Significance
The spindle assembly checkpoint (SAC) works as a surveillance mechanism to ensure accurate segregation of genetic materials during cell division. Protein kinase monopolar spindle 1 (Mps1) plays a key role in SAC, but the mechanism of Mps1 action in chromosome segregation remains elusive. In this study, we identified a previously undefined structural determinant of Mps1 named “IRK” (internal region for kinetochore localization) and demonstrated its functional importance in accurate kinetochore–microtubule attachment. Mechanistically, a dynamic hierarchical interaction between Mps1 and the nuclear division cycle 80 complex (Ndc80C) orchestrates accurate mitosis, because persistent association of inactive Mps1 with Ndc80C via the IRK perturbs correct kinetochore–microtubule attachment. Our results provide a new mechanistic insight into the spatiotemporal dynamics of Mps1 activity at the kinetochore in mitosis.
Keywords: Mps1 kinase, spindle assembly checkpoint, chromosome alignment, kinetochore–microtubule attachment, reversine
Abstract
The spindle assembly checkpoint (SAC) is a conserved signaling pathway that monitors faithful chromosome segregation during mitosis. As a core component of SAC, the evolutionarily conserved kinase monopolar spindle 1 (Mps1) has been implicated in regulating chromosome alignment, but the underlying molecular mechanism remains unclear. Our molecular delineation of Mps1 activity in SAC led to discovery of a previously unidentified structural determinant underlying Mps1 function at the kinetochores. Here, we show that Mps1 contains an internal region for kinetochore localization (IRK) adjacent to the tetratricopeptide repeat domain. Importantly, the IRK region determines the kinetochore localization of inactive Mps1, and an accumulation of inactive Mps1 perturbs accurate chromosome alignment and mitotic progression. Mechanistically, the IRK region binds to the nuclear division cycle 80 complex (Ndc80C), and accumulation of inactive Mps1 at the kinetochores prevents a dynamic interaction between Ndc80C and spindle microtubules (MTs), resulting in an aberrant kinetochore attachment. Thus, our results present a previously undefined mechanism by which Mps1 functions in chromosome alignment by orchestrating Ndc80C–MT interactions and highlight the importance of the precise spatiotemporal regulation of Mps1 kinase activity and kinetochore localization in accurate mitotic progression.
Faithful distribution of the duplicated genome into two daughter cells during mitosis depends on proper kinetochore–microtubule (MT) attachments. Defects in kinetochore–MT attachments result in chromosome missegregation, causing aneuploidy, a hallmark of cancer (1, 2). To ensure accurate chromosome segregation, cells use the spindle assembly checkpoint (SAC) to monitor kinetochore biorientation and to control the metaphase-to-anaphase transition. Cells enter anaphase only after the SAC is satisfied, requiring that all kinetochores be attached to MTs and be properly bioriented (3, 4). The core components of SAC signaling include mitotic arrest deficient-like 1 (Mad1), Mad2, Mad3/BubR1 (budding uninhibited by benzimidazole-related 1), Bub1, Bub3, monopolar spindle 1 (Mps1), and aurora B. The full SAC function requires the correct centromere/kinetochore localization of all SAC proteins (5).
Among the SAC components, Mps1 was identified originally in budding yeast as a gene required for duplication of the spindle pole body (6). Subsequently, Mps1 orthologs were found in various species, from fungi to mammals. The stringent requirement of Mps1 for SAC activity is conserved in evolution (6–13). Human Mps1 kinase (also known as “TTK”) is expressed in a cell-cycle–dependent manner and has highest expression levels and activity during mitosis. Its localization is also dynamic (8, 14). Although the molecular mechanism remains unclear, Mps1 is required to recruit Mad1 and Mad2 to unattached kinetochores, supporting its essential role in SAC activity (15–18). It also is clear that aurora B kinase activity and the outer-layer kinetochore protein nuclear division cycle 80 (Ndc80)/Hec1 are required for Mps1 localization to kinetochores, as evidenced by recent work, including ours (17, 19–24). How Mps1 activates the SAC is now becoming clear. Mps1 recruits Bub1/Bub3 and BubR1/Bub3 to kinetochores through phosphorylation of KNL1, the kinetochore receptor protein of Bub1 and BubR1 (25–30).
Despite much progress in understanding Mps1 functions, it remains unclear how Mps1 is involved in regulating chromosome alignment. In budding yeast mitosis, Mps1 regulates mitotic chromosome alignment by promoting kinetochore biorientation independently of Ipl1 (aurora B in humans) (31), but in budding yeast meiosis Mps1 must collaborate with Ipl1 to mediate meiotic kinetochore biorientation (32). In humans, Mps1 regulates chromosomal alignment by modulating aurora B kinase activity (33), but recent chemical biology studies show that Mps1 kinase activity is important for proper chromosome alignment and segregation, independently of aurora B (22, 34–36). Therefore whether Mps1 regulates chromosome alignment through modulation of aurora B kinase activity is still under debate (37).
In this study, we reexamined the function of human Mps1 in chromosome alignment. We found that chromosomal alignment is largely achieved in Mps1 knockdown cells, provided that cells are arrested in metaphase in the presence of MG132, a proteasome inhibitor. However, disrupting Mps1 activity via small molecule inhibitors perturbs chromosomal alignment, even in the presence of MG132. This chromosome misalignment is caused by the abnormal accumulation of inactive Mps1 in the kinetochore and the subsequent failure of correct kinetochore–MT attachments. Further, we demonstrate that inactive Mps1 does not depend on the previously reported tetratricopeptide repeat (TPR) domain for localizing to kinetochores, and we identify a previously unidentified region adjacent to the C terminus of the TPR domain that is responsible for localizing inactive Mps1 to kinetochores. Thus, our work highlights that Mps1 kinase activity is necessary in regulating chromosome alignment and that it must be tightly regulated in space and time to ensure proper localization of Mps1 at kinetochores.
Results
Mps1 Plays a Minor Role in Promoting Chromosome Alignment.
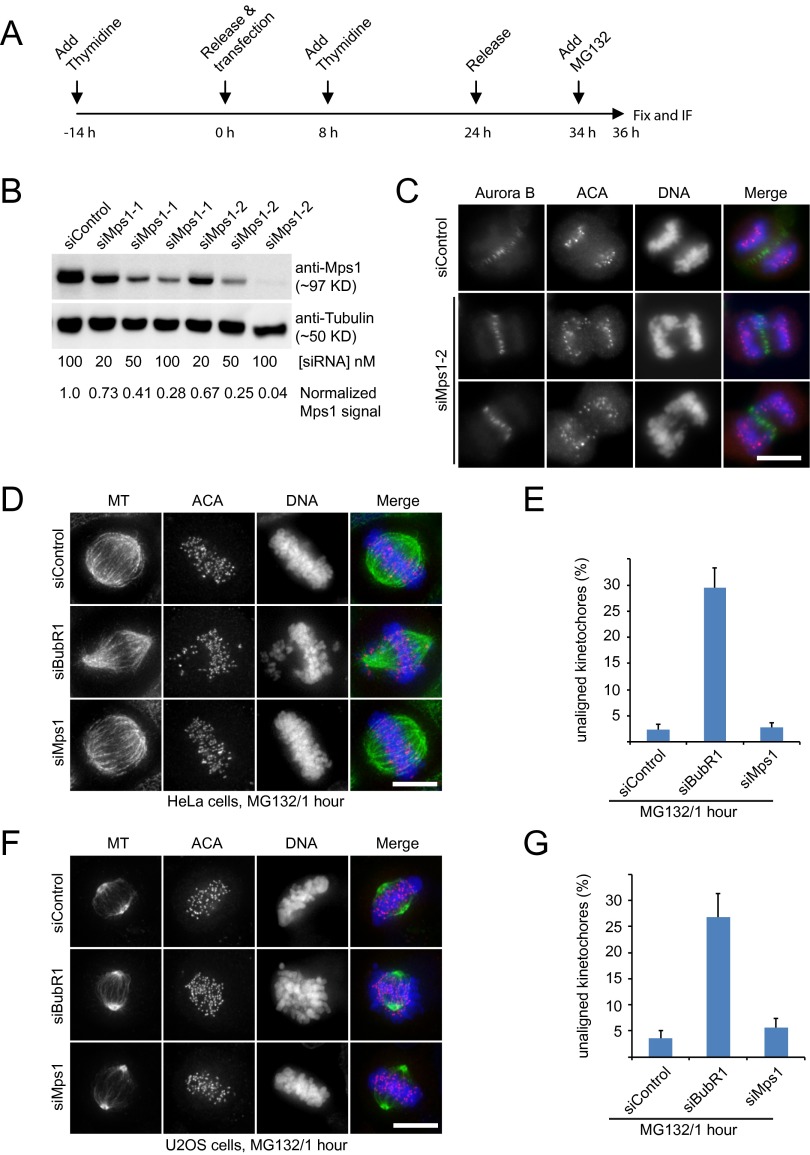
Mps1 has been shown to be required for chromosome alignment, likely through regulating aurora B kinase activity (33). However, later studies demonstrated that inhibiting Mps1 activity did not perturb aurora B kinase activity (22, 35, 36). To address this discrepancy, we aimed to reassess the role of Mps1 in chromosome alignment. We used the well-established MG132 treatment approach, in which cells were synchronized at G2/M by double-thymidine block release, followed by the addition of MG132 to arrest cells in metaphase (Fig. S1A) (38). Immunofluorescence staining indicated that the chromosomes aligned properly to the spindle equator in shMock-transfected cells (Fig. 1 A and B). Consistent with published results, knocking down BubR1 disrupted normal chromosome alignment (38–40). However, when Mps1 was knocked down by shRNAs (designated “shMps1-1” and “shMps1-2”), most chromosomes aligned correctly. Full chromosome alignment was achieved in about 80% of Mps1-knockdown cells, and one or two pairs of chromosomes remained unaligned in less than 20% of cells (Fig. 1 A and B). We next examined Mps1 function using chemically synthesized siRNA with different targeting sequences. siMps1-2 siRNA knocked down endogenous Mps1 more effectively than siMps1-1, and more than 90% of anaphase cells contained lagging chromosomes (Fig. S1 B and C). Therefore, we used siMps1-2 to investigate Mps1 function. Consistently, most chromosomes aligned properly in siMps1-2–transfected cells, whereas severe defects in chromosome alignment were found in cells transfected with siBubR1 (Fig. S1 D and E). To confirm our findings, we carried out similar experiments using untransformed diploid hTERT-RPE1 cells (Fig. 1 C and D) and p53-positive U2OS cells (Fig. S1 F and G) and obtained nearly the same results.
