Significance
Nonhomologous end joining (NHEJ) is a DNA double-strand break repair pathway required for development of the adaptive immune response, maintenance of cellular proliferative capacity, and the response to several commonly used cancer treatments. A major challenge faced by this pathway is that chromosome breaks can have dirty end structures, making them difficult to repair. We show here that two mammalian DNA polymerases have an unexpectedly pivotal role in helping resolve such ends. Each is proficient in different contexts and has a differing impact on repair fidelity. This work sheds light on how NHEJ has evolved to be flexible during repair and identifies two polymerases as critical for this process.
Keywords: nonhomologous end joining, double-strand break repair, Pol X, polymerase mu, polymerase lambda
Abstract
Nonhomologous end joining (NHEJ) repairs chromosome breaks and must remain effective in the face of extensive diversity in broken end structures. We show here that this flexibility is often reliant on the ability to direct DNA synthesis across strand breaks, and that polymerase (Pol) μ and Pol λ are the only mammalian DNA polymerases that have this activity. By systematically varying substrate in cells, we show each polymerase is uniquely proficient in different contexts. The templating nucleotide is also selected differently, with Pol μ using the unpaired base adjacent to the downstream 5′ phosphate even when there are available template sites further upstream of this position; this makes Pol μ more flexible but also less accurate than Pol λ. Loss of either polymerase alone consequently has clear and distinguishable effects on the fidelity of repair, but end remodeling by cellular nucleases and the remaining polymerase helps mitigate the effects on overall repair efficiency. Accordingly, when cells are deficient in both polymerases there is synergistic impact on NHEJ efficiency, both in terms of repair of defined substrates and cellular resistance to ionizing radiation. Pol μ and Pol λ thus provide distinct solutions to a problem for DNA synthesis that is unique to this pathway and play a key role in conferring on NHEJ the flexibility required for accurate and efficient repair.
Chromosome breaks are most often repaired by nonhomologous end joining (NHEJ) in mammals. This pathway is thus essential for assembly of the antigen receptor genes (Ig and TcR) required for adaptive immunity, normal cellular replicative capacity and nervous system development (1) and shapes the effectiveness and safety of break-inducing cancer therapies (ionizing radiation and certain chemotherapeutic agents) (2). Repair is accomplished by directly ligating broken ends together. Such a mechanism makes NHEJ unique among the major DNA repair pathways, as an unbroken template cannot easily be used to guide replacement of lost or damaged nucleotides by DNA polymerases. Polymerase-dependent synthesis might thus be expected to play a lesser role in NHEJ, relative to the other DNA transactions (excision and ligation) required for DNA repair.
Nevertheless, at least three different members of the mammalian X family of DNA polymerases can be specifically implicated in NHEJ; polymerase (Pol) λ, Pol μ, and terminal deoxynucleotidyl transferase (TdT) (3). All three of these polymerases contain N-terminal BRCT (breast cancer carboxy terminal associated) domains, which confers on the polymerases the ability to form stable complexes with the core NHEJ factors Ku, X-ray cross-complementing gene 4 (XRCC4), and ligase IV. All three polymerases are also active in synthesis during NHEJ in vitro, but there is an abundance of evidence for structural elements unique to each polymerase that account for striking differences in these in vitro activities (4–8). Most clearly, the general character of synthesis activities have a gradient of decreasing dependence on an intact template strand, Pol λ> Pol μ > TdT (9, 10), that in part can be attributed to differences in a variable insert in the palm subdomain (Loop1) (9, 11, 12). However, evidence that these different in vitro activities effects the biological role of these polymerases is definitive only for TdT, which adds template-independent nucleotides (N additions) to broken intermediates during V(D)J recombination. Accordingly, TdT is expressed only in cells active in V(D)J recombination (13).
By comparison, both Pol μ and Pol λ are widely expressed (14, 15), and it has been difficult to clearly link phenotypes of deficiency in these polymerases to an important role for them in NHEJ. For example, resistance to ionizing radiation, a hallmark of proficiency in NHEJ, is not affected by deficiency in Pol λ (16, 17), whereas the effects of deficiency in Pol μ on resistance to ionizing radiation depends partly on cell type (16, 18, 19). Effects of these polymerases on the fidelity of NHEJ have also been observed (9, 10, 16, 20–23), although interpretations vary as to whether differences in catalytic activities are important (i.e., they are redundant), or even if their catalytic activities are relevant at all. Much of the uncertainty can be attributed to changes in relative expression level comparing different cell types and especially to difficulties in unambiguously defining the substrates on which the polymerases acted in these cellular experiments.
We therefore systematically varied both substrate and polymerase in a cellular setting. We identify contexts where only Pol μ or Pol λ, among all of the DNA polymerases expressed in mouse embryo fibroblasts, are active—their cognate substrates—and clarify the mechanistic basis for their specific activities. We show how their differing activities promotes accurate repair and that the ability of either to perform synthesis in this challenging context is surprisingly essential.
Results
Pol μ and Pol λ Are Preferentially Active on Different Substrates.
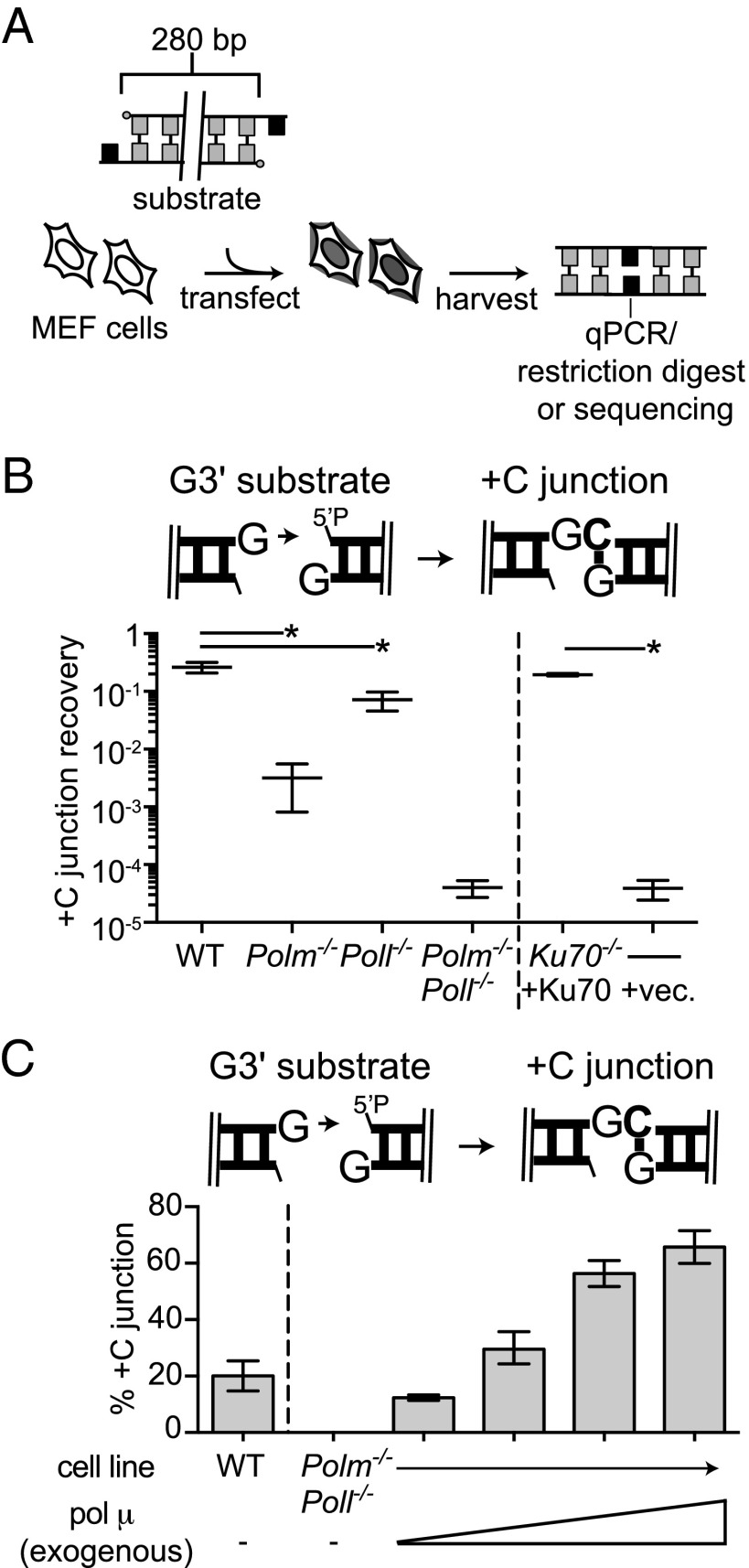
Substrates were introduced into C57BL/6 mouse embryo fibroblasts (MEFs) that were WT or deficient in Pol μ (Polm−/−), Pol λ (Poll−/−), or both. Conditions were optimized to reflect joining by classically defined NHEJ, as is evident from parallel experiments performed in mouse dermal fibroblasts lines with and without a factor essential for NHEJ (Ku70; Figs. 1–5). Head to tail joined products were characterized by quantitative PCR (qPCR) to assess overall product recovery and then sequenced to assess differences in product structures (Fig. 1A). We considered first only those products consistent with activity of a polymerase and ligase alone (Figs. 1–4) to more clearly evaluate polymerase specificity, but then also addressed the significance of polymerase activity after nucleolytic remodeling of the original substrate’s end structure (Fig. 5).
