Abstract
Comparative studies of the mitochondrial proteome have identified a conserved core of proteins descended from the α-proteobacterial endosymbiont that gave rise to the mitochondrion and was the source of the mitochondrial genome in contemporary eukaryotes. A surprising result of phylogenetic analyses is the relatively small proportion (10–20%) of the mitochondrial proteome displaying a clear α-proteobacterial ancestry. A large fraction of mitochondrial proteins typically has detectable homologs only in other eukaryotes and is presumed to represent proteins that emerged specifically within eukaryotes. A further significant fraction of the mitochondrial proteome consists of proteins with homologs in prokaryotes, but without a robust phylogenetic signal affiliating them with specific prokaryotic lineages. The presumptive evolutionary source of these proteins is quite different in contending models of mitochondrial origin.
Keywords: mitochondria, proteome, endosymbiont, phagotrophy, syntrophy
Understanding the origin and evolution of the mitochondrion remains a challenge, despite the flood of relevant biochemical, cell and molecular biological, and phylogenetic data and insights that have accumulated in the almost five decades since the modern resurrection (1, 2) of the long-standing endosymbiont hypothesis: the idea that this organelle is a tamed and highly reworked endosymbiotic bacterium. The abundance of information bearing on eukaryotic cell evolution (particularly and most recently sequence data) and differences over how the data are analyzed and interpreted have prompted a plethora of often-conflicting ideas about when and how, within an endosymbiotic context, the mitochondrion originated (3, 4). Considering the spate of recent papers dealing with various aspects of mitochondrial origin and evolution either explicitly (e.g., refs. 5–10) or as part of a broader view of eukaryotic cell evolution (e.g., refs. 11–14), interest in this subject is unlikely to wane any time soon.
That the mitochondrial genome is of bacterial (specifically α-proteobacterial) origin is now indisputable (15, 16), and that fact has underpinned the current wide acceptance of a xenogenous/exogenous (endosymbiotic) origin of the mitochondrion (17, 18), effectively vanquishing autogenous/endogenous (nonsymbiotic) origin hypotheses (e.g., refs. 19–21). However, the mitochondrial proteome, most of which is nucleus-encoded, tells another, murkier story. It was expected that many mitochondrial proteins would not have obvious bacterial homologs because they would have emerged specifically within eukaryotes, as part of the endosymbiont-to-organelle retailoring process. What was not anticipated was how relatively few mitochondrial proteins with bacterial homologs would group specifically with α-Proteobacteria in phylogenetic reconstructions: At most, only 10–20% of any of the mitochondrial proteomes examined so far display a robust α-proteobacterial signal. This minority group includes proteins encoded by the mitochondrial genome as well as nucleus-encoded mitochondrial proteins whose genes we infer were transferred to the nucleus via endosymbiotic gene transfer (EGT) during mitochondrial evolution. This essay briefly highlights the results of studies that revealed the mosaic nature of the mitochondrial proteome and discusses the implications of that observation for models of mitochondrial origin and evolution.
Proteins Encoded by Mitochondrial DNA
The mitochondrial genome specifies a miniscule but essential portion of the mitochondrial proteome, ranging from a low of 3 proteins of defined function in apicomplexans such as Plasmodium to a high of 66 in the excavate Andalucia godoyi, a member of the core jakobids. The latter assemblage of protists contains the most ancestral (bacteria-like) mitochondrial genomes known (22, 23), with mitochondrial-gene content varying only slightly within the group. Among them, jakobid mtDNAs encode 67 proteins: 25 components of the mitochondrial respiratory chain, 29 translation proteins (mostly ribosomal proteins), 9 proteins involved in protein import and maturation, and 4 proteins constituting a multisubunit bacteria-like α2ββ′σ RNA polymerase. Mitochondrial-gene content, although extremely variable among eukaryotes, is largely a subset of that in the core jakobids (15), whose mitochondrial genomes lack only genes for small subunit ribosomal protein S16 (rps16), recently identified in the mtDNAs of Acanthamoeba castellanii and Vermamoeba vermiformis (6, 24) and large subunit ribosomal protein L36 (rpl36), annotated in the mtDNA sequence of Malawimonas jakobiformis (www.ncbi.nlm.nih.gov/gene?cmd=Retrieve&dopt=full_report&list_uids=2695583). On that basis, we can infer that the mitochondrial genome of the last eukaryotic common ancestor (LECA) encoded a minimum of ∼70 proteins (6, 23). In extant eukaryotes, the role of the mitochondrial genome is a highly circumscribed and critical one: It supports the translation and maturation of a small number of proteins that are components of complexes I to IV (CI–CIV) of the electron transport chain (ETC) as well as complex V (CV; ATP synthase). These mtDNA-encoded proteins are assembled with nucleus-encoded partner proteins to generate a functional respiratory chain capable of carrying out coupled electron transport–oxidative phosphorylation.