Fig. S1.
Mps1 plays a minor role in facilitating chromosome alignment during mitosis. (A) Schematic representation of the protocol used to analyze the chromosome alignment functions of differently treated cells. (B) Western blot for Mps1 and α-tubulin of differently treated cells. Cells were transfected with siControl, siMps1-1, or siMps1-2 at the indicated concentration. Thirty-six hours post transfection, mitotic cell lysates were probed with anti-Mps1 and anti-tubulin antibodies, respectively. (C) Representative immunofluorescence images of HeLa cells treated with different siRNAs as indicated. Thirty-six hours post transfection, cells were fixed and costained for aurora B (green), ACA (red), and DNA (blue). (Scale bar, 10 μm.) (D and F) Representative immunofluorescence images of HeLa cells (D) or U2OS cells (F) treated with different siRNAs as indicated. Thirty-five hours post transfection, cells were treated with MG132 for 1 h. Then cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bars, 10 μm.) (E and G) Bar graphs illustrating the percentage of unaligned kinetochores in cells treated as in D (E) or F (G). In each experiment, five cells were measured (>60 kinetochores per cell). Values are mean ± SE of three independent experiments.
Fig. 1.
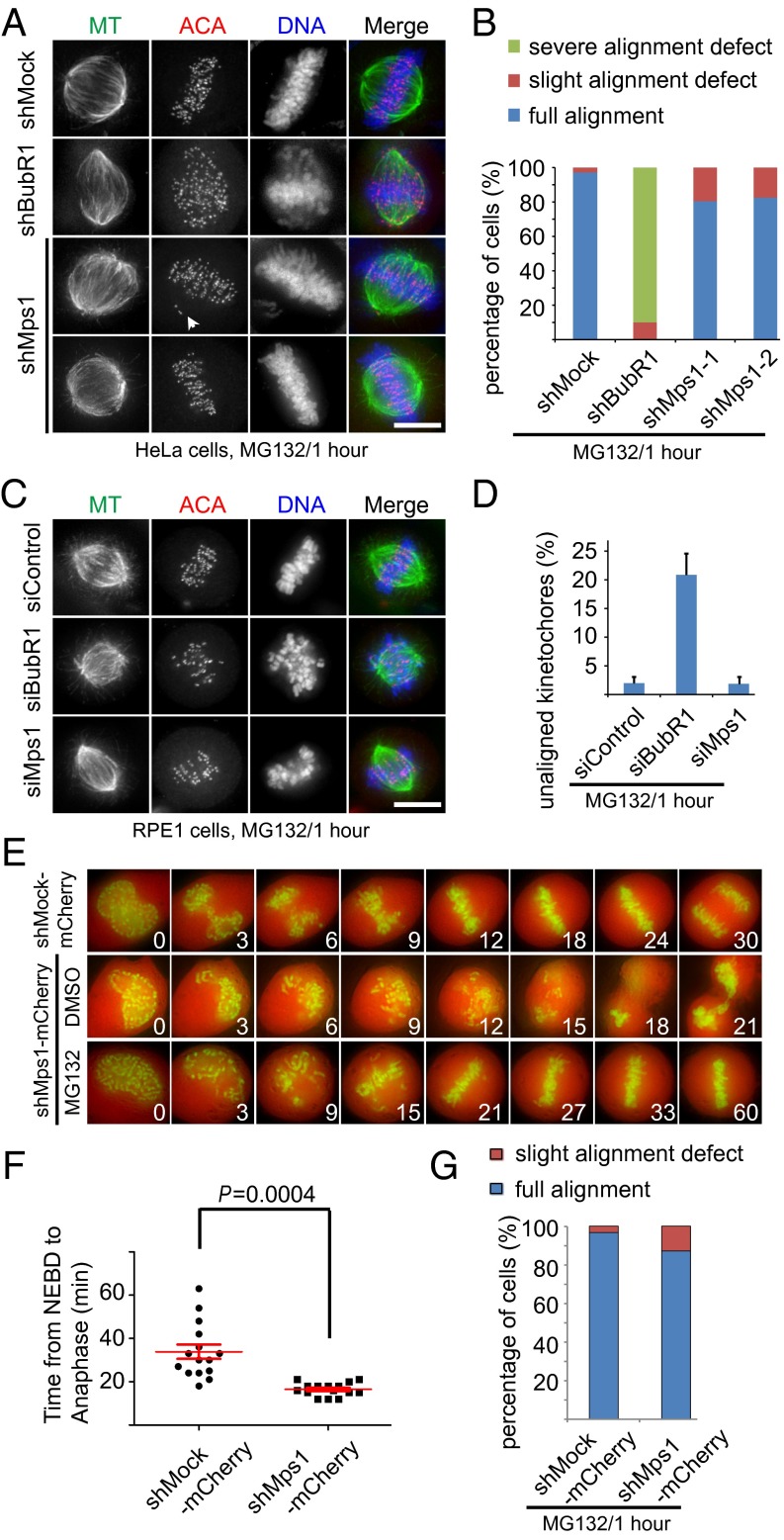
Mps1 plays a minor role in facilitating chromosome alignment during mitosis. (A and C) Representative immunofluorescence images of HeLa cells (A) and RPE1 cells (C) treated with different shRNAs/siRNAs, as indicated. At 36 h after transfection, cells were treated with MG132 for 1 h. Then cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bars, 10 μm.) (B) Bar graph illustrating percentages of cells with fully aligned kinetochores, with 1–10 unaligned kinetochores (slight alignment defect), and with more than 10 unaligned kinetochores (severe alignment defect), treated as in A. Values are means ± SE of three independent experiments. (D) Bar graph illustrating the percentage of unaligned kinetochores in cells treated as in C. Values are means ± SE of three independent experiments. (E) Representative still photographs illustrating mitotic progression in H2B-GFP–expressing cells transfected with plasmid coexpressing mCherry (as readout for shMps1 transfection) and shMps1 or with plasmid coexpressing mCherry and shMock. At 36 h after transfection, cells were treated with DMSO and/or MG132. Images were acquired at the indicated time points after the start of NEBD. (F) Scatter plots of the time from NEBD to the beginning of anaphase in cells treated as in E (n = 15 cells). Bars indicate means ± SE. Student’s t test was used to calculate P values for comparison of shMock and shMps1 samples. (G) Bar graph illustrating percentages of cells photographed in E with fully aligned chromosomes or with a few unaligned chromosomes within 1 h after treatment.
Next, we carried out live-cell imaging to examine chromosome alignment in cells expressing shMps1. In shMock-transfected cells, most chromosomes congressed to the metaphase plate ∼12 min after nuclear envelope breakdown (NEBD) and entered anaphase about 30 min after NEBD. shMps1-transfected cells, however, appeared to enter mitosis normally but entered anaphase prematurely with unaligned chromosomes, indicating that the SAC is defective (Fig. 1 E and F). Examination showed that most control cells formed a metaphase plate at 12 min after NEBD, but Mps1-depleted cells lacked a defined metaphase plate at the same time point. These data suggest that Mps1 promotes efficient chromosome alignment. When the cells were arrested at metaphase with MG132, 88% of these shMps1-expressing cells achieved chromosome alignment within 1 h (25 cells were examined) (Fig. 1G). We conclude that Mps1 promotes efficient chromosome alignment during unperturbed mitosis, but Mps1 is dispensable for chromosome alignment in the presence of MG132.
Mps1 Is Not Required for Correcting Errors in the Kinetochore–MT Attachment.
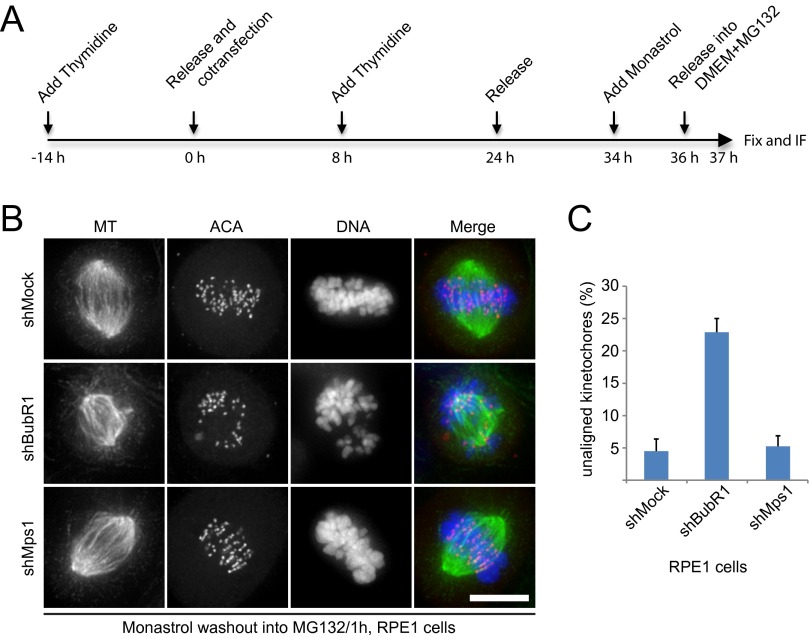
Next, we asked whether Mps1 plays a role in correcting erroneous kinetochore–MT attachments. We used the well-established monastrol washout approach as illustrated in Fig. S2A (41), because cells treated with monastrol contain a large number of erroneous kinetochore–MT attachments. In shMock- and shMps1-transfected HeLa cells, chromosome alignment appeared to be normal. In shBubR1-transfected HeLa cells, however, chromosome misalignment was apparent (Fig. 2 A and B). Live-cell imaging confirmed the findings (Fig. 2C). Consistently, experiments carried out using RPE1 cells showed similar results (Fig. S2 B and C). Thus, we concluded that Mps1 is not involved in correcting errors in kinetochore–MT attachments.
Fig. S2.
Mps1 is dispensable for the correction of errors in kinetochore–MT attachment. (A) Schematic representation of the protocol used to analyze the functions of kinetochore–MT attachment error correction in differently treated cells. (B) Representative immunofluorescence images of RPE1 cells treated with different shRNAs as indicated. Thirty-four hours post transfection, cells were treated with monastrol for 2 h and then were released into medium containing MG132 for 1 h. Then cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bar, 10 μm.) (C) Bar graph illustrating the percentage of unaligned kinetochores in cells treated as in B. Values are mean ± SE of three independent experiments.
Fig. 2.
