Abstract
Many eukaryotes have obligate associations with microorganisms that are transmitted directly between generations. A model for heritable symbiosis is the association of aphids, a clade of sap-feeding insects, and Buchnera aphidicola, a gammaproteobacterium that colonized an aphid ancestor 150 million years ago and persists in almost all 5,000 aphid species. Symbiont acquisition enables evolutionary and ecological expansion; aphids are one of many insect groups that would not exist without heritable symbiosis. Receiving less attention are potential negative ramifications of symbiotic alliances. In the short run, symbionts impose metabolic costs. Over evolutionary time, hosts evolve dependence beyond the original benefits of the symbiosis. Symbiotic partners enter into an evolutionary spiral that leads to irreversible codependence and associated risks. Host adaptations to symbiosis (e.g., immune-system modification) may impose vulnerabilities. Symbiont genomes also continuously accumulate deleterious mutations, limiting their beneficial contributions and environmental tolerance. Finally, the fitness interests of obligate heritable symbionts are distinct from those of their hosts, leading to selfish tendencies. Thus, genes underlying the host–symbiont interface are predicted to follow a coevolutionary arms race, as observed for genes governing host–pathogen interactions. On the macroevolutionary scale, the rapid evolution of interacting symbiont and host genes is predicted to accelerate host speciation rates by generating genetic incompatibilities. However, degeneration of symbiont genomes may ultimately limit the ecological range of host species, potentially increasing extinction risk. Recent results for the aphid–Buchnera symbiosis and related systems illustrate that, whereas heritable symbiosis can expand ecological range and spur diversification, it also presents potential perils.
Keywords: Buchnera, aphid, Muller's ratchet, selection levels, coevolution
Obligate symbiotic relationships shape the evolution of partner lineages. In symbioses that are mutually beneficial, partners evolve traits that enable and stabilize the symbiosis: this cooperative coevolution is emphasized in most studies of symbioses. Genomic work has also revealed that obligate symbiosis produces unusual genome modifications, including extreme reduction, rapid protein evolution, and codon reassignments, all of which are evident in ancient obligate symbionts of insects (1, 2). Recent studies suggest that host genomes also have acquired unusual modifications that are linked to symbiosis, including acquisition of genes from bacterial donors that seem to play a role in controlling or supporting symbionts (3–5). Below, we explore why lineages entering into obligate heritable symbiosis undergo strange patterns of genome evolution and display features that are difficult to interpret simply as adaptations for improving symbiotic function. We refer to the commitment to obligate, inherited symbiosis as the evolutionary “rabbit hole” of obligate symbiosis, implying a generally irreversible journey into a very odd world where the usual rules do not apply.
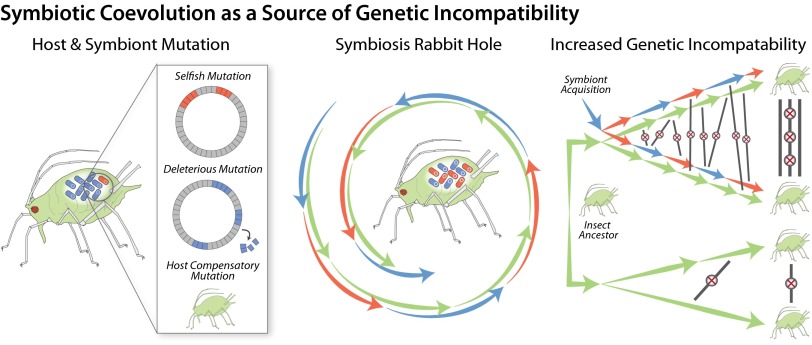
Broadly, the symbiosis rabbit hole refers to the confluence of selection and neutral evolution in generating the extreme patterns of genomic evolution observed in symbiotic partners. As we argue, these extremes are driven by three main forces: deleterious symbiont evolution due to genetic drift, within-host selection leading to symbiont selfishness, and adaptive compensation on the part of hosts (Fig. 1). The interaction of these forces results in rapid and ongoing evolutionary change in both symbiotic partners, with profound evolutionary consequences. Symbiont degeneration coupled with host compensation is a defining characteristic of heritable symbiosis. A salient feature of this relationship is that the host must maintain a viable symbiosis with a partner that has a rapidly evolving genome due to the nonadaptive fixation of mutations through drift. In sum, the host must keep pace with its symbiont as multiple forces draw it ever further down the rabbit hole.
Fig. 1.
Causes and consequences of symbiotic coevolution. Mutations that negatively impact the symbiosis can be fixed through genetic drift due to clonality and small population size (shown in blue) or through within-host selection for selfish symbionts that favor their own fitness over that of the host (red). In response, the host is selected to buffer these mutations (green), leading to a spiral down the symbiosis rabbit hole. This symbiont–host coevolution may drive the rapid accumulation of genetic incompatibilities between host lineages and between hosts and symbiont strains. Lineage-specific symbiont–host coevolution may lead to accelerated reproductive isolation and speciation, which could further reduce the effective size of genetically compatible host populations.
In this perspective, we focus on insect–bacterial symbioses, especially the symbiosis of pea aphid (Hemiptera: Acyrthosiphon pisum) and Buchnera aphidicola (Gammaproteobacteria), for which recent experimental studies have yielded new insights into the integration of symbiotic partners. These ideas are potentially applicable to a broad range of heritable symbioses in which the symbiont is strictly clonal and restricted to living in hosts.
Evolutionary Opportunities from Symbiosis: Ecological Benefit and Lineage Expansion
On macroevolutionary time scales, symbiont acquisition has often enabled evolutionary diversification and ecological expansion. By acquiring maternally transmitted bacterial symbionts, many insect lineages have succeeded in unlocking new ecological niches, particularly ones that present nutritionally unbalanced diets. Aphids and other sap-feeding insects rely on phloem sap or xylem sap as their only food, and these diets are extremely limited in essential amino acids and some vitamins (6). Use of these unbalanced diets is possible because symbionts supply missing nutrients (7–10).
The macroevolutionary and ecological consequences of acquiring symbionts can be immense. Continuing with the same example, symbiont-dependent sap-feeding insects were among the first herbivores to exploit vascular plants (11, 12) and include highly successful clades such as aphids (5,000 described species), whiteflies (1,600 species), psyllids (3,000 species), scale insects (8,000 species), leafhoppers (>20,000 species), cicadas (2,500 species), spittlebugs (3,000 species), and planthoppers (13,000 species) (11). All possess needle-like mouthparts, or stylets, used to access plant fluid and sap diets. These groups exhibit diverse plant–parasitic lifestyles and are critical players in terrestrial ecosystems as vectors of plant disease, food to diverse predators and parasites, and mutualists to other insects including ants. In each of these sap-feeding insect groups, phylogenetic analyses show that obligate symbionts have been vertically transmitted for millions of years, in many cases from the late Permian [>250 Mya (million years ago)] (8, 10). The diversity and abundance of insects feeding on xylem or phloem sap reflect the dominance of vascular plants in terrestrial ecosystems: symbionts provided the entry to a vast and expanding new niche that spread across the globe. As a counterexample, the Coleorrhyncha, which resembles other sap-feeding groups in originating around the same time (the Permian) and having an obligate symbiont, contains only about 24 species, reflecting ties to specific nonvascular plants (certain mosses) (13, 14).
