoci, and APCC. Foci and megafoci expressed CYP11B2 and DAB2, whereas APCCs were defined as clusters that expressed CYP11B2 but not DAB2. It is likely that APCCs from our study would include some CYP11B2-expressing foci and megafoci based on the criteria by Boulkroun et al. (30). With the detailed transcriptome analysis, we have identified additional potential protein markers for APCCs other than CYP11B2. For example, we found that SLC35F1, MC2R, and PPP4R4 also had significantly higher transcript expression in APCCs than in ZG or ZF/ZR (Fig. 1B). Although the specific function of SLC35F1 is unknown, given the known role of SLC35 transporter family members (SLC35A–SLC35F) in glucose transport, SLC35F1 may have a metabolic role in APCCs (31). The up-regulation of MC2R in APCCs is of interest, because this gene encodes the adrenocorticotropic hormone receptor, which regulates aldosterone production along with angiotensin II and serum K+ (32). The increased MC2R in APCCs may imply an enhanced role for adrenocorticotropic hormone in aldosterone production in APCCs. PPP4R4 (also known as PP4R4 or KIAA1622) binds to protein phosphatase 4, catalytic subunit (PPP4C), and the PP4R4–PPP4C complex has a role in the regulation of intracellular phosphorylation/dephosphorylation cycles, which may relate to aldosterone production in APCCs (33). Thus, up-regulated genes in APCCs might play a role in aldosterone production, which can be studied in future functional studies.
A key aspect of this study was our NGS analysis of a targeted panel of genes known for somatic mutation in APA. This approach is considerably more sensitive and requires less DNA for mutation detection than standard Sanger sequencing. NGS analysis identified several known and previously unreported variants as somatic acquired events in APCCs. Across 23 total informative APCC samples from FFPE and frozen tissues subjected to NGS, 8 (35%) harbored known aldosterone driver mutations identified in APA. Although insufficient DNA remained after NGS to confirm the presence of these variants by conventional Sanger sequencing, the enrichment of known APA somatic mutations at high variant frequencies in APCC strongly supports these same events as frequent alterations in APCC. Although isolating minute lesions can be challenging for macrodissection, our targeted NGS approach (which can be expanded with additional APA drivers as identified) is applicable to FFPE tissue cohorts. FFPE provides considerable utility for researchers interested in adrenal neoplasia that have archival collections of tissue blocks. Likewise, given rapid technological advances, we anticipate that exome or whole-genome sequencing from APCCs may be possible in the near future. Such approaches may illuminate drivers in ∼65% of APCCs that did not harbor candidate drivers by our panel-based approach.
Of interest, no well-supported somatic KCNJ5 mutations were observed in our APCCs subjected to NGS, although mutations in KCNJ5 occur in at least 30% of APAs (7, 8, 17, 21). Given that APCCs in our cohort harbored somatic mutations in CACNA1D (26%) and ATP1A1 (9%), at residues previously reported in APA, APCCs may represent a precursor population of cells that lead to APA with these mutations through unknown mechanisms. Alternatively, APCCs with KCNJ5 mutations may rapidly progress to APA, and hence, KCNJ5 mutations may only be rarely detected in APCCs. Assessment of adrenal cohorts with well-annotated clinical status from ethnically diverse groups with a range of adrenal pathology will be needed to further understand the mutation spectrum observed in APCCs and the relationship to APA.
Recently, PA was found to associate with germ-line mutations in armadillo repeat containing 5 (ARMC5) (34). To determine if such germ-line variants may predispose to APCC accumulation, APCC score, or mutation status of genes somatically altered in APA, we performed Sanger sequencing of the ARMC5 coding sequence on germ-line DNA isolated from five adrenals with the highest APCC scores (DAN8, -11, -15, -21, and -26). Only one adrenal (DAN15) had a nonsynonymous alteration (P507L), which was previously reported as benign based on in silico analysis (34). Hence, although our results do not support a role for germ-line variants in ARMC5 driving APCC accumulation, determining genetic associations with APCC development is an important area for future research. Although ARMC5 was not included in our NGS panel, it can be included in future panel iterations for evaluating additional cohorts.