Mps1 is not essential for the correction of errors in kinetochore–MT attachment. (A) Representative immunofluorescence images of HeLa cells treated with different shRNAs as indicated. At 34 h after transfection, cells were treated with monastrol for 2 h and then were released for 1 h in medium containing MG132. Then cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bar, 10 μm.) (B) Bar graph illustrating the percentage of unaligned kinetochores in cells treated as in A. Values are means ± SE of three independent experiments. (C) Representative still photographs illustrating mitotic progression in H2B-GFP–expressing cells transfected with different plasmids coexpressing mCherry (as a readout for shRNA transfection) and shMock/shMps1/shBubR1. At 36 h after transfection, cells were treated with monastrol for 2 h and then were released in DMEM containing MG132. Images were acquired at the indicated time points. (Scale bars, 10 μm.)
Inhibition of Mps1 Kinase Activity Prevents Kinetochore–MT Attachments.
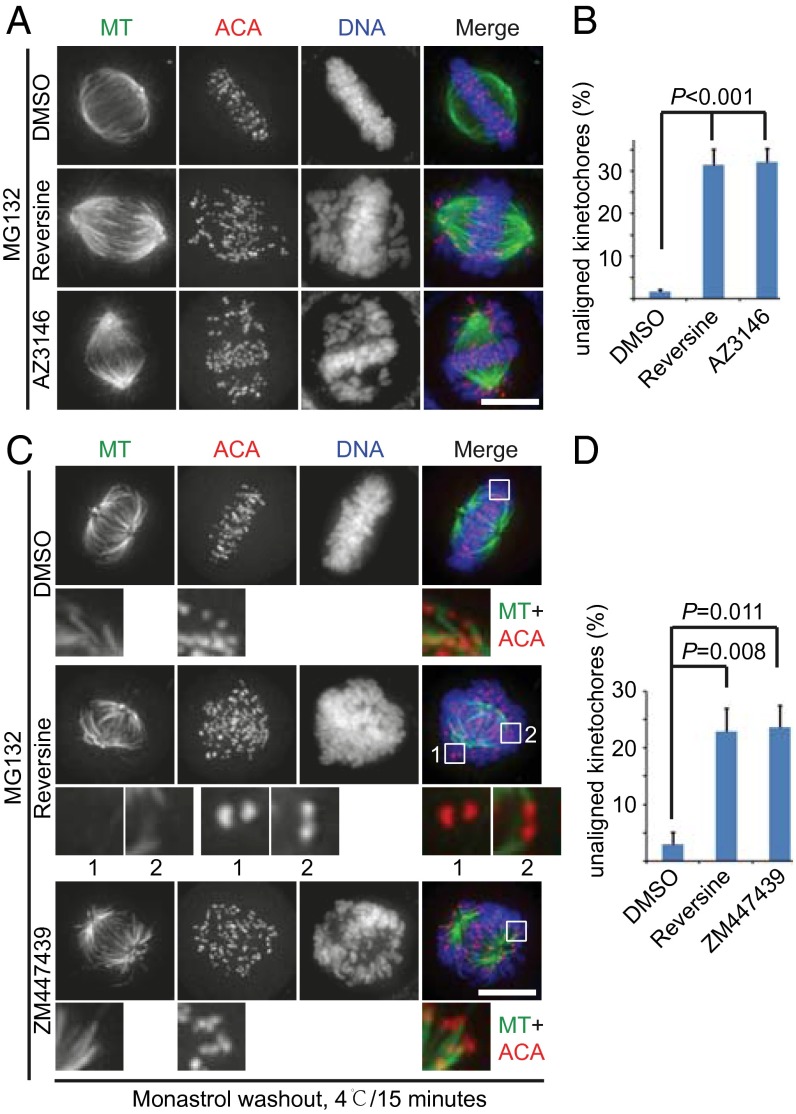
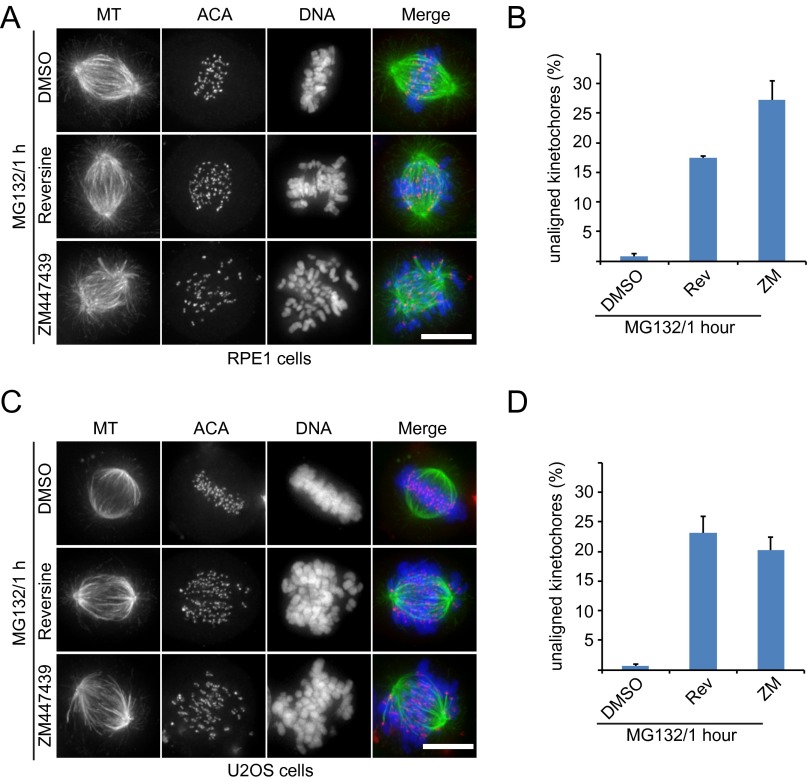
Previous RNA interference and chemical biology studies found that human Mps1 is essential for chromosome alignment and error correction (22, 33, 35, 36). However, our observations so far could not fully confirm those results. Therefore, we tested whether Mps1 kinase activity is important for regulating chromosome alignment. HeLa cells treated with both reversine and AZ3146, two potent chemical inhibitors for inhibiting Mps1 kinase activity, displayed remarkable chromosome misalignment (Fig. 3 A and B), suggesting that Mps1 kinase activity is essential for proper alignment. We next performed a monastrol washout assay to examine the correction of errors in chromosome attachment. When released in DMEM (Dulbecco's modified eagle medium) containing MG132 for 1 h, cells established bipolar spindles, and most chromosomes aligned correctly (Fig. 3C, Top). When cells were released in DMEM containing MG132 plus reversine, there were numerous unaligned chromosomes (Fig. 3 C, Middle and D). Consistent with the previous finding (42), after cells were released in DMEM containing MG132 plus the aurora B inhibitor ZM447439 (referred to as “ZM” hereafter), numerous chromosomes remained unaligned, and syntelic kinetochore–MT attachment was clear (Fig. 3 C, Bottom and D). However, examination of the kinetochore–MT attachment revealed that, unlike the stable syntelic kinetochore–MT attachment in the aurora B inhibition group, all unaligned kinetochores lost the MT attachment in the Mps1 inhibition group (enlarged inset 1 in Fig. 3C). This observation suggests that, although inhibiting Mps1 disrupts chromosome alignment, it interferes with a pathway other than aurora B-dependent error correction. We next examined the phenotype of Mps1 inhibition in RPE1 cells and U2OS cells. Consistent with the data for HeLa cells, inhibition of Mps1 also perturbed chromosome alignment severely in RPE1 (Fig. S3 A and B) and U2OS (Fig. S3 C and D) cells. Taken together, these findings demonstrate that inhibiting Mps1 kinase activity disrupts correct chromosomal alignment but does so through a mechanism other than aurora B inhibition.
Fig. 3.
Inhibiting Mps1 kinase activity causes a severe defect in chromosome alignment through a mechanism other than aurora B inhibition. (A) Representative immunofluorescence images of HeLa cells treated as indicated. Cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bar, 10 μm.) (B) Quantification of the degree of chromosome misalignment in cells treated as in A. Kinetochore position was measured as a function of the distance from the pole to the spindle equator. Bars represent the means ± SE of three independent experiments. In each experiment, five cells were measured (>60 kinetochores per cell). (C) Representative immunofluorescence images of HeLa cells treated with monastrol for 2 h and then released for 1 h in medium containing DMSO/MG132, reversine/MG132, or ZM/MG132. Cells then were incubated at 4 °C for 15 min and fixed for immunofluorescence staining for MT (green), ACA (red), and DNA (blue). Magnified views of the boxed areas are shown in the lower panels. (Scale bar, 10 μm.) (D) Quantification of the degree of chromosome misalignment in cells treated as in C. Bars represent the means ± SE of three independent experiments. In each experiment, five cells were measured (>60 kinetochores per cell). Student’s t test was used to calculate P values.
Fig. S3.
Inhibiting Mps1 kinase activity causes a severe defect in chromosome alignment. (A and C) Representative immunofluorescence images of RPE1 cells (A) and U2OS cells (C) treated as indicated. Cells were fixed and costained for MT (green), ACA (red), and DNA (blue). (Scale bars, 10 μm.) (B and D) Quantification of the degree of chromosome misalignment in cells treated as in A (B) or C (D). Kinetochore position was measured as a function of the distance from pole to spindle equator. Bars represent the mean ± SE of three independent experiments. In each experiment, five cells were measured (>60 kinetochores per cell).
Inactive Mps1 Accumulates Abnormally at Kinetochores, Leading to Defects in the Establishment of Kinetochore–MT Attachments.