Parallel cases of symbiont-driven ecological expansion have been documented in other insects, including cockroaches (15, 16), ants (17), lice (18), and beetles (19–21). Examples extend into other animal hosts although, in some, transmission may be partly or wholly horizontal rather than strictly vertical. Examples include vesicomyid clams (22, 23), corals (24), earthworms (25), sponges (26), tunicates (24), and flashlight fish (27). In most, phylogenetic analyses demonstrate that symbiont acquisitions occurred in a shared ancestor of a major clade. By freeing hosts from specific nutritional requirements or by providing new protection against pathogens or predators, symbiosis has enabled novel lifestyles and increased long-term fitness.
Acquiring a heritable symbiont is effectively a mutation of major effect, increasing host fitness at the population and clade level. In many, although not all, identified cases, these acquisitions have resulted in a proliferation of descendant lineages, usually comprised of species restricted to a particular dietary niche. Thus, long-term, heritable symbiosis underlies many dominant insect lifestyles and has shaped macroevolutionary and ecological patterns.
Evolutionary Hazards of Symbiosis
Becoming Irreversibly Obligate.
The continuous presence of a vertically transmitted symbiont leads to the evolution of developmental dependence beyond the symbiont’s original contribution; that is, hosts become addicted to their symbionts. In aphids, elimination of Buchnera through antibiotic treatment interferes with development, which typically stalls if Buchnera fails to colonize (28). Aphid females deprived of Buchnera, due to heat, antibiotics, or old age, produce few or no progeny, even when dietary nutrition is sufficient (29). This dependence on Buchnera for development reflects 150 million years of fixation of aphid mutations that are beneficial or neutral in the presence of Buchnera but potentially deleterious in its absence. Thus, adoption of symbionts for nutrient provisioning is a gateway to developmental dependence even when those nutrients are not needed. Indeed, so long as the symbiont is continuously present, addiction can evolve even to deleterious microbes, such as the reproductive parasite Wolbachia (30).
Reflecting their reliance on symbionts, hosts have evolved specialized mechanisms and tissues for housing and supporting symbionts and for transferring them from mother to progeny. In aphids, cells that are specified to become bacteriocytes show distinctive gene expression in early developmental stages, and their cellular fate is determined before Buchnera colonization (31, 32). Bacteriocyte expression of genes underlying amino acid metabolism complements Buchnera pathways for amino acid biosynthesis, reflecting extensive host–symbiont collaboration in this central nutritional function (33–35). Certain aphid genes seem to function solely in controlling or supporting Buchnera. For example, some highly expressed peptides are confined to bacteriocytes or surrounding sheath cells (36). An amino acid transporter expressed in bacteriocytes has altered substrate affinity that imposes negative feedback regulation of essential amino acid production by Buchnera (37). Finally, an aphid-encoded protein, originally of bacterial origin (but not Buchnera), has been shown to be localized within Buchnera cells although its function is not yet known (5). Taken together, these findings for the Buchnera–aphid symbiosis point to extensive genomic and metabolic integration of symbiotic partners and blur the distinction between symbiont and organelle.
Accommodation of symbionts may require that hosts suppress or modify immune responses (38, 39), potentially elevating risk of pathogen invasion. In aphids, many genes underlying responses to Gram-negative bacteria have been eliminated, including the immune deficiency signaling pathway (IMD), peptidoglycan receptor proteins, and antimicrobial peptides (40, 41). Potentially, these losses facilitated the evolution of symbioses with Buchnera, and with numerous facultative symbionts, as supported by the observation that Buchnera cells elicit the IMD pathway in other insects (42). This reduction in immunity seems to have consequences because aphids are susceptible to infections by bacterial pathogens during feeding and during nutritional stress (43, 44). The prospect that immune-system reduction paved the way for the elaborate symbioses in sap-feeding insects generally will likely be resolved from ongoing genome sequencing of additional insect species that vary in symbiotic associations.
As these examples illustrate, once a symbiont is required for development, hosts may become locked in, even when the original symbiotic benefit is reduced or eliminated due to changing ecological conditions or deterioration of symbiont functionality. Evidence that such deterioration indeed occurs is discussed in the next sections.
Symbiont Decay.
A well-documented force affecting heritable symbionts is genetic drift leading to the fixation of neutral or deleterious mutations that cause gene inactivation, gene loss, or inefficiency of gene products (45, 46). The basis for elevated genetic drift is the drastic shift in population genetic structure that occurs when a free-living microorganism adopts an obligate symbiotic lifestyle. The genetic population size becomes largely dependent on the host population size (47), and free-living bacteria have much larger populations than do animals (48). Furthermore, most heritable symbionts are strictly clonal, being transmitted only through host matrilines. This radical change in population structure results in less efficient selection genome-wide, leading to elevated rates of fixation of deleterious mutations (45, 46, 49, 50).
Symbiont genome decay affects genes in all functional categories (8). The first obligate symbiont genome sequenced, that of Buchnera of the pea aphid, was most notable for the fact that it had undergone extensive gene loss and contained no novel genes yet did retain genes for biosynthesis of essential amino acids needed by hosts (51). With more genome sequencing, it became apparent that Buchnera genomes in different aphid lineages continue to undergo irreversible gene loss, over long and short time scales (52–56). Similar ongoing gene loss is evident in every obligate symbiont clade for which multiple genomes have been sequenced (16, 57, 58). Many show far more extreme genome reduction than does Buchnera. Indeed, symbiont genomes have repeatedly evolved to be the very smallest genomes known in cellular organisms (aside from organelles), with total gene counts often <300 and sometimes <150 (1, 3, 10, 58–62). Continuing losses from established obligate symbionts include genes underlying central cellular functions and cell-envelope production, as well as genes underlying symbiotic benefits such as nutrient biosynthesis.
Essential genes that are retained are subject to elevated burdens of slightly deleterious mutations in heritable symbionts. Compared with homologs in free-living relatives, gene products have lower efficiencies and reduced thermal stability (53, 63). Symbionts also exhibit genome-wide accelerated sequence evolution and mutation-driven biases in nucleotide base composition (8, 45, 46, 64, 65). This mutation-driven bias generally favors A+T nucleotides and has extreme effects on polypeptide composition; all encoded proteins in most insect symbionts are strongly shifted toward amino acids that enable higher A+T in the DNA sequence. The negative effects of these mutations are partially masked by constitutively high expression of chaperones that help to stabilize impaired proteins (66–68), but high chaperone expression is itself metabolically costly.