Based on our findings from microarray sequencing and NGS, we propose that APCCs represent a precursor population of cells that can progress to APA. In this study, we confirmed that the majority of APCCs consist of subcapsular small ZG-like cells and inner lipid-rich large ZF-like cells (15). However, both of these ZG/ZF-like cells strongly express CYP11B2, consistent with autonomous aldosterone production in APCCs. Recurrent mutations observed in APA are known to cause aldosterone overproduction in in vitro experiments (8, 12); therefore, the existence of these mutations in APCCs supports autonomous aldosterone production. Of note, we identified distinct somatic mutations in individual APCCs from the same adrenal gland, which is in contrast to previously studied APAs that have been reported to harbor a single mutation driving aldosterone production. There are multiple ways in which APCCs might contribute to PA, which include APCC progression to APA as a result of the single-somatic mutation events described in this paper. Alternatively, expansion to APA may require second-hit mutations within the APCC that increase cell proliferation. Finally, some APCCs may be dead-end lesions that do not have the capability to progress to a macroscopic adenoma. Additional studies will be needed to track potential progression from APCC to APA, which will likely require assessment of incidentally resected adrenals with lesions intermediate between APCC and APA.
In summary, our study shows the presence of somatic mutations known to impact aldosterone synthesis in many adrenal APCCs. These mutations would explain the higher expression of CYP11B2 in APCCs and suggest that they would produce aldosterone in a renin-independent manner. The role played by APCCs in PA resulting from both unilateral (potentially as a precursor to APA) and bilateral adrenal aldosterone production warrants additional research.
Materials and Methods
Human Adrenal Samples.
All experimental procedures carried out in this study were reviewed and approved by the Institutional Review Boards of Georgia Reagents University and the University of Michigan. Human adrenal samples were obtained from 44 renal transplantation donors at Georgia Reagents University: DAN samples 2–53 (Table 1). Adrenal pieces of 3 mm were either fixed in 10% (vol/vol) formaldehyde (FFPE) or frozen in optimum cutting temperature compound (O.C.T. block; Sakura Finetek). Adrenal histology was evaluated by a board-certified Anatomic Pathologist with subspecialty expertise in endocrine pathology (T.J.G).
FFPE Sections for APCC Scoring and NGS.
Paraffin blocks of DAN samples without overt pathology by histologic analysis were used to prepare six 5-µm serial sections (FFPE slides 1–6) for immunohistochemistry and mutation analysis. For each adrenal sample, slides 1 and 6 were immunostained with a monoclonal mouse antibody selective to human CYP11B2 as previously reported (15, 35). The remaining sections (slides 2–5) were used for sample preparation for NGS (see below). CYP11B2-stained sections were used for independent blinded estimation of size and frequency of APCCs by K. Nishimoto, A.R.S., K. Nanba, W.E.R., and one additional adrenal researcher (Adina Turcu). APCC scoring was based on the following: a score of 0 was given to adrenals with no APCC; a score of 1 was given to adrenals with no large APCCs where the number of small APCCs per centimeter capsular length was less than 0.5; a score of 2 was given to adrenals with no large APCCs where the number of small APCCs per centimeter capsular length was greater than 0.5; a score of 3 was given to adrenals where large APCCs were found but the number of large APCCs per centimeter capsular length was less than 0.5; a score of 4 was given to adrenals where the number of large APCCs per centimeter capsular length was between 0.5 and 1; and a score of 5 was given to adrenals where the number of large APCCs per centimeter capsular length was greater than 1. Of note, large APCCs were defined as follows: ≥100 µm along the capsule length with a thickness of ≥20 µm (dimension at the farthest point perpendicular to the capsule). Small APCCs were defined as follows: APCCs that were smaller than the size criterion of a large APCC. For NGS, APCCs were identified by CYP11B2 immunostaining and subsequently isolated by macrodissection on intervening unstained FFPE sections (samples marked P in Table 1).