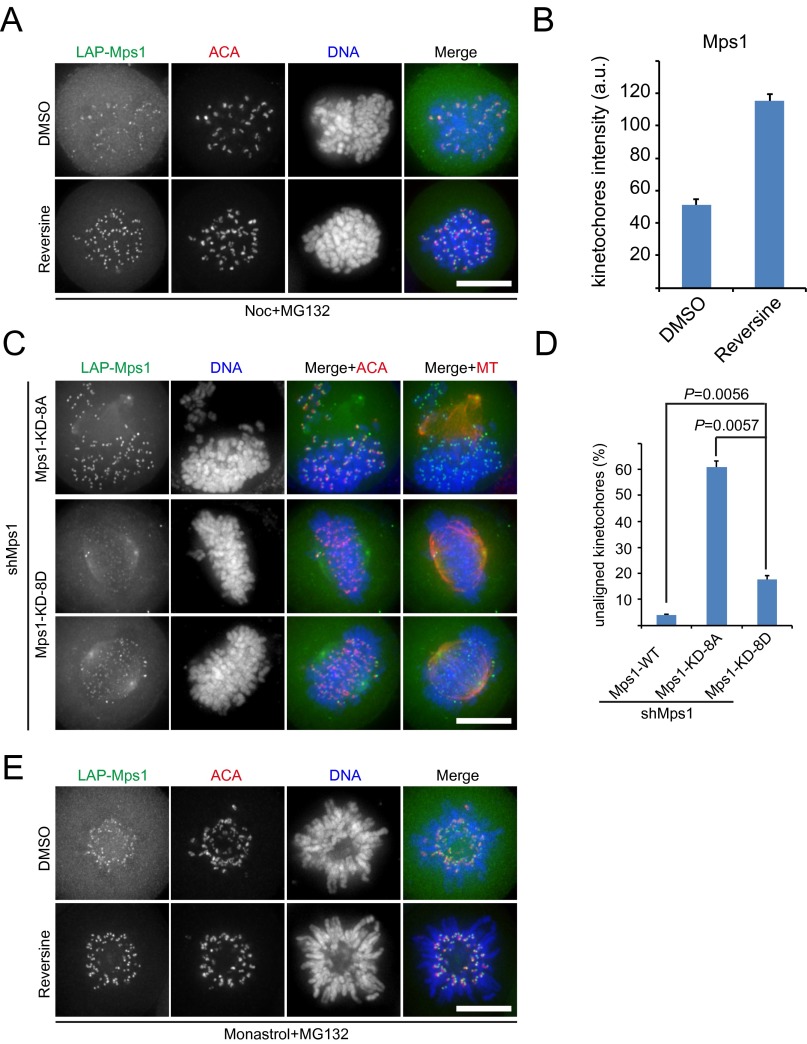
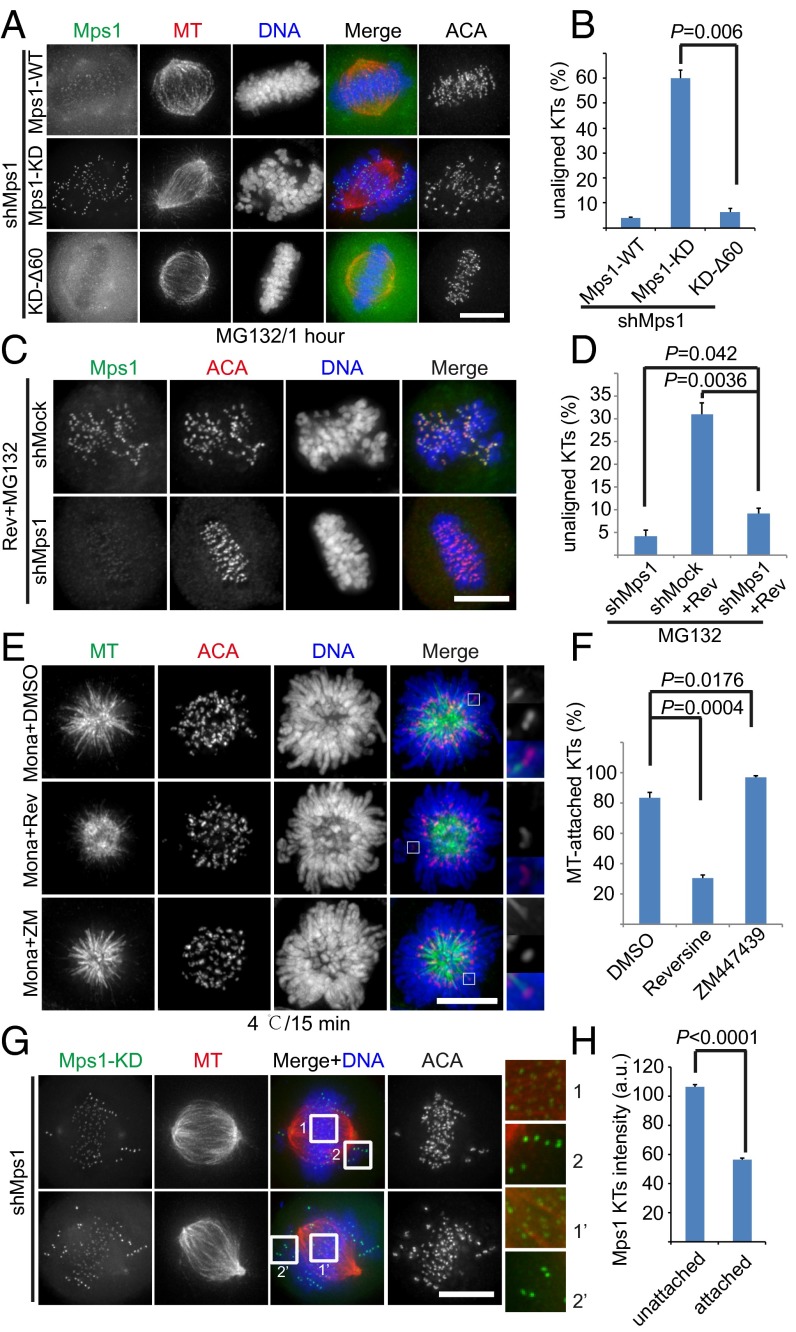
Why does the absence of Mps1 cause insignificant chromosome misalignment, whereas inhibiting Mps1 kinase activity severely disrupts chromosome alignment? Normally, Mps1 at kinetochores with inappropriate MT attachment exchanges dynamically with the cytoplasmic pool of Mps1 (43, 44). Upon inhibition of kinase activity, Mps1 localization on kinetochores is elevated significantly (Fig. S4 A and B) (22, 35, 44, 45). Therefore, we hypothesized that the abnormal accumulation of inactive Mps1 in kinetochores causes defects in kinetochore–MT attachments. To test this hypothesis, we used a kinase-dead Mps1 mutant (Mps1KD), which has significantly elevated kinetochore localization compared with WT Mps1. Indeed, we observed massive chromosome misalignment in cells expressing Mps1KD (Fig. 4 A and B). Consistently, removal of the N-terminal extension (NTE) of Mps1, which is responsible for its localization to kinetochores (designated as Mps1KD-Δ60), reduced the level of Mps1 at kinetochores but did not affect chromosome alignment (Fig. 4 A and B) (21). Similarly, the previously reported phospho-mimicking Mps1 mutant in Mps1KD (designated as Mps1KD-8D) also induced a slight chromosome alignment defect because of its weaker localization on kinetochores (Fig. S4 C and D) (45). These findings are consistent with the hypothesis that an abnormal accumulation of inactive Mps1 at kinetochores causes chromosome misalignment. We further predicted that, when endogenous Mps1 was knocked down, reversine treatment would not disrupt chromosome alignment. Consistent with our prediction, knocking down endogenous Mps1 decreased reversine-induced defects in chromosome alignment (Fig. 4 C and D). These observations further support the concept that Mps1 is not essential for chromosome alignment. Taken together, our data demonstrate that the chromosome alignment defect caused by Mps1 inhibitors is caused mainly by the elevated localization of an inactive form of Mps1 in the kinetochore.
Fig. S4.
The kinetochore localization of inactive Mps1 perturbs stable kinetochore–MT attachment. (A) Representative immunofluorescence images of HeLa cells cotransfected with shMps1 and shRNA-resistant constructs as indicated. Thirty-six hours post transfection, cells were treated with MG132 for 1 h and then were immunostained for MT (shown as red in merged images), ACA (shown as red in merged images), and DNA (blue). (Scale bar, 10 μm.) (B) Quantification of the degree of chromosome misalignment in cells treated as in A. Kinetochore position was measured as a function of the distance from pole to spindle equator. Bars represent the mean ± SE of three independent experiments. In each experiment, five cells were measured (>60 kinetochores per cell). (C) Weakening the kinetochore localization of Mps1KD can rescue the chromosome misalignment phenotype induced by the expression of Mps1KD. Shown are representative immunofluorescence images of cells cotransfected with shMps1 and shRNA-resistant Mps1 constructs as indicated. Thirty-six hours post transfection, cells were treated with MG132 for 1 h and then were immunostained for MT (red), ACA (shown as black and white images), and DNA (blue). (Scale bar, 10 μm.) (D) Bar graph illustrating the percentage of unaligned kinetochores in cells treated as in C. Values are mean ± SE of three independent experiments. (E) Representative immunofluorescence images of LAP-Mps1 HeLa cells treated with the inhibitors as indicated and then immunostained for ACA (red) and DNA (blue). (Scale bar, 10 μm.)
Fig. 4.
Kinetochore localization of inactive Mps1 prevents the establishment of stable MT attachment. (A) Representative immunofluorescence images of HeLa cells cotransfected with Mps1 shRNA and different shRNA-resistant, GFP-tagged Mps1 constructs. At 36 h after transfection, cells were treated with MG132 for 1 h and then were fixed and costained for MT (red), ACA (shown as black and white images), and DNA (blue). (Scale bar, 10 μm.) (B) Bar graph illustrating the degree of chromosome misalignment in cells treated as in A. Bars represent the means ± SE of three independent experiments. In each experiment, five cells were measured (>60 kinetochores per cell). (C) HeLa cells were transfected with different shRNAs as indicated. At 36 h after transfection, cells were treated with reversine plus MG132 for 1 h and then were fixed and stained for Mps1 (green), ACA (red), and DNA (blue). (Scale bar, 10 μm.) (D) Bar graph illustrating the degree of chromosome misalignment in cells treated as in C. Bars represent the means ± SE of five measured cells. (E) Representative immunofluorescence images of HeLa cells treated for 2 h with monastrol in combination with DMSO, reversine, or ZM. Cells then were incubated at 4 °C for 15 min and fixed for immunofluorescence staining for MT (green), ACA (red), and DNA (blue). Magnified views of the boxed areas are shown in the panels on the right. (Scale bar, 10 μm.) (F) Bar graph showing the percentage of kinetochores with MT attachment in cells treated as in E. Bars represent the means ± SE of kinetochores with attachment from five cells. (G) Representative immunofluorescence images of HeLa cells cotransfected with shMps1 and shRNA-resistant LAP-Mps1KD constructs. At 36 h after transfection, cells were treated with MG132 for 1 h and then were immunostained for MT (red), ACA (shown as black and white images), and DNA (blue). Magnified views of the boxed areas are shown in the panels at the right. (Scale bar, 10 μm.) (H) Bar graph representing the intensity of the Mps1 signal at unattached kinetochores and attached kinetochores (average of five cells ± SE). The y axis shows the intensity of the kinetochore signal. Student’s t test was used to calculate P values. a.u., arbitrary units.