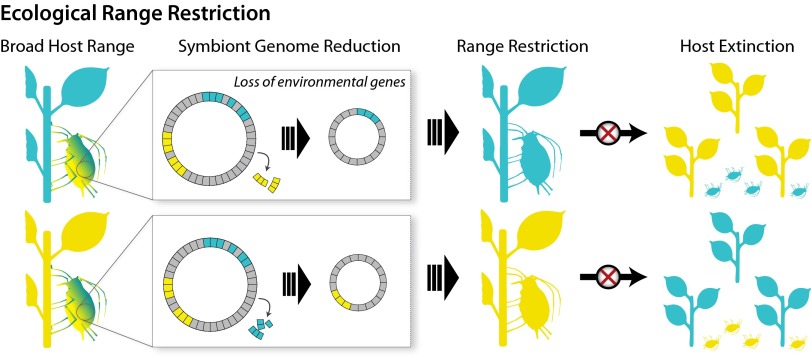
These observations raise a question: How can seemingly deleterious mutations that eliminate or hinder useful symbiont functions become fixed? One explanation depends on the fluctuations in nutrient availability in environments. Host insects encounter varying ecological conditions, such as changes in host plants that affect nutrient availability. If the symbiont provisions nutrients, but the diet sometimes is enriched for those nutrients, selection to maintain the corresponding symbiont pathway will be relaxed, opening the way for inactivation of the underlying genes. The result is that the host now requires the dietary supply, leading to a long-term narrowing of its ecological range (Fig. 2). For example, in some gall-feeding aphids, Buchnera has lost biosynthetic pathways for some nutrients (53), probably because gall formation results in enrichment of ingested sap. This unidirectional loss of Buchnera capabilities potentially prevents the aphid lineage from returning to a broader feeding niche.
Fig. 2.
Ecological range restriction by symbiont gene loss. In a specific environment, some symbiont genes may not be needed, resulting in relaxed selection for their maintenance and inactivation. In sap-feeding insects with obligate symbionts, using a food plant with abundant levels of a particular nutrient can lead to irreversible loss of symbiont genes for making that nutrient. A consequence is permanent restriction of the host’s ecological range: for example, confinement to a smaller set of food plant species. As available resources change over time (e.g., due to climate change), a possible consequence of a narrower ecological niche is smaller population size or eventual extinction.
Loss and decay of symbiont functionality result in selection on hosts to compensate. Hosts require symbionts for nutritional benefits or for proper development so ongoing symbiont decay forces hosts to continually adapt. Host compensatory adaptations are reflected in the elaborate support systems that are beginning to be revealed from studies of symbioses of sap-feeding insects. For example, in mealybugs and psyllids, symbionts have lost most of the genetic machinery for generating cell-envelope components, and genes underlying these functions are instead found within host genomes and are highly expressed in bacteriocytes (3, 4). As hosts evolve to shore up symbiont shortcomings, the latter are able to lose even more functionality, leading to increasingly intricate host support systems. This pressure on hosts to compensate for symbiont decay explains why obligate symbionts have the smallest genomes, by far, of any cellular organisms: Their hosts evolve to compensate for symbiont gene losses, facilitating further loss of symbiont function over time. Thus, the lineage descends into the symbiosis rabbit hole, driven by genetic drift in the symbionts and compensatory adaptation by hosts (Fig. 1).
Symbiont Selfishness.
Fitness of a maternally inherited symbiont is closely aligned with that of its hosts. Thus, natural selection generally favors symbiont features that benefit hosts, such as mechanisms for efficient nutrient provisioning at economical metabolic costs. Indeed, some symbiont features seem to be specific adaptations for increasing host-level fitness: e.g., in Buchnera, pathways for production of some essential amino acids are amplified and located on plasmids as mechanisms for overproducing or regulating the production of these nutrients (9). Nonetheless, the fitness interests of host and symbiont are not identical. The potential for the spread of “selfish” symbiont mutations persists even in the most intimate codependent associations (69). A symbiont mutation that speeds replication of the mutant cell line within a host–and thereby increases its proportional representation in the progeny–can increase in frequency even if it lowers her overall fecundity. Although mutualistic symbioses are often considered as fully cooperative, in fact, we should expect elements of a coevolutionary arms race, or Red Queen evolution, of the sort widely demonstrated for host–pathogen coevolution (70). Accordingly, the machinery underlying symbiont–host integration may represent not a stable solution to host–symbiont integration, but the current status of an ongoing struggle, driven by both conflict and concordance in evolutionary interests.
Among the clear examples of selfishness on the part of maternally inherited symbionts are mechanisms that favor female over male progeny. In most cases, males are a dead end for symbionts (but see ref. 71), and symbionts are not expected to support male reproduction (72). Numerous cases of symbionts manipulating reproduction to favor production of infected matrilines, at male expense, have been documented, including many within the widely known Wolbachia (Alphaproteobacteria) (73), as well as Cardinium (Bacteroidetes) (74). In ancient obligate symbiosis, mechanisms for the propagation of symbionts have been fixed, but ongoing mutations can favor the proliferation and transmission of selfish symbionts within matrilines.
In general, a mutant symbiont cell lineage that replicates faster but does not provision nutrients to hosts might increase proportionally within progeny of a female. However, once fixed, its hosts will have lower fecundity than hosts in which all symbionts provision nutrients (47). Thus, matrilines in which selfish symbionts become fixed are negatively selected within the host population, implying strict limits on symbiont selfishness. Nonetheless, selection at the host level will not eliminate selfish tendencies of symbionts, and hosts are expected to evolve counteradaptations. Several observations do suggest elements of arms race coevolution in intimate, heritable insect symbioses. In both aphids and Sitophilus grain weevils, peptides resembling the classic antimicrobial peptides that are effectors of the innate immune system seem to be key in the containment of maternally inherited nutritional symbionts (36, 75). Likewise, in the obligate symbiosis of tsetse, immune components play a part in regulating symbionts (38). Thus, hosts seem to control symbionts using mechanisms related to those that limit pathogen invasion. However, host control of heritable symbiont proliferation has been investigated in only a few systems.
In addition to favoring direct controls on selfish symbionts, selection on hosts could lead to mechanisms that limit the potential for symbiont-level selection. Such adaptations could involve separation of a distinct symbiont pool used for transmission to progeny (69) or enforcing small inoculum size (47). Host controls seem most likely to evolve when symbionts replicate many times per host generation. Potentially, hosts can eliminate a bacteriocyte along with its resident bacteria if the bacteriocyte is underperforming by not provisioning sufficient nutrients or if proliferating symbiont cells become cancerous. During the life of an aphid female, bacteriocytes are lost; speculatively, this elimination could be selective, functioning as a means of disfavoring retention and transmission of selfish Buchnera cell lines. Although hosts might police their symbionts so as to minimize selfish tendencies and promote cooperation (76), such policing is not yet known from insect symbioses. Most likely, some selfish mutations occur and are countered by hosts. Thus, along with genomic decay through drift, symbiont selfishness is an additional pressure that ultimately tightens the specificity of host–symbiont associations.
Consequences of Symbiosis for Host Evolution
Speciation Rates.
