Significance
Chronic inflammation is a significant risk factor for cancer and other human diseases. During chronic inflammation, cells exposed to neutrophil-derived hypochlorous acid accumulate in their genomes the DNA lesion 5-chlorocytosine (5ClC). Using a battery of chemical, structural, and genetic tools, the present study demonstrates that 5ClC is a mutagenic lesion, suggesting that genomic 5ClC accumulation could have very serious biological consequences. 5ClC induces C→T transitions, a type of mutation commonly observed in tissues under inflammatory stress as well as in the genomes of inflammation-driven cancers. Thus, the mutagenic properties of 5ClC represent an appealing molecular mechanism by which chronic inflammation induces the genetic changes that potentially enable and stimulate carcinogenesis.
Keywords: 5-chloro-deoxycytidine, hypochlorite, myeloperoxidase, inflammatory bowel disease, carcinogenesis
Abstract
During chronic inflammation, neutrophil-secreted hypochlorous acid can damage nearby cells inducing the genomic accumulation of 5-chlorocytosine (5ClC), a known inflammation biomarker. Although 5ClC has been shown to promote epigenetic changes, it has been unknown heretofore if 5ClC directly perpetrates a mutagenic outcome within the cell. The present work shows that 5ClC is intrinsically mutagenic, both in vitro and, at a level of a single molecule per cell, in vivo. Using biochemical and genetic approaches, we have quantified the mutagenic and toxic properties of 5ClC, showing that this lesion caused C→T transitions at frequencies ranging from 3–9% depending on the polymerase traversing the lesion. X-ray crystallographic studies provided a molecular basis for the mutagenicity of 5ClC; a snapshot of human polymerase β replicating across a primed 5ClC-containing template uncovered 5ClC engaged in a nascent base pair with an incoming dATP analog. Accommodation of the chlorine substituent in the template major groove enabled a unique interaction between 5ClC and the incoming dATP, which would facilitate mutagenic lesion bypass. The type of mutation induced by 5ClC, the C→T transition, has been previously shown to occur in substantial amounts both in tissues under inflammatory stress and in the genomes of many inflammation-associated cancers. In fact, many sequence-specific mutational signatures uncovered in sequenced cancer genomes feature C→T mutations. Therefore, the mutagenic ability of 5ClC documented in the present study may constitute a direct functional link between chronic inflammation and the genetic changes that enable and promote malignant transformation.
Chronic inflammation is an established risk factor for many human cancers (1–6). By generating reactive chemical entities (e.g., reactive oxygen, nitrogen, and halogen species) intended to neutralize invading pathogens, inflammatory responses may also inflict chemical collateral damage on nucleic acids and proteins in surrounding healthy tissues (7–10). When the immune response aberrantly persists over months and years, the chemical damage may exceed the repair capabilities of the exposed cells, leading to cell death or to genetic and epigenetic modifications that, in principle, can set the stage for malignant transformation (7–10).
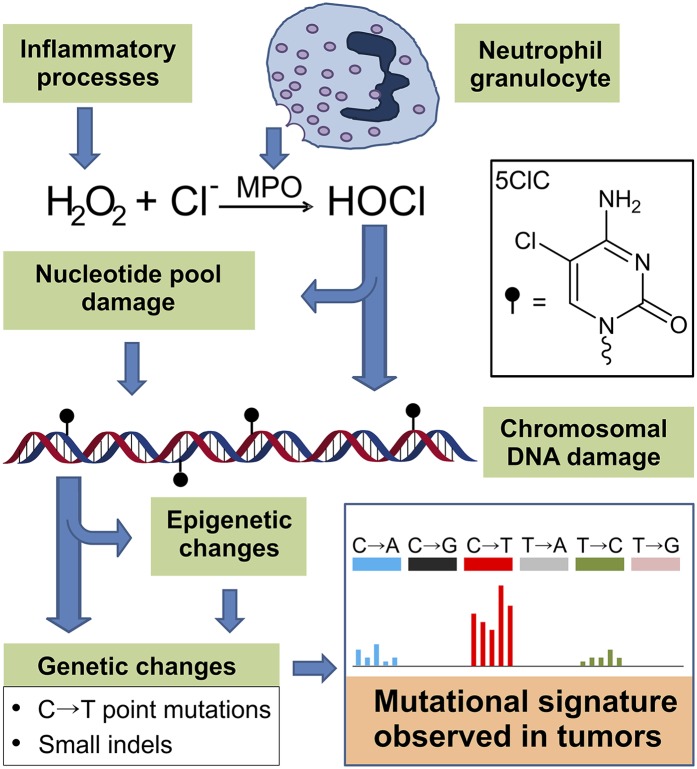
One type of chemical damage associated with inflammation is chlorination of cellular macromolecules by hypochlorous acid (HOCl). This type of damage is the result of the infiltration and activation of neutrophil granulocytes (the most abundant leukocyte), which produce myeloperoxidase, an enzyme capable of synthesizing HOCl from hydrogen peroxide and chloride anions (Fig. 1) (11–14). The main biomarkers found in cells exposed to HOCl are chlorinated tyrosines (in proteins) and chlorinated nucleobases (in nucleic acids and nucleotide pools) (15, 16). Among the chlorinated nucleobases, 5-chlorocytosine (5ClC) is the most abundant (16–19) and recently has been recognized as a biomarker associated with chronic inflammation (20, 21). In one study, the levels of 5ClC in both DNA and RNA were elevated in the colon of Rag2−/− mice infected with Helicobacter hepaticus, a model of chronic inflammation in the gut (20). In a follow-up study, the same biomarkers were found at similarly high levels in colonic tissue samples from human patients with inflammatory bowel disease (Crohn’s disease or ulcerative colitis) (21). Surprisingly, these studies did not find significantly increased levels of classical DNA biomarkers of inflammation and oxidative stress, such as 8-oxo-7,8-dihydroguanine (8oxoG) and other oxidative DNA lesions (20, 21). These findings, together with the observed persistence and accumulation of genomic 5ClC in chronically inflamed tissues suggested that 5ClC is not efficiently repaired and could have a functional role in disease initiation or progression.
Fig. 1.
A model of the 5ClC biomarker as a functional link between chronic inflammation and the genetic changes observed in tumors. During inflammation, activated neutrophil granulocytes produce the enzyme myeloperoxidase (MPO); other inflammatory processes produce hydrogen peroxide, among other reactive oxygen species. Using hydrogen peroxide and chloride, MPO catalyzes the formation of HOCl. Exposure of the surrounding cells to HOCl can generate 5ClC in their genomic DNA, a type of DNA damage occurring either by direct chlorination of cytosines or by the incorporation of damaged nucleotides from the pool. The present study showcases the mutagenic ability of 5ClC, which can directly induce C→T mutations. Additionally, 5ClC is known to induce epigenetic changes that could lead to the inactivation of tumor-suppressor genes such as MLH1 by hypermethylation, in turn possibly leading to additional mutagenic outcomes such as small indels and microsatellite instability. Together, the biological effects of 5ClC generate a characteristic mutational signature (the relative distribution of all possible point mutations and indels over all possible sequence contexts), which could account for certain mutational signatures found in malignancies associated with chronic inflammation. The mutational signature shown is a cartoon representation of signature 6 from Alexandrov et al. (56), highlighting the preponderance of C→T transitions.
So far, research on 5ClC has focused on the lesion’s ability to interact with DNA methyl transferases (22–24) when 5ClC is part of a CpG island; this interaction was shown to lead to changes in DNA methylation patterns (25). However, there are no reports affirming the ability of 5ClC to induce genetic changes directly during the replication or repair of genomic DNA. Knowledge of the mutagenicity of 5ClC would be especially relevant for the cells surrounding the inflammation site, which have both an increased level of DNA repair (e.g., removing the aforementioned oxidative stress lesions that do not persist in inflamed tissues) and a higher propensity to replicate (to compensate for the inflammation-induced cell death). Both of these processes put the cells in the inflamed tissues at an increased risk for progressing toward cancer, particularly if the 5ClC they accumulate is a mutagenic lesion.
In the present work, we have used cellular, biochemical, and structural approaches to show that 5ClC is indeed mutagenic, inducing C→T transitions at a significant rate; when the levels of genomic 5ClC are taken into account, the mutagenesis of 5ClC translates into an anticipated 1–2 orders of magnitude increase in the cellular mutation rate in the inflamed tissues. The molecular basis for the mutagenic property of 5ClC was elucidated by crystallizing human polymerase β (pol β) as it incorporated dGTP or dATP analogs opposite the lesion. In conjunction with the previously known epigenetic effects of 5ClC, the mutagenic properties of 5ClC establish this biomarker as a promising functional link between chronic inflammation and the genetic and transcriptional changes observed in inflammation-associated tumors (Fig. 1).
Results
5ClC Is a Mutagenic Base That Is Easily Bypassed by Polymerases in Vivo.