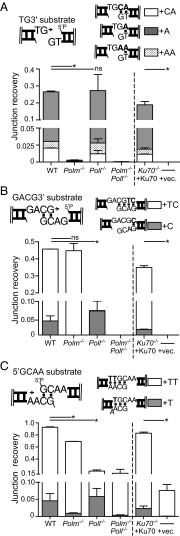
Fig. 1.
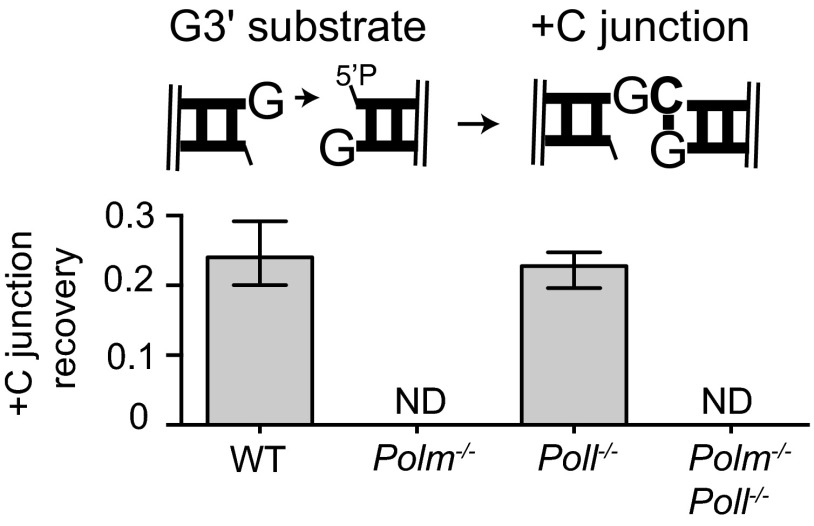
Synthesis primed from ends with an unpaired 3′ terminus. (A) Description of the extrachromosomal DSB repair assay. (B) The G3′ substrate (described in cartoon) was introduced into the noted fibroblast cell lines. For each cell line, the efficiency of recovery for junctions formed after addition of a single complementary nucleotide (+C junction recovery) is expressed as a fraction of the total junctions recovered from WT cells. Error bars reflect the range of means from two triplicate transfections, performed on different days. Mean recovery efficiencies were assessed as significantly different with confidence P < 0.05 (*) or not significantly different (ns). (C) The percentage of junctions formed after accurate synthesis was determined after introduction into the Polm−/−Poll−/− cell line of GCG3′ substrate together with 0, 1, 10, 100, or 1,000 ng of Pol μ. The means and error bars were determined as in B.
Fig. 5.
Effect of polymerase deficiency on the efficiency and fidelity of NHEJ. Shown are efficiencies of junction recovery determined as in previous figures, but now for all junction types. (A) Junctions formed with the G3′ substrate are categorized according to whether (i) their formation is consistent with activity of polymerase and ligase alone (dark blue sectors), (ii) overhangs were remodeled by nuclease activity (light blue), or (iii) overhang sequences were removed. Grayed positions in the cartoons at left are nucleotides we can infer are removed before joining. Products where overhang sequence is missing (iii) were grouped according to whether the original overhangs were precisely removed (tan), deletions extend to sites of >1 nt flanking sequence identity (microhomologies”; red), or other dsDNA deletions (orange). (B and C) Sector areas are determined by recovery efficiencies for corresponding junction types, relative to matched WT control cells, with the angle of rotation for each sector fixed for all pies within an isogenic cell line series. Overall joining efficiencies are listed (Jeff) and assessed as significantly different from the matched WT control with confidence P < 0.05 (*) or not significantly different (ns). The resulting junction spectra were generated after introduction of G3′ substrate into (B) WT and matched polymerase-deficient cells or (C) cells with and without Ku. Junction spectra generated after introduction of the substrate GACG3′ (D; see also Fig. S5) into (E) WT and matched polymerase-deficient cells or (F) cells with and without Ku70.
Fig. 4.
Evaluation of structural elements that distinguish polymerase activities. Polm−/−Poll−/− cells were complemented by introducing the indicated human WT or mutant polymerases together with (A) G3′ and GCG3′, (B) GACG3′, or (C) GCG3′ and GCAG3′ substrates. Either 10 ng (Pol μ constructs) or 1 ng (Pol λ constructs) was used for complementation. The percentage of the indicated synthesis-associated products was determined by sequencing, with error bars derived as in Fig. 1.
Synthesis across a broken DNA backbone poses unique problems for DNA polymerases. The most extreme example of this is occurs when ends with 3′ noncomplementary overhangs pair, as a complementary sequence cannot help align the primer with template. We therefore introduced such a substrate—a DNA fragment with a single 3′ overhanging G at both head and tail ends (G3′)—into the MEF panel described above. Head-to-tail junctions formed after incorporation of a single C, followed by end ligation (+C products), accounted for 26% of those recovered from WT MEFs, whereas this product was recovered 83-fold less frequently in Pol μ-deficient cells (Fig. 1B and Dataset S1). Similar results were also observed using an independently generated panel of MEF lines (Fig. S1). Incorporation of one or more nucleotides other than C (+N) was rare; such products accounted for only 0.5% of those recovered from WT MEFs, and their recovery was equally affected by loss of Pol μ and Pol λ (Dataset S1).
Fig. S1.
Synthesis primed from ends with an unpaired 3′ terminus. The G3′ substrate (described in cartoon) was introduced into MEFs as described in Fig. 1B, except using a panel of MEFs generated independently, derived from mice from different litters. Joining efficiency was determined by qPCR as in Fig. 1B. The proportion of recovered junctions formed after synthesis of a single complementary base (+C junction recovery) was approximated here by assessing the fraction of products sensitive to a restriction enzyme specific for this junction (NsiI). Error bars reflect the range of means from two triplicate transfections, performed on different days. ND, not detectable (recovery efficiency < 0.01).
We additionally complemented Polm−/−Poll−/− MEFs with increasing amounts of Pol μ, resulting in corresponding increases in recovery of the +C junction up to more than double that observed in WT MEFs (63% compared with 26%; Fig. 1C). Participation of Pol μ during NHEJ in this cell type is thus limited by subsaturating expression. Nevertheless, on this class of substrate (noncomplementary 3′ overhangs; Figs. 1 and 3A), even very low levels of Pol μ account for far more activity than the other polymerases expressed in MEFs.
Fig. 3.
Synthesis at ends that align to generate two nucleotide gaps. The recovery efficiencies for junctions formed after synthesis were determined as in Fig. 1B for cells transfected with (A) GT3′, (B) GACG3′, or (C) 5′GCAA substrates. Mean recoveries of the most abundant product were assessed as significantly different with confidence P < 0.05 (*) or not significantly different (ns).
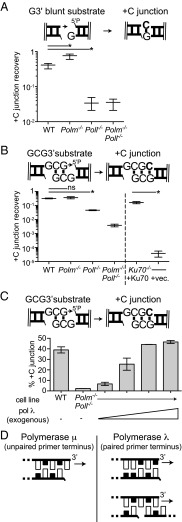
We considered two possible ways the lack of sequence complementarity at ends restricts activity to Pol μ; it is the only polymerase active when aligned ends lack complementary sequence to assist end-bridging or it is the only polymerase active in such a context, but when the primer terminus is additionally single stranded (unpaired). To distinguish between these possibilities, we generated a substrate with one blunt end and one 3′ G overhang (Blunt/G3′); synthesis must now initiate from a blunt end (double-stranded primer), but there is still no complementary sequence to promote end-bridging and primer/template alignment. Surprisingly, the contribution of the two polymerases to synthesis during NHEJ was inverted for this new variant substrate; recovery of the +C junction now relied primarily on Pol λ (Fig. 2A and Dataset S1), even though this new substrate differed from that described in in Fig. 1 by only a single nucleotide (Fig. 1B compared with Fig. 2A).
Fig. 2.
Synthesis primed from ends with a paired 3′ terminus. The recovery efficiencies for junctions formed after synthesis were determined as in Fig. 1B for cells transfected with (A) G3′-blunt or (B) GCG3′ substrates. Error bars and statistics were determined as in Fig. 1B. (C) The percentage of junctions associated with accurate synthesis was determined after introduction of the GCG3′ substrate together with 0, 0.1, 1, 10, or 100 ng of Pol λ into Polm−/−Poll−/− cells, and mean frequencies and error bars derived as in Fig. 1. (D) Cartoon describing the primer contexts preferred by Pol μ and Pol λ for synthesis activity associated with cellular NHEJ.
A similar reduction in junction recovery in Poll−/− cells is also apparent when using a substrate with partly complementary 3′ overhangs (GCG3′; Fig. 2B). In this context, there is again a complementary sequence opposite the primer terminus, but only after alignment of a pair of ends. Additionally, on this substrate, there is little synthesis associated with NHEJ in cells deficient in both Pol μ and Pol λ; thus, other polymerases expressed in MEFs are much less active on GCG3′ than even Pol μ. Introduction of Pol λ into Polm−/−Poll−/− MEFs was also sufficient to recover activity on this substrate, but in contrast to Pol μ with G3′ (Fig. 1C), it was not possible to significantly exceed the levels of Pol λ-dependent synthesis observed in WT MEFs by addition of increasing amounts of Pol λ (Fig. 2C).