Frozen Sections for Microarray Analysis and NGS.
Frozen O.C.T blocks were cut into two sets of serial sections. One set was used for RNA microarray analysis (marked M in Table 1), whereas the other was used for isolating genomic DNA for NGS (marked F in Table 1).
Transcriptome Analysis Using Frozen Sections.
Frozen adrenal glands in O.C.T. compound from DAN samples 45, 46, 48, and 50 (marked M in Table 1) were cut into 7-µm sections and mounted onto Superfrost Plus Microscope Slides (Thermo Fisher Scientific). To recognize enriched populations of aldosterone-producing ZG or APCC, slides 1, 10, and 20 were immunostained for CYP11B2. For immunohistochemistry, sections were fixed with 100% acetone and incubated with primary/secondary antibodies without antigen retrieval. The remaining tissue sections (slides 2–9 and 11–19) were stained with cresyl violet and used for LCM as previously reported (36, 37). APCC and ZG cells were captured from CYP11B2-positive cells based on CYP11B2-stained sections (slides 1, 10, and 20). ZF and ZR were captured for transcriptome comparison from lipid-rich cells in the middle layer and compact cells outside of the medulla, respectively. RNA from APCC, ZG, ZF, and ZR cells was isolated using a PicoPure RNA Isolation Kit (Molecular Devices). Total RNA (1–10 ng) from APCC, ZG, ZF, and ZR samples was submitted to the University of Michigan DNA Sequencing and Microarray Cores for reverse transcription and amplification using the Ovation Pico WTA System V2 (NuGEN Technologies). The cDNA was purified using the QIAquick PCR Purification Kit (Qiagen) and biotin-labeled using the Encore Biotin Module (NuGEN Technologies) followed by hybridization to the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix). This array was designed to interrogate 21,702 genes, including 18,802 distinct unambiguous genes on the human genome with 54,675 probe sets comprised of 11 perfect match probes per probe set. Expression values for each probe set were calculated using a robust multiarray average method (38). We fit two-way ANOVA models with terms for four tissues (APCC, ZG, ZF, and ZR) and four subjects (DAN samples 45, 46, 48, and 50) to log-transformed data for each probe set and used the resulting F tests to compare tissue pairs. Results of all probe sets on the array are available as GEO accession no. GSE68889.
qPCR for CYP11B2.
Residual RNA, which was prepared in the transcriptome analysis, was used for confirmation qPCR. This RNA was again reverse-transcribed to cDNA and amplified as described above. For qPCR, 1 ng prepared cDNA was mixed with Fast Universal PCR Master Mix (Applied Biosystems) and TaqMan primer/probe mix specific for CYP11B2 as previously reported (39). TaqMan primer/probe mix for peptidylprolyl isomerase A (cyclophilin A) transcript was purchased from Applied Biosystems and used for normalization. The delta–delta threshold cycle (ΔΔCt) method was used to calculate fold changes in expression (40).
NGS of Adrenal DNA.
DNA for NGS to identify mutations was isolated using two methods: manual macrodissection from FFPE sections and LCM from frozen sections of O.C.T.-embedded adrenals.
For FFPE samples, macrodissection was accomplished using a scalpel on intervening unstained sections (FFPE slides 2–5) by localizing APCCs using CYP11B2-stained slides (FFPE slides 1 and 6) from 11 DAN cases with large APCCs (marked P in Table 1). In each case, an area of normal adrenal cortex and/or medulla ∼10 times the size of the largest isolated APCC was similarly dissected. DNA was isolated using the Allprep FFPE DNA/RNA Kit (Qiagen) according to the manufacturer’s instructions. The isolation protocol was modified by extending the xylene incubation to 5 min and the centrifugation during deparaffinization to 5 min and eluting in a volume of 20 µL. DNA was quantified using the Qubit 2.0 Fluorometer (Life Technologies). If APCC DNA was not sufficient for quantification, the quantity was estimated at 1/10th the quantity in the matched normal tissue.