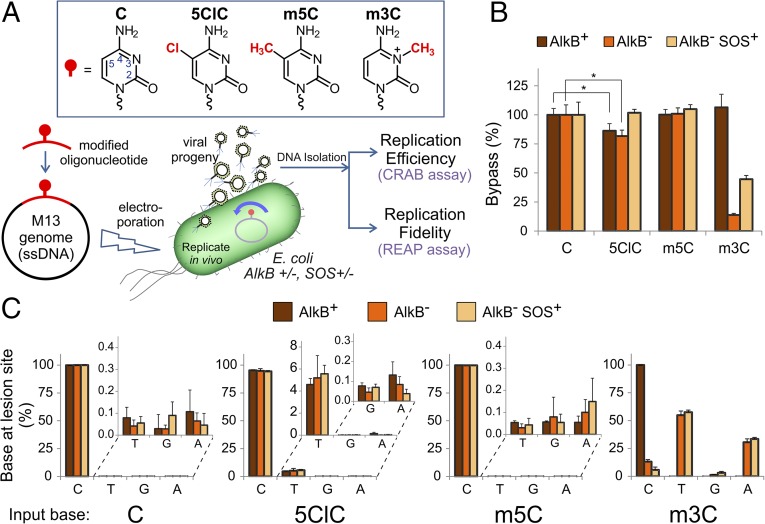
The mutagenic properties of 5ClC, as well as the lesion’s ability to block replication, were evaluated by introducing the lesion site-specifically on a viral vector, replicating the vector in living cells, and analyzing the resulting viral progeny DNA using the established REAP (Restriction Endonuclease and Postlabeling) and CRAB (Competitive Replication of Adduct Bypass) assays (26–28). The 5ClC nucleoside was incorporated site-specifically into a 16mer oligonucleotide, which subsequently was ligated into an M13 single-stranded DNA vector (Fig. 2A and Fig. S1). Similarly, control vectors were constructed containing the negative controls cytosine (C) and 5-methylcytosine (m5C) and the positive control 3-methylcytosine (m3C). The m3C-containing vector was used primarily to validate the assays: in the absence of the Fe(II)/α-ketoglutarate-dependent dioxygenase AlkB, a direct reversal DNA repair enzyme, m3C is a strong block to replication and leads to a characteristic mutational pattern, whereas in wild-type cells m3C is repaired efficiently and completely, thus behaving like an unmodified cytosine (27, 29). Additionally, m3C was used to confirm the successful induction of the SOS response (see below).
Fig. 2.
The biological consequences of 5ClC in vivo. (A) General strategy for characterizing the in vivo properties of 5ClC. Oligodeoxynucleotides containing 5ClC, the negative controls cytosine (C) or 5-methylcytosine (m5C), or the positive control 3-methylcytosine (m3C) were ligated into M13 genomes, which were then introduced into E. coli cells of different genetic backgrounds and allowed to replicate. DNA from the resulting viral progeny was analyzed to determine the replication efficiency (a measure of how easily the lesion is bypassed) and replication fidelity (the ability of the lesion to induce mutations). A detailed graphical overview of the methods is shown in Fig. S1. (B) In vivo bypass efficiency of 5ClC and controls. The colors correspond to the three E. coli strains used, as indicated. Each graphed value corresponds to the mean of three independent biological replicates; error bars indicate one SD. *P < 0.05. Data are also tabulated in Table S1. (C) The mutagenicity of 5ClC in vivo. Each graph shows the base composition at the lesion site in the progeny phage coming from the constructed genomes containing the indicated input base at the lesion site. Each graphed value corresponds to the mean of three independent biological replicates; error bars indicate one SD. Data are also tabulated in Table S2.
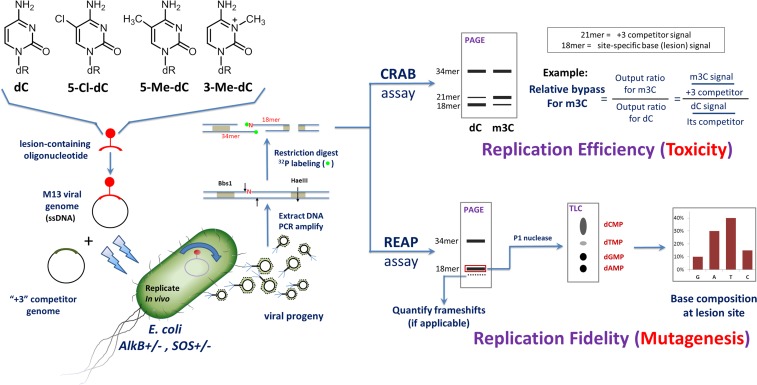
Fig. S1.
Outline of the in vivo experimental strategy. 16mer oligonucleotides containing 5ClC or controls at a defined site were used to construct M13 ssDNA viral genomes. The lesion-containing genomes were mixed with a “+3” competitor genome in a fixed ratio and were transformed into several strains of E. coli cells by electroporation. The resulting phage progeny were collected and reamplified. DNA was extracted from the phage progeny, and a region of interest was PCR amplified with assay-specific primers. Restriction endonucleases and 32P end-labeling were used to radiolabel an 18mer piece containing the lesion site at its 5′ end. The CRAB assay evaluated the ratio between the 18mer band (coming from the lesion-containing genome) and the 21mer band (given by the “+3” competitor genome) and normalized it to the equivalent ratio of the unmodified dC control to obtain the relative bypass percentage. The lower the bypass percentage, the stronger is the replication-blocking ability of the lesion and therefore its toxic effect on the cell. In the REAP assay, the 18mer oligonucleotide was isolated using PAGE and was digested with P1 nuclease to release the radioactive monophosphate nucleotide from its 5′ end. TLC separation and phosphorimagery then provided the identity and amount of the base at the lesion site.
Two strains of Escherichia coli AB1157 denoted AlkB+ (wild type) and AlkB− (AlkB deficient) were used to replicate the constructed M13 vectors. Additionally, an SOS-induced version of the AlkB− strain (denoted AlkB−SOS+) was used. The induction of the SOS response, accomplished by briefly exposing the cells to UV radiation, increases the levels of cellular Y-class translesion polymerases such as Pol IV and Pol V. Following in vivo replication, the progeny phage DNA was extracted and analyzed to determine for each lesion (or controls) and cell condition the percentage bypass (a measure of how readily a lesion is bypassed during replication) and the percentage base at the lesion site, from which the type and amount of mutagenesis can be determined (26). A detailed graphical overview of the method is shown in Fig. S1.
The bypass experiments revealed that 5ClC is a mild block to replication; bypass relative to cytosine was ∼75% in both wild-type and AlkB− strains and increased to ∼100% in the SOS-induced cell line (Fig. 2B and Table S1). The negative control m5C showed a level of bypass indistinguishable from that of cytosine in all cell strains investigated. The behavior of the positive control m3C was consistent with previous studies (27, 29): in the AlkB− strain, m3C was a very strong block to replication (15% bypass relative to dC); the amount of bypass increased with SOS induction to 35%, whereas in the wild-type strain, because of its efficient repair by AlkB, m3C showed a bypass of ∼100% (Fig. 2B and Table S1).
Table S1.
In vivo bypass efficiency of 5ClC and controls
| Input base | AlkB+ | AlkB− | AlkB− SOS+ |
| Cytosine | 100 ± 6 | 100 ± 9 | 100 ± 11 |
| 5ClC | 86 ± 6 | 82 ± 5 | 102 ± 3 |
| m5C | 100 ± 4 | 101 ± 5 | 105 ± 4 |
| m3C | 106 ± 11 | 14 ± 1 | 45 ± 3 |
Data are from Fig. 2B and are shown as % average ± SD.
The mutagenesis assay revealed that 5ClC is a mutagenic lesion, coding as a C ∼95% of the time and as a T ∼5% of the time, in all three cell strains investigated (Fig. 2C and Table S2). This result therefore suggests that 5ClC base-paired with guanine most of the time (∼95%), but it also base-paired with adenine (∼5%), an unexpected result. To put this value in perspective, 5ClC is approximately as mutagenic as 8oxoG, evaluated with a similar assay in a wild-type E. coli strain (i.e., a strain with normal levels of the 8oxoG repair enzymes MutM and MutY) (30, 31). The positive control m3C behaved as previously reported (27, 29). When fully repaired in the wild-type (AlkB+) strain, m3C coded exclusively as C; however, in the AlkB− strains, m3C was highly mutagenic, coding primarily as a T or an A, with smaller amounts of C and G also observed. Additionally, SOS induction exacerbated the mutagenesis of m3C by increasing the percentage of non-C base detected at lesion site (Fig. 2C and Table S2). In contrast, the negative controls cytosine and m5C coded exclusively as C in all cell lines tested; the non-C levels shown in the inset graphs in Fig. 2C and in Table S2 were <0.5%, in good agreement with the expected sensitivity of the assay.
Table S2.
In vivo mutagenesis of 5ClC and controls
| Input base | Base at lesion site | AlkB+ | AlkB− | AlkB− SOS+ |
| Cytosine | C | 99.8 ± 0.1 | 99.9 ± 0.6 | 99.8 ± 0.5 |
| T | 0.08 ± 0.05 | 0.04 ± 0.03 | 0.05 ± 0.03 | |
| G | 0.03 ± 0.06 | 0.03 ± 0.02 | 0.09 ± 0.06 | |
| A | 0.11 ± 0.1 | 0.06 ± 0.04 | 0.05 ± 0.05 | |
| 5ClC | C | 95.2 ± 0.6 | 94.7 ± 2.0 | 94.3 ± 0.8 |
| T | 4.6 ± 0.6 | 5.2 ± 2.0 | 5.6 ± 0.8 | |
| G | 0.08 ± 0.02 | 0.05 ± 0.02 | 0.07 ± 0.02 | |
| A | 0.13 ± 0.07 | 0.08 ± 0.04 | 0.04 ± 0.02 | |
| m5C | C | 99.8 ± 0.0 | 99.8 ± 0.2 | 99.8 ± 0.2 |
| T | 0.05 ± 0.01 | 0.03 ± 0.02 | 0.04 ± 0.03 | |
| G | 0.06 ± 0.01 | 0.08 ± 0.09 | 0.05 ± 0.04 | |
| A | 0.05 ± 0.03 | 0.1 ± 0.06 | 0.15 ± 0.11 | |
| m3C | C | 99.9 ± 0.0 | 13.2 ± 1.8 | 5.8 ± 2.6 |
| T | 0.02 ± 0.01 | 54.9 ± 3.7 | 57.5 ± 1.9 | |
| G | 0.01 ± 0.00 | 1.3 ± 0.6 | 3.1 ± 1.3 | |
| A | 0.03 ± 0.00 | 30.6 ± 3.0 | 33.6 ± 1.4 |
Data are from Fig. 2C and are shown as % average ± SD.