We have argued that symbionts are prone to evolve in directions detrimental to hosts, due both to genetic drift in clonal symbiont populations and to selection favoring selfish traits. An implication is that hosts are continually selected to compensate. Under this scenario, the host–symbiont interface is predicted to rely on rapidly evolving genes that quickly acquire incompatibilities between populations (Fig. 1). In effect, host and symbiont coevolution will drag each symbiotic lineage deeper into its own unique rabbit hole. Thus, incompatibilities between symbiont and host loci, or between different host loci involved in symbiotic control, are expected to emerge quickly and to accelerate the emergence of postzygotic isolating mechanisms, reinforcing reproductive isolation at early stages of lineage divergence. Incompatibilities involving loci functioning in symbioses might arise even for host loci and symbiont genotypes circulating within a population, as seems to occur for nuclear loci within populations (77).
If symbiont–host incompatibilities emerge rapidly, insect clades with obligate symbionts might have higher speciation rates than similarly aged clades without obligate symbionts. In theory, this prediction is testable using a comparative phylogenetic framework to evaluate speciation rates in host clades with and without symbionts. In practice, such tests would be difficult because we still have poor estimates of species diversity in many insect clades, and comprehensive phylogenies are nonexistent. A further prediction, and one that might be tested more readily, is that reproductive isolation in insects with symbionts will often be enforced by incompatibilities between host and symbiont loci or between different host loci that contribute to the regulation, support, and transmission of symbionts. Understanding the role of symbiosis in generating reproductive isolation can be approached through experimental investigations of symbiosis using hybridization or transfection to produce novel host–symbiont combinations (78).
If symbiosis does facilitate speciation, one of the driving forces, genetic drift affecting symbiont genomes, is exacerbated. A major determinant of the rate of fixation of deleterious mutations in symbionts is host population size (47, 50), and each speciation event generates two smaller populations. As the host strives to keep pace with its symbiont, we expect an accelerated descent into the symbiosis rabbit hole.
Ecological Range.
Symbiont evolution can lead to restricted ecological range of hosts by limiting tolerance of both biotic and abiotic factors, such as nutritional availability and temperature. As discussed in Symbiont Decay, if a symbiont loses genes underlying pathways for provisioning its host with nutrients, due to relaxation of purifying selection during periods of temporary nutrient abundance, then the host lineage becomes permanently dependent on environmental sources (or must acquire a new symbiont) (Fig. 2). Losses of nutrient provisioning capabilities are ongoing in all groups of obligate insect symbionts for which genome comparisons within a symbiont clade are available (2, 52–55). By enforcing dietary requirements, these losses are expected to narrow the range of suitable environments for host-insect lineages (Fig. 2).
Host insects cannot completely buffer their symbionts’ environment, and symbionts incur mutations that impact their environmental tolerance, particularly to heat. Obligate symbionts in insects are heat-sensitive and can be killed by temperatures that do not kill their hosts. For example, carpenter ants are limited by the heat sensitivity of their obligate symbiont, Blochmannia (79). Likewise, Buchnera numbers plummet after heat exposure (80, 81). In pea aphid populations that experience continuous cool temperatures, Buchnera evolves to become even more heat sensitive, due to the spread of a mutation inactivating a heat shock promoter (82, 83).
Although symbiont heat sensitivity will be constrained by prevailing temperatures, symbionts generally seem to have narrow thermal range relative to that of hosts (84). The major effect of deleterious amino acid replacements is to lower protein stability. As a general compensation for protein instability, chaperone expression in obligate symbionts is high even under nonstress conditions (68). In Buchnera, chaperonin (GroEL) is produced constitutively at levels equivalent to those during extreme heat shock in Escherichia coli (66). Other Buchnera chaperones are also overexpressed constitutively, and only a few genes retain any transcriptional response to heat (67). Thus, ability to compensate for environmental stressors that destabilize proteins seems to be compromised. The net effect of these tendencies in symbionts is to reduce ecological range and, thus, host population size.
Escaping the Hazards of Symbiosis: Acquiring Novel Symbioses Through Replacement and Supplementation
Once a host lineage has proceeded down the irreversible path into obligate symbiosis, it seems that there is little opportunity to exit. In a few cases, such as the leafhopper subfamily Typhlocybinae, symbionts may have been lost in connection with dietary shifts: e.g., phloem sap to parenchyma. More often, the only escape from degenerate partners seems to be to supplement or replace them with new symbionts. Numerous clear examples of an ancient obligate symbiont being joined or replaced by a newer one are evident, including several in the sap-feeding insects of suborder Auchenorrhyncha (Fig. 3). A likely driver for adding a new symbiont is the degradation of functions in an ancient one: A new symbiont can replace or supplement functions that are lost or inefficient in the older partner.
Fig. 3.
Summary of the gains and losses of heritable symbionts across sap-feeding insects in the order Hemiptera. The phylogenies show evolutionary relationships of host insect groups (gray) and heritable symbionts. For color-coded lines, see Inset legend. Host phylogeny represents the most recent understanding, but placement of certain lineages (e.g., the Coleorrhyncha and Heteroptera) is uncertain (12, 89). Ancestral symbiont names are in boxes along their lineage; in some cases, the same symbiont lineage has different names in different insect clades. Names of acquired symbionts are shown where the symbiont is acquired on the host phylogeny. Dashed lines represent hypothetical relationships and possible origins of symbiosis deep in the evolution of the Hemiptera. The white-dashed lineage represents an ancestral symbiont that permitted the initial diversification of the Hemiptera; its identity is not yet known. Lineages that terminate at a question mark remain uncertain; host–symbiont relationships in these clades are diverse and their origins unclear. See text for citations regarding specific symbionts presented on the tree.
Initially a newly acquired symbiont has a large set of biosynthetic capabilities, including some that are redundant with those of the existing symbiont. Over evolutionary time, this redundancy is eliminated, as illustrated by the perfectly complementary and nonredundant combinations of biosynthetic pathways repeatedly observed for genomes of coresident symbionts in sap-feeding insects (1, 85, 86). Depending on which genome initially loses specific biosynthetic capabilities, a likely outcome is that both old and new symbionts become obligate for the host, each maintaining distinct and complementary contributions.
New symbionts can take on functions previously carried out by more ancient symbionts. For example, the aphid Cinara cedri contains Buchnera along with a second obligate symbiont, Serratia symbiotica, which lives in a distinct type of bacteriocyte (87–89). The acquisition of S. symbiotica coincides with further gene loss in Buchnera: The C. cedri Buchnera genome is substantially smaller and lacks several amino acid biosynthetic genes present in other Buchnera (55). The missing pathways are retained by S. symbiotica, despite its genome also being reduced (87, 90). In this case and others (86), a new symbiont has replaced or supplemented capabilities of an older one. However, the new symbiont embarks on the same evolutionary path of genome decay, driven by mutation and drift.