For each sample, 4.2 µL isolated DNA (containing an estimated 0.7–7.0 ng) was used for barcoded library generation by multiplexed PCR using a custom Ion AmpliSeq Panel and the Ion AmpliSeq Library Kit 2.0 (Life Technologies) according to the manufacturer’s instructions, except that 30 amplification cycles were used. The custom Ion AmpliSeq Panel was designed to target genes previously shown to be mutated in APA or other adrenal hyperplasias/neoplasms (APA_v1 Panel). The APA_v1 Panel contains 310 independent pairs of forward and reverse primers targeting the entire coding regions of ATP1A1, ATB2B3, KCNJ5, and CACNA1D as well as genes shown to harbor germ-line or somatic variants associated with adrenal hyperplasia (PDE11A, PDE8B, and PRKAR1A) (7, 10, 13, 18–26). Templates were prepared using the Ion PI Template OT2 200 Kit v2 on the Ion One Touch 2 according to the manufacturer’s instructions (Life Technologies). NGS of multiplexed templates was performed on an Ion Proton P1 Chip using the Ion PI Sequencing 200 Kit v2 according to the manufacturer’s instructions on the Ion Proton Sequencer (Life Technologies).
Data analysis was performed in Torrent Suite 4.0 essentially as described (41), with alignment by the Torrent Mapping Alignment Program (version 4.0; Life Technologies) using default parameters and variant calling by the Torrent Variant Caller plugin (version 4.0) using default low-stringency somatic variant settings. Variants were annotated using Annovar (42). Called variants were filtered to identify potential driving somatic mutations by removing synonymous or noncoding variants and those with frequencies >0.01 in normal populations from Exome Sequencing Project 6500 or 1000 Genomes, flow-corrected read depths <50, flow variant allele-containing reads <10, variant allele fractions (flow variant allele-containing reads/flow-corrected read depths) <0.10, or flow variant allele calling forward to reverse read ratio <0.2 or >5. Variants occurring exclusively in reads with other variants [single-nucleotide variants or indels (insertion or deletion of bases) or those occurring in the last mapped base of a read] were excluded. Variants passing filtering in APCCs that were called as variants (regardless of filtering status) in any normal tissue were considered germ line/artifacts and excluded from additional analysis unless occurring at a previously reported residue associated with APA. These filtering criteria are more stringent than our previously validated criteria for calling single-nucleotide/indel variants from AmpliSeq data (41, 43). All somatic APCC variants were visually confirmed in Integrative Genomics Viewer (Broad Institute), and paired normal samples were inspected in Integrative Genomics Viewer to confirm lack of substantial read support for the called variant.
Frozen sections from DAN samples 45 and 50 (marked F in Table 1) were cut onto Membrane Slides PEN-Membrane (Leica) at 7-µm thickness. All of these sections were immunostained for CYP11B2 as previous reported (15, 44) with the following modifications. Sections were fixed with 70% ethanol and incubated with primary/secondary antibodies without antigen retrieval. Populations of APCC cells were laser-captured using the Leica LMD 600 from CYP11B2-stained sections. ZF was captured as a control from lipid-rich cells below the captured ZG cells. DNA was isolated using the Pico Pure DNA Extraction Kit (Thermo Fisher) according to the manufacturer’s instruction. NGS was performed on 9 µL isolated DNA per sample for barcoded library generation by multiplexed PCR using APA_v1 as described above, except that 27 amplification cycles were used. Templates were prepared using the Ion Personal Genome Machine Template OT2 Kit v2 (Life Technologies) according to the manufacturer’s instructions. Sequencing of multiplexed templates was performed using the Ion Torrent Personal Genome Machine (Life Technologies) on an Ion 314 Chip using the Ion Personal Genome Machine 200 Sequencing Kit v2 (Life Technologies) according to the manufacturer’s instructions. Data analysis was performed as described above.
Sanger Sequencing of ARMC5.