Establishing the Purity of 5ClC-Containing Oligonucleotides and Genomes.
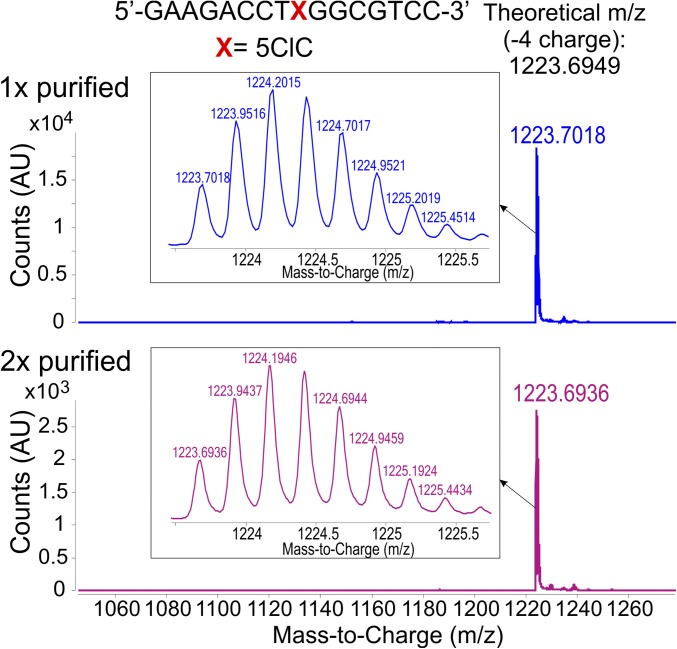
A series of validation experiments was performed to rule out any potential experimental artifacts (e.g., impurities in the starting materials) that could confound the in vivo experiment described above, which indicated that 5ClC was mutagenic. The 5ClC-containing oligonucleotide used to construct the M13 genome was thoroughly scrutinized using mass spectrometry and was confirmed to be free of residual protecting groups and/or failure sequences (Fig. S2). As a further safeguard, a portion of the 5ClC-containing oligonucleotide was repurified with an independent HPLC method (Materials and Methods and Figs. S2 and S3) and was used to construct a genome that was analyzed side by side with the one prepared initially in the experiments outlined below. Finally, a lesion integrity assay was performed (described in Materials and Methods and Fig. S4A), which directly established the identity and purity of the 5ClC base present site-specifically in the constructed M13 genomes (Fig. S4B). Taken together, these analyses confirmed the high purity (>99%) of the 5ClC-containing oligonucleotides and M13 genomes, engendering confidence in the reported genetic results.
Fig. S2.
Oligonucleotide purity analysis. ESI Q-TOF mass spectrometry traces of the 16mer oligodeoxynucleotides of the sequence 5′-GAAGACCTXGGCGTCC-3′ (in which X = 5ClC) after one round (1×) and two rounds (2×) of HPLC purification described in Materials and Methods. Insets show the parent peaks, highlighting their characteristic isotope distribution envelopes.
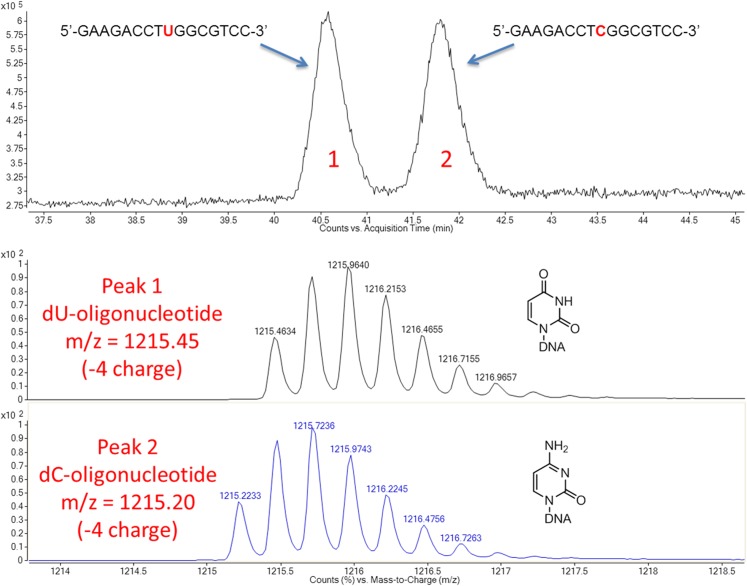
Fig. S3.
HPLC separation of 16mer oligonucleotides that differ by only one mass unit. Oligodeoxynucleotides of the sequence 5′-GAAGACCTXGGCGTCC-3′, in which X is either deoxyuridine (dU) or deoxycytidine (dC), were separated on an Acclaim Polar Advantage (Thermo Scientific) column using 10 mM ammonium acetate and acetonitrile as solvents. The gradient used was 2–12% acetonitrile over 100 min. The peaks then were analyzed on an ESI Q-TOF mass spectrometer (Agilent). The isotope distribution envelope and the theoretical mass (for the −4-charged species) of each oligonucleotide are shown.
Fig. S4.
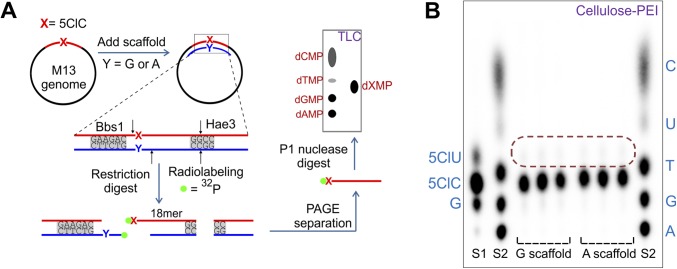
Purity analysis of the 5ClC-containing genome. (A) Lesion integrity assay. Scaffolds containing G or A opposite the lesion site and restriction enzymes (with the restriction sites indicated) were used to isolate a 32P-radiolabeled monophosphate of the base X (base at the lesion site, i.e., 5ClC) and evaluate its identity and purity on TLC (Cellulose-PEI). (B) The result of the lesion integrity assay applied to an M13 genome containing 5ClC. Two mixtures of deoxynucleotide monophosphate standards, S1 (5ClU, 5ClC, G) and S2 (C, U, T, G, A) were loaded on the TLC plate alongside three independent replicates for each scaffold. The area encircled by the red dotted line indicates the location where any putative 5ClU contaminant would appear. Quantification revealed that the constructed genomes contained >99% 5ClC at the lesion site.
The Intrinsic Mutagenic Properties of 5ClC.
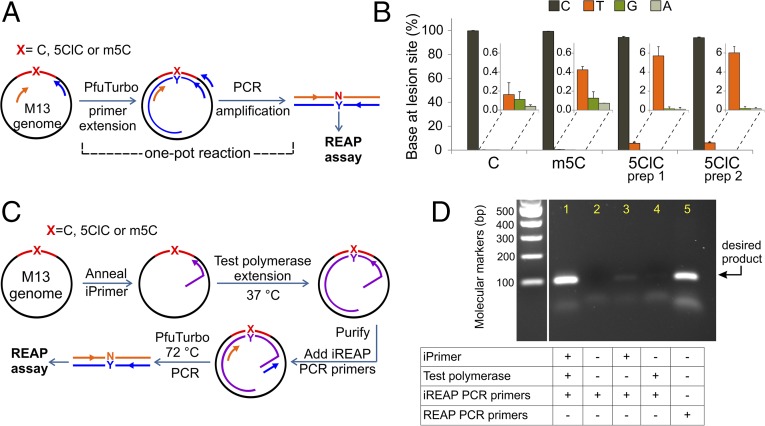
Two possibilities could explain the in vivo mutagenic properties of 5ClC: either 5ClC is intrinsically mutagenic, or a cellular enzymatic process could act on 5ClC to convert it into a secondary, more mutagenic product. For example, 5ClC could be the substrate of a glycosylase, generating an abasic site, or it could be enzymatically deaminated to 5-chlorouracil (5ClU). Both scenarios, even if inefficient, could cause a portion of 5ClC to code as a T. To rule out a contribution of the intracellular milieu to the observed mutagenic properties of 5ClC, the modified M13 genomes were used as templates for an in vitro primer extension assay. The high-fidelity polymerase PfuTurbo was used to extend a primer and then generate a PCR product that can be analyzed directly with the REAP assay (Fig. 3A). The results of the in vitro primer extension were comparable to those obtained in the in vivo assays. Specifically, 5ClC was found to be mutagenic, coding both as a C (∼94.2%) and as a T (∼5.8%), whereas the controls (cytosine and m5C) coded, as expected, only as Cs (Fig. 3B and Table S3). Additionally, the results obtained with the repurified oligonucleotide were indistinguishable from the results obtained with the initial 5ClC oligonucleotide (Fig. 3B and Table S3). Thus, the qualitative and quantitative concordance of the in vivo and in vitro results strongly supports the view that 5ClC is intrinsically mutagenic and base-pairs with A at a rate of ∼5%.
Fig. 3.