The sequential acquisition of multiple symbionts that retain complementary biosynthetic capabilities can be reconstructed for several lineages in the sap-feeding suborder Auchenorrhyncha [e.g., cicadas, spittlebugs, leafhoppers, and sharpshooters (10, 58, 91, 92)] and also some aphids, adelgids, and scale insects (87, 93–96) (Fig. 3). Most Auchenorrhyncha lineages contain the widespread ancestral symbiont Sulcia muelleri, plus a coresident partner, which varies among lineages. In each case, Sulcia and its partner have complementary amino acid biosynthetic pathways. The original symbiotic pair in Auchenorrhyncha was Sulcia plus a Betaproteobacterial symbiont; this pair originated >270 Mya and is retained by some descendant lineages (10, 92). In other lineages, including cicadas, sharpshooters, and one tribe of spittlebugs, Sulcia is retained, but the other symbiont is replaced by a new symbiont type (91). These replacements potentially expand the ecological niche of the host insect. For example, in the sharpshooters, a clade within the large leafhopper family Cicadellidae, Baumannia replaced Nasuia (the betaproteobacterium) and may have facilitated the dietary transition from phloem sap to xylem sap. Baumannia has many more biosynthetic pathways than does Nasuia, possibly compensating for the lack of nutrients in xylem sap (97).
Relative to the time scale of host species diversification, symbiont replacements are relatively rare. Examining the morphology of bacteriocytes in symbiont replacements gives some insight into why replacements might be so few. The Sodalis-like symbiont that replaced Zinderia in spittlebugs of tribe Philaenini (Fig. 3) has a reduced genome but retains pathways complementary to those of its Sulcia partner (86). This new symbiont occupies a distinct cell type from the bacteriocytes that house Zinderia in other spittlebugs (91). The occupation of distinct cell types by each coresident symbiont suggests that the Sodalis-like symbiont initially coexisted with Zinderia by invading separate cells of the same host. Indeed, some relatives with the Sodalis group are opportunistic facultative symbionts that invade multiple cell types of insects using invasion machinery closely homologous to that found in pathogenic bacteria (98). In Philaenini and some other hosts (19), Sodalis lineages have become obligate symbionts restricted to specialized host cells. Strikingly, the evolution of novel bacteriocytes for new symbiont acquisitions is the norm among the Auchenorrhyncha (91).
In some insect groups, multiple gains and losses of symbionts have resulted in a confusing mosaic of symbiont combinations in different host clades. For example, scale insect (Coccoidea) families display varied associations, reflecting repeated symbiont acquisitions, replacements, and losses (96, 99). Mealybugs (Pseudococcidae), one clade of scale insects, host an ancestral betaproteobacterium, Tremblaya spp., which coresides with a variety of partners (95). In the mealybug Planococcus citri, this pair is so codependent that Tremblaya has eliminated parts of its own translational machinery, apparently depending on gene products of its partner, which lives within the Tremblaya cytoplasm (3). Similarly psyllids (Psylloidea) and whiteflies (Aleyrodoidea) host ancient gammaproteobacterial symbionts (Carsonella ruddii and Portiera aleyrodidarum) that seem to descend from a single colonization of an ancestor of these related insect groups (Fig. 3). Often, this ancestral symbiont coresides with more recently acquired symbionts, such as symbionts from the Sodalis group or the polyketide-producing symbiont Profftella armatura (100). In some psyllids, Carsonella shows metabolic interdependence with coresident symbionts (57). In each case, the newer obligate symbiont is subject to the same genome decay process as the older symbiont (87, 90, 94, 100).
Outside the Hemiptera, one of the best-studied cases of symbiont replacement is in weevils, one of the most species-rich animal clades. Phylogenetic reconstructions for hosts and symbionts show that an ancestor of weevils was colonized 125 Mya by the symbiont clade Nardonella (gammaproteobacteria), which was retained in many weevil lineages but replaced in several (20, 101, 102). In Sitophilus grain weevils, Nardonella was replaced with a Sodalis-like symbiont that has undergone genome rearrangement and decay. Thus, an evolutionary succession of heritable symbionts may be more widespread than previously appreciated.
The Long-Term Fate of Heritable Symbiosis
Understanding Host–Symbiont Interactions.
We have argued that obligate, heritable symbionts present a moving target requiring ongoing counteradaptation on the part of hosts. This view parallels the proposal that prominent features of genomes, such as size and number of introns and abundance of nongenic DNA, reflect the interplay of natural selection and genetic drift and are therefore governed by population size, in addition to natural selection (48, 103). Similarly, features of intimate symbioses must be considered in the light of the evolutionary processes that govern them, including conflicts between selection on symbionts and selection on hosts, clonality of many symbiont lineages, and genetic population sizes of hosts and symbionts. Both deleterious mutations and selfish mutations are expected to recur in symbionts, and we expect hosts to continually adapt by controlling and supporting their symbionts, and sometimes by admitting novel symbionts. These expectations are consistent with recent findings on the molecular mechanisms acting at the host–symbiont interface, from aphids and Buchnera symbiosis and from other insect symbioses. It is interesting to speculate on the long-term fate of heritable symbiosis in which symbiont genomes are continually declining. Potentially, these processes sometimes limit host distribution so severely that extinction results. However, most obligate insect symbioses are millions of years old so speciation rates must often outnumber extinction rates in these clades.
Differences Between Symbionts and Organelles.
The most evolutionarily successful of heritable symbioses are those that gave rise to mitochondria and plastids, raising the question of why organelles have not been limiting baggage for eukaryotic hosts. Although numerous studies have documented excesses of deleterious mutations circulating within organelle genomes, these mutations are generally recent, remain at low frequencies, and do not become fixed within populations (104). An apparent reason, at least for animal mitochondria, that drift does not more often bring deleterious mutations to fixation is potent selection within the female germ line against mutations that affect mitochondrial function (105). Thus, hosts have evolved mechanisms for preventing transmission of symbionts with harmful mutations. Such mechanisms have the short-term advantage of increasing fitness of offspring and the long-term effect of limiting the accumulation of harmful mutations within lineages. The extent of mechanisms for selective symbiont transmission in heritable symbioses such as those of sap-feeding insects is unknown.
Another difference between insect symbionts and organelles is that genomes of the latter encode little of their own machinery for self-replication and depend on import of needed gene products from the host. One consequence is that they typically have fewer genes and thus present a smaller mutational target. More importantly, organelle genes that are retained are subject to strong host-level selection, limiting their deterioration or selfish tendencies. In contrast, highly reduced genomes of insect symbionts contain mostly genes involved with cell replication, transcription, and translation: Mutations in these genes will impact fitness of individual symbiont cells, where selfishness can originate. Thus, eukaryotic organelles may have escaped the symbiosis rabbit hole primarily because genes controlling symbiont replication are transferred to the host genome.