Bidirectional Sanger sequencing of ARMC5 coding sequence was performed from germ-line DNA using previously reported primer sequences (45).
Acknowledgments
S.A.T. is supported by the A. Alfred Taubman Medical Research Institute. S.A.T. and R.K. are supported by the National Cancer Institute Grant CA46592 (to the Michigan Cancer Center Core). This work was also supported by fellowships from the Federation of National Public Service Personnel Mutual Aid Associations and the Tachikawa Hospital, Japan (to K. Nishimoto); National Heart, Lung, and Blood Institute Grant R01HL27255 (to C.E.G.-S.); American Heart Association Fellowship 14POST20020003 (to K. Nanba); and National Institutes of Diabetes and Digestive and Kidney Diseases Grant DK43140 (to W.E.R.).
Footnotes
Conflict of interest statement: S.A.T has a separate sponsored research agreement with Compendia Bioscience/Life Technologies. No part of the study described herein was supported by Compendia Bioscience/Life Technologies, and they had no role in the data collection, interpretation, or analysis, and did not participate in the study design or decision to submit for publication.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE68889).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505529112/-/DCSupplemental.
References
- 1.Hannemann A, Wallaschofski H. Prevalence of primary aldosteronism in patient’s cohorts and in population-based studies--a review of the current literature. Horm Metab Res. 2012;44(3):157–162. doi: 10.1055/s-0031-1295438. [DOI] [PubMed] [Google Scholar]
- 2.Husebye ES, et al. Consensus statement on the diagnosis, treatment and follow-up of patients with primary adrenal insufficiency. J Intern Med. 2014;275(2):104–115. doi: 10.1111/joim.12162. [DOI] [PubMed] [Google Scholar]
- 3.Lacroix A, Feelders RA, Stratakis CA, Nieman LK. Cushing’s syndrome. Lancet. May 21, 2015 doi: 10.1016/S0140-6736(14)61375-1. [DOI] [PubMed] [Google Scholar]
- 4.Speiser PW, et al. High frequency of nonclassical steroid 21-hydroxylase deficiency. Am J Hum Genet. 1985;37(4):650–667. [PMC free article] [PubMed] [Google Scholar]
- 5.Chao CT, et al. Diagnosis and management of primary aldosteronism: An updated review. Ann Med. 2013;45(4):375–383. doi: 10.3109/07853890.2013.785234. [DOI] [PubMed] [Google Scholar]
- 6.Lifton RP, et al. Hereditary hypertension caused by chimaeric gene duplications and ectopic expression of aldosterone synthase. Nat Genet. 1992;2(1):66–74. doi: 10.1038/ng0992-66. [DOI] [PubMed] [Google Scholar]
- 7.Choi M, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science. 2011;331(6018):768–772. doi: 10.1126/science.1198785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monticone S, et al. Effect of KCNJ5 mutations on gene expression in aldosterone-producing adenomas and adrenocortical cells. J Clin Endocrinol Metab. 2012;97(8):E1567–E1572. doi: 10.1210/jc.2011-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulkroun S, et al. KCNJ5 mutations in aldosterone producing adenoma and relationship with adrenal cortex remodeling. Mol Cell Endocrinol. 2013;371(1-2):221–227. doi: 10.1016/j.mce.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Scholl UI, et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet. 2013;45(9):1050–1054. doi: 10.1038/ng.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monticone S, et al. A novel Y152C KCNJ5 mutation responsible for familial hyperaldosteronism type III. J Clin Endocrinol Metab. 2013;98(11):E1861–E1865. doi: 10.1210/jc.2013-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oki K, Plonczynski MW, Luis Lam M, Gomez-Sanchez EP, Gomez-Sanchez CE. Potassium channel mutant KCNJ5 T158A expression in HAC-15 cells increases aldosterone synthesis. Endocrinology. 2012;153(4):1774–1782. doi: 10.1210/en.2011-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azizan EA, et al. Somatic mutations in ATP1A1 and CACNA1D underlie a common subtype of adrenal hypertension. Nat Genet. 2013;45(9):1055–1060. doi: 10.1038/ng.2716. [DOI] [PubMed] [Google Scholar]