The mutagenesis of 5ClC in vitro. (A) Strategy for analyzing the mutagenesis of 5ClC in vitro. The modified M13 genomes, used as templates, were incubated with the REAP PCR primers (blue and orange) and PfuTurbo polymerase. The first incubation extended only the blue primer, generating a double-stranded product, which was then PCR amplified with the aid of the orange primer. The identity and amount of base N is determined by the REAP assay. (B) In vitro mutagenesis of 5ClC and controls. The graphed values are averages of three replicates, with error bars representing one SD. Data are also tabulated in Table S3. (C) Schematic of the iREAP strategy. The M13 genome containing a lesion (5ClC) or a control base (cytosine or m5C) at a defined site was used as a template for a primer extension reaction. The primer (iPrimer) contained a unique 5′ overhang that did not anneal to the M13 genome. The primer extension was carried out with a chosen test polymerase at 37 °C. Subsequently, the extended product was purified to remove the polymerase and unextended iPrimer, and a PCR was performed so that only the newly synthesized strand was amplified. The PCR product was then analyzed using the REAP assay to identify the type and amount of base (denoted N) present at the lesion site. The results of this assay with eight different test polymerases are summarized in Table 1. (D) A control experiment demonstrating the specificity of the primers used in the iREAP assay. The PCR products obtained from control iREAP experiments 1–5 with a C genome have been separated on a 2% agarose gel and stained with ethidium bromide. In each experiment, the iPrimer, test polymerase (Klenow fragment), the iREAP PCR primers (or REAP PCR primers) were added (+) or left out (−), as indicated.
Table S3.
In vitro mutagenesis of 5ClC and controls
| Input base | Base at lesion site | % average ± SD |
| Cytosine | C | 99.7 ± 0.2 |
| T | 0.16 ± 0.13 | |
| G | 0.11 ± 0.08 | |
| A | 0.04 ± 0.02 | |
| m5C | C | 99.3 ± 0.12 |
| T | 0.48 ± 0.04 | |
| G | 0.14 ± 0.08 | |
| A | 0.08 ± 0.01 | |
| 5ClC prep1 | C | 94.2 ± 1.0 |
| T | 5.7 ± 0.9 | |
| G | 0.05 ± 0.01 | |
| A | 0.04 ± 0.01 | |
| 5ClC prep2 | C | 93.9 ± 0.6 |
| T | 6.0 ± 0.6 | |
| G | 0.04 ± 0.02 | |
| A | 0.04 ± 0.02 |
Data are from Fig. 3B and are shown as % average ± SD.
Next, we evaluated the mutagenic properties of 5ClC when traversed by different, but functionally well-understood, polymerases. A panel of eight different polymerases was chosen, covering all known classes of DNA polymerases (A, B, C, D, X, Y, and RT) and all domains of life (Table 1). The inclusion of three different human DNA polymerases (pol β, pol γ, and pol η) allowed exploration of the possible relevance of the 5ClC lesion in human cells and, by extension, to human disease.
Table 1.
Mutagenic consequences of traversing 5ClC with several different DNA polymerases
| Family | Polymerase | Domain | Species | Proofreading | % C→T induced by 5ClC |
| A | Pol I (Klenow fragment Exo+) | Bacteria | E. coli | Yes | 4.4 ± 0.3 |
| A | Pol I (Klenow fragment Exo−) | Bacteria | E. coli | No | 4.4 ± 0.3 |
| A | T4 DNA polymerase | Viruses | T4 phage | Yes | 4.1 ± 0.2 |
| A | Pol γ (gamma) | Eukaryota | H. sapiens | Yes | 3.5 ± 0.3 |
| B | Pol ζ (zeta) | Eukaryota | S. cerevisiae | No | 3.4 ± 0.2 |
| C | Pol III holoenzyme (in vivo)* | Bacteria | E. coli | Yes | 4.6 ± 0.6 |
| D | PfuTurbo† | Archaea | P. furiosus | Yes | 5.8 ± 0.7 |
| X | Pol β (beta) | Eukaryota | H. sapiens | No | 9.1 ± 0.6 |
| Y | Pol η (eta) | Eukaryota | H. sapiens | No | 3.7 ± 0.7 |
| RT | HIV RT | Viruses | HIV | No | 4.9 ± 1.2 |
A panel of DNA polymerases spanning all known polymerase families (A, B, C, D, X, Y, RT) and all life domains was used to extend a primer annealed to an M13 ssDNA genome containing one 5ClC lesion. Subsequently, the newly synthesized strand was specifically PCR amplified (Fig. 3C), and the PCR product was analyzed with the REAP assay to determine the amount of C→T transitions induced by 5ClC.
The result from Fig. 2C.
The average of the two results from Fig. 3B.
Once again, the M13 genome site-specifically modified with 5ClC was used as a template in a modified primer extension assay denoted “iREAP” (in vitro REAP), which coupled the extension product of any test polymerase with the downstream quantitative analysis of the REAP assay (Fig. 3C). Here, the primer used (denoted “iPrimer”) contained a unique 5′ overhang, which allowed a subsequent strand-specific PCR amplification of the newly synthesized strand (Fig. 3C). One key feature of this design was the low annealing temperature of the iPrimer (∼40 °C), sufficient for a 37 °C primer extension reaction but inadequate for PCR. This feature prevented any unextended iPrimer from “jump-starting” the PCR and ensured that the outcome of the assay—the type and amount of the base present at the lesion site—reflected only the base-pairing preference and fidelity of the test polymerase used in the primer extension reaction and not that of the PCR polymerase. The strand-specific amplification approach permitted subsequent mutagenic analysis regardless of the yield of the primer extension reaction, and thus the assay could also provide data about the fidelity of low processivity polymerases. A set of control experiments was performed to demonstrate the primer design features. As shown in Fig. 3D, only when the iPrimer was successfully extended in the primer extension reaction did the subsequent PCR process yield the necessary amplicon for REAP analysis.
We found that 5ClC was mutagenic regardless of the polymerase used to replicate across the lesion, inducing a characteristic C→T transition at a frequency ranging from ∼3.5% (pol ζ and pol γ) to ∼9.1% (pol β) (Table 1). No clear correlation between the proofreading ability of the polymerase and the extent of miscoding was observed (Table 1). Together, these data strengthen the notion that 5ClC is an intrinsically mutagenic lesion being decoded as a base-pairing partner for both G and A by all classes of DNA polymerases found in nature.
Structural Analysis of 5ClC Replicative Bypass.
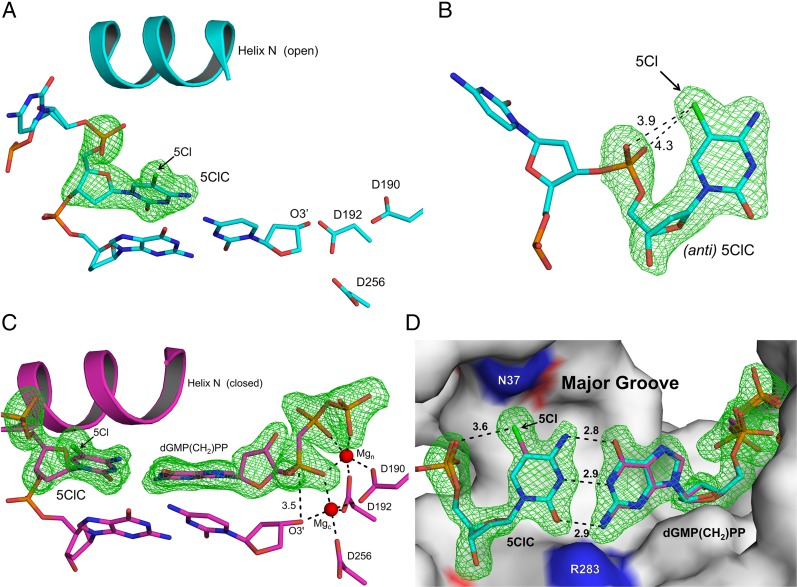
To investigate the molecular basis for the mutagenic ability of 5ClC, a structural analysis was performed on a polymerase caught in the process of replicating across a templated 5ClC. To this end, we determined binary (pol/DNA) and ternary (pol/DNA/dNTP) X-ray crystal structures of human DNA polymerase β (pol β) with 5ClC in the templating position of a 1-nt–gapped DNA duplex (Table 2). Pol β is a model mammalian DNA polymerase involved in base excision repair and may encounter DNA lesions in the templating strand during repair of the genome (32–34). The binary DNA complex structure with 5ClC in the templating position diffracted to 2.0 Å. Fig. 4A shows pol β in an open conformation with 5ClC in an anti-conformation. The 5Cl atom is 3.9 and 4.3 Å away from the backbone phosphate oxygen atoms (Fig. 4B) and is accommodated within the major groove of the DNA, in contrast to other known mutagenic lesions, such as 8oxoG, which occupies both the syn- and anti-conformations in the open binary complex because of the steric clash of the O8 with the phosphate backbone (35).
Table 2.
Data collection and refinement statistics of pol β binary and ternary complexes with 5ClC as the templating base
| Binary | Ternary incoming dGPCPP | Ternary incoming dAPCPP | |
| Data collection | |||
| Wavelength | 1.54 | 1.54 | 1.54 |
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c, Å | 55.9, 79.5, 55.1 | 50.6, 79.1, 55.7 | 54.6, 78.6, 55.1 |
| α, β, γ, ° | 90, 109.2, 90 | 90, 107.6, 90 | 90, 113.2, 90 |
| Resolution, Å | 50–2.0 | 50–1.98 | 50–2.0 |
| Rsym or Rmerge*, % | 10.7 (48.1) | 7.5 (50.3) | 7.2 (51.1) |
| I/sI | 14.1 (2.1) | 16.3 (2.2) | 19.8 (2.4) |
| Completeness, % | 98.9 (90.8) | 99.3 (95.9) | 98.9 (90.9) |
| Redundancy | 4.2 (2.5) | 3.5 (2.2) | 4.7 (2.8) |
| Refinement | |||
| Resolution, Å | 2.0 | 1.98 | 2.0 |
| No. reflections | 51,361 | 52,969 | 50,142 |
| Rwork/Rfree | 20.3/25.3 | 19.7/24.0 | 21.7/27.3 |
| No. atoms | |||
| Protein | 2,593 | 2,674 | 2,670 |
| DNA | 644 | 644 | 644 |
| Water | 284 | 320 | 191 |
| B-factors, Å | |||
| Protein | 37.1 | 28.1 | 41.4 |
| DNA/5ClC/dNTP | 33.6/45.8/– | 35.4/24.5/19.4 | 40.8/54.1/33.4 |
| Water | 37.7 | 33.3 | 39.1 |
| rms deviations | |||
| Bond length, Å | 0.01 | 0.01 | 0.01 |
| Bond angles, ° | 1.1 | 1.1 | 1.07 |
| Protein Data Bank ID | 5BOM | 5BOL | 5BPC |
Highest-resolution shell is shown in parentheses.