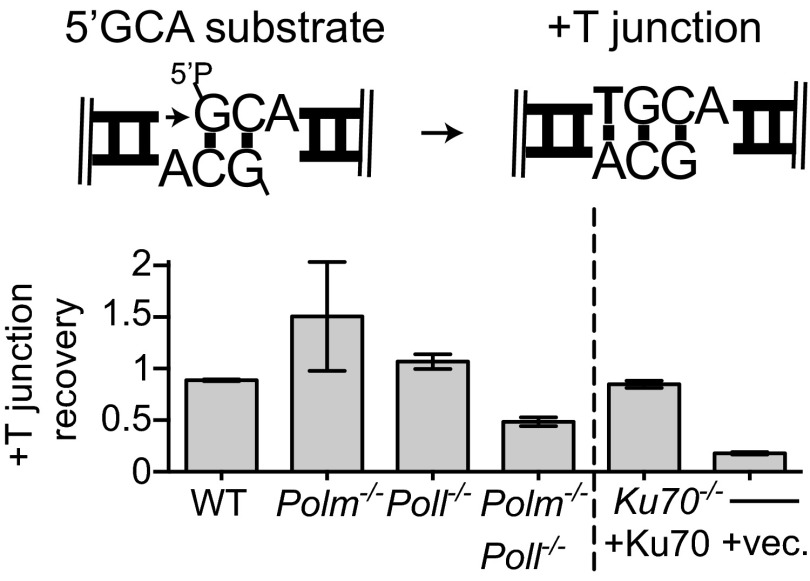
When ends with 5′ overhangs align, gap-filling synthesis does not have to transit a strand break, and this less-challenging reaction has been shown to be largely independent of polymerase specialization in Saccharomyces cerevisiae (10). Consistent with this result, synthesis is 2.7-fold more efficient when comparing analogous 5′ vs. 3′ overhang substrates (Fig. S2 compared with Fig. 2B), and at least when the gap is a single nucleotide, there was no effect of deficiency in Pol λ or Pol μ (Fig. 3C).
Fig. S2.
Synthesis at ends with partly complementary 5′ overhangs. The 5′GCA substrate (described in cartoon) was introduced into MEFs as described in Fig. 1B. Joining efficiency was determined by qPCR. The proportion of recovered junctions formed after synthesis of a single complementary base (+T junction recovery) was approximated by assessing the fraction of products sensitive to restriction enzyme specific for this junction (HpyCH4V). Error bars reflect the range of means from two triplicate transfections, performed on different days. ND, not detectable (<0.01).
Specialized polymerases are thus required during NHEJ in any context where synthesis must transit a strand break; i.e., aligned ends where one or both ends have a 3′ overhang. Importantly, the context of the 3′ terminus then defines which of these two polymerases is most active. Pol μ alone can efficiently initiate synthesis during NHEJ from an unpaired 3′ terminus, whereas this polymerase is less active than Pol λ when there is complementary sequence opposite the 3′ terminus (Fig. 2D).
Pol μ and Pol λ Use Different Mechanisms to Locate Templating Base.
These polymerases are thus essential for activity when the juxtaposition of primer and template must rely on interactions between these DNA molecules and the higher-order protein complex (i.e., NHEJ core factors and the polymerase) that mediates end alignment. This strategy for assembling the substrates for these polymerases raises questions as to how Pol μ and Pol λ locate the templating base, and whether they use the same mechanism for this. We addressed this issue using DNA ends that, in contrast to those described above, are expected to align such that they require synthesis of more than a single nucleotide before ligation can occur.
We addressed this question first with noncomplementary overhangs. Synthesis and ligation of a substrate with two nucleotide noncomplementary overhangs (TG3′) relied on Pol μ (Fig. 3A), similar to results with the comparable single nucleotide overhang substrate (G3′; Fig. 1A). However, the majority of these products involved synthesis of only a single A, the nucleotide complementary to the template base immediately adjacent to the site of ligation. By comparison, junctions with addition of nucleotides complementary to both overhang template positions (+CA) were 20-fold less frequent than +A and comparable in frequency to junctions with repeated addition of A (+AA) (Fig. 3A). Pol μ thus efficiently “skips ahead” to find template during synthesis of this substrate.
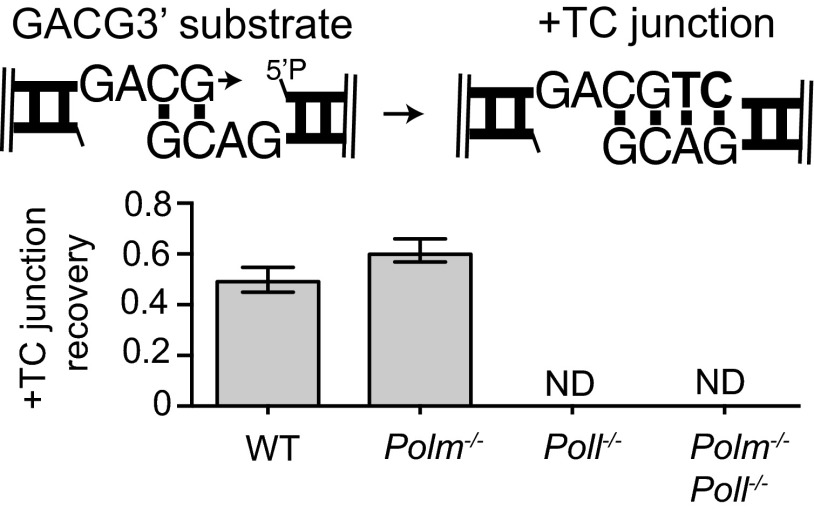
We next addressed synthesis of longer gaps when the ends are partly complementary (GACG3′); again, this is comparable to a substrate described above (GCG3), except the gap after alignment is now two nucleotides instead of one. Strikingly, we were readily able to recover products where both nucleotides were added (+TC) for this substrate—in contrast to the two nucleotide noncomplementary overhang substrate—but this product is dependent on Pol λ (Fig. 3B and Dataset S1). Similar results were also observed using an independently generated panel of MEF lines (Fig. S3). However, a less frequent product of skip-ahead synthesis (+C) was also observed, and similar to the noncomplementary overhang substrate, this skip-ahead product required Pol μ (Fig. 3B). In the absence of both Pol μ and Pol λ, junctions were reduced 3,000-fold. Thus, Pol λ is uniquely effective on this substrate and synthesizes both nucleotides of the two nucleotide gap, and although Pol μ is the only significant alternative, it is more than 10-fold less efficient and, when active, skips ahead (Fig. 3B).
Fig. S3.
Synthesis at ends that align to generate two nucleotide gaps. The GCAG3′ substrate (described in cartoon) was introduced into MEFs as described in Fig. 1B, but using a panel of MEFs generated independently, derived from mice from different litters. Joining efficiency was determined by qPCR, but the proportion of recovered junctions formed after synthesis of TC was approximated by assessing the fraction of products sensitive to a restriction enzyme specific for this junction (AatII). Error bars reflect the range of means from two triplicate transfections, performed on different days. ND, not detectable (recovery efficiency < 0.01).
As noted above, synthesis at ends with partly complementary 5′ overhangs has no specific requirement for either Pol μ or Pol λ, at least when end alignment generates a one nucleotide gap. However, the unique effectiveness of Pol λ on two nucleotide gaps in the context of 3′ overhangs led us to readdress this question. Consistent with this idea, we show Pol λ is uniquely effective in synthesizing both nucleotides of a two-nucleotide gap even on 5′ overhang substrates (+TT; Fig. 3C and Dataset S1). A skip-ahead product (+T) is again observed for this substrate and depleted in Pol μ-deficient cells.
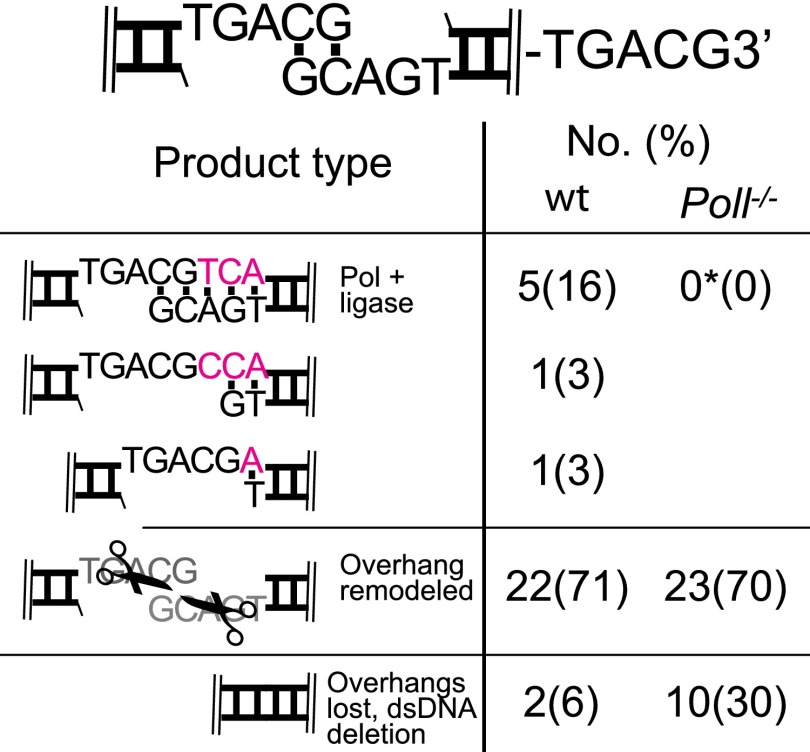
We also assessed repair using a substrate (TGACG3′) comparable to the ones described in Figs. 2C and 3B, except the gap generated after alignment is now three nucleotides (instead of two or one). Complete fill in of this longer gap still requires Pol λ (Fig. S4). However, this reaction is inefficient even in WT cells (16%, Fig. S4, compare with 41% for a two nucleotide gap, Fig. 3B), and the frequency of products generated after remodeling of the initial end structure by a nuclease increases to compensate [71% (Fig. S4) compared with 26% (Fig. 5E)]. Importantly, overhang remodeling often generates gaps less than three nucleotides, and polymerase activity on these shorter gaps can now be inferred to occur at least as frequently as complete fill-in of the unremodeled end structure (Fig. 5 and Fig. S4). Thus, the end alignment configurations expected to be most important for activity of specialized polymerases in NHEJ—paired and unpaired primer, and gaps of one to two nucleotides—can be accounted for by the substrates described in Figs. 1–3.