Composition of the Mitochondrial Proteome
The basic approach to identifying bona fide mitochondrial proteins currently comprises a combination of direct subcellular proteomics techniques [tandem mass spectrometry (MS/MS) applied to purified mitochondria or submitochondrial fractions] in combination with indirect bioinformatics-based prediction of proteins exhibiting N-terminal mitochondrial targeting sequences (25–27). The two techniques have their complementary strengths and limitations, with MS/MS capable of revealing novel mitochondrial proteins (ones having no known homologs) but often missing low-abundance proteins that may be identified by in silico analyses, which, however, fail to retrieve mitochondrial proteins that lack well-defined N-terminal mitochondrial targeting signals. Delineation of the mitochondrial proteome in any organism requires a list of cellular proteins predicted from complete nuclear and mitochondrial genome sequences, as well as assessment of possible nonmitochondrial contamination. Given these requirements and constraints, it is hardly surprising that a relatively small number of largely complete and robustly validated mitochondrial proteomes have been determined to date, primarily from multicellular animals (notably human and mouse), fungi (e.g., the yeast Saccharomyces cerevisiae), and plants (e.g., Arabidopsis thaliana, rice). Although eukaryotic microbes (protists) constitute most of the evolutionary diversity within the eukaryotic domain, comprehensive proteomic analyses of protist mitochondria are still relatively few (24, 28–30). Nevertheless, with the increasing availability of substantially complete eukaryotic-genome sequences, valuable comparative information about mitochondrial proteomes can be gleaned by in silico data-mining methods alone.
Although our knowledge of complete mitochondrial proteomes is patchy at present, a great deal of compositional heterogeneity is already evident, both in the total number of proteins that constitute the organelle in a given organism and in their range of functional activities (31, 32). Multicellular animals and plants seem to have the largest repertoire of mitochondrial proteins (>1,000) although, as demonstrated in recent studies, the compositional complexity of certain unicellular protists may rival that of their multicellular eukaryotic relatives (24, 30). As detailed below, much of the variability in mitochondrial-proteome composition can be understood as lineage-specific additions to a basic protein core that is conserved among eukaryotes, although lineage-specific losses also come into play in shaping the mitochondrial proteome.
Comparative mitochondrial proteomics (comprehensive comparison of mitochondrial-proteome data across the range of eukaryotes) has helped to define the composition of what we may term the “last mitochondrial common ancestor” (LMCA): effectively, the mitochondrion of the LECA. In particular, comparative analysis of individual mitochondrial-protein complexes has highlighted the evolutionary process by which a basic α-proteobacterial core has been modified (typically by acquisition of additional components) during eukaryogenesis. For example, ETC CI (NADH: ubiquinone oxidoreductase) has expanded from 17 subunits in an α-proteobacterium, Paracoccus denitrificans (33), to as many as 45 subunits in animals (34). Although early studies had suggested a lineage-specific distribution of novel CI subunits (34, 35), with 13 of them emerging specifically within metazoan animals, a broader survey of eukaryotic CI using enhanced bioinformatics search techniques has revealed that virtually all of the eukaryote-specific CI subunits were already present in the LMCA (36, 37). ETC CV (ATP synthase) presents a somewhat different picture, with evidence in this case of species-specific subunits in addition to a eukaryote-specific core (38–40).