Fig. 4.
Structure of pol β binary and nonmutagenic ternary complex with 5ClC-containing oligonucleotide. Fo-Fc omit maps (green) are contoured at 3σ. Helix N of pol β, primer terminus (O3′), 5Cl of 5ClC, and key active-site residues are indicated. (A) The binary pol β complex shown in the open conformation. (B) A different viewing angle of the structure highlights the anti-geometry of the 5ClC base and the distances (in Ångstroms) between the 5Cl atom and the oxygen atoms of the phosphate backbone. (C) The pol β ternary complex with an incoming dGMP(CH2)PP base-pairing with 5ClC. Mg2+ ions are shown as red spheres in the catalytic and nucleotide-binding sites. The distance (in Ångstroms) between O3′ and Pα is shown. (D) The 5ClC and dGMP(CH2)PP are shown in a stick representation (purple), and the protein is shown in surface representation (gray). The distances correspond to the Watson–Crick hydrogen bonding between 5ClC and the dGTP analog. The DNA major groove edge is indicated. For reference the dC:dGTP (PDB ID code 4UB4) ternary complex is superimposed (light blue).
Soaking pol β binary DNA complex crystals in a cryosolution with MgCl2 and the nonhydrolyzable dGMP(CH2)PP analog promoted the open-to-closed conformational change without catalysis. Using this approach, we determined the nonmutagenic closed ternary preinsertion complex to 1.98 Å (Table 2). 5ClC remained in the anti-conformation and formed a good Watson–Crick base pair with the incoming dGTP analog (Fig. 4C). Mg2+ was bound both in the catalytic and nucleotide metal-binding sites, and O3′ of the primer terminus was in a catalytically competent position 3.5 Å to Pα of the bound nucleotide (Fig. 4C). Overlaying this structure with the nondamaged dC:dGTP ternary complex structure trapped with calcium showed they were nearly identical, with an rmsd of 0.17 Å; both structures formed identical Watson–Crick hydrogen-bonding interactions (Fig. 4D). The major groove can accommodate the 5Cl atom, with the closest active site residue being Asn37 (3.8 Å away). Additionally, the phosphate backbone did not have to alter its position and remained 3.6 Å away. In contrast to the major groove, the DNA minor groove restricts the geometry of the nascent base pair. Arg283 was within hydrogen bonding distance of O2 for both templating dC and 5ClC. These results demonstrate that the major groove accommodates the 5Cl atom, facilitating Watson–Crick base-pairing between 5ClC and dGTP within the active site. This accommodation explains the propensity for error-free lesion bypass (G being the major base-pairing partner for 5ClC) observed in the cellular studies.
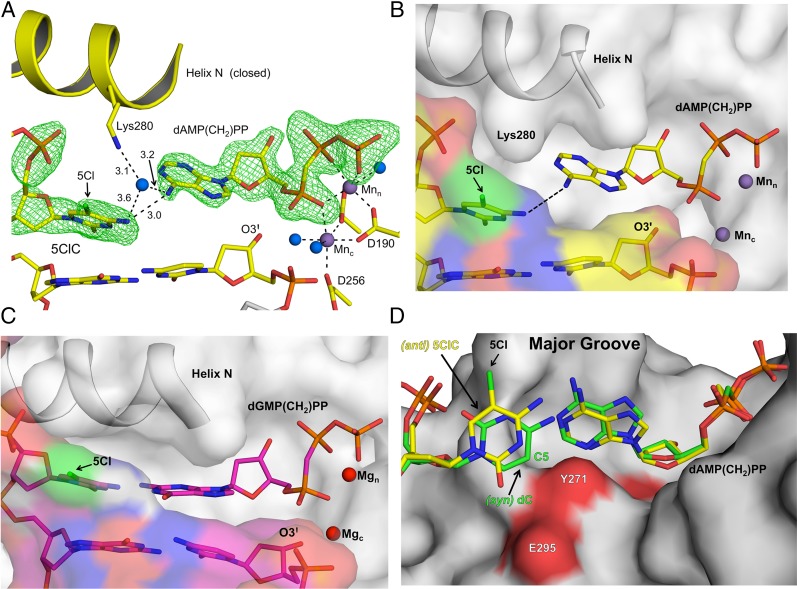
The in vivo and in vitro results reported here suggest that 5ClC promotes C:G→T:A transition mutations by base-pairing with adenine in addition to the expected base-pairing partner guanine. To elucidate the mechanistic basis for the 5ClC mutagenesis, we obtained the closed ternary complex of a templating 5ClC with an incoming dAMP(CH2)PP to 2.0 Å (Table 2). Because we were unable to obtain a closed ternary structure in the presence of MgCl2, we instead used MnCl2 during the soak step. The need for a more accommodating metal cation likely reflects the reduced binding affinity of dATP for the templating 5ClC. Fig. 5A shows the resulting closed ternary complex forming a mismatch intermediate conformation with Mn2+ in the catalytic and nucleotide-binding sites (36). This conformation resulted in the coding template base being shifted upstream by more than 3 Å and generating an abasic-like templating pocket within the active site that is occluded by Lys280 (Fig. 5 A and B). In this conformer, the only unique interaction of the templating 5ClC with the incoming dATP analog was between N4 of 5ClC and N6 of the incoming dATP analog. This interaction was facilitated by a well-ordered bridging water molecule near N4 of 5ClC, N6 of dATP, and Lys280 (Fig. 5A). In addition, the primer terminus sugar rotated away from the catalytic metal, generating a suboptimal O3′ position that did not coordinate the catalytic metal (Fig. 5 A and B). Accordingly, misinsertion of dATP would require a minor shift of O3′ to coordinate the catalytic metal and initiate catalysis, as has been proposed previously during mismatch insertion (37). These structural rearrangements are in contrast to the ordered active site and Watson–Crick base-pairing interactions observed between the templating 5ClC and dGTP analog (Fig. 5C).
Fig. 5.
Structures of the pol β closed ternary insertion complex during mutagenic 5ClC lesion bypass. Fo-Fc omit maps (green) are contoured at 3σ. Helix N, primer terminus (O3′), 5Cl of 5ClC, and key active-site residues are indicated. (A) The pol β mismatch ternary complex with an incoming dAMP(CH2)PP opposite 5ClC with putative hydrogen-bonding interaction distances (in Ångstroms) shown. Mn2+ ions are shown as purple spheres in the catalytic and nucleotide-binding sites. Water molecules are shown in blue. (B) The view in A with the protein (gray) and DNA (yellow) shown in surface representation. Incoming dAMP(CH2)PP is shown in stick representation. (C) The nonmutagenic ternary complex with an incoming dGMP(CH2)PP base-pairing with 5ClC is shown with the protein (gray) and DNA (purple) in surface representation. The Mg2+ ions are shown in red, and the incoming dGMP(CH2)PP is shown in stick representation. (D) The anti-5ClC and dAMP(CH2)PP are shown in stick representation (yellow), and the protein is shown in surface representation (gray). The DNA major groove edge is indicated. For reference, the mismatch syn-dC:dATP (PDB ID code 3C2L) ternary complex is superimposed (green). The C5 position of the nondamaged cytosine is indicated.
During the mutagenic insertion of the dATP analog, 5ClC remained in the anti-conformation, and N4 of 5ClC was within hydrogen-bonding distance to N6 of the incoming dATP analog (Fig. 5A). An overlay of the pol β dAMP(CH2)PP insertion opposite 5ClC or natural cytosine is shown in Fig. 5D. In contrast to 5ClC, the nondamaged templating cytosine adopted a syn-conformation with no hydrogen-bonding interactions between the templating base and the incoming dATP analog. This result indicates that the 5Cl substituent on 5ClC was promoting the anti-conformation of the base by favoring the major over the minor DNA groove position. Fig. 5D shows a surface representation highlighting the unrestrained space in the major groove that accommodates the 5Cl group. If 5ClC were to adopt a syn-conformation, similar to the nondamaged cytosine, the 5Cl atom would sterically clash with Tyr271 and Glu295 (Fig. 5D). Therefore, the anti-conformation of the 5ClC base, adopted to accommodate the 5Cl bulky substituent in the DNA major groove, is a key factor that promotes a favorable hydrogen-bonding interaction between 5ClC and the incoming dATP, and likely constitutes the molecular basis for the mutagenic mispairing ability of 5ClC, given the constrained minor groove of most DNA polymerases.
Discussion
The Molecular Basis of 5ClC Mutagenesis.
The present work unveils the mutagenic properties of 5ClC, a modified base previously shown to accumulate in cells and tissues exposed to chronic inflammation (20, 21). We found that when present in a template strand, 5ClC has an increased probability (compared with the replication fidelity of canonical bases) of mispairing with adenine and thus miscoding as a T. The structural evidence reported here confirms the existence of a direct interaction between 5ClC and incoming dATP in the active site of a polymerase. This unique interaction and the formation of an apparent abasic site templating pocket are sufficient to promote the mutagenic insertion of dATP opposite 5ClC by an “enhanced A-rule” mechanism whereby a polymerase incorporates an A opposite a structure resembling an abasic site (38).