Conclusion
Symbiosis opens new ecological niches for hosts and could accelerate speciation rates. However, it can also impose long-term fitness costs. The potential negative repercussions of obligate symbiosis raise the possibility that it can limit the ecological range of hosts, reduce population sizes, or even cause extinction of some symbiont-dependent host lineages (along with their symbionts). We have argued that acquiring a maternally inherited obligate symbiont thrusts lineages into a peculiar irreversible coevolutionary relationship that potentially increases speciation rate as well as extinction risk. Genomics-based analyses provide some supportive evidence for these disparate evolutionary consequences of obligate symbiosis.
A main driving force for this process is the genomic decay in symbionts that results from strict clonality and small genetic population size. Therefore, we emphasize that these same expectations do not apply when the symbionts undergo horizontal or environmental transmission or when they are transmitted biparentally. In such cases, the opportunity for continued DNA uptake from the environment or for homologous recombination (sex) persist, circumventing the ratchet-like loss of symbiont function and genes. This point is illustrated by the nephridial symbioses of earthworms. The ancient vertically transmitted symbiont, Verminephrobacter, is inherited biparentally, continues to incorporate foreign DNA, and does not undergo genome reduction (106, 107). Conversely, entrance to the symbiosis rabbit hole does not require that symbionts be intracellular: Genome decay is observed in maternally transmitted extracellular symbionts, exemplified by Ishikawaella capsulata in plataspid stinkbugs (108). The evolutionary rabbit hole does require that the symbiosis be beneficial to hosts, driving them to coadapt. Hosts do not adapt to maintain pathogens, which therefore must retain sufficient capabilities to function independently. Although genomic reduction occurs in host-restricted pathogens, gene loss is far more extreme in obligate symbionts (2), implying that reduction is facilitated by host adaptation. Finally, acquiring a novel symbiont can slow the descent into the symbiosis rabbit hole, but new symbionts ultimately undergo the same drastic genome decay, requiring compensatory evolution in the host.
Acknowledgments
We thank members of the N.A.M. and Howard Ochman laboratory group for feedback. Funding came from a US Department of Agriculture, Agriculture and Food Resource Initiative postdoctoral fellowship TEXW-2013-03420 (to G.M.B.) and National Science Foundation Award IOS1347116.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission.
References
- 1.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 3.Husnik F, et al. Horizontal gene transfer from diverse bacteria to an insect genome enables a tripartite nested mealybug symbiosis. Cell. 2013;153(7):1567–1578. doi: 10.1016/j.cell.2013.05.040. [DOI] [PubMed] [Google Scholar]
- 4.Sloan DB, et al. Parallel histories of horizontal gene transfer facilitated extreme reduction of endosymbiont genomes in sap-feeding insects. Mol Biol Evol. 2014;31(4):857–871. doi: 10.1093/molbev/msu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakabachi A, Ishida K, Hongoh Y, Ohkuma M, Miyagishima S-Y. Aphid gene of bacterial origin encodes a protein transported to an obligate endosymbiont. Curr Biol. 2014;24(14):R640–R641. doi: 10.1016/j.cub.2014.06.038. [DOI] [PubMed] [Google Scholar]
- 6.Sandström J, Moran N. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 1999;91(1):203–210. [Google Scholar]
- 7.Douglas AE. Nutritional interactions in insect-microbial symbioses: Aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 8.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 9.Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 10.Bennett GM, Moran NA. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a Phloem-feeding insect. Genome Biol Evol. 2013;5(9):1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimaldi D, Engel MS. Evolution of the Insects. Cambridge Univ Press; New York: 2005. [Google Scholar]
- 12.Misof B, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–767. doi: 10.1126/science.1257570. [DOI] [PubMed] [Google Scholar]
- 13.Santos-Garcia D, et al. Small but powerful, the primary endosymbiont of moss bugs, Candidatus Evansia muelleri, holds a reduced genome with large biosynthetic capabilities. Genome Biol Evol. 2014;6(7):1875–1893. doi: 10.1093/gbe/evu149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuechler SM, Gibbs G, Burckhardt D, Dettner K, Hartung V. Diversity of bacterial endosymbionts and bacteria-host co-evolution in Gondwanan relict moss bugs (Hemiptera: Coleorrhyncha: Peloridiidae) Environ Microbiol. 2013;15(7):2031–2042. doi: 10.1111/1462-2920.12101. [DOI] [PubMed] [Google Scholar]
- 15.Lo N, Bandi C, Watanabe H, Nalepa C, Beninati T. Evidence for cocladogenesis between diverse dictyopteran lineages and their intracellular endosymbionts. Mol Biol Evol. 2003;20(6):907–913. doi: 10.1093/molbev/msg097. [DOI] [PubMed] [Google Scholar]
- 16.Sabree ZL, Degnan PH, Moran NA. Chromosome stability and gene loss in cockroach endosymbionts. Appl Environ Microbiol. 2010;76(12):4076–4079. doi: 10.1128/AEM.00291-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauer C, Stackebrandt E, Gadau J, Hölldobler B, Gross R. Systematic relationships and cospeciation of bacterial endosymbionts and their carpenter ant host species: Proposal of the new taxon Candidatus Blochmannia gen. nov. Int J Syst Evol Microbiol. 2000;50(Pt 5):1877–1886. doi: 10.1099/00207713-50-5-1877. [DOI] [PubMed] [Google Scholar]
- 18.Allen JM, Reed DL, Perotti MA, Braig HR. Evolutionary relationships of “Candidatus Riesia spp.,” endosymbiotic enterobacteriaceae living within hematophagous primate lice. Appl Environ Microbiol. 2007;73(5):1659–1664. doi: 10.1128/AEM.01877-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heddi A, et al. Molecular and cellular profiles of insect bacteriocytes: Mutualism and harm at the initial evolutionary step of symbiogenesis. Cell Microbiol. 2005;7(2):293–305. doi: 10.1111/j.1462-5822.2004.00461.x. [DOI] [PubMed] [Google Scholar]
- 20.Lefèvre C, et al. Endosymbiont phylogenesis in the dryophthoridae weevils: Evidence for bacterial replacement. Mol Biol Evol. 2004;21(6):965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- 21.Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T. Diversification of endosymbiosis: Replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J. 2013;7(7):1378–1390. doi: 10.1038/ismej.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart FJ, Young CR, Cavanaugh CM. Lateral symbiont acquisition in a maternally transmitted chemosynthetic clam endosymbiosis. Mol Biol Evol. 2008;25(4):673–687. doi: 10.1093/molbev/msn010. [DOI] [PubMed] [Google Scholar]
- 23.Newton ILG, et al. The Calyptogena magnifica chemoautotrophic symbiont genome. Science. 2007;315(5814):998–1000. doi: 10.1126/science.1138438. [DOI] [PubMed] [Google Scholar]
- 24.Kwan JC, et al. Genome streamlining and chemical defense in a coral reef symbiosis. Proc Natl Acad Sci USA. 2012;109(50):20655–20660. doi: 10.1073/pnas.1213820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinel N, Davidson SK, Stahl DA. Verminephrobacter eiseniae gen. nov., sp. nov., a nephridial symbiont of the earthworm Eisenia foetida (Savigny) Int J Syst Evol Microbiol. 2008;58(Pt 9):2147–2157. doi: 10.1099/ijs.0.65174-0. , and erratum (2013) 63(Pt 2):796. [DOI] [PubMed] [Google Scholar]