Next, we examined the stability of the kinetochore–MT attachment in cells treated with or without reversine. The recruitment of active Mps1 to the kinetochore was evident in HeLa cells treated with monastrol (Fig. S4E) and was enhanced when Mps1 was inhibited by reversine. To monitor the stability of the kinetochore fiber (K-fiber), we incubated the cells at 4 °C for 15 min. As expected, the kinetochore–MT attachment was stable in the control situation after cold treatment (Fig. 4 E and F), but numerous kinetochores treated with reversine lost their interaction with MTs (Fig. 4 E and F). Once again, this effect is specific for Mps1, and not aurora B, inhibition, because the K-fibers were stable in cells treated with ZM (Fig. 4E, Bottom Row). This observation indicates that localization of inactive, as opposed to active, Mps1 to the kinetochore impairs the establishment of stable MT attachments.
We further observed that the signal intensity of Mps1KD was weaker at the kinetochores attached to MTs than at the unattached kinetochores (Fig. 4 G and H). Preferred localization on unattached kinetochores is a common feature for SAC proteins (46). However, SAC was inactive in this assay because endogenous Mps1 was knocked down. Thus, these data indicate that Mps1KD has a higher affinity for the unattached kinetochore and that this higher affinity is independent of the SAC. Taken together, these findings demonstrate that the elevated and stable kinetochore recruitment of inactive Mps1 perturbs the establishment of stable kinetochore–MT attachments.
Inactive and Active Mps1 Kinases Exhibit Distinctly Different Structural Requirements for Localization to the Kinetochore.
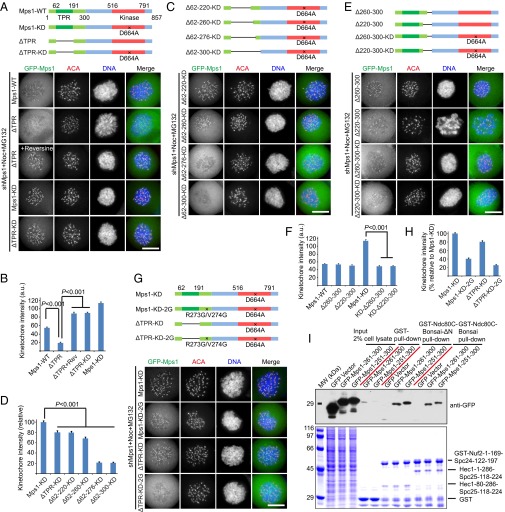
Inactivating Mps1 kinase activity causes strongly elevated kinetochore localization of Mps1 (22, 35, 44, 45, 47). However, the mechanism remains unknown. Several observations suggest that inactive Mps1 may localize to kinetochores via a mechanism different from that used by Mps1WT. First, inactive Mps1 has a much slower turnover rate at kinetochores than Mps1WT (22, 35, 44). Second, although two Mps1 mutants with a disrupted TPR domain (a domain shown to be responsible for the kinetochore localization of Mps1WT) fail to localize to kinetochores, inhibition of their kinase activity with reversine restores their normal kinetochore localization (48). Third, Mps1 mutants lacking the entire TPR domain localize to kinetochore in the presence of reversine (21). Therefore, it is likely that Mps1 localizes to kinetochores independently of its TPR domain. To confirm this notion, we examined the localization of an Mps1 mutant lacking the TPR domain (Mps1ΔTPR) (see Fig. 5A for the schematic representation). In contrast to the clear kinetochore localization of Mps1WT, the kinetochore signal of Mps1ΔTPR was nearly undetectable (Fig. 5 A and B), supporting the concept that the TPR domain is essential for the localization of Mps1WT. In the presence of reversine, however, the Mps1ΔTPR signals increased significantly at kinetochores. The same was true when Mps1ΔTPR was mutated further to create its kinase-dead mutant (Fig. 5 A and B). These findings indicate that a region other than the TPR domain is responsible for localizing inactive Mps1 to kinetochores.
Fig. 5.
Inactive Mps1 has a kinetochore localization mechanism distinct from that of Mps1WT. (A, C, E, and G) Representative immunofluorescence images of HeLa cells cotransfected with Mps1 shRNA and different shRNA-resistant plasmids as indicated. At 36 h after transfection, cells were treated with the indicated drugs for 1 h. Then cells were fixed and costained for ACA (red) and DNA (blue). (Scale bars, 10 μm.) A schematic representation of Mps1 deletion constructs is shown above the images in each panel. (B and F) Bar graphs illustrating the kinetochore intensity of different Mps1 deletions treated as in A (B) and E (F). The y axis shows the kinetochore signal intensity in arbitrary units. Bars represent the means ± SE of five measured cells (>20 kinetochores per cell). Student’s t test was used to calculate P values. (D and H) Bar graphs illustrating relative kinetochore intensity of different Mps1 deletions treated as in C (D) and G (H). Bars represent the means ± SE of five measured cells (>20 kinetochores per cell). Student’s t test was used to calculate P values. (I) GST-, GST-Ndc80CBonsai–, or GST-Ndc80CBonsai-ΔN–bound agarose beads were used as affinity matrices to absorb cell lysates expressing GFP-tagged Mps1251–300 or Mps1261–300 fusion protein. Pull-downs were analyzed by SDS/PAGE and probed by anti-GFP blot.
One candidate region could be the NTE (amino acids 1–61) of Mps1, because it is important for targeting Mps1 to kinetochores (21). Therefore we tested the kinetochore localization of GFP-tagged Mps11–61. In the absence of endogenous Mps1, Mps11–61 did not localize to kinetochores at all, but weak kinetochore localization of Mps11–61 appeared in the presence of endogenous Mps1 (Fig. S5A). These data indicate that the NTE is not sufficient for localizing Mps1 to kinetochores.
Fig. S5.
Inactive Mps1 has a kinetochore localization mechanism distinct from that of Mps1WT. (A) Representative immunofluorescence images of HeLa cells cotransfected with GFP-Mps11–61 together with shMps1 or shMock. Thirty-six hours post transfection cells were fixed and costained for ACA (red) and DNA (blue). (Scale bar, 10 μm.) (B and C) Representative immunofluorescence images of HeLa cells cotransfected with Mps1 shRNA and different shMps1-resistant plasmids as indicated. Thirty-six hours post transfection cells were treated with indicated drugs for 1 h. Then cells were fixed and costained for ACA (red) and DNA (blue). (Scale bars, 10 μm.) (D) Bar graph illustrating kinetochore intensity of different Mps1 deletions treated as in C. Bars represent the mean ± SE of five cells (>20 kinetochores per cell). (E) Multiple sequence alignment of Mps1 proteins from different species as indicated identifies the conserved RVPV motif within the Mps1 251–300 fragment. The sequence alignment was done by ClustalW2 software.
To pinpoint the domain(s) essential for the localization of Mps1KD to the kinetochore, we constructed a series of Mps1 deletion mutants and examined their localization in transiently transfected HeLa cells (Fig. 5C, Upper). As shown in Fig. 5C, Mps1KD-Δ62–220 localized on kinetochores with an intensity similar to that for Mps1KD-ΔTPR. The localization of Mps1KD-Δ62–260 at kinetochores was readily apparent. Strikingly, Mps1KD-Δ62–276 and Mps1KD-Δ62–300 exhibit no apparent kinetochore localization (Fig. 5 C and D). Nevertheless, the Mps1Δ301–514 is readily apparent at the kinetochores (Fig. S5B), indicating that Mps1301–514 is not a determinant for Mps1 localization to the kinetochore. These data suggest that the Mps1261–300 (hereafter referred as the “internal region for kinetochore localization,” IRK) is the structural determinant for Mps1KD localization to the kinetochores. In particular, the sequence encompassing amino acids 261–276 plays a key role. Consistently, the kinetochore localization of a series of Mps1 deletion mutants lacking the catalytic domain also showed that the IRK region contributes to the kinetochore localization of inactive Mps1 (Fig. S5 C and D).
An Mps1KD construct lacking both the TPR domain and the IRK lost kinetochore localization capacity (Fig. 5 C and D). We next examined the localization of recombinant Mps1 proteins lacking only the IRK. Removing the amino acids 260–300 or 220–300 did not affect the localization of Mps1 proteins with a WT kinase domain. Compared with full-length Mps1KD, however, the Mps1KD lacking amino acids 260–300 or 220–300 exhibited greatly decreased kinetochore localization, suggesting that the IRK is involved in the recruitment of inactive Mps1 to kinetochores (Fig. 5 E and F). Further mapping experiments showed that the residues R273 and V274 within the IRK are two key residues responsible for targeting inactive Mps1 to kinetochores (Fig. 5 G and H and Fig. S5E). In addition, pull-down assays showed that GFP-tagged Mps1 fragments containing the IRK were coprecipitated by the recombinant Ndc80CBonsai complex (Fig. 5I), suggesting that the newly identified IRK is indeed the region responsible for targeting Mps1 to kinetochores.
Our data demonstrate that inactive Mps1 uses a structural determinant distinct from that of Mps1WT for kinetochore targeting. The TPR domain is essential for mediating the localization of Mps1WT to the kinetochore but is dispensable for localizing inactive Mps1. Thus, we conclude that IRK is the structural determinant responsible for localizing inactive Mps1 to the kinetochore and that Mps1 uses two distinct structural modules for its localization to kinetochores, depending on its kinase activity.
The Localization of Inactive Mps1 to Unattached Kinetochores Is Independent of Aurora B Activity.