Fig. S4.
Synthesis at ends that align to generate three nucleotide gaps. Repair junctions recovered from WT and Poll−/− cells are categorized as consistent with formation by polymerase and ligase activity alone (e.g., as in Figs. 1–4), formation after nuclease activity limited to overhang sequence (overhang remodeled), or formation after nuclease activity removes all overhang sequence (dsDNA deletion and overhangs removed). The number of sequences for each junction type and representation as a percentage of the total analyzed is shown. Accurate fill-in synthesis was significantly reduced in Poll−/− relative to WT cells (*confidence level, P < 0.05 by Fisher’s exact test).
We conclude the two polymerases use different cues for locating the templating nucleotide. Pol λ locates template nucleotides in normal sequence, progressing from 3′ to 5′ across any gaps present after end alignment. Pol μ instead identifies template primarily through a spacing rule, where it uses as template only the nucleotide adjacent to the site of ligation, even when one or more nucleotides of template is available further upstream.
Structural Elements Essential for Polymerase Specialization in Cellular NHEJ.
We next addressed structural elements that could help distinguish the activities of Pol μ and Pol λ. When considering Pol μ, we and others have emphasized an extended loop insert (Loop1) in its palm subdomain (9, 11, 12). Deletion of this loop (Pol μΔloop1), or substitution of a residue thought to help limit its mobility (Pol μF385A), specifically reduces activity on noncomplementary ends (its cognate substrate) in vitro (7). To determine whether the requirement for this element is maintained during cellular NHEJ, we introduced the substrate with noncomplementary 3′ overhangs (G3′) into Polm−/−Poll−/− MEFs and then complemented these cells with various Pol μ constructs. As expected, introduction of WT Pol μ was essential for recovery of products of template-dependent synthesis and ligation (+C product); by comparison, Pol μF385A was one-third as effective as the WT construct, and Pol μΔloop1 was more than 10-fold less effective (Fig. 4A, Left, and Dataset S2).
Activities for all three constructs (WT, Pol μF385A, and Pol μ Δloop1) were nevertheless indistinguishable on a substrate with a complementary sequence opposite the primer (Fig. 4A, Right); deleting or disabling the loop1 element thus impairs Pol μ activity on its cognate substrate, but does not make it generally unable to perform synthesis on other substrates. Deletion or mutation of the loop also does not alter how the polymerase identifies template, as all three Pol μ constructs remained unable to fill in both nucleotides (+TC; Fig. 4B and Dataset S2) when end align generates a two nucleotide gap. Instead, all three Pol μ constructs still skip ahead; indeed, when loop1 is deleted, the most frequent product involves repeated use of this template position (+CC product; Fig. 4B). The loop is thus an essential component of Pol μ specialization—it is required for activity on its cognate substrate—but deletion of the loop neither enables the polymerase on preferred substrates for Pol λ nor reduces the tendency of this polymerase to skip ahead on longer gaps.
Prior structural studies also suggest Pol λ possesses an element that makes it uniquely suited for its cognate substrate (4). When ends align to generate a two nucleotide gap, Pol λ can “scrunch” the next-to-be copied template nucleotide by burying it in a pocket generated by amino acids L277, H112, and R514. We addressed whether this pocket was important for activity on two nucleotide gaps in cells by complementing Polm−/−Poll−/− cells with WT Pol λ, as well as a Pol λ construct where these residues were substituted with alanine (Pol λ 3A). This mutant construct had generally reduced activity, but the degree of reduction was much greater on the substrate with ends that align to generate a two nucleotide gap vs. a one nucleotide gap, in accordance with a central role for this pocket in promoting synthesis on the longer gap (Fig. 4C and Dataset S2).
Polymerase Specialization Is Essential for NHEJ.
We identified cognate substrates for each polymerase above by excluding products where it was clear the initial substrate’s end structure had been remodeled by nuclease activity (Figs. 1–4). Here we address the impact of polymerase deficiency on NHEJ overall by extending analysis to all products. We represent in Fig. 5 the relative recoveries of different products or product classes as different sectors of a pie first for WT cells. For polymerase-deficient cells, the areas of each pie sector are adjusted to reflect the recovery efficiencies, relative to WT cells, for corresponding products (or product classes). The angle of rotation for each sector is kept constant to emphasize how the effects of polymerase deficiency vary for different products, and the overall joining efficiency (i.e., summing together all products; Jeff) for each line, relative to the matched WT control, is listed.
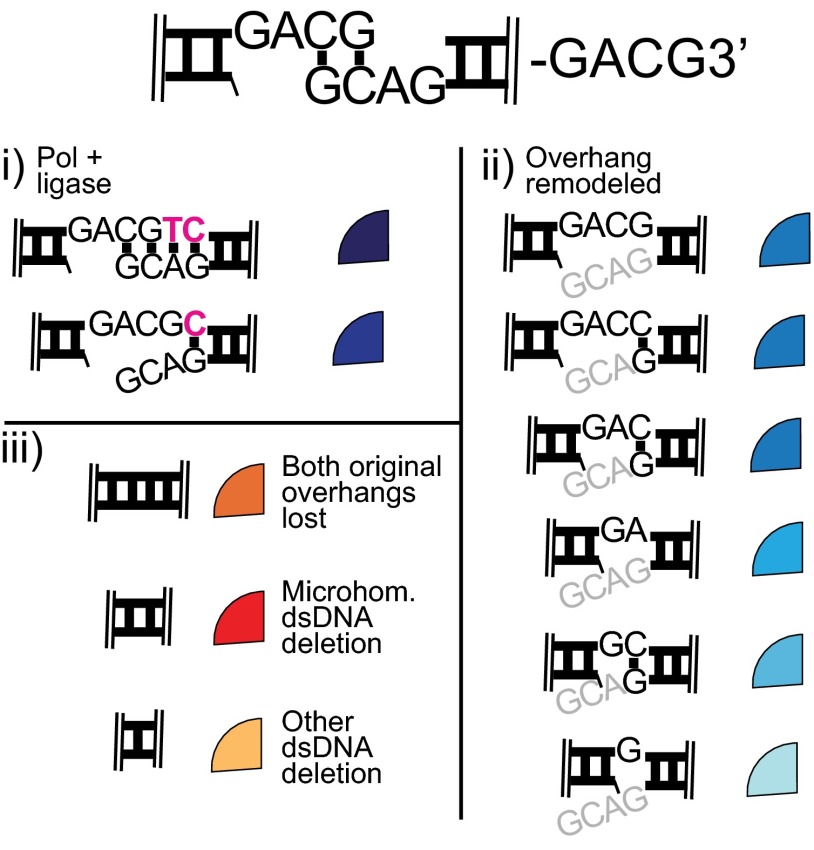
As previously emphasized (Fig. 1), G3′ originates as a cognate substrate for Pol μ; this is apparent by the 83-fold reduction in the area of the dark blue sector in Polμ−/− cells, relative to the WT control line (Fig. 5 A, i and B, compare top left and top right quadrants). However, there is an inverted dependency on polymerase for a second major product of this substrate (Fig. 5 A, ii, light blue); recovery of this product is reduced 23-fold in cells deficient in Pol λ and not significantly impacted by deficiency in Pol μ (Fig. 5B). Taking into account the product structure and previous analysis (Fig. 2A), we infer that the initial substrate, a cognate substrate for Pol μ, was remodeled by nuclease activity into a cognate substrate for Pol λ (Fig. 5 A, ii). Using a substrate initially “cognate” for Pol λ (GACG3′), we again observed a pattern of alternating dependency on one or the other polymerase, although there are many more permutations of products with partial overhang loss (sectors with different blue shades, Fig. 5 D–F and Fig. S5). Remodeling of overhangs by nuclease activity thus adds to the flexibility of end processing during NHEJ. However, the specificity of each polymerase is retained after overhang remodeling, and their presence remains integral to efficient joining of the remodeled end structures.
Fig. S5.
Legend including junction sequences. Synthesized nucleotides in pink. Overhang sequence retained in each junction sequence is listed in black text. Gray text refers to opposite strand sequence that was lost from the product.
The remaining products—those where both overhangs are lost, including deletions that extend dsDNA flanking sequence—are also reduced in polymerase-deficient cells, but for these products, the degree of reduction is similar regardless which polymerase is missing. This result can be explained if nuclease activity generates heterogeneous intermediates, where some are cognate substrates for Pol μ and others are cognate substrates for Pol λ; however, as diagrammed for a product where both overhangs are lost (Fig. 5 B, iii), the same product can then be generated by activity of either polymerase on their different cognate substrates. Moreover, for these product classes, it is also possible to generate intermediates that can be directly ligated, independent of polymerase activity (i.e., when by chance, nuclease activity precisely removes both overhangs). This polymerase-independent mechanism is especially relevant for microhomology-directed deletions (red segment), where the intermediates potentially have complementary overhanging sequence (sticky ends). These products consequently account for a much larger fraction of the residual products recovered from polymerase-deficient cells.