- 26.Hentschel U, Piel J, Degnan SM, Taylor MW. Genomic insights into the marine sponge microbiome. Nat Rev Microbiol. 2012;10(9):641–654. doi: 10.1038/nrmicro2839. [DOI] [PubMed] [Google Scholar]
- 27.Hendry TA, de Wet JR, Dunlap PV. Genomic signatures of obligate host dependence in the luminous bacterial symbiont of a vertebrate. Environ Microbiol. 2014;16(8):2611–2622. doi: 10.1111/1462-2920.12302. [DOI] [PubMed] [Google Scholar]
- 28.Koga R, Tsuchida T, Sakurai M, Fukatsu T. Selective elimination of aphid endosymbionts: Effects of antibiotic dose and host genotype, and fitness consequences. FEMS Microbiol Ecol. 2007;60(2):229–239. doi: 10.1111/j.1574-6941.2007.00284.x. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson TL, Ishikawa H. Injection of essential amino acids substitutes for bacterial supply in aposymbiotic pea aphids (Acyrthosiphon pisum) Entomol Exp Appl. 2000;94(1):85–91. [Google Scholar]
- 30.Dedeine F, et al. Removing symbiotic Wolbachia bacteria specifically inhibits oogenesis in a parasitic wasp. Proc Natl Acad Sci USA. 2001;98(11):6247–6252. doi: 10.1073/pnas.101304298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Braendle C, et al. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1(1):E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koga R, Meng X-Y, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109(20):E1230–E1237. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102(15):5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poliakov A, et al. Large-scale label-free quantitative proteomics of the pea aphid-Buchnera symbiosis. Mol Cell Proteomics. 2011;10(6):007039. doi: 10.1074/mcp.M110.007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108(7):2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shigenobu S, Stern DL. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc Biol Sci. 2013;280(1750):20121952. doi: 10.1098/rspb.2012.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price DRG, et al. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111(1):320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106(29):12133–12138. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ratzka C, Gross R, Feldhaar H. Gene expression analysis of the endosymbiont-bearing midgut tissue during ontogeny of the carpenter ant Camponotus floridanus. J Insect Physiol. 2013;59(6):611–623. doi: 10.1016/j.jinsphys.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Gerardo NM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11(2):R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8(2):e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Douglas AE, Bouvaine S, Russell RR. How the insect immune system interacts with an obligate symbiotic bacterium. Proc Biol Sci. 2011;278(1704):333–338. doi: 10.1098/rspb.2010.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stavrinides J, McCloskey JK, Ochman H. Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl Environ Microbiol. 2009;75(7):2230–2235. doi: 10.1128/AEM.02860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakabachi A, Ishikawa H, Kudo T. Extraordinary proliferation of microorganisms in aposymbiotic pea aphids, Acyrthosiphon pisum. J Invertebr Pathol. 2003;82(3):152–161. doi: 10.1016/s0022-2011(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 45.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1996;93(7):2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3(11):850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 47.Rispe C, Moran NA. Accumulation of deleterious mutations in endosymbionts: Muller’s ratchet with two levels of selection. Am Nat. 2000;156(4):425–441. doi: 10.1086/303396. [DOI] [PubMed] [Google Scholar]
- 48.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 49.Rispe C, Delmotte F, van Ham RCHJ, Moya A. Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res. 2004;14(1):44–53. doi: 10.1101/gr.1358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersson ME, Berg OG. Muller’s ratchet in symbiont populations. Genetica. 2007;130(2):199–211. doi: 10.1007/s10709-006-9007-7. [DOI] [PubMed] [Google Scholar]
- 51.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407(6800):81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 52.Tamas I, et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science. 2002;296(5577):2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- 53.van Ham RCHJ, et al. Reductive genome evolution in Buchnera aphidicola. Proc Natl Acad Sci USA. 2003;100(2):581–586. doi: 10.1073/pnas.0235981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323(5912):379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 55.Pérez-Brocal V, et al. A small microbial genome: The end of a long symbiotic relationship? Science. 2006;314(5797):312–313. doi: 10.1126/science.1130441. [DOI] [PubMed] [Google Scholar]
- 56.Jiang Z, et al. Comparative analysis of genome sequences from four strains of the Buchnera aphidicola Mp endosymbion of the green peach aphid, Myzus persicae. BMC Genomics. 2013;14:917. doi: 10.1186/1471-2164-14-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sloan DB, Moran NA. Genome reduction and co-evolution between the primary and secondary bacterial symbionts of psyllids. Mol Biol Evol. 2012;29(12):3781–3792. doi: 10.1093/molbev/mss180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. MBio. 2014;5(5):e01697–e14. doi: 10.1128/mBio.01697-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rio RVM, et al. Insight into the transmission biology and species-specific functional capabilities of tsetse (Diptera: glossinidae) obligate symbiont Wigglesworthia. MBio. 2012;3(1):e00240–e11. doi: 10.1128/mBio.00240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sabree ZL, Huang CY, Okusu A, Moran NA, Normark BB. The nutrient supplying capabilities of Uzinura, an endosymbiont of armoured scale insects. Environ Microbiol. 2013;15(7):1988–1999. doi: 10.1111/1462-2920.12058. [DOI] [PubMed] [Google Scholar]
- 61.Williams LE, Wernegreen JJ. Sequence context of indel mutations and their effect on protein evolution in a bacterial endosymbiont. Genome Biol Evol. 2013;5(3):599–605. doi: 10.1093/gbe/evt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314(5797):267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 63.Lambert JD, Moran NA. Deleterious mutations destabilize ribosomal RNA in endosymbiotic bacteria. Proc Natl Acad Sci USA. 1998;95(8):4458–4462. doi: 10.1073/pnas.95.8.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herbeck JT, Funk DJ, Degnan PH, Wernegreen JJ. A conservative test of genetic drift in the endosymbiotic bacterium Buchnera: Slightly deleterious mutations in the chaperonin groEL. Genetics. 2003;165(4):1651–1660. doi: 10.1093/genetics/165.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wernegreen JJ. Reduced selective constraint in endosymbionts: Elevation in radical amino acid replacements occurs genome-wide. PLoS ONE. 2011;6(12):e28905. doi: 10.1371/journal.pone.0028905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baumann P, Baumann L, Clark MA. Levels of Buchnera aphidicola chaperonin GroEL during growth of the aphid Schizaphis graminum. Curr Microbiol. 1996;32:279–285. [Google Scholar]
- 67.Wilcox JL, Dunbar HE, Wolfinger RD, Moran NA. Consequences of reductive evolution for gene expression in an obligate endosymbiont. Mol Microbiol. 2003;48(6):1491–1500. doi: 10.1046/j.1365-2958.2003.03522.x. [DOI] [PubMed] [Google Scholar]
- 68.Kupper M, Gupta SK, Feldhaar H, Gross R. Versatile roles of the chaperonin GroEL in microorganism-insect interactions. FEMS Microbiol Lett. 2014;353(1):1–10. doi: 10.1111/1574-6968.12390. [DOI] [PubMed] [Google Scholar]