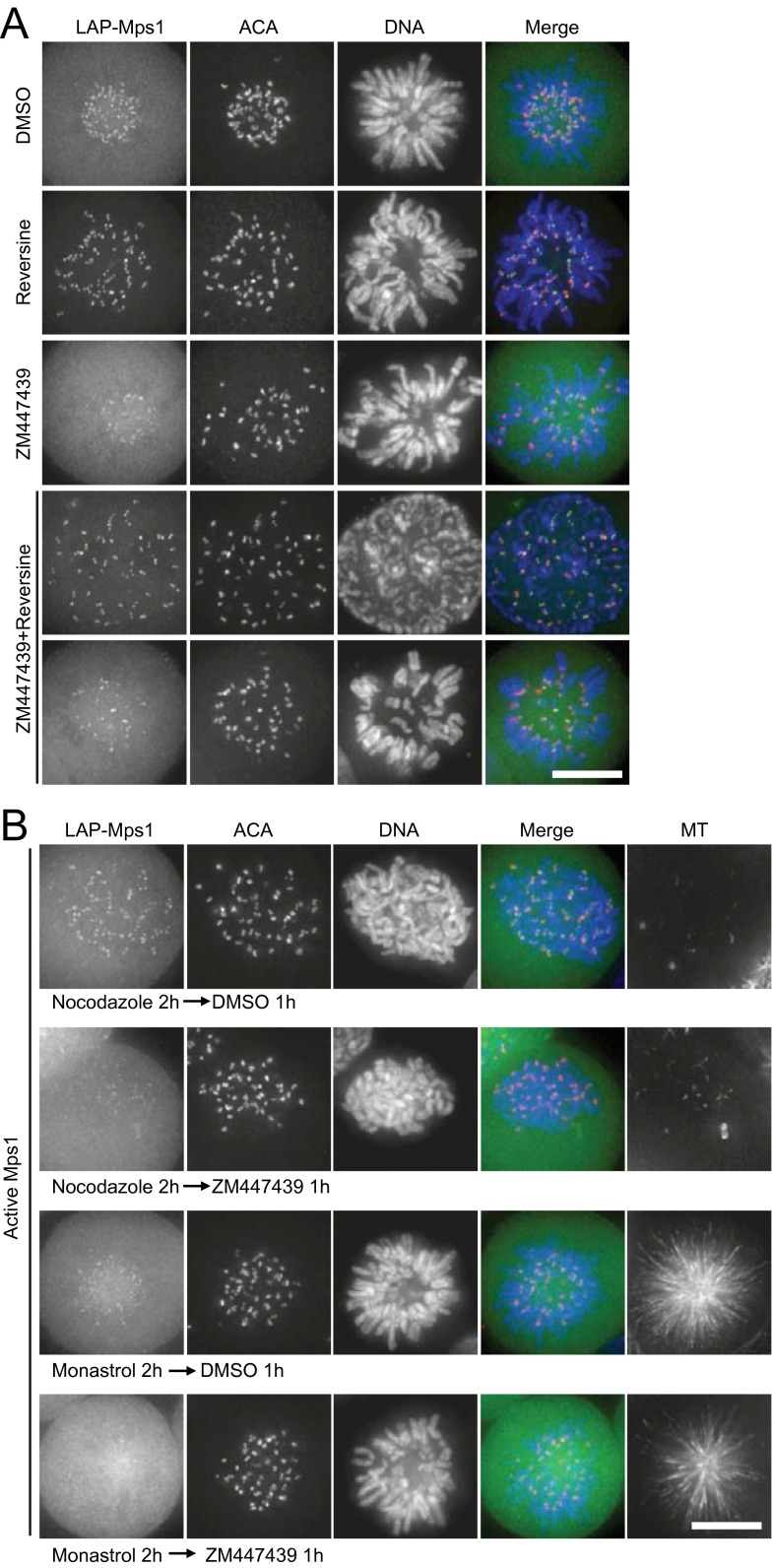
The kinetochore localization of Mps1WT depends on aurora B kinase activity (19, 20, 22, 23). We next asked whether the recruitment of inactive Mps1 to the kinetochore also depends on aurora B. For this purpose, we examined the kinetochore localization of Mps1 in the presence of reversine and ZM. Relative to cells treated with reversine, there was a heterogeneous result in cells double-treated with reversine and ZM: The kinetochore signal of Mps1 decreased in some cells and remained unchanged in other cells (Fig. S6A). Examination indicated that the cells with an unchanged Mps1 signal were in prophase, and the cells with a decreased Mps1 signal were in prometaphase. One difference between prophase and prometaphase is that all kinetochores are unattached to MTs in prophase, whereas in prometaphase most kinetochores show MT attachment, although not necessarily in a correct manner. Therefore we thought that inactive Mps1 might adopt either aurora B-dependent or aurora B-independent localization depending on the state of MT attachment. To challenge our hypothesis, we treated cells with nocodazole plus reversine for 2 h to depolymerize MT and then treated these cells with DMSO or ZM for 1 h. In this experiment we observed that the Mps1 kinetochore signal in ZM-treated cells was as bright as that in DMSO-treated cells (Fig. 6 A and B). This result suggests that, in the absence of a kinetochore–MT interaction, aurora B is not required to recruit inactive Mps1 at the kinetochores. On the contrary, when cells were treated first with monastrol plus reversine, the Mps1 kinetochore signal decreased sharply in cells subsequently treated with ZM (Fig. 6 C and D). This result suggests that the recruitment of inactive Mps1 onto MT-bound kinetochores depends upon aurora B. Taken together, these data suggest that aurora B is required to localize inactive Mps1 only to kinetochores that are already bound to MTs but not to the “naked” kinetochores in prophase. However, aurora B activity is required, in both unattached and attached situations, for the recruitment of active Mps1 to the kinetochore (Fig. S6B).
Fig. S6.
The kinetochore localization of inactive Mps1 does not stringently require aurora B kinase activity. (A) Representative immunofluorescence images of LAP–Mps1 HeLa cells treated as indicated. The cells were fixed and costained for ACA (red) and DNA (blue). (Scale bar, 10 μm.) (B) The kinetochore localization of WT Mps1 depends on aurora B activity in both the no MT-attachment situation and the with MT-attachment situation. Cells were treated with nocodazole or monastrol for 2 h. Then one group of nocodazole/monastrol-treated cells was treated with ZM447439 for 1 h, and the other group of cells was treated with DMSO for 1 h. Cells were fixed and costained for ACA (red) and DNA (blue). (Scale bar, 10 μm.)
Fig. 6.
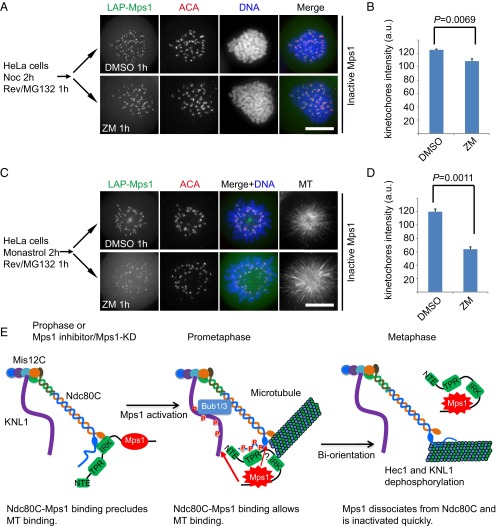
The kinetochore localization of inactive Mps1 is insensitive to aurora B activity. (A and C) Representative immunofluorescence images of LAP–Mps1 HeLa cells treated with different drugs as indicated. At 8 h after G1/S release, cells were treated with nocodazole (A) or monastrol (C) for 2 h. Reversine and MG132 were added to the medium for 1 h. Then cells were treated with or without ZM for 1 h. Cells were fixed and costained for MT (shown as black-and-white images), ACA (red), and DNA (blue). (Scale bars, 10 μm.) (B and D) Bar graphs illustrating Mps1 kinetochore intensity of cells treated as in A (B) or C (D). The y axis shows the kinetochore signal intensity in arbitrary units. Values are means ± SE of kinetochores from five cells. Student’s t test was used to calculate P values. (E) A model illustrating dynamic localization of Mps1 at the kinetochore before and after its activation. During early prophase, inactive Mps1 accumulates quickly at kinetochores through its binding with Ndc80C using its IRK (mainly) and TPR domain. Inactivation of Mps1, either by chemical inhibition or an engineered kinase-death mutation, promotes Mps1 localization to the kinetochore via binding to Ndc80C; this binding prevents the establishment of a stable Ndc80C–MT attachment. In an accurate mitosis, Mps1 is activated by autophosphorylation in trans when the local concentration of Mps1 at kinetochores is above a critical concentration. Upon activation, Mps1 exhibits a conformational change by which the IRK is released from binding to Ndc80C. In this activity-elicited domain-switching model, active Mps1 binds to kinetochores using its TPR domain in an aurora B-dependent manner. The binding of active Mps1 to Ndc80C is compatible with an initial establishment of kinetochore–MT attachments. After the establishment of a correct end-on MT attachment, Mps1 is liberated from kinetochores by dephosphorylation of Hec1 and competitive Ndc80C–MT binding. Mps1 is inactivated by PP1/PP2A promptly after its liberation from the kinetochore, and the SAC signaling then is satisfied. For simplicity, Mps1 is not shown as a dimer.
Discussion
Mps1 is an evolutionarily conserved SAC kinase (3, 49). A prevailing hypothesis accounting for Mps1 function is that it has a role in accurate chromosome alignment during eukaryotic cell division (31, 33). Our results have identified a structural determinant underlying the localization of inactive Mps1 to the kinetochore and have revealed a dual mode of kinetochore localization of Mps1, sensing the temporal dynamics of Mps1 kinase activity at the kinetochore for accurate SAC execution (modeled in Fig. 6E). First, Mps1 localizes to the kinetochore of prophase cells via an interaction mediated by the IRK and nuclear division cycle 80 complex (Ndc80C). This IRK-mediated Ndc80C–Mps1 interaction competes for Ndc80C–MT association, preventing premature end-on capture of kinetochores. After phosphorylation by Cdk1 and/or its autophosphorylation at the kinetochore, Mps1 undergoes a conformational change that switches its binding interface with Ndc80C from the IRK to the TPR domain to facilitate a dynamic molecular turnover of Mps1 at the kinetochore. This spatiotemporal dynamics of Mps1 acts as a fine-tuning mechanism to facilitate accurate kinetochore biorientation and subsequent end-on MT capture during the prometaphase–metaphase transition. Thus, phosphorylation-induced conformational rearrangement of the Mps1 molecule provides a spatial cue to ensure accurate Ndc80C–MT attachment during chromosome congression and alignment, integrating its enzymatic catalysis-coupled mechanosensing into SAC activity control.
Mps1 kinase promotes efficient chromosome alignment in unperturbed mitosis. Most chromosomes in Mps1-suppressed cells achieved full alignment in the presence of MG132, suggesting that chromosomes could achieve alignment via a less efficient manner when the Mps1 level is low for a prolonged period. Mps1 is required for the maximal kinetochore recruitment of BubR1 and centromere-associated protein E (CENP-E), two proteins that play dual roles in checkpoint signaling and chromosome alignment (6). Approximately 30% of BubR1 and CENP-E remains bound to kinetochores in Mps1-suppressed cells (36), suggesting that reduced but sufficient levels of BubR1 and CENP-E are essential to regulate chromosome alignment, particularly during the prolonged mitosis in cells treated with MG132. Consistent with this notion, we also noticed that a few chromosomes fail to align to the spindle equator and remain at the spindle poles in ∼20% of Mps1 knockdown cells. We interpret this phenotype as a consequence of insufficient CENP-E localization at kinetochores, because the phenotype is similar to that seen in cells lacking CENP-E motor activity (50–52). It is possible that an alternative back-up mechanism drives chromosome alignment in Mps1-suppressed cells when mitotic exit is blocked by MG132. In the future, it would be of great interest to elucidate the precise kinetics of chromosome alignment in Mps1-deficient cells. Such kinetics can be understood by visualizing and quantifying the temporal dynamics of the Mps1 kinase gradient during chromosome congression and alignment in normal mitosis and when alignment is perturbed (53).