The structural snapshots show that during the mutagenic bypass of the lesion, 5ClC adopts a conformation in which the 5Cl substituent is placed in the major groove of DNA. Because, for most DNA polymerases, the DNA major groove of the nascent base pair can generally accommodate bulky substituents, this phenomenon is likely universal, as supported by the mutagenesis data across all families of DNA polymerases (Table 1). In contrast, the minor groove edge often contacts protein side chains within the polymerase active site, thereby limiting base conformations that protrude in the DNA minor groove. By favoring the major groove position for the 5Cl group, the 5ClC base primarily adopts an anti-conformation, thereby presenting its Watson–Crick hydrogen-bonding edge to the incoming nucleotide (Figs. 4 and 5). In contrast, a nondamaged cytosine is known to occupy both the anti- and syn-conformations, and thus the probability of forming a stable base pair with incoming dATP is greatly reduced (Fig. 5D). Furthermore, our crystal structures reveal an unusual and rather unique interaction between 5ClC and the incoming adenine, with the N4 of 5ClC and N6 of adenine being within hydrogen-bonding distance (3.0 Å). Although the interaction has only one putative hydrogen bond (and/or a water bridge), it is sufficient to promote the conformational change in the polymerase from the open form to the prechemistry closed form. In fact, mutagenic mispairings of damaged bases that rely on only one hydrogen bond have been reported before [e.g., N2,3-ethenoguanine mispairing with T (39)]. Moreover, the base-pairing we observe between 5ClC and A is strikingly different from the base-pairing between cytosine and adenine previously reported to occur across multiple polymerase families as either a wobble base pair or a base pair involving minor tautomeric forms of either base (40–42). Nevertheless, we cannot rule out the possibility that during the nucleotidyl transfer reaction, the pairing between 5ClC and A transitions toward a wobble base pair or a tautomeric pair. Importantly, both of these possibilities will be stabilized by the presence of the 5Cl substituent, which promotes the anti-conformation of the 5ClC base, whereas an alternative syn-conformer would not form stable wobble base pairs.
Interestingly, by extending the steric argument, m5C, similarly to 5ClC, is expected to adopt a primarily anti-conformation. However, 5ClC is much more mutagenic than m5C, suggesting that factors other than sterics must be involved. For example, the presence of the chlorine atom on the base is known to affect the electron density and distribution on the aromatic ring; as a consequence, the pKa of the N3 position of 5ClC (2.65) is significantly lower than the pKa of cytosine at the same atom (4.30) (23). In comparison, m5C has a pKa similar to that of cytosine (4.35) (23). The electronic effect of chlorine was shown to be strong enough to influence the energy of the hydrogen bonds involved in base-pairing; thus, the estimated base-pairing energy of 5ClC:G is slightly less than the energy of the C:G pair (43). Future studies probing the electronic configuration of 5ClC will be of interest to provide a more comprehensive understanding of the way that the chlorine substituent alters the base-pairing potential of 5ClC.
5ClC joins a substantial group of other substituted cytosines or cytosine analogs that are known to be mutagenic and induce C→T mutations. 5-Hydroxycytosine, an oxidative stress-induced lesion, is believed to mispair with adenine when it adopts an imine minor tautomeric form (44). Tautomerism is also the explanation for the mutagenic properties of 5-aza-5,6-dihydrocytosine, a cytosine analog used as an antiviral therapeutic (28). Cytosine analogs such as bicyclocytosine (45), N4-hydroxycytosine (46), and N4-aminocytosine (47) are mutagenic because of their ability to form Watson–Crick or wobble base pairs with both purines. Although 5-methylcytosine, an important epigenetic marker, is not a mutagenic base (as confirmed by the data in the present study), it has an increased potential to deaminate to thymine; if the resulting mismatch is not repaired, the deamination of 5-methylcytosine thus can lead to C→T mutations (48, 49). This mechanism is believed to account for the C→T mutations occurring in the CpG islands, where the cytosine is most likely to be methylated (48, 49). Additionally, 5-methylcytosine is also oxidatively processed by the TET enzymes to generate sequentially 5-hydroxymethylcytosine, 5-formylcytosine, and 5-carboxycytosine; of these 5-substituted cytosines, only 5-formylcytosine can induce C→T mutations in vitro, at a frequency of ∼1% (50, 51).
The Mutagenic Impact of 5ClC in Mammalian Cells.
Our data convincingly suggest that 5ClC is a mutagenic lesion, inducing about 3–9% C→T transition mutations depending on the polymerase at play. Although by itself this number seems relatively low, the impact of 5ClC and, by extension, the impact of chlorination damage on mammalian cell biology will depend on the total number of lesions present in the genome. The upper bound of the extent of 5ClC formation in DNA was reported in a study in which physiologically achievable concentrations of HOCl transformed in vitro about 1–10 of every 1,000 cytosines in 5ClC (19). At this level of incorporation, the mutagenesis of 5ClC would be catastrophic for a mammalian-sized genome. More typically, the levels of genomic 5ClC in chronically inflamed tissues have been measured at ∼1–30 lesions per 108 bases (20, 21) or 66–1,980 lesions per diploid human genome. Given the inherent mutagenicity of 5ClC described here, when a cell having this lesion burden replicates, it would acquire about 3.3–99 new mutations. This number represents a notable increase over the spontaneous mutation rate, estimated for mammalian cells to be on average 1.1–1.5 mutations per cell division (52, 53). Given that no repair pathway specific for 5ClC has yet been described (perhaps explaining the accumulation and persistence of this lesion in the genome), it is plausible that 5ClC is an important contributor to the mutagenic burden previously observed in tissues subjected to chronic inflammation (54, 55) and thus increases the probability that such tissues progress toward malignancy.
The Mechanistic Link Between Chronic Inflammation and Genetic Modifications in Tumors.
The patterns of genetic changes seen in the genomes of cancer cells are indicative of the mutagenic processes that contributed to the conversion of normal cells to tumors (48, 56, 57). Thirty such patterns, called “mutational signatures,” have been identified; each consists of a histogram of relative frequencies of all possible point mutations occurring within all possible trinucleotide sequence contexts (48, 56). However, only a fraction of mutational signatures have been ascribed to specific mutagenic processes (56). Among all possible base substitution mutations, C→T (and correspondingly G→A) transitions are the most common, and for certain mutational signatures, they have been attributed to well-known mechanisms such as m5C deamination and AICDA (activation-induced cytidine deaminase)/APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like)-dependent deamination (48, 56). Nevertheless, other mutational signatures, including many of unknown etiology, include C→T mutations (48, 56). Given our present study, we suspect that 5ClC could also be a contributor to the C→T mutations observed in sequenced tumors as part of one or several different mutational signatures. The well-established infiltration of tumors with immune cells, including neutrophils (2, 9, 58), provides a rich opportunity for HOCl production and, hence, 5ClC formation.
We postulate that cells exposed to chronic inflammatory processes, more specifically those mediated by activated neutrophils and HOCl production, would bear in their DNA the mutagenic legacy of chlorination damage (Fig. 1). Such a mutagenic signature would feature C→T mutations characteristic of 5ClC formation and also other mutations corresponding to the variety of oxidative and chlorinated lesions induced by HOCl (18). Some features of the mutagenic spectrum generated by chronic inflammation already have been reported in mouse models, and they include increased C→T mutations as well as small DNA base insertions and deletions (indels) (54). Intriguingly, in tumor sequence data, both the mutational signature 6 of Alexandrov et al. (56), and the H signature of Jia et al. (57) feature extensive C→T transitions (but also smaller levels of other mutations such as T→C and C→A) and are strongly associated with colorectal cancers, malignancies that often develop in chronically inflamed tissues. It has been proposed that these mutational signatures reflect the microsatellite instability phenotype of many colorectal tumors (48, 56, 57), explaining well the indels that also have been reported to associate with these signatures (56). However, microsatellite instability does not fully explain the observed C→T-dominated point mutation pattern. Interestingly, one of the mechanisms that leads to microsatellite instability is inhibition of MLH1 expression, a key mismatch repair protein, by hypermethylation (59). As mentioned earlier, one biochemical feature of 5ClC, when present in CpG islands, is the ability to induce DNA hypermethylation (25, 60). Therefore, we speculate that signature 6 (and/or signature H) may reflect both the mutagenic and the epigenetic features of genomic 5ClC accumulation and thus represent the mutational signature of neutrophil-driven chronic inflammation (Fig. 1). Further work is needed to investigate this hypothesis.
Materials and Methods
Enzymes.
Polymerase pol β used for in vitro extension reactions was from Novoprotein, polymerases γ and ζ were from Enzymax LLC, HIV-RT was from Worthington Biochemical Corp., and PfuTurbo was from Agilent Technologies. Polymerase η was a generous gift from Wei Yang (National Institute of Diabetes and Digestive and Kidney Diseases/NIH, Bethesda). The other polymerases and restriction enzymes were from New England Biolabs.
5ClC Nucleoside.
5ClC deoxynucleoside was from Alchem Laboratories Corporation; its identity and purity was confirmed by NMR. The aqueous stability of 5ClC nucleoside was also evaluated. 5ClC was found to be stable to deamination at biologically relevant pHs. No appreciable deamination (evaluated on reverse-phase HPLC) was detected after incubation with 2 mM NaOH, 2 mM NaOMe, or 30% (wt/vol) ammonia for 24 h.
Oligonucleotides.
All unmodified oligonucleotides were from Integrated DNA Technologies. 5ClC, m3C, and m5C modified bases were incorporated into 16mer oligonucleotides (5′-GAAGACCTXGGCGTCC-3′ and 5′CCGACXGCGCATCAGC-3′, where X is the modified nucleobase) by using phosphoramidite solid-phase methods described before (26, 29). The 5ClC nucleoside was protected at N4 with a benzoyl group and then was used to prepare the 5ClC phosphoramidite (at ChemGenes). Oligonucleotide syntheses containing the 5ClC phosphoramidite were done at ChemGenes or Boston Open Labs.