- 69.Frank SA. Models of symbiosis. Am Nat. 1997;150(Suppl 1):S80–S99. doi: 10.1086/286051. [DOI] [PubMed] [Google Scholar]
- 70.Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15(6):379–393. doi: 10.1038/nrg3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Watanabe K, Yukuhiro F, Matsuura Y, Fukatsu T, Noda H. Intrasperm vertical symbiont transmission. Proc Natl Acad Sci USA. 2014;111(20):7433–7437. doi: 10.1073/pnas.1402476111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frank SA. Evolution: Mitochondrial burden on male health. Curr Biol. 2012;22(18):R797–R799. doi: 10.1016/j.cub.2012.07.066. [DOI] [PubMed] [Google Scholar]
- 73.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6(10):741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 74.Penz T, et al. Comparative genomics suggests an independent origin of cytoplasmic incompatibility in Cardinium hertigii. PLoS Genet. 2012;8(10):e1003012. doi: 10.1371/journal.pgen.1003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Login FH, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334(6054):362–365. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 76.Frank SA. Perspective: Repression of competition and the evolution of cooperation. Evolution. 2003;57(4):693–705. doi: 10.1111/j.0014-3820.2003.tb00283.x. [DOI] [PubMed] [Google Scholar]
- 77.Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. Genetic incompatibilities are widespread within species. Nature. 2013;504(7478):135–137. doi: 10.1038/nature12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran NA, Yun Y. Experimental replacement of an obligate insect symbiont. Proc Natl Acad Sci USA. 2015;112(7):2093–2096. doi: 10.1073/pnas.1420037112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan Y, Wernegreen JJ. Can’t take the heat: High temperature depletes bacterial endosymbionts of ants. Microb Ecol. 2013;66(3):727–733. doi: 10.1007/s00248-013-0264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burke G, Fiehn O, Moran N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 2010;4(2):242–252. doi: 10.1038/ismej.2009.114. [DOI] [PubMed] [Google Scholar]
- 81.Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27(2):189–195. [Google Scholar]
- 82.Dunbar HE, Wilson ACC, Ferguson NR, Moran NA. Aphid thermal tolerance is governed by a point mutation in bacterial symbionts. PLoS Biol. 2007;5(5):e96. doi: 10.1371/journal.pbio.0050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burke GR, McLaughlin HJ, Simon J-C, Moran NA. Dynamics of a recurrent Buchnera mutation that affects thermal tolerance of pea aphid hosts. Genetics. 2010;186(1):367–372. doi: 10.1534/genetics.110.117440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wernegreen JJ. Mutualism meltdown in insects: Bacteria constrain thermal adaptation. Curr Opin Microbiol. 2012;15(3):255–262. doi: 10.1016/j.mib.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106(36):15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koga R, Moran NA. Swapping symbionts in spittlebugs: Evolutionary replacement of a reduced genome symbiont. ISME J. 2014;8(6):1237–1246. doi: 10.1038/ismej.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lamelas A, et al. Serratia symbiotica from the aphid Cinara cedri: A missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7(11):e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gómez-Valero L, et al. Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J Bacteriol. 2004;186(19):6626–6633. doi: 10.1128/JB.186.19.6626-6633.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Burke GR, Normark BB, Favret C, Moran NA. Evolution and diversity of facultative symbionts from the aphid subfamily Lachninae. Appl Environ Microbiol. 2009;75(16):5328–5335. doi: 10.1128/AEM.00717-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koga R, Bennett GM, Cryan JR, Moran NA. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 2013;15(7):2073–2081. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 92.Urban JM, Cryan JR. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea) BMC Evol Biol. 2012;12:87. doi: 10.1186/1471-2148-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Toenshoff ER, Gruber D, Horn M. Co-evolution and symbiont replacement shaped the symbiosis between adelgids (Hemiptera: Adelgidae) and their bacterial symbionts. Environ Microbiol. 2012;14(5):1284–1295. doi: 10.1111/j.1462-2920.2012.02712.x. [DOI] [PubMed] [Google Scholar]
- 94.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21(16):1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thao ML, Gullan PJ, Baumann P. Secondary (gamma-Proteobacteria) endosymbionts infect the primary (beta-Proteobacteria) endosymbionts of mealybugs multiple times and coevolve with their hosts. Appl Environ Microbiol. 2002;68(7):3190–3197. doi: 10.1128/AEM.68.7.3190-3197.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rosenblueth M, Sayavedra L, Sámano-Sánchez H, Roth A, Martínez-Romero E. Evolutionary relationships of flavobacterial and enterobacterial endosymbionts with their scale insect hosts (Hemiptera: Coccoidea) J Evol Biol. 2012;25(11):2357–2368. doi: 10.1111/j.1420-9101.2012.02611.x. [DOI] [PubMed] [Google Scholar]
- 97.Wu D, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4(6):e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dale C, Plague GR, Wang B, Ochman H, Moran NA. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc Natl Acad Sci USA. 2002;99(19):12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosas-Pérez T, Rosenblueth M, Rincón-Rosales R, Mora J, Martínez-Romero E. Genome sequence of “Candidatus Walczuchella monophlebidarum” the flavobacterial endosymbiont of Llaveia axin axin (Hemiptera: Coccoidea: Monophlebidae) Genome Biol Evol. 2014;6(3):714–726. doi: 10.1093/gbe/evu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nakabachi A, et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 2013;23(15):1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 101.Toju H, et al. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol. 2010;76(1):275–282. doi: 10.1128/AEM.02154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Conord C, et al. Long-term evolutionary stability of bacterial endosymbiosis in curculionoidea: Additional evidence of symbiont replacement in the dryophthoridae family. Mol Biol Evol. 2008;25(5):859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- 103.Kuo C-H, Moran NA, Ochman H. The consequences of genetic drift for bacterial genome complexity. Genome Res. 2009;19(8):1450–1454. doi: 10.1101/gr.091785.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nachman MW. Deleterious mutations in animal mitochondrial DNA. Genetica. 1998;102-103(1-6):61–69. [PubMed] [Google Scholar]
- 105.Hill JH, Chen Z, Xu H. Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat Genet. 2014;46(4):389–392. doi: 10.1038/ng.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kjeldsen KU, et al. Purifying selection and molecular adaptation in the genome of Verminephrobacter, the heritable symbiotic bacteria of earthworms. Genome Biol Evol. 2012;4(3):307–315. doi: 10.1093/gbe/evs014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lund MB, Kjeldsen KU, Schramm A. The earthworm-Verminephrobacter symbiosis: An emerging experimental system to study extracellular symbiosis. Front Microbiol. 2014;5:128. doi: 10.3389/fmicb.2014.00128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 2011;3:702–714. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]