It is worth noting that chemical inhibition of Mps1 induces much more severe defects in chromosome alignment than those seen in cells treated with Mps1 shRNA, which are caused mainly by an accumulation of enzymatic inactive Mps1 (Mps1KD or chemically inhibited Mps1). Thus, we reason that the persistent accumulation of inactive Mps1 at kinetochores, caused by a stable Ndc80C–IRK interaction, prevents the establishment of stable kinetochore–MT attachments and results in an aberrant chromosome alignment. How then does the accumulated inactive Mps1 at kinetochores affect chromosome alignment? Because the Hec1 MT-binding domain also is required for recruiting Mps1 to kinetochores (21, 23), it is likely that the Hec1–MT binding and Hec1–Mps1 binding are mutually exclusive. In support of this view, our study showed that the kinetochore localization of inactive Mps1, but not active Mps1, prevented the establishment of stable kinetochore–MT attachments. Conversely, stable MT attachment also attenuated the kinetochore localization of inactive Mps1 (Fig. 4 G and H). During the revision of our manuscript, two groups reported the competition between Mps1 and MT for Ndc80C binding (54, 55), supporting our conclusion that the kinetochore localization of inactive Mps1 induces chromosome misalignment by interfering with MT attachment to Ndc80C.
Interestingly, the recruitment of Mps1WT to the kinetochores strictly depends on the TPR domain, whereas the kinetochore localization of inactive Mps1 requires the IRK. Removal of the IRK impaired the capacity of inactive Mps1 to localize to the kinetochores (Fig. 5 E and F), whereas deletion of both the TPR domain and the IRK abolished the kinetochore localization of Mps1 (Fig. 5 C and D). These observations suggest that inactive Mps1 depends mainly on the IRK for its kinetochore recruitment. Although not playing a major role, the TPR domain also contributes to the kinetochore localization of inactive Mps1. We also present evidence that inactive Mps1 localizes to kinetochores in two different regulatory manners: in an aurora B-independent manner in the absence of MT attachment and in an aurora B-dependent manner otherwise. What is the biological significance of using two distinct regions for Mps1 localization to the kinetochore before and after Mps1 full activation? We speculate that this highly orchestrated localization of Mps1 has several benefits. First, the localization of inactive Mps1 via the high-affinity binding between its IRK and Ndc80C allows a quick increase in the local concentration of Mps1 at kinetochores in prophase and promotes its activation through autophosphorylation in trans and further by Cdk1-dependent phosphorylation (56, 57). Previous studies show that Mps1 is active in interphase and promotes the formation of an interphase Cdc20 inhibitory complex (36, 58). However, mounting evidence demonstrates that Mps1 kinase activity peaks in mitosis (8, 56). Therefore, we argue that the predominant pool of cytoplasmic Mps1 is inactive and that kinetochore localization is essential for the full activation of Mps1. Second, the localization of active Mps1 depends on aurora B activity in a TPR domain-dependent manner and promotes Mps1 to phosphorylate its key kinetochore substrate, KNL1, which is essential for the downstream SAC signaling (59). Numerous reports demonstrate that active Mps1 has a higher turnover rate and lower signal intensity at kinetochores than does inactive Mps1 (22, 35, 43–45). We envision that low-affinity localization ensures a prompt silencing of SAC when biorientation is achieved. The activation of Mps1 may switch its binding interface with Ndc80C in several ways. First, it is well known that kinase conformation changes after the activation by phosphorylation of sites within the activation loop (60). In addition, the active conformation of Mps1 kinase may block the access of IRK to Ndc80C. The other possibility is that the direct phosphorylation of Ndc80/Hec1 by Mps1 changes the binding interface between Mps1 and Ndc80C (24). Finally, autophosphorylation sites outside the activation loop (and also outside the IRK) may participate in the regulation via an as yet uncharacterized mechanism (45). Thus, it would be of great interest to quantify the dynamics of Mps1 kinase activity at the kinetochore using a FRET-based optical sensor (53) and to establish the relationship between the temporal dynamics of Mps1 kinase activity and accurate attachment during mitosis.
The results of previous publications combined with our current findings enable us to propose a two-module model for the Mps1 kinetochore localization. In early prophase, inactive Mps1 associates with Ndc80C in an aurora B-independent manner and remains stable at kinetochores. During this stage, the IRK region serves as the major kinetochore-targeting module (Fig. 6E). The accumulated Mps1 at the kinetochore then undergoes autophosphorylation to activate Mps1 via a conformational change, promoting Mps1 dynamics and the establishment of Ndc80C–MT binding (43, 44, 56). It is likely that, upon activation, the conformation of Mps1 changes to block the interaction of the IRK with Ndc80C. This blocking then would allow the TPR domain (together with the NTE) to act solely and to bias Mps1 binding to Hec1 in a dynamic manner (21, 23). We emphasize that the localization of active Mps1 to the kinetochore is compatible with the incorrect MT attachment (e.g., monastrol treatment; Fig. S4E and Fig. S6B). In our opinion, the competition between Mps1 and MT for binding with Ndc80C would not be sufficient to discriminate between correct and incorrect attachment (54, 55). Upon the establishment of proper bioriented kinetochore–MT attachment, maximal intrakinetochore tension pulls aurora B away from outer kinetochores (61). Together with the localized feedback loop between PP1 and PP2A-B56 (62), this event leads further to the liberation of Mps1 from kinetochores (checkpoint satisfaction) and stable MT attachment concurrently with the dephosphorylated Knl1–Mis12–Ndc80 network. Alternatively, end-on MT attachment separates Mps1 from KNL1/Spc105 and enables SAC silencing (63). The dynamic association of Mps1 with Ndc80C provides exquisite temporal regulation to prevent premature stable association between Hec1/Nuf2 and MT at early prometaphase, and prompt activation of Mps1 promotes its release for an accurate kinetochore–MT attachment (Fig. 6E). Thus, the task ahead is to delineate the mechanism of action underlying the conformational changes of Mps1 molecules at the kinetochore.
In summary, we have identified a dynamic, hierarchical interaction between Mps1, Ndc80C, and MT at kinetochores that orchestrates accurate mitosis. The persistent association of inactive Mps1 with Ndc80C via the IRK perturbs faithful kinetochore–MT attachment and mitotic progression. Our results provide a novel mechanistic insight into the spatiotemporal dynamics of Mps1 activity at the kinetochore and in accurate mitosis.
Methods
Cell Culture and Drug Treatments.
HeLa cells were routinely maintained in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS and penicillin-streptomycin (100 IU/mL and 100 mg/mL, respectively; GIBCO). BAC TransgeneOmics LAP–Mps1 stable HeLa cells were kindly provided by A. Hyman (Max Planck Institute, Dresden, Germany) and were maintained in DMEM containing G418 (0.5 μg/μL) (64). hTERT-RPE1 cells were maintained in DMEM/F12 medium containing 0.01 mg/mL hygromycin supplemented with 10% FBS. Thymidine was used at 2 mM, nocodazole at 100 ng/mL, the Eg5 inhibitor monastrol at 100 μM, the Mps1 inhibitor reversine at 0.5 μM, AZ3146 at 2 μM, the aurora B inhibitor ZM447439 at 2.5 μM, and MG132 at 20 μM. In some cases, the CENP-E inhibitor syntelin was used at 1 μM (51).
Plasmids, RNAi, and Transfection.
Wild-type and kinase-dead LAP–Mps1 constructs and pSuper-Mps1 (shMps1-1) and pSuper-Mock shRNA constructs were described previously and were a kind gift from G. Kops, University Medical Center Utrecht, Utrecht, The Netherlands (33). pSuper-BubR1 and pSuper-Mps1-2 (shMps1-2) were constructed as described (65) by using the target sequences 5′-ATGAGACTTCAGAAAACCC-3′ and 5′-GAACAAAGTGAGAGACATT-3′, respectively. To construct plasmids coexpressing shRNA and mCherry, pmCherry-C2 plasmid (based on pEGFP-C2) was cut by BglII and BamHI and then was ligated again to eliminate the multiple restriction enzyme sites. The PCR-amplified fragment encompassing the whole mCherry expression cassette, including the CMV promoter, mCherry cDNA, and polyadenylation sequence, was inserted into the pSuper vector via BamHI and EcoRI sites. GFP-tagged Mps1 and truncation Mps1KD-Δ60, Mps11–61, and Mps11–303 were generated by inserting the corresponding PCR-amplified fragments into the pEGFP-C1 vector via BglII and SalI sites. The GFP-tagged Mps1 deletion construct Mps1ΔTPR was generated by inserting Mps1192–857 PCR-amplified fragments into the GFP-Mps11–61 plasmid via SalI and BamHI sites. The other deletion constructs, Mps1Δ62–220, Mps1Δ62–260, Mps1Δ62–276, and Mps1Δ62–300, were generated similarly.
Mutagenesis was performed using Mut Express II fast mutagenesis kits (Vazyme Biotech Co. Ltd) according to the manufacturer’s instructions. All constructs were verified by sequencing. The target sequences of different siRNAs are siBubR1 (GGAGATCCTCTACAAAGGG, nucleotides 1126–1144), siMps1-1 (CTTGAATCCCTGTGGAAAT, nucleotides 2627–2646), and siMps1-2 (CGGAATTCATTGAGACAAA, nucleotides 766–784), which have been described previously and synthesized by Qiagen (48, 66). All the plasmids and siRNAs were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the user’s manual. To enrich mitotic cells, cells were treated with thymidine for 14–16 h, starting 8 h after transfection. Then cells were released into normal DMEM. At 10 h after release, cells were treated with the Eg5 motor inhibitor monastrol for 2 h and then were fixed for immunofluorescence staining. For rescue experiments, Mps1 shRNA was cotransfected with different rescue plasmids (or empty vector) at a 3:1 ratio.
Antibodies.
Monoclonal anti–hMps1-N1 (8), anti-BubR1 (66), and anti-Mad2 (67) antibodies have been described previously. Anti–α-tubulin (DM1A; Sigma), anti-aurora B (anti–AIM-1; BD), and anti-centromere antibodies (ACA; ImmunoVision) were obtained commercially. For all Western blotting, signals were detected using HRP-conjugated anti-mouse or anti-rabbit antibodies (Pierce).
Immunofluorescence Microscopy, Image Processing, and Quantification.