We thus observe very striking differences in product spectra when comparing cells deficient in a single polymerase. At the same time, nuclease activity often converts end structures that are a cognate substrate for one polymerase into the cognate substrate for the other, so there is typically only a modest impact (less than threefold) on overall joining efficiency (Jeff; Fig. 5) in these cells. In contrast, the efficiency of joining is reduced ∼20-fold in cells deficient in both. Residual products are also almost entirely comprised of those with both overhangs lost and microhomology-directed deletions. This impact of deficiency in both polymerases on overall joining efficiencies, as well as the spectrum of products generated, is similar to that observed after complete loss of NHEJ (i.e., due to deficiency in Ku; Fig. 5 C and F).
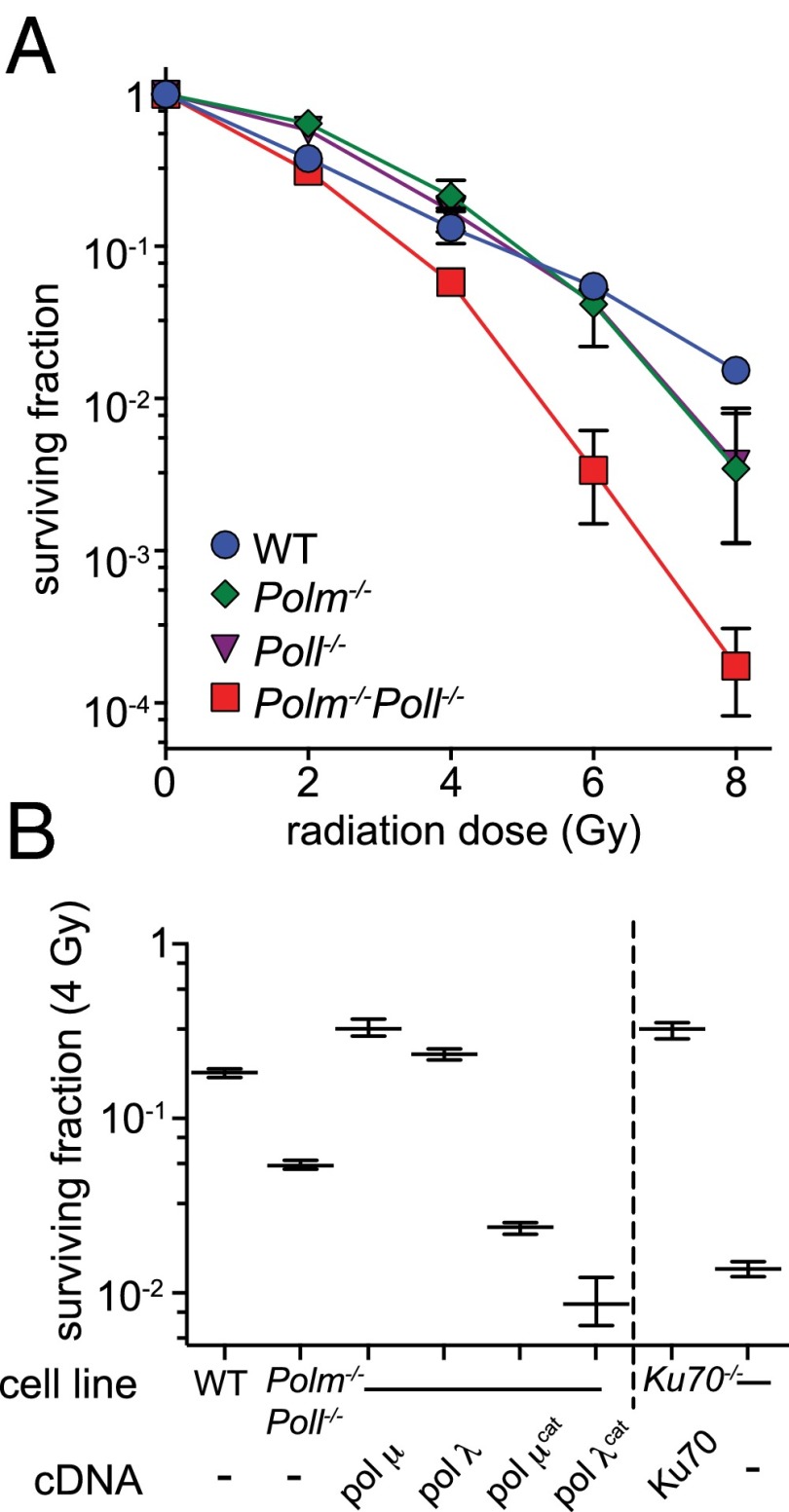
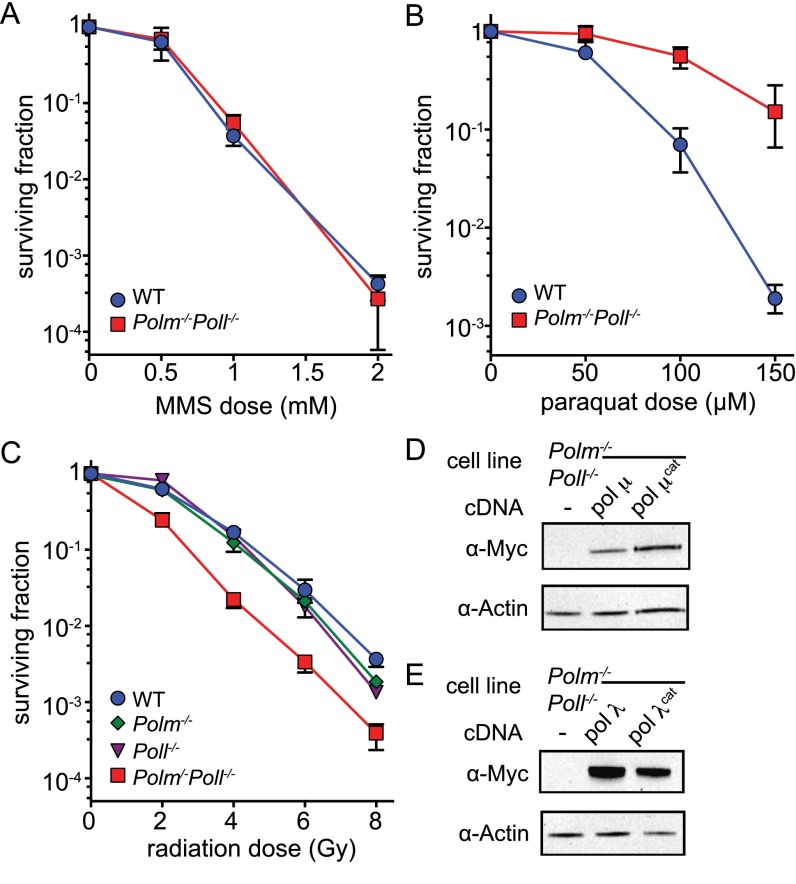
These results are readily reconciled with an assay of gross phenotype. Both of the cell lines deficient in a single polymerase were at best mildly sensitive to a double-strand break-inducing agent, ionizing radiation (IR), whereas cells deficient in both polymerases were much more clearly IR sensitive (Fig. 6A). Doubly deficient cells were not sensitive to sources of DNA damage that do not directly introduce DSBs [methyl-methane sulfonate (Fig. S6A) and paraquat (Fig. S6B)], and a similarly specific sensitivity to IR was observed using an independently generated panel of MEF lines (Fig. S6C). Moreover, overexpression of either WT Pol μ or Pol λ alone was sufficient to complement the radio-sensitivity observed in cells deficient in both (Fig. 6B). We were also able to confirm catalytic activities were essential for this complementation, using catalytically defective variants of these polymerases. Indeed, expression of either catalytically inactive polymerase resulted in a severe hypersensitivity to IR, relative to even the parental doubly deficient line.
Fig. 6.
Effect of polymerase deficiency on resistance to ionizing radiation. (A and B) Resistance to ionizing radiation was determined by evaluating colony-forming proficiency after exposure to X-irradiation, relative to untreated controls. Error bars represent the SD from the mean of triplicate experiments. (B) Resistance to 4 Gr of ionizing radiation for WT and Polm−/−Poll−/− lines or stable subclones of the latter line engineered to express mouse cDNAs that were either WT, Pol μ cat (D330E + D332E), or Pol λ cat (D425E + D427E).
Fig. S6.
Sensitivity of polymerase-deficient cells to agents that damage DNA. (A) Resistance to ionizing radiation as in Fig. 6, except using a panel of MEFs generated independently, derived from mice from a different litter. (B) Cellular resistance to methyl methanesulfonate (MMS) was determined after a 4-h exposure and (C) resistance to paraquat after exposure for 18 h. Error bars represent the SD from the mean survival at each dose. (D and E) Western blot analysis of Polm−/−Poll−/− subclones used in Fig. 6B that were engineered to express WT or catalytically inactive Pol μ (D) or Pol λ (E) proteins.
Discussion
Polymerase Specialization.
Seventeen different DNA polymerases are widely expressed in mammals, arguing that disparate needs for DNA synthesis has driven a high degree of specialization. We show here how 2 of these 17 polymerases are specifically designed to direct synthesis across a broken DNA backbone: synthesis in a context that is unique to, and we show here is essential for, efficient repair of double-strand breaks by NHEJ.