Mitochondrial ribosomes exhibit a similar pattern, with the mitoribosome of the LCMA inferred to have had an α-proteobacterial–contributed core consisting of 20 small-subunit (SSU) and 33 large-subunit (LSU) proteins supplemented, respectively, by 10 and 9 additional, eukaryote-specific subunits (41, 42). As in the case of ETC CI, retailoring of the endosymbiont ribosome was essentially complete before divergence of the main eukaryotic supergroups, with novel nucleus-encoded subunits in place and many of the original endosymbiont-encoded ribosomal protein genes having already undergone EGT to the nucleus. Subsequent evolution involved additional mitochondrion-to-nucleus gene relocation, as well as lineage-specific gains and losses (41, 42). The most extreme examples of mitoribosome retailoring are seen in the kinetoplastid protists, Trypanosoma brucei (43) and Leishmania tarentole (44). In T. brucei, the ribosomal SSU and LSU contain 56 and 77 proteins, compared with 21 and 34, respectively, in a bacterial (e.g., Escherichia coli) ribosome, with the novel mitochondrial proteins displaying very limited sequence similarity with their counterparts within Kinetoplastidae and no detectable similarity outside this lineage. These focused studies highlight the dynamics of mitochondrial-proteome evolution, as well as emphasizing the importance of direct MS/MS analysis in revealing novel mitochondrial proteins that cannot be identified through sequence comparisons.
As the database for mitochondrial-proteome comparisons has expanded, it has become apparent that the LMCA was already as complex in its key functions as modern mitochondria; importantly, for example, it possessed an essentially complete protein-import apparatus (45). Indeed, molecular reconstructions demonstrate that LECA itself was a structurally complex cell with a sophisticated metabolism, already containing the hallmarks of a typical eukaryotic cell (46). Evidently, basic endosymbiont-to-mitochondrion retailoring was essentially complete before the divergence of the main eukaryotic lineages, indicating that the initial endosymbiotic event must have occurred at an earlier point in eukaryogenesis.
Proto-Mitochondrion: An α-Proteobacterial Perspective
Comparisons of the experimentally determined or inferred mitochondrial proteomes of extant eukaryotes allow the reconstruction of a minimal proteome of the LMCA. Conversely, by exploiting whole-genome sequence searches to extract eukaryotic proteins having a close evolutionary relationship with α-proteobacterial homologs, and assuming that such proteins were contributed by the α-proteobacterial ancestor of mitochondria, one can gain insight into the composition of the ancestral (proto-) mitochondrial proteome. Within the set of retrieved α-proteobacteria-like proteins will be ones that are bona fide components of the contemporary mitochondrial proteome (including proteins encoded by mtDNA and nucleus-encoded proteins targeted to and imported into mitochondria), as well as α-proteobacteria–like proteins that are localized elsewhere in the cell. In fact, the latter group comprises the largest proportion of proteins assumed to have been transferred from the α-proteobacterial endosymbiont to the nuclear genome via EGT, effectively having been recruited to function outside the evolving mitochondrion.
In an initial study along these lines, Gabaldón and Huynen (47) used a database of 77 genome sequences, including 6 α-proteobacterial and 9 eukaryotic, and identified 630 clusters of orthologous groups (COGs) that displayed a close α-proteobacterial/eukaryotic evolutionary relationship. This number represents a minimal estimate of the proteins that were likely present in the proto-mitochondrion (this approach obviously fails to account for proteins present in the proto-mitochondrion but subsequently lost from all of the α-proteobacterial or all of the eukaryotic genomes under consideration, as well as proteins that do not have a sufficiently strong phylogenetic signal to be detected). In a subsequent phylogenetic analysis of an expanded database (144 genomes, including 16 eukaryotic and 11 α-proteobacterial), Gabaldón and Huynen (32) identified 842 α-proteobacteria–like eukaryotic proteins. Of this set, only 74 (8.8%) and 130 (15.4%) could be identified, respectively, in the yeast and human mitochondrial proteome sets available at the time.