- 14.Beuschlein F, et al. Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat Genet. 2013;45(4):440–444e2. doi: 10.1038/ng.2550. [DOI] [PubMed] [Google Scholar]
- 15.Nishimoto K, et al. Adrenocortical zonation in humans under normal and pathological conditions. J Clin Endocrinol Metab. 2010;95(5):2296–2305. doi: 10.1210/jc.2009-2010. [DOI] [PubMed] [Google Scholar]
- 16.Nanba K, et al. Histopathological diagnosis of primary aldosteronism using CYP11B2 immunohistochemistry. J Clin Endocrinol Metab. 2013;98(4):1567–1574. doi: 10.1210/jc.2012-3726. [DOI] [PubMed] [Google Scholar]
- 17.Monticone S, et al. Immunohistochemical, genetic and clinical characterization of sporadic aldosterone-producing adenomas. Mol Cell Endocrinol. 2015;411:146–154. doi: 10.1016/j.mce.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Åkerström T, et al. Comprehensive re-sequencing of adrenal aldosterone producing lesions reveal three somatic mutations near the KCNJ5 potassium channel selectivity filter. PLoS One. 2012;7(7):e41926. doi: 10.1371/journal.pone.0041926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Salameh A, Cohen R, Desailloud R. Overview of the genetic determinants of primary aldosteronism. Appl Clin Genet. 2014;7:67–79. doi: 10.2147/TACG.S45620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azizan EA, et al. Somatic mutations affecting the selectivity filter of KCNJ5 are frequent in 2 large unselected collections of adrenal aldosteronomas. Hypertension. 2012;59(3):587–591. doi: 10.1161/HYPERTENSIONAHA.111.186239. [DOI] [PubMed] [Google Scholar]
- 21.Dekkers T, et al. Adrenal nodularity and somatic mutations in primary aldosteronism: One node is the culprit? J Clin Endocrinol Metab. 2014;99(7):E1341–E1351. doi: 10.1210/jc.2013-4255. [DOI] [PubMed] [Google Scholar]
- 22.Fernandes-Rosa FL, et al. Genetic spectrum and clinical correlates of somatic mutations in aldosterone-producing adenoma. Hypertension. 2014;64(2):354–361. doi: 10.1161/HYPERTENSIONAHA.114.03419. [DOI] [PubMed] [Google Scholar]
- 23.Murthy M, Azizan EA, Brown MJ, O’Shaughnessy KM. Characterization of a novel somatic KCNJ5 mutation delI157 in an aldosterone-producing adenoma. J Hypertens. 2012;30(9):1827–1833. doi: 10.1097/HJH.0b013e328356139f. [DOI] [PubMed] [Google Scholar]
- 24.Rothenbuhler A, et al. Identification of novel genetic variants in phosphodiesterase 8B (PDE8B), a cAMP-specific phosphodiesterase highly expressed in the adrenal cortex, in a cohort of patients with adrenal tumours. Clin Endocrinol (Oxf) 2012;77(2):195–199. doi: 10.1111/j.1365-2265.2012.04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vezzosi D, et al. Phosphodiesterase 11A (PDE11A) gene defects in patients with acth-independent macronodular adrenal hyperplasia (AIMAH): Functional variants may contribute to genetic susceptibility of bilateral adrenal tumors. J Clin Endocrinol Metab. 2012;97(11):E2063–E2069. doi: 10.1210/jc.2012-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams TA, et al. Somatic ATP1A1, ATP2B3, and KCNJ5 mutations in aldosterone-producing adenomas. Hypertension. 2014;63(1):188–195. doi: 10.1161/HYPERTENSIONAHA.113.01733. [DOI] [PubMed] [Google Scholar]