Oligonucleotide Purification and Characterization.
The oligonucleotides containing 5ClC were deprotected with aqueous ammonium hydroxide and purified by reverse-phase HPLC on a Varian Microsorb-MV 100-5 250 × 4.6 mm C18 column (Varian) or on an Acclaim Polar Advantage 3-µm 250 × 3.0 mm C18 column (Thermo Scientific) using 10 mM ammonium acetate and acetonitrile as solvents. The 2× purified oligonucleotide was purified on both columns sequentially. Purity was evaluated on an Agilent 6510 electrospray ionization (ESI) quadrupole time-of-flight (Q-TOF) mass spectrometer (Fig. S2). The purification conditions allowed the separation of 16mer oligonucleotides that differ by only one mass unit (Fig. S3) and therefore were expected to separate the oligonucleotide containing 5ClC from one containing the putative contaminant 5ClU.
M13 Genomes Construction.
Oligonucleotides containing lesions and controls were ligated into an M13mp7(L2) single-stranded viral genome by using reported methods (26, 29). The constructed genomes were purified using phenol-chloroform extraction and QIAquick columns (Qiagen).
Lesion Integrity Assay.
A lesion integrity assay, adapted from ref. 29, was performed to establish directly the identity and purity of the site-specific base present in the constructed M13 genomes. Although the HPLC purification of the oligonucleotides should have removed any sequences containing 5ClU, detecting small levels of 5ClU in the MS analysis was challenging because 5ClU is only 1 Da heavier than 5ClC. It was important to detect and quantify the amount of 5ClU still remaining in the oligonucleotide sample or that may have formed during genome construction, because 5ClU would likely code as a T and contribute to the mutagenic signal. To that end, a scaffold was used to frame the lesion on the single-stranded M13 genome, after which the site of interest was analyzed using REAP. To account for any bias the scaffold might introduce, the experiment was performed independently with two scaffolds, one containing G and the other containing A opposite the lesion. To 150 fmol of M13 genome containing 5ClC prepared above, 250 fmol of a scaffolding oligonucleotide was added and incubated at 80 °C for 5 min, followed by a slow cooling (0.1 °C/s) for annealing. The scaffolds (5′-CCAGTGAATTGGACGCCYAGGTCTTCCACTGAATCA-3′, where Y is either G or A) spanned the lesion site, being complementary to 15 or 16 bases on either side of the lesion. This construct was subjected directly to the REAP assay, starting with the BbsI digest step (Fig. S4A) (26). To prepare 5CldCMP and 5CldUMP standards, an oligonucleotide containing 5ClC at the 5′ end was radiolabeled, isolated by PAGE, and digested to monophosphates with P1 nuclease. The mixture then was incubated in 0.1 M HCl for 30 min to promote partial deamination of 5ClC to 5ClU, and the spot for 5CldUMP was confirmed by a cold 5′ monophosphorylated standard. With either scaffold, the constructed M13 genome was found to contain >99% 5ClC at the site of inquiry, whereas any putative 5ClU present (as judged from the TLC standard) was <1% (Fig. S4B).
In Vivo Lesion Bypass and Mutagenesis Assays.
The constructed M13 viral genomes (mixed with a competitor genome) were transfected into E. coli strains by electroporation. The E. coli strains used were AlkB+ (wild type) (AB1157, nalA) and AlKB− (AlkB deficient) (AB1157, nalA, alkB22). Lesion bypass was evaluated with the CRAB assay; lesion mutagenesis was measured with the REAP assay (26). Briefly, the resulting progeny phage were collected and reamplified; their DNA then was extracted using the QIAprep M13 kit (Qiagen). The region of interest (where the lesion used to reside) was PCR amplified with assay-specific primers. The CRAB primers amplified both input and competitor sequences and thus allowed the estimation of the relative bypass (Fig. S1). A genome normalization step (26) also was used to estimate the relative concentrations of the different M13 genomes accurately. The REAP primers amplified only the progeny coming from the input genomes (lesion-containing genomes) to maximize the signal obtained from the lesion site. Restriction digests and radiolabeling yielded an18mer fragment that contained the base at the lesion site at its 5′ end. PAGE isolation of this fragment and the subsequent P1 nuclease digest produced the radioactive monophosphate of the base at the lesion site, which was subsequently analyzed by TLC (Cellulose-PEI) and quantified by phosphorimagery (Fig. S1).
In Vitro Mutagenesis Assay.
The M13 genomes constructed as described above were used as templates for the in vitro mutagenesis assay (iREAP). An extension primer (5′-CGTGACGATGCGCAGACTGATATCATGTGTAAAACGACGGC-3′) was added in equimolar amount and annealed by heating the mixture at 85 °C for 5 min, followed by a slow (0.1 °C/s) cooling to 4 °C. The primer was extended using 1 U of polymerase (as indicated) in NEBuffer1 (New England Biolabs) supplemented with 1 mM DTT and 200 µM of each of the four dNTPs (New England Biolabs) at 37 °C for 1 h. The polymerase was inactivated at 95 °C for 10 min; then the reaction was transferred quickly on ice to minimize reannealing of unextended primer. The extension product was purified from unextended primer and polymerase using QIAquick columns (Qiagen) and an additional wash with 0.5× PE buffer (Qiagen). The eluent was then PCR amplified (32 cycles of 95/70/72 °C for 30/15/60 s, respectively) with PfuTurbo polymerase, using the primers 5′-HGGAAACAGCTATGACCATGATTCAGTGGAAGAC-3′ (forward) and 5′-HCGTGACGA TGCGCAGACTGATATCATGT-3′ (reverse), where H is a phospho-hexylamino linker which prevents 5′ phosphorylation during subsequent steps. The PCR products were then purified and analyzed with the REAP assay as previously described (26). For the in vitro mutagenesis assay using PfuTurbo, the M13 genomic template containing a site-specific lesion was directly PCR amplified with PfuTurbo with the following primers: 5′-HCAGCTATGACCATGATTCAG-TGGAAGAC-3′ (forward) and 5′-HTGTAAAACGACGG CCAGTGAATTGGACG-3′ (reverse), because in this case the first PCR step is essentially a primer extension reaction using PfuTurbo. The resulting PCR products were subsequently purified and analyzed as above.
Structural Studies of 5ClC with DNA pol β.
Human wild-type pol β was overexpressed in E. coli and purified as described previously (61). Binary complex crystals with a templating 5ClC in a 1-nt–gapped DNA were grown as previously described (62). The 16mer oligonucleotide containing 5ClC was synthesized and thoroughly purified on reverse-phase HPLC as described above. The binary complex crystals were transferred to a cryosolution containing 12% ethylene glycol, 50 mM imidazole (pH 7.5), 20% PEG3350, 90 mM sodium acetate, and 5 mM nonhydrolyzable dNTP analogs dGMP(CH2)PP or dAMP(CH2)PP with 200 mM MgCl2 or MnCl2, thus resulting in the ternary complex.
Data were collected at 100 K on an in-house Saturn92 CCD detector system mounted on a MiraMax-007HF rotating anode generator at a wavelength of 1.54 Å. Data were processed and scaled using the HKL2000 software package (63). Initial models were determined using molecular replacement with the open [Protein Data Bank (PDB) ID code 3ISB] or closed (PDB ID code 2FMS) structures of pol β as a reference. All Rfree flags were taken from the starting model, and refinement was carried out using PHENIX (64) and model building using Coot (65). The figures were prepared using PyMOL and AxPyMOL Molecular Graphics plugin (v1.0, Schrodinger LLC), and all density maps were generated after simulated annealing was performed. Ramachandran analysis determined that 100% of nonglycine residues lie in the allowed regions.
Acknowledgments
We thank Allison Simi, Maria Kulikova, and Nicole Zatorski for their assistance; Koli Taghizadeh for technical advice; and Charles Knutson, Steven Tannenbaum, and Gerald Wogan for valuable feedback on the manuscript. We also thank the MIT Center for Environmental Health Sciences (MIT CEHS) for providing access to the Instrumentation Core facility. This work was supported by NIH Grants P01 CA26731 and R37 CA080024 (both to J.M.E), NIH Center Grant P30 ES002109 (to the MIT CEHS), and by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences, Project Nos. Z01-ES050158 (S.H.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates of the X-ray structures reported in this paper have been deposited at the Research Collaboratory for Structural Bioinformatics Protein Data Bank [RCSB PDB ID codes 5BOM (binary complex), 5BOL (ternary complex with dGTP analog), and 5BPC (ternary complex with dATP analog)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507709112/-/DCSupplemental.