In a more recent analysis of this type, Wang and Wu (8) used a greatly expanded genome database (1,613 total, 30 eukaryotic, 171 α-proteobacterial), retrieving 394 COGs that they consider to represent mitochondrion-derived nuclear genes: a substantially lower number than the 842 reported by Gabaldón and Huynen (32). Wu and Wang (8) claim that their analysis is more specific and more sensitive than that of Gabaldón and Huynen (32), resulting in a “leaner” reconstructed metabolism in the proto-mitochondrion, but one in which a number of gaps in the latter study are filled by the identification of additional components of various pathways. In their reconstruction, Gabaldón and Huynen (32) included as mitochondrial proteins ones that clustered with β- and γ-proteobacteria, a concession necessary to recover the bulk of Reclinomonas americana mtDNA-encoded ribosomal proteins. However, this problem proved not to be an issue with the increased organismal sampling in the Wu and Wang (8) study. In fact, Gabaldón and Huynen (32) noted that the number of COGs they recovered varied substantially, depending on the parameters they selected for phylogenetic analysis, and could be reduced substantially by application of more selective criteria (e.g., by setting more stringent cutoffs). The results of these reconstructions are obviously highly influenced by the database used (both number and phylogenetic coverage of the genome sequences examined) as well as the methods used and parameters chosen for tree construction.
Despite these qualifications, these studies do provide a minimalist picture of the proto-mitochondrial metabolism, retrieving virtually complete pathways for pyruvate metabolism, TCA cycle, coupled electron transport-oxidative phosphorylation, fatty-acid biosynthesis, β-oxidation, branched-chain amino acid degradation, and biosynthesis of ubiquinone, biotin, 1-carbon units, and iron-sulfur clusters, most of which metabolism is localized in contemporary mitochondria. Proteins involved in mitochondrial-ribosome biogenesis and translation are prominent in these reconstructions, as are metabolite transporters, but replication and transcription proteins are virtually absent, with heterotrophic carbohydrate metabolism (e.g., glycolysis) entirely missing. Notable α-proteobacterial contributions to nonmitochondrial metabolism include several ER-localized enzymes involved in lipid (steroid) metabolism, as well as enzymes participating in purine and UDP-sugar biosynthesis (8).
The overall impression from these studies is that of a (facultatively) aerobic endosymbiont exhibiting a streamlined metabolism with a bias toward energy conversion, amino acid biosynthesis, and protein synthesis. This α-proteobacteria–derived portion of the mitochondrial proteome constitutes much of the functional core that is conserved throughout eukaryotes.
Origin and Evolution of the Mitochondrial Proteome
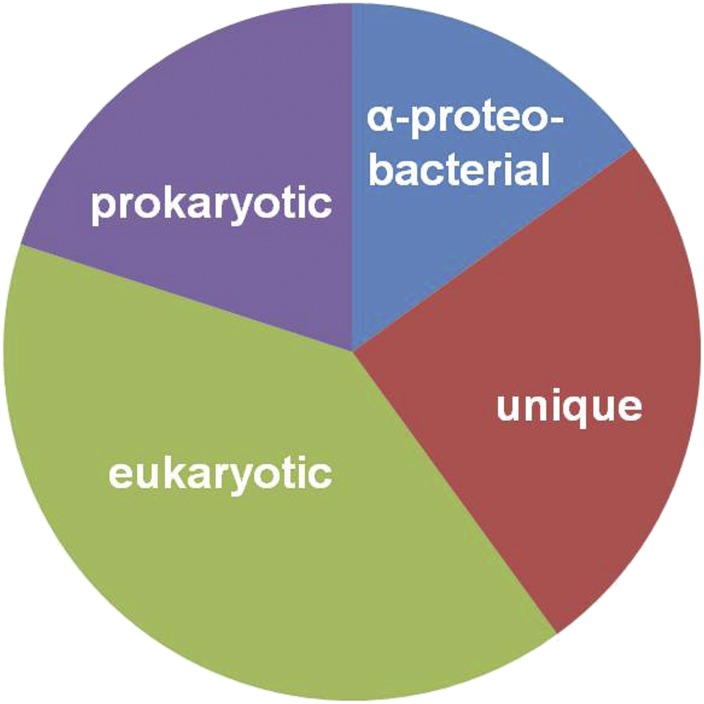
Acceptance that the mitochondrial genome is of α-proteobacterial origin naturally prompted the question: How much of the total mitochondrial proteome is also of α-proteobacterial ancestry? Initial phylogenetic analyses provided a surprising answer: relatively less than might have been anticipated (47–49). These studies revealed that mitochondrial proteins can largely be binned into four categories: category I, strong relationship with α-proteobacteria; category II, non–α-proteobacterial homologs in prokaryotes, often without a strong affiliation to any particular