Pol μ alone had significant activity on all of the substrates tested. This flexibility was largely attributable to its ability to prime synthesis from a single-stranded terminus (Fig. 1), but a consequence of this characteristic activity is that it is not possible to use canonical means for identifying template. Accordingly, accompanying work [Moon et al. (24)] and we show this polymerase typically chooses its template during cellular NHEJ by following a “one nucleotide gap rule,” where the unpaired base adjacent to the downstream 5′ phosphate is used as template, even when there are available template sites further upstream of this position: the polymerase skips ahead (Fig. 3 A–C and Fig. S7C). Although deletion or mutation of the loop1 motif of Pol μ impairs its ability to prime synthesis from single-stranded termini, the mutant polymerases still locate template by skipping ahead (Fig. 4B), indicating these two facets of Pol μ specialization can surprisingly be uncoupled from each other.
Fig. S7.
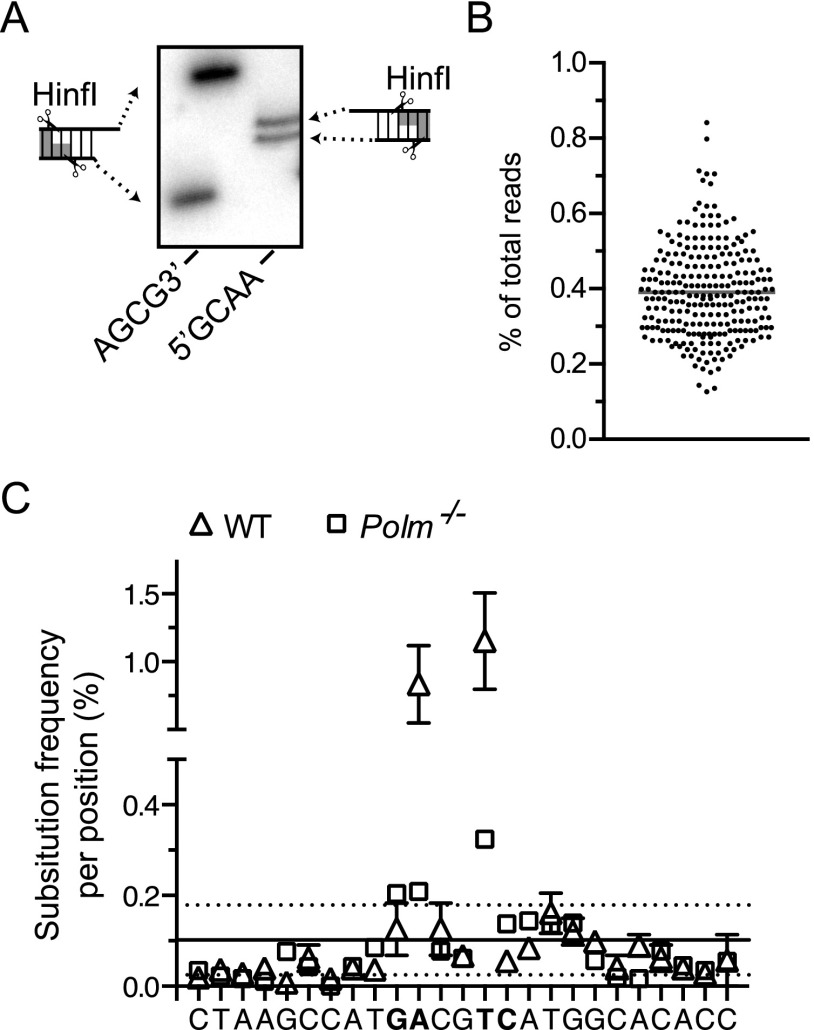
Validation of the cellular NHEJ assay. (A) Representative substrates were HinfI digested, 32P-labeled, and resolved by denaturing polyacrylamide gel electrophoresis. (B) A library of a control oligonucleotide with an embedded degenerate tetramer was prepared for sequencing from the same number of input molecules and using an identical workflow as experimental samples. The frequency of reads for each different sequence, from a total of 11,776 reads, is plotted for each of the predicted 256 sequences. (C) Mean frequency of single nucleotide substitutions for joined products of GACG3′ recovered from WT (triangles) and Pol μ-deficient cells (squares) over a portion of the junction flanking the ends. Sites of cellular polymerase-dependent gap filling are bolded. Error frequencies at each position of the experimental samples fall within 1 SD (dashed lines) of the mean substitution error (solid line) derived from sample processing (as determined for a control amplicon generated by T4 ligation of 5′GATC in vitro), except at two positions in which substitution rate is higher and largely attributable to repeated C addition by Pol μ (see also Fig. 3A).
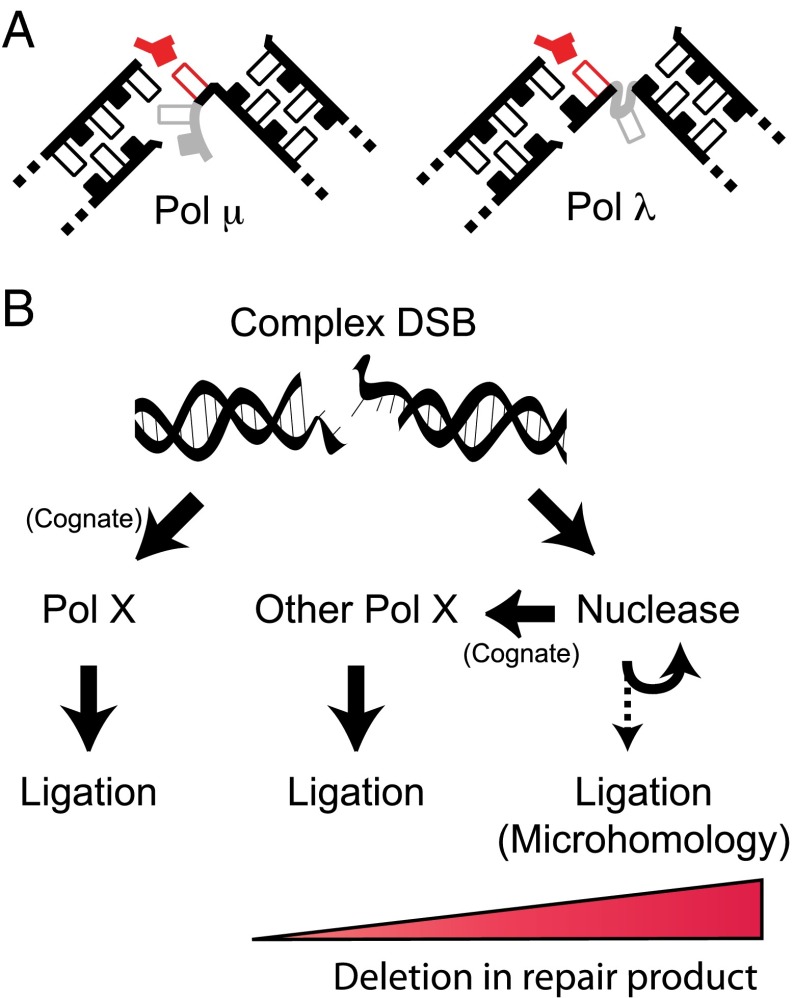
Like Pol μ, Pol λ is able to synthesize over broken template, but Pol λ accounts for the majority of synthesis activity when there is complementary sequence opposite the primer (dsDNA primer; Fig. 2). Pol λ also solves the problem of locating template in such contexts differently than does Pol μ; when ends align to generate a two nucleotide gap, only Pol λ has the significant ability to incorporate both nucleotides (Fig. 3). We show that Pol λ is uniquely proficient on the longer gap because it uses a variation of the one nucleotide gap rule; like Pol μ, it maintains the arrangement of strand break termini as if it were a single nucleotide gap, but now does so by scrunching the downstream template base within a pocket unique to this enzyme (4) (Figs. 4C and 7A).
Fig. 7.
Mechanism by which specialized polymerases contribute to NHEJ. (A) Diverse end structures relevant to synthesis during NHEJ may be addressed by these polymerases similarly, as if alignments always generate one-nucleotide gaps, by adjusting template (grayed positions) upstream (Pol μ) or down stream (Pol λ) of the nascent base pair (red) when gaps are longer than one nucleotide. (B) Complex DSBs are ligated after synthesis by a Pol X polymerase on its cognate substrate or after nuclease activity remodels the initial end structure to a cognate substrate for the other polymerase. In the absence of polymerase activity ligation is more reliant on microhomologies, likely uncovered by repeated cycles of nuclease activity, and is associated with increased deletion.
Indeed, for ternary structures of either polymerase on a one nucleotide gap (6, 25), Pol λ scrunched on a two nucleotide gap (4), and Pol μ skipping ahead on a two nucleotide gap [Moon et al. (24)], both the distances between the two-strand-break termini and the angles of upstream and downstream strands are nearly the same [the one nucleotide gap rule; figures 7A and 6 of Moon et al. (24)]. As noted above (Figs. 1–3 and Fig. S4), our work argues these polymerases are important for NHEJ primarily for filling in short one to two nucleotide gaps; these four structures are thus sufficient to model the majority of end alignment configurations expected to be relevant to the activity of specialized polymerases during NHEJ. We suggest the single spatial arrangement of strand-break termini observed in these structures is driven by a need to incorporate polymerase activity within the NHEJ paired-end complex (including Ku, XRCC4, ligase IV, XLF, and PAXX), which assumes the burden of preassembling the substrates for these polymerases (Fig. 7B). However, the paired-end complex cannot reasonably sample enough different end alignment configurations to keep pace with substrate diversity. Accordingly, the two polymerases elegantly address this problem by adjusting only the template strand and only immediately upstream (Pol μ) [Moon et al. (24)] or downstream (Pol λ) (4) of the nascent base pair binding site (Fig. 7A).