References
- 1.Brenner DR, et al. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol Biomarkers Prev. 2014;23(9):1729–1751. doi: 10.1158/1055-9965.EPI-14-0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elinav E, et al. Inflammation-induced cancer: Crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 3.Kundu JK, Surh Y-J. Inflammation: Gearing the journey to cancer. Mutat Res. 2008;659(1-2):15–30. doi: 10.1016/j.mrrev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Hussain SP, Harris CC. Inflammation and cancer: An ancient link with novel potentials. Int J Cancer. 2007;121(11):2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 5.Balkwill F, Charles KA, Mantovani A. Smoldering and polarized inflammation in the initiation and promotion of malignant disease. Cancer Cell. 2005;7(3):211–217. doi: 10.1016/j.ccr.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mangerich A, Dedon PC, Fox JG, Tannenbaum SR, Wogan GN. Chemistry meets biology in colitis-associated carcinogenesis. Free Radic Res. 2013;47(11):958–986. doi: 10.3109/10715762.2013.832239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: Reconciling chemical mechanisms and biological fates. Int J Cancer. 2011;128(9):1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic Biol Med. 2010;49(11):1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantovani A, Cassatella MA, Costantini C, Jaillon S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol. 2011;11(8):519–531. doi: 10.1038/nri3024. [DOI] [PubMed] [Google Scholar]
- 12.van der Veen BS, de Winther MPJ, Heeringa P. Myeloperoxidase: Molecular mechanisms of action and their relevance to human health and disease. Antioxid Redox Signal. 2009;11(11):2899–2937. doi: 10.1089/ars.2009.2538. [DOI] [PubMed] [Google Scholar]
- 13.Winterbourn CC, Hampton MB, Livesey JH, Kettle AJ. Modeling the reactions of superoxide and myeloperoxidase in the neutrophil phagosome: Implications for microbial killing. J Biol Chem. 2006;281(52):39860–39869. doi: 10.1074/jbc.M605898200. [DOI] [PubMed] [Google Scholar]
- 14.Henderson JP, Byun J, Heinecke JW. Molecular chlorine generated by the myeloperoxidase-hydrogen peroxide-chloride system of phagocytes produces 5-chlorocytosine in bacterial RNA. J Biol Chem. 1999;274(47):33440–33448. doi: 10.1074/jbc.274.47.33440. [DOI] [PubMed] [Google Scholar]
- 15.Winterbourn CC. Biological reactivity and biomarkers of the neutrophil oxidant, hypochlorous acid. Toxicology. 2002;181-182:223–227. doi: 10.1016/s0300-483x(02)00286-x. [DOI] [PubMed] [Google Scholar]
- 16.Winterbourn CC, Kettle AJ. Biomarkers of myeloperoxidase-derived hypochlorous acid. Free Radic Biol Med. 2000;29(5):403–409. doi: 10.1016/s0891-5849(00)00204-5. [DOI] [PubMed] [Google Scholar]
- 17.Kawai Y, et al. Endogenous formation of novel halogenated 2′-deoxycytidine. Hypohalous acid-mediated DNA modification at the site of inflammation. J Biol Chem. 2004;279(49):51241–51249. doi: 10.1074/jbc.M408210200. [DOI] [PubMed] [Google Scholar]
- 18.Whiteman M, Jenner A, Halliwell B. Hypochlorous acid-induced base modifications in isolated calf thymus DNA. Chem Res Toxicol. 1997;10(11):1240–1246. doi: 10.1021/tx970086i. [DOI] [PubMed] [Google Scholar]
- 19.Kang JI, Jr, Sowers LC. Examination of hypochlorous acid-induced damage to cytosine residues in a CpG dinucleotide in DNA. Chem Res Toxicol. 2008;21(6):1211–1218. doi: 10.1021/tx800037h. [DOI] [PubMed] [Google Scholar]
- 20.Mangerich A, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci USA. 2012;109(27):E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knutson CG, et al. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc Natl Acad Sci USA. 2013;110(26):E2332–E2341. doi: 10.1073/pnas.1222669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valinluck V, Liu P, Kang JI, Jr, Burdzy A, Sowers LC. 5-halogenated pyrimidine lesions within a CpG sequence context mimic 5-methylcytosine by enhancing the binding of the methyl-CpG-binding domain of methyl-CpG-binding protein 2 (MeCP2) Nucleic Acids Res. 2005;33(9):3057–3064. doi: 10.1093/nar/gki612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valinluck V, Wu W, Liu P, Neidigh JW, Sowers LC. Impact of cytosine 5-halogens on the interaction of DNA with restriction endonucleases and methyltransferase. Chem Res Toxicol. 2006;19(4):556–562. doi: 10.1021/tx050341w. [DOI] [PubMed] [Google Scholar]
- 24.Valinluck V, Sowers LC. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res. 2007;67(3):946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 25.Lao VV, et al. Incorporation of 5-chlorocytosine into mammalian DNA results in heritable gene silencing and altered cytosine methylation patterns. Carcinogenesis. 2009;30(5):886–893. doi: 10.1093/carcin/bgp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- 27.Shrivastav N, et al. A chemical genetics analysis of the roles of bypass polymerase DinB and DNA repair protein AlkB in processing N2-alkylguanine lesions in vivo. PLoS One. 2014;9(4):e94716. doi: 10.1371/journal.pone.0094716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li D, et al. Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2′-deoxycytidine. Proc Natl Acad Sci USA. 2014;111(32):E3252–E3259. doi: 10.1073/pnas.1405635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine, and 3-methylthymine in alkB Escherichia coli. Proc Natl Acad Sci USA. 2004;101(39):14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46(5):1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- 31.Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29(30):7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- 32.Freudenthal BD, Beard WA, Wilson SH. DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 2013;41(3):1848–1858. doi: 10.1093/nar/gks1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kothandapani A, et al. Novel role of base excision repair in mediating cisplatin cytotoxicity. J Biol Chem. 2011;286(16):14564–14574. doi: 10.1074/jbc.M111.225375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beard WA, Batra VK, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat Res. 2010;703(1):18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH. Binary complex crystal structure of DNA polymerase β reveals multiple conformations of the templating 8-oxoguanine lesion. Proc Natl Acad Sci USA. 2012;109(1):113–118. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol Cell. 2008;30(3):315–324. doi: 10.1016/j.molcel.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freudenthal BD, Beard WA, Shock DD, Wilson SH. Observing a DNA polymerase choose right from wrong. Cell. 2013;154(1):157–168. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zahn KE, Averill A, Wallace SS, Doublié S. The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site. Biochemistry. 2011;50(47):10350–10358. doi: 10.1021/bi201219s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao L, et al. Basis of miscoding of the DNA adduct N2,3-ethenoguanine by human Y-family DNA polymerases. J Biol Chem. 2012;287(42):35516–35526. doi: 10.1074/jbc.M112.403253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA. 2011;108(43):17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia S, Wang J, Konigsberg WH. DNA mismatch synthesis complexes provide insights into base selectivity of a B family DNA polymerase. J Am Chem Soc. 2013;135(1):193–202. doi: 10.1021/ja3079048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, et al. Mechanism of somatic hypermutation at the WA motif by human DNA polymerase η. Proc Natl Acad Sci USA. 2013;110(20):8146–8151. doi: 10.1073/pnas.1303126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Theruvathu JA, Yin YW, Pettitt BM, Sowers LC. Comparison of the structural and dynamic effects of 5-methylcytosine and 5-chlorocytosine in a CpG dinucleotide sequence. Biochemistry. 2013;52(47):8590–8598. doi: 10.1021/bi400980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suen W, Spiro TG, Sowers LC, Fresco JR. Identification by UV resonance Raman spectroscopy of an imino tautomer of 5-hydroxy-2′-deoxycytidine, a powerful base analog transition mutagen with a much higher unfavored tautomer frequency than that of the natural residue 2′-deoxycytidine. Proc Natl Acad Sci USA. 1999;96(8):4500–4505. doi: 10.1073/pnas.96.8.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negishi K, et al. The mechanism of mutation induction by a hydrogen bond ambivalent, bicyclic N4-oxy-2′-deoxycytidine in Escherichia coli. Nucleic Acids Res. 1997;25(8):1548–1552. doi: 10.1093/nar/25.8.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sledziewska E, Janion C. Mutagenic specificity of N4-hydroxycytidine. Mutat Res. 1980;70(1):11–16. doi: 10.1016/0027-5107(80)90053-6. [DOI] [PubMed] [Google Scholar]
- 47.Nomura A, Negishi K, Hayatsu H, Kuroda Y. Mutagenicity of N4-aminocytidine and its derivatives in Chinese hamster lung V79 cells. Incorporation of N4-aminocytosine into cellular DNA. Mutat Res. 1987;177(2):283–287. doi: 10.1016/0027-5107(87)90012-1. [DOI] [PubMed] [Google Scholar]
- 48.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15(9):585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutsenko E, Bhagwat AS. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437(1):11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 50.Münzel M, et al. Improved synthesis and mutagenicity of oligonucleotides containing 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine. Chemistry. 2011;17(49):13782–13788. doi: 10.1002/chem.201102782. [DOI] [PubMed] [Google Scholar]
- 51.Xing X-W, et al. Mutagenic and cytotoxic properties of oxidation products of 5-methylcytosine revealed by next-generation sequencing. PLoS One. 2013;8(9):e72993. doi: 10.1371/journal.pone.0072993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513(7518):422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99(2):803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato Y, et al. IL-10 deficiency leads to somatic mutations in a model of IBD. Carcinogenesis. 2006;27(5):1068–1073. doi: 10.1093/carcin/bgi327. [DOI] [PubMed] [Google Scholar]
- 55.Sheh A, et al. Mutagenic potency of Helicobacter pylori in the gastric mucosa of mice is determined by sex and duration of infection. Proc Natl Acad Sci USA. 2010;107(34):15217–15222. doi: 10.1073/pnas.1009017107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alexandrov LB, et al. Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia P, Pao W, Zhao Z. Patterns and processes of somatic mutations in nine major cancers. BMC Med Genomics. 2014;7:11. doi: 10.1186/1755-8794-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lakritz JR, et al. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget. 2015;6(11):9387–9396. doi: 10.18632/oncotarget.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weisenberger DJ, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 60.Valinluck V, Sowers LC. Inflammation-mediated cytosine damage: A mechanistic link between inflammation and the epigenetic alterations in human cancers. Cancer Res. 2007;67(12):5583–5586. doi: 10.1158/0008-5472.CAN-07-0846. [DOI] [PubMed] [Google Scholar]
- 61.Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase beta. Methods Enzymol. 1995;262:98–107. doi: 10.1016/0076-6879(95)62013-3. [DOI] [PubMed] [Google Scholar]
- 62.Batra VK, et al. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14(4):757–766. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Otwinowski Z, Minor W. Processsing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Emsley P, Cowtan K. 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2126–2132.