bacterial or archaeal group; category III, homologs exclusively in other eukaryotes; and category IV, proteins unique to the organism in question, lacking evident homologs in other eukaryotes (Fig. 1). This pattern, initially deduced for yeast, S. cerevisiae (48, 49), has been observed for the mitochondrial proteome of other organisms, although with variation in the proportions of the different categories (e.g., ref. 28). Consistently, only ∼10–20% of the mitochondrial proteome in diverse eukaryotes exhibits a strong α-proteobacterial phylogenetic signal (8, 32).
Fig. 1.
Representation of the evolutionary ancestry of the mitochondrial proteome, deduced from phylogenetic-tree reconstructions. Proportions of the four categories vary depending on the organism in question, but the α-proteobacterial component typically constitutes ∼10–20% of the total mitochondrial proteome (8, 32).
The mosaic character of the mitochondrial proteome raises a number of questions about the acquisition of the individual proteins. As detailed above, we can safely infer that the α-proteobacterial component (APC) of the mitochondrial proteome was introduced into the evolving eukaryotic cell by the endosymbiont that also brought in the mitochondrial genome, with many endosymbiont genes subsequently either lost or undergoing EGT to the nucleus. The origin of the non–α-proteobacterial component (NPC) of the mitochondrial proteome is less clear. With some exceptions, NPC proteins were most likely acquired subsequently by the evolving mitochondrion and presumably might have had one of three origins: (i) acquisition from the host of preexisting proteins (i.e., proteins already present in the host); (ii) elaboration de novo within the eukaryotic cell; or (iii) lateral gene transfer (LGT) from diverse prokaryotes (bacteria other than α-proteobacteria or archaea) or viruses. Acquisition of proteins via LGT must have occurred during the period of eukaryogenesis leading up to LECA (after the establishment of the proto-mitochondrion) although examples of lineage-specific, post-LECA LGT are certainly evident. Contending models for the origin of mitochondria have different implications for the origin of the mitochondrial NPC, in particular the prokaryotic cohort that does not cluster with α-proteobacteria.
Phagotrophic vs. Syntrophic: Two Contending Scenarios
Although many specific symbiotic models for the origin of mitochondria have been proposed over the past five decades (see, e.g., ref. 50), these hypotheses can be broadly grouped into one of two scenarios: phagotrophic, in which an α-proteobacterial endosymbiont is taken up by a host cell that already has some degree of eukaryotic character, in particular the ability to carry out phagocytosis (ingestion of other organisms as a food source); and syntrophic, where intimate metabolic interdependence between two different kinds of prokaryotic cell leads eventually to one cell (the host, assumed to be an archaeon) subsuming the second cell (the symbiont, assumed to be a eubacterium) (see, e.g., refs. 3, 4, 14, 46, 51, and 52 for recent synopses and critiques). [Although this categorization is useful for discussion purposes, it should be emphasized that phagotrophy and syntrophy, as biological processes, are not mutually exclusive: It is conceivable, for example, that a syntrophic association might have occurred involving a host cell that already had some eukaryotic characteristics, rather than an archaeon per se (see ref. 4 for a pertinent discussion of this point).]