Significance of Polymerase Specialization to NHEJ.
The ligation step during NHEJ is much less tolerant of mispairs and other distortions when these are 3′ of strand breaks, relative to 5′ of strand breaks (2). As a consequence, synthesis that initiates from 3′ overhangs or blunt ends—and that then is sufficient to generate paired termini 3′ of strand breaks—is especially significant to this pathway. Pol μ and Pol λ alone among mammalian polymerases address this problem but use different solutions.
Pol μ allows retention of activity on a wider range of substrates, relative to Pol λ; it can make any end with a 3′ overhang, regardless of sequence (and possibly associated damage as well), somewhat of a “universal donor” for the ligation step. This flexibility in activity might explain why phenotypes of Pol μ deficiency, including immunodeficiency, impaired hematopoiesis, and cellular radio-sensitivity, can be more severe than phenotypes of Pol λ deficiency (16, 19, 26). Pol λ is nevertheless more active than Pol μ for some substrates, and on a subset of these (2nt gaps), Pol λ allows for retention of an extra nucleotide in the product and thus is more accurate.
The increased accuracy of Pol λ in this context is most clearly significant to NHEJ as a means to further enable ligation. Pol μ activity on the same substrates indeed generates a paired 3′ terminus (a universal donor), but this is not an ideal solution, as the one or more mispairs that are upstream of the site of ligation still interferes with the ligation step. There may be additional negative consequences of using only Pol μ for repair; for example, it is prone to generating mononucleotide repeats (26, 27) (Fig. 3A and Fig. S7C), and, indeed, Pol μ−/− mice have been characterized as healthier than their matched WT counterparts in some respects (28). As a means to possibly limit these negative consequences, the levels of Pol μ, but not Pol λ, expressed in MEFs is subsaturating with respect to its ability to contribute to NHEJ (Fig. 1C compared with Fig. 2C), and high levels of Pol μ expression are restricted to a very short window in B-cell development (16). We suggest that there is a hierarchy of polymerase use, where the more accurate Pol λ is typically given priority in most cell types: this is similar to the hierarchy of polymerase use described in the bypass of UV photoproducts.
Together these two polymerases make up a comprehensive toolbox that is essential for repair of diverse end structures by NHEJ. The results described in Fig. 5D provide a particularly striking example of this; we observed significant amounts of eight different permutations of product that included overhang loss (blue sectors), and one or the other polymerase was typically essential for each one. However, the ability of this pathway to also use nuclease(s) to process ends means the cognate substrate for one polymerase can frequently be altered to generate a cognate substrate for the other (Fig. 7B). Therefore, loss of one or the other polymerase alone has a mild impact on overall NHEJ efficiency, whereas the impact of deficiency in both polymerases is severe, with levels indistinguishable from those observed in Ku-deficient cells for some substrates (Fig. 5). Deficiencies in these specialized polymerases also have a synergistic impact on overall NHEJ efficiency using a gross measure of chromosomal double-strand break repair (sensitivity to ionizing radiation; Fig. 6).
We show here that Pol μ and Pol λ are uniquely effective when synthesis must transit a strand break and thereby play a key role in classically defined NHEJ. Strikingly, one other eukaryotic polymerase has a related activity—Pol θ also directs synthesis across double-strand break ends, although it needs a short patch of terminal complementary sequence, or microhomology, to initiate synthesis—and it is essential for an alternate NHEJ pathway (29–31). More complex and longer-lived organisms are expected to need more flexible and efficient versions of end joining; it is clear that the use of specialized polymerases is an integral part of how this has been achieved.
Methods
Cell Lines.
Polm−/− mice were generated as previously described (32), Poll−/− mice were generated by disruption of exon 5, and these two mice were crossed to generate Polm−/−Poll−/− double KO mice. MEFs were generated from E14.5-d embryos from WT (C57BL/6), Poll−/−, Polm−/−, and Poll−/−Polm−/− mice and were immortalized by infection with a retrovirus that expresses SV-40 large T antigen (Addgene; #1779). Genotypes were confirmed using PCR and Western blot analysis. Cells overexpressing either myc-tagged WT or catalytically inactive polymerase variants were generated by infecting the Polm−/−/Poll−/− cells with retrovirus derived from pBabe-puro constructs containing the appropriate murine cDNAs, and comparable levels of expression were verified by Western blot (Fig. S6 D and E). Ku70−/−p53−/− cells were obtained from P. Hasty (The University of Texas Health Science Center at San Antonio) and then complemented by expression of the mouse Ku70 cDNA (+Ku70) or the pBabe-puro empty vector (vec). All cell lines were maintained in DMEM supplemented with 10% (vol/vol) FBS (Sigma), 5 mM N-acetyl-l-cysteine (Sigma), 2 mM l-glutamine (Gibco), 1× penicillin-streptomycin, and 2 μg/mL puromycin (if necessary), at 37 °C and 5% CO2.
Generation of DSB Repair Substrates.
Substrates were prepared by PCR amplification of a common 285-bp DNA segment with primer pairs containing embedded restriction enzyme digest sites chosen to generate the desired end structures. The primer pairs and restriction enzymes used for each substrate are listed in Table S1. Substrates were purified using the QIAquick PCR purification kit (Qiagen), and complete digestion of each substrate was validated by native gel electrophoresis. Two substrates were selected for further validation that this procedure is sufficient to generate the intended end structures at nucleotide-level resolution. These substrates were sequentially treated with HinfI, shrimp alkaline phosphatase, and T4-kinase in the presence γ-[32P]ATP and analyzed by denaturing gel electrophoresis (Fig. S7A).
Table S1.
Primers and restriction enzymes used to generate substrates
| Substrate | Primers | Restriction enzymes |
| G3′ | 5′-CAAGTGGACCACATGTCTTAGCTGTATAGTCAGGGAAATC-3′ | AhdI |
| 5′-CCGCCGACGCCATGTCACACCCATCTCAGACTGGCTACCC-3′ | ||
| G3′-Blunt | 5′-TCAGTATCCTGTGACTGCACACCCATCTCA-3′ | BciVI/PvuII |
| 5′-GTGGTCAGCTGGCTTAGCTGTATAGTCAG-3′ | ||
| GCG3′ | 5′-TTTTTGCCACGCTGGCTTAGCTGTATAGTCAGGGA-3′ | BglI |
| 5′-CACCTGCCTCGCTGGCACACCCATCTCAGACTGGC-3′ | ||
| TG3′ | 5′-TTTTTGCAATGCAATCGCTTAGCTGTATAG-3′ | BsrDI |
| 5′-TCACGCAATGCAATCACACCCATCTCA-3′ | ||
| GACG3′ | 5′-CAAGTGGTCCACCGTCATGGCTTAGCTGTATAG-3′ | BstXI |
| 5′-GCCGACCAGCGTCATGGCACACCCATCTCA-3′ | ||
| 5′GCAA | 5′-CAAGTGGTCTCCGCAAATCGCTTAGCTGTATAG-3′ | BsaI |
| 5′-GCCGAGGTCTCAGCAAATCACACCCATCTCA-3′ | ||
| 5′GCA | 5′-TCATGCTCTTCCGCAATCACAGAAAATCGCTTAGCTG-3′ | BspQI |
| 5′-CAAGGCTCTTCAGCAATCTGGACGAATCACACCCATCT-3′ |
Extrachromosomal DSB Repair Assay.
Extrachromosomal DNA substrates (20 ng) and pMAX-GFP plasmid (600 ng) were introduced into the MEF cell lines (2 × 105 cells per transfection) by electroporation using a 1,350-V, 30-ms pulse in a 10-μL chamber (Neon; Invitrogen). After electroporation, cells were incubated in fresh media without antibiotics for 1 h at 37 °C to allow time for the cells to carry out end joining. Cells were washed with PBS, and then the total cellular DNA was harvested using a QIAamp DNA mini kit as per the manufacturer’s instructions (Qiagen). Each electroporation was performed in triplicate and repeated in triplicate on a second day. The complementation of cells with purified proteins was performed as above with the addition of 1–1,000 ng of protein to the substrate transfection solution immediately before electroporation. The proteins used in this study were expressed and purified as described previously (4, 7).
Recovery of joined products was quantified by real-time PCR (qPCR) using an ABI 7900HT or QuantStudio 6 System (Applied Biosystems), primers that amplify head-to-tail junctions (5′-CTTACGTTTGATTTCCCTGACTATACAG and 5′-GCAGGGTAGCCAGTCTGAGATG), and VeriQuest SYBR Green qPCR Master Mix (Affymetrix). This assay has previously been validated as efficient, reproducible, and linear over the range relevant to these experiments (2). The number of junctions recovered from cells was determined by comparison of the threshold cycles (CT) to a standard curve run in parallel, generated by serial dilution of a model amplicon into DNA harvested from mock-transfected DNA.
Joining efficiency (Jeff) was determined by comparing the number of junctions recovered from cells deficient in the polymerases or Ku to the number of junctions recovered from a matched WT cell line electroporated in parallel (WT Jeff ≡ 1). The proportion of specific junction sequences in each sample (P) was then typically determined (Figs. 1B, 2 A and B, and 3–5) by amplification and sequencing on the Illumina Miseq platform (see below). In selected cases (Figs. 1C and 2C and Figs. S1–S3), junctions were characterized by diagnostic restriction enzyme digests on repair products that were amplified using Cy-5–labeled versions of the primers described above. The restriction enzymes used for each substrate were as follows: NsiI for products of G3′, AatII for products of GCAG3′, or AfeI for products of GCG3′. Digestion products were resolved on a 5% (wt/vol) native polyacrylamide gel and visualized using a Typhoon Imager (GE Healthcare). Quantification of relative band intensities was carried out using ImageJ software.