- 27.Azizan EA, et al. Microarray, qPCR, and KCNJ5 sequencing of aldosterone-producing adenomas reveal differences in genotype and phenotype between zona glomerulosa- and zona fasciculata-like tumors. J Clin Endocrinol Metab. 2012;97(5):E819–E829. doi: 10.1210/jc.2011-2965. [DOI] [PubMed] [Google Scholar]
- 28.Loh KC, Koay ES, Khaw MC, Emmanuel SC, Young WF., Jr Prevalence of primary aldosteronism among Asian hypertensive patients in Singapore. J Clin Endocrinol Metab. 2000;85(8):2854–2859. doi: 10.1210/jcem.85.8.6752. [DOI] [PubMed] [Google Scholar]
- 29.Rossi E, et al. High prevalence of primary aldosteronism using postcaptopril plasma aldosterone to renin ratio as a screening test among Italian hypertensives. Am J Hypertens. 2002;15(10 Pt 1):896–902. doi: 10.1016/s0895-7061(02)02969-2. [DOI] [PubMed] [Google Scholar]
- 30.Boulkroun S, et al. Adrenal cortex remodeling and functional zona glomerulosa hyperplasia in primary aldosteronism. Hypertension. 2010;56(5):885–892. doi: 10.1161/HYPERTENSIONAHA.110.158543. [DOI] [PubMed] [Google Scholar]
- 31.Ishida N, Kawakita M. Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35) Pflugers Arch. 2004;447(5):768–775. doi: 10.1007/s00424-003-1093-0. [DOI] [PubMed] [Google Scholar]
- 32.Hattangady NG, Olala LO, Bollag WB, Rainey WE. Acute and chronic regulation of aldosterone production. Mol Cell Endocrinol. 2012;350(2):151–162. doi: 10.1016/j.mce.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen GI, et al. PP4R4/KIAA1622 forms a novel stable cytosolic complex with phosphoprotein phosphatase 4. J Biol Chem. 2008;283(43):29273–29284. doi: 10.1074/jbc.M803443200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zilbermint M, et al. Primary Aldosteronism and ARMC5 Variants. J Clin Endocrinol Metab. 2015;100(6):E900–E909. doi: 10.1210/jc.2014-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomez-Sanchez CE, et al. Development of monoclonal antibodies against human CYP11B1 and CYP11B2. Mol Cell Endocrinol. 2014;383(1-2):111–117. doi: 10.1016/j.mce.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimoto K, Harris RB, Rainey WE, Seki T. Sodium deficiency regulates rat adrenal zona glomerulosa gene expression. Endocrinology. 2014;155(4):1363–1372. doi: 10.1210/en.2013-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishimoto K, et al. Transcriptome analysis reveals differentially expressed transcripts in rat adrenal zona glomerulosa and zona fasciculata. Endocrinology. 2012;153(4):1755–1763. doi: 10.1210/en.2011-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 39.Ye P, Mariniello B, Mantero F, Shibata H, Rainey WE. G-protein-coupled receptors in aldosterone-producing adenomas: A potential cause of hyperaldosteronism. J Endocrinol. 2007;195(1):39–48. doi: 10.1677/JOE-07-0037. [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 41.Warrick JI, et al. Tumor evolution and progression in multifocal and paired non-invasive/invasive urothelial carcinoma. Virchows Arch. 2015;466(3):297–311. doi: 10.1007/s00428-014-1699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang X, Wang K. wANNOVAR: Annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDaniel AS, et al. HRAS mutations are frequent in inverted urothelial neoplasms. Hum Pathol. 2014;45(9):1957–1965. doi: 10.1016/j.humpath.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freedman BD, et al. Adrenocortical zonation results from lineage conversion of differentiated zona glomerulosa cells. Dev Cell. 2013;26(6):666–673. doi: 10.1016/j.devcel.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Assié G, et al. ARMC5 mutations in macronodular adrenal hyperplasia with Cushing's syndrome. N Engl J Med. 2013;369(22):2105–2114. doi: 10.1056/NEJMoa1304603. [DOI] [PMC free article] [PubMed] [Google Scholar]