HeLa cells grown on coverslips were fixed and permeabilized simultaneously with PTEMF buffer [50 mM Pipes (pH 6.8), 0.2% Triton X-100, 10 mM EGTA, 1 mM MgCl2, 4% formaldehyde] at room temperature and were processed for indirect immunofluorescence microscopy. Samples were examined on a DeltaVision microscope (Applied Precision) with a 60× objective lens, NA = 1.42, with optical sections acquired 0.2 μm apart in the z axis. Deconvoluted images from each focal plane were projected into a single picture using Softworx (Applied Precision). Images were taken at identical exposure times within each experiment and were acquired as 16-bit gray-scale images. After deconvolution, the images were exported as 24-bit RGB images and processed in Adobe Photoshop. Images shown in the same panel have been identically scaled. Kinetochore intensities were measured in ImageJ (rsb.info.nih.gov/ij/) on nondeconvoluted images. The levels of kinetochore-associated proteins were quantified as described previously (19). In brief, the average pixel intensities from at least 100 kinetochore pairs from five cells were measured, and background pixel intensities were subtracted. The pixel intensities at each kinetochore pair then were normalized against ACA pixel values to account for any variations in staining or image acquisition. Unless otherwise specified, the values for treated cells then were plotted as a percentage of the values obtained from cells of the control groups.
Live-Cell Imaging.
HeLa cells were cultured in glass-bottomed culture dishes (MatTek). During imaging, cells were cultured at 37 °C in CO2-independent medium (Invitrogen) containing 10% FBS and 2 mM glutamine and were observed with the DeltaVision RT system (Applied Precision). Images were prepared for publication using Adobe Photoshop software.
GST Pull-Down Assay.
GST-Ndc80CBonsai and GST-Ndc80CBonsaiΔN fusion proteins were purified as previously described (68). GST-tagged Ndc80CBonsai or Ndc80CBonsaiΔN fusion protein-bound glutathione beads were incubated with 293T cell lysates expressing GFP-Mps1251–300 fusion proteins in Hepes lysis buffer containing 0.1% Triton X-100 for 2 h at 4 °C. After the incubation, the beads were washed three times with PBS containing 0.1% Triton X-100 and once with PBS and were boiled in SDS/PAGE sample buffer. The bound proteins then were separated on 10% SDS/PAGE.
Acknowledgments
We thank Dr. A. Hyman for providing BAC TransgeneOmice LAP–Mps1 cells; Dr. Xueliang Zhu for providing RPE1 cells; Dr. Minhao Wu for help on structural biology analysis; and all members of the X.Y. laboratory for helpful discussions. This work was supported by Chinese Natural Science Foundation Grants 31320103904, 31430054, 31071184, 31371363, 91313303, 31471275, and 31301099; Chinese 973 Project Grants 2012CB945002, 2014CB964803, and 2013CB911203; Anhui Provincial Natural Science Foundation Grants 1408085MKL09 and 1508085SMC213; China Postdoctoral Science Foundation Grants 2014M560517 and IRT13038; and NIH Grants CA146133, DK56292, and U54 CA118948.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508791112/-/DCSupplemental.
References
- 1.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holland AJ, Cleveland DW. Losing balance: The origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13(6):501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 4.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol. 2013;14(1):25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jia L, Kim S, Yu H. Tracking spindle checkpoint signals from kinetochores to APC/C. Trends Biochem Sci. 2013;38(6):302–311. doi: 10.1016/j.tibs.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem. 2012;81:561–585. doi: 10.1146/annurev-biochem-061611-090435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 8.Stucke VM, Silljé HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. EMBO J. 2002;21(7):1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu ST, et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14(4):1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poss KD, Nechiporuk A, Stringer KF, Lee C, Keating MT. Germ cell aneuploidy in zebrafish with mutations in the mitotic checkpoint gene mps1. Genes Dev. 2004;18(13):1527–1532. doi: 10.1101/gad.1182604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer MG, Heeger S, Häcker U, Lehner CF. The mitotic arrest in response to hypoxia and of polar bodies during early embryogenesis requires Drosophila Mps1. Curr Biol. 2004;14(22):2019–2024. doi: 10.1016/j.cub.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Hardwick KG, Weiss E, Luca FC, Winey M, Murray AW. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273(5277):953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- 13.He X, Jones MH, Winey M, Sazer S. Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J Cell Sci. 1998;111(Pt 12):1635–1647. doi: 10.1242/jcs.111.12.1635. [DOI] [PubMed] [Google Scholar]
- 14.Dou Z, et al. Dynamic distribution of TTK in HeLa cells: Insights from an ultrastructural study. Cell Res. 2003;13(6):443–449. doi: 10.1038/sj.cr.7290186. [DOI] [PubMed] [Google Scholar]
- 15.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297(5590):2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 16.Vigneron S, et al. Kinetochore localization of spindle checkpoint proteins: Who controls whom? Mol Biol Cell. 2004;15(10):4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrich S, Windecker H, Hustedt N, Hauf S. Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores. J Cell Sci. 2012;125(Pt 20):4720–4727. doi: 10.1242/jcs.110387. [DOI] [PubMed] [Google Scholar]
- 18.Sliedrecht T, Zhang C, Shokat KM, Kops GJ. Chemical genetic inhibition of Mps1 in stable human cell lines reveals novel aspects of Mps1 function in mitosis. PLoS One. 2010;5(4):e10251. doi: 10.1371/journal.pone.0010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dou Z, et al. Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS One. 2011;6(4):e18793. doi: 10.1371/journal.pone.0018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saurin AT, van der Waal MS, Medema RH, Lens SM, Kops GJ. Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat Commun. 2011;2:316. doi: 10.1038/ncomms1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201(2):217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190(1):73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu T, et al. Phosphorylation of microtubule-binding protein Hec1 by mitotic kinase Aurora B specifies spindle checkpoint kinase Mps1 signaling at the kinetochore. J Biol Chem. 2013;288(50):36149–36159. doi: 10.1074/jbc.M113.507970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemmler S, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28(8):1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14(7):746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 26.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22(10):900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepperd LA, et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22(10):891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vleugel M, et al. Sequential multisite phospho-regulation of KNL1-BUB3 interfaces at mitotic kinetochores. Mol Cell. 2015;57(5):824–835. doi: 10.1016/j.molcel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Primorac I, et al. Bub3 reads phosphorylated MELT repeats to promote spindle assembly checkpoint signaling. eLife. 2013;2:e01030. doi: 10.7554/eLife.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overlack K, et al. A molecular basis for the differential roles of Bub1 and BubR1 in the spindle assembly checkpoint. eLife. 2015;4:e05269. doi: 10.7554/eLife.05269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maure JF, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007;17(24):2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer RE, et al. Mps1 and Ipl1/Aurora B act sequentially to correctly orient chromosomes on the meiotic spindle of budding yeast. Science. 2013;339(6123):1071–1074. doi: 10.1126/science.1232518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132(2):233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6(5):359–368. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190(1):25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190(1):89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan W, Cleveland DW. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J Cell Biol. 2010;190(1):21–24. doi: 10.1083/jcb.201006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7(1):93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 39.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev Cell. 2012;23(4):745–755. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Elowe S, et al. Uncoupling of the spindle-checkpoint and chromosome-congression functions of BubR1. J Cell Sci. 2010;123(Pt 1):84–94. doi: 10.1242/jcs.056507. [DOI] [PubMed] [Google Scholar]
- 41.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6(3):232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 42.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161(2):267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14(11):953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 44.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJ. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol. 2010;191(2):281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang X, et al. Dynamic autophosphorylation of mps1 kinase is required for faithful mitotic progression. PLoS One. 2014;9(9):e104723. doi: 10.1371/journal.pone.0104723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoffman DB, Pearson CG, Yen TJ, Howell BJ, Salmon ED. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell. 2001;12(7):1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, et al. Characterization of spindle checkpoint kinase Mps1 reveals domain with functional and structural similarities to tetratricopeptide repeat motifs of Bub1 and BubR1 checkpoint kinases. J Biol Chem. 2012;287(8):5988–6001. doi: 10.1074/jbc.M111.307355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thebault P, et al. Structural and functional insights into the role of the N-terminal Mps1 TPR domain in the SAC (spindle assembly checkpoint) Biochem J. 2012;448(3):321–328. doi: 10.1042/BJ20121448. [DOI] [PubMed] [Google Scholar]
- 49.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22(22):R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 50.Yao X, Abrieu A, Zheng Y, Sullivan KF, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2(8):484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- 51.Ding X, et al. Probing CENP-E function in chromosome dynamics using small molecule inhibitor syntelin. Cell Res. 2010;20(12):1386–1389. doi: 10.1038/cr.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapoor TM, et al. Chromosomes can congress to the metaphase plate before biorientation. Science. 2006;311(5759):388–391. doi: 10.1126/science.1122142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chu Y, et al. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol Cell Biol. 2011;3(4):260–267. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji Z, Gao H, Yu H. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science. 2015;348(6240):1260–1264. doi: 10.1126/science.aaa4029. [DOI] [PubMed] [Google Scholar]
- 55.Hiruma Y, et al. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348(6240):1264–1267. doi: 10.1126/science.aaa4055. [DOI] [PubMed] [Google Scholar]
- 56.Kang J, Chen Y, Zhao Y, Yu H. Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc Natl Acad Sci USA. 2007;104(51):20232–20237. doi: 10.1073/pnas.0710519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morin V, et al. CDK-dependent potentiation of MPS1 kinase activity is essential to the mitotic checkpoint. Curr Biol. 2012;22(4):289–295. doi: 10.1016/j.cub.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Bravo V, et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell. 2014;156(5):1017–1031. doi: 10.1016/j.cell.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ghongane P, Kapanidou M, Asghar A, Elowe S, Bolanos-Garcia VM. The dynamic protein Knl1 - a kinetochore rendezvous. J Cell Sci. 2014;127(Pt 16):3415–3423. doi: 10.1242/jcs.149922. [DOI] [PubMed] [Google Scholar]
- 60.Bayliss R, Fry A, Haq T, Yeoh S. On the molecular mechanisms of mitotic kinase activation. Open Biol. 2012;2(11):120136. doi: 10.1098/rsob.120136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323(5919):1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nijenhuis W, Vallardi G, Teixeira A, Kops GJ, Saurin AT. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat Cell Biol. 2014;16(12):1257–1264. doi: 10.1038/ncb3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aravamudhan P, Goldfarb AA, Joglekar AP. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat Cell Biol. 2015;17(7):868–879. doi: 10.1038/ncb3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poser I, et al. BAC TransgeneOmics: A high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5(5):409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296(5567):550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 66.Elowe S, Hümmer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21(17):2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 2011;30(16):3322–3336. doi: 10.1038/emboj.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133(3):427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]