The efficiency of recovery for each junction was then defined as Jeff × P. Whether differences in means of recovery efficiencies were significant was assessed by one-way ANOVA test, with the P values adjusted to account for multiple comparisons by the Bonferroni method.
Next-Generation Sequencing.
Sequencing was performed in two separate runs. Template DNA for each sequencing library was prepared by pooling the three independent electroporations performed on a single day. Amplification of 5 × 105 input molecules (calculated from qPCR) was performed using Phusion DNA polymerase (NEB) and variants of qPCR primers possessing six-nucleotide barcode sequences appended to their 5′-ends for 21 cycles. Amplified DNA (15.5 ng/library) were pooled into groups of 8–12 libraries, 5′phosphorylated, and treated with Klenow exo- to add dA to the 3′ termini (NEB). The ends were then ligated to adapters for paired-end sequencing (Illumina). Agarose gel purification was used to remove free adapter. A final enrichment amplification of 10 cycles was performed, and products again werew purified (Agencourt Ampure XP; Beckman Coulter). Either 180 (sequencing run 1) or 70 ng (sequencing run 2) of the purified DNA from each group of libraries was then combined. These libraries were submitted for a 2 × 80-bp (run 1) or 2 × 250-bp (run 2) sequencing run (MiSeq; Illumina). A PhiX174 DNA “spike” was included in each run. We additionally validated that sample diversity was maintained after amplification and sequencing by amplifying and sequencing a control template with an embedded degenerated tetramer, using the same input number of molecules as we used for experimental samples (5 × 105; Fig. S7B).
Reads of PhiX174 DNA were removed, adapter sequences were stripped, read pairs were merged, and libraries were de-indexed using Genomics workbench v7.5.1 (CLC-Bio). Sequences that were improperly de-indexed were identified by exact match to common sequences in other libraries and removed. Substitution error due to sample processing (1.2 × 10−3) was assessed by determining the average frequency of substitution at each position using a control library generated from the in vitro ligation of a 5′GATC substrate with T4 ligase. The single nucleotide substitution rate for sequences from experimental samples was not significantly greater than the control, except as noted in Fig. S7C; thus, analysis of experimental samples was restricted to counting exact matches to a list of uniquely identifiable sequences generated from all combinations of sequential terminal deletions. We also included in our analysis “N-additions,” defined as those junctions with sequence of any length or composition inserted between left and right ends. To compensate for reduced counts of exact matches due to processing-dependent sequence substitutions, we applied the formula y = x[1 − (a × n)], where y is the corrected count, x is the observed count of exact matches, a = 1.2 × 10−3 (average substitution frequency of control library), and n is the length of the sequence.
Cytotoxicity Studies.
Experiments where cells were treated with IR were carried out by seeding 100–100,000 cells per 10-cm dish in fresh media lacking antibiotic 4 h before treatment with varying doses of radiation using a RS 2000 biological irradiator (Rad Source Technologies). Colonies formed 10 d after treatment were strained using a crystal violet [0.5% (wt/vol)] solution. Experiments where cells were treated with methyl methanesulfonate (Sigma) and paraquat dicholoride (Ultra Scientific) were carried out similarly, but the cells were dosed for 4 or 18 h, respectively, before seeding. N-acetyl-l-cysteine was omitted from the media for cells treated with paraquat dichloride. Plates containing a minimum of 50 colonies were counted by hand, and at least three plates were counted for each dose.
Supplementary Material
Acknowledgments
We thank Andrea Moon, Lars Pedersen, Miguel Garcia Diaz, and Rajendrakumar A. Gosavi (The National Institute of Environmental Health Sciences) for purified proteins and Dr. Paul Hasty (The University of Texas Health Science Center at San Antonio) for fibroblasts lines deficient in Ku70 and p53. We thank the laboratory for helpful discussions. This research was supported by National Cancer Institute Grant CA097096 (to D.A.R.), Postdoctoral Fellowship PF-14-0438-01-DMC from the American Cancer Society (to J.M.P.), Ministerio de Ciencia y Tecnologia Grant BFU2012-37969 (to L.B.), and a European Molecular Biology Organization fellowship (to A.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505805112/-/DCSupplemental.
References
- 1.O’Driscoll M, Jeggo PA. The role of double-strand break repair: Insights from human genetics. Nat Rev Genet. 2006;7(1):45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 2.Waters CA, et al. The fidelity of the ligation step determines how ends are resolved during nonhomologous end joining. Nat Commun. 2014;5:4286. doi: 10.1038/ncomms5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsden DA, Asagoshi K. DNA polymerases in nonhomologous end joining: Are there any benefits to standing out from the crowd? Environ Mol Mutagen. 2012;53(9):741–751. doi: 10.1002/em.21725. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Diaz M, et al. Template strand scrunching during DNA gap repair synthesis by human polymerase lambda. Nat Struct Mol Biol. 2009;16(9):967–972. doi: 10.1038/nsmb.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin MJ, Blanco L. Decision-making during NHEJ: A network of interactions in human Polμ implicated in substrate recognition and end-bridging. Nucleic Acids Res. 2014;42(12):7923–7934. doi: 10.1093/nar/gku475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moon AF, et al. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol. 2007;14(1):45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 7.Moon AF, et al. Sustained active site rigidity during synthesis by human DNA polymerase μ. Nat Struct Mol Biol. 2014;21(3):253–260. doi: 10.1038/nsmb.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romain F, Barbosa I, Gouge J, Rougeon F, Delarue M. Conferring a template-dependent polymerase activity to terminal deoxynucleotidyltransferase by mutations in the Loop1 region. Nucleic Acids Res. 2009;37(14):4642–4656. doi: 10.1093/nar/gkp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nick McElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19(3):357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 10.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280(32):29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 11.Delarue M, et al. Crystal structures of a template-independent DNA polymerase: Murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21(3):427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juárez R, Ruiz JF, Nick McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34(16):4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- 14.Aoufouchi S, et al. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 2000;28(18):3684–3693. doi: 10.1093/nar/28.18.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domínguez O, et al. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19(7):1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertocci B, De Smet A, Weill JC, Reynaud CA. Nonoverlapping functions of DNA polymerases mu, lambda, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity. 2006;25(1):31–41. doi: 10.1016/j.immuni.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 17.Vermeulen C, Bertocci B, Begg AC, Vens C. Ionizing radiation sensitivity of DNA polymerase lambda-deficient cells. Radiat Res. 2007;168(6):683–688. doi: 10.1667/RR1057R.1. [DOI] [PubMed] [Google Scholar]
- 18.Chayot R, Danckaert A, Montagne B, Ricchetti M. Lack of DNA polymerase μ affects the kinetics of DNA double-strand break repair and impacts on cellular senescence. DNA Repair (Amst) 2010;9(11):1187–1199. doi: 10.1016/j.dnarep.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Lucas D, et al. Altered hematopoiesis in mice lacking DNA polymerase mu is due to inefficient double-strand break repair. PLoS Genet. 2009;5(2):e1000389. doi: 10.1371/journal.pgen.1000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capp J-P, et al. Involvement of DNA polymerase mu in the repair of a specific subset of DNA double-strand breaks in mammalian cells. Nucleic Acids Res. 2007;35(11):3551–3560. doi: 10.1093/nar/gkm243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Capp J-P, et al. The DNA polymerase lambda is required for the repair of non-compatible DNA double strand breaks by NHEJ in mammalian cells. Nucleic Acids Res. 2006;34(10):2998–3007. doi: 10.1093/nar/gkl380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chayot R, Montagne B, Ricchetti M. DNA polymerase μ is a global player in the repair of non-homologous end-joining substrates. DNA Repair (Amst) 2012;11(1):22–34. doi: 10.1016/j.dnarep.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 23.Ogiwara H, Kohno T. Essential factors for incompatible DNA end joining at chromosomal DNA double strand breaks in vivo. PLoS One. 2011;6(12):e28756. doi: 10.1371/journal.pone.0028756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon AF, Gosavi RA, Kunkel TA, Pedersen LC, Bebenek K. Creative template-dependent synthesis by human polymerase mu. Proc Natl Acad Sci USA. 2015;112:E4530–E4536. doi: 10.1073/pnas.1505798112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12(1):97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 26.Escudero B, et al. Polμ deficiency increases resistance to oxidative damage and delays liver aging. PLoS One. 2014;9(4):e93074. doi: 10.1371/journal.pone.0093074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aza A, Martin MJ, Juarez R, Blanco L, Terrados G. DNA expansions generated by human Polμ on iterative sequences. Nucleic Acids Res. 2013;41(1):253–263. doi: 10.1093/nar/gks1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas D, et al. Increased learning and brain long-term potentiation in aged mice lacking DNA polymerase μ. PLoS One. 2013;8(1):e53243. doi: 10.1371/journal.pone.0053243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan SH, Yu AM, McVey M. Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila. PLoS Genet. 2010;6(7):e1001005. doi: 10.1371/journal.pgen.1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roerink SF, van Schendel R, Tijsterman M. Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans. Genome Res. 2014;24(6):954–962. doi: 10.1101/gr.170431.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousefzadeh MJ, et al. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014;10(10):e1004654. doi: 10.1371/journal.pgen.1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucas D, et al. Polymerase mu is up-regulated during the T cell-dependent immune response and its deficiency alters developmental dynamics of spleen centroblasts. Eur J Immunol. 2005;35(5):1601–1611. doi: 10.1002/eji.200526015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.