In perhaps the best-known and most widely discussed syntrophic model, the hydrogen hypothesis of Martin and Müller (53), the host is a strictly anaerobic methanogenic archaeon, which is dependent on hydrogen produced by the symbiont (a facultatively aerobic α-proteobacterium). Integration of the α-proteobacterial partner into the archaeal partner followed by massive gene transfer from symbiont to host genome effectively initiates the process of eukaryogenesis, with the symbiont undergoing subsequent conversion into the mitochondrion. In syntrophic models, the origin of the mitochondrion is coincident with the origin of the eukaryotic cell, per se, and is the conditio sine qua non for eukaryogenesis.
In phagotrophic models, the origin of the mitochondrion is placed later in eukaryogenesis and may in fact be one of the last major eukaryotic features to be elaborated. Cavalier-Smith, in particular, has long championed a phagotrophic model of mitochondrial origin (11, 54–57), with detailed schema of eukaryogenesis developed particularly from a cell-biological perspective.
In both scenarios, the emergence of the mitochondrion is a singular event. In the syntrophic scenario, it is seen as a radical innovation resulting from an extremely rare, even unlikely, event; in the phagotrophic scenario, it is regarded as a graduated innovation resulting as an almost inevitable consequence of a process (phagocytosis) that is now common among microbial eukaryotes. [It is interesting to consider, as O’Malley (3) has pointed out, the way in which proponents of syntrophic and phagotrophic scenarios use the monophyletic origin of mitochondria to bolster their particular viewpoint. Syntrophic proponents ask why, if phagocytosis is so common, mitochondria emerged only once whereas phagotrophic proponents ask why, if a syntrophic scenario is so “unspeakably rare” (11), it happened at all.]
Within these two scenarios, how can we account for those mitochondrial proteins that seem to have a prokaryotic but not specifically α-proteobacterial ancestry (referred to below as “prokaryotic NPC proteins”)? Trivially, some mitochondrial proteins that score as NPC in phylogenetic reconstructions may have diverged too far to be readily recognizable as α-proteobacterial. However, this possibility is unlikely to account for the entirety of this cohort. Martin and coworkers (58, 59) have argued for a fluid prokaryotic-chromosome model in which LGT among free-living prokaryotes preceding and subsequent to the origin of mitochondria could explain the observed pattern. However, although contemporary α-proteobacterial (and other prokaryotic) genomes almost certainly differ in gene content from their ancestral genomes existing at the time of the mitochondrial endosymbiosis, and accepting that LGT has played a major role in prokaryotic genome evolution, this explanation would seem to demand that the genes in question were preferentially lost from the α-proteobacterial genomes under consideration, or were so widely transferred among non–α-proteobacterial genomes that evidence of their α-proteobacterial provenance has effectively been erased. Although such a scenario might apply in isolated cases, it is unlikely, in my view, that it could rationalize the entirety of prokaryotic NPC mitochondrial proteins. Considering that the mitochondrial APC is relatively constant, whereas the NPC subsets are quite diverse, one would have to imagine a very large proto-mitochondrial genome if the bulk of the NPC were assumed to have been present in, and contributed by, an α-proteobacterial genome in a syntrophic scenario. Lang and Burger (14, 52) have presented other arguments in strongly argued critiques of a syntrophic scenario.
An alternative, straightforward explanation for lack of phylogenetic signal in these cases is that the proteins in question are of ancient origin. The idea here of “ancient origin” is an origin that considerably predates that of the mitochondrial endosymbiosis: i.e., proteins that had existed for a long time in the host since their acquisition (e.g., by LGT) from a prokaryotic source, or that were present in LUCA and inherited vertically, before the advent of the mitochondrial endosymbiosis. Thus, lack of affiliation with a specific bacterial or archaeal group could simply reflect progressive sequence divergence over a long evolutionary history. A similar issue arises with the peroxisome: In a phylogenetic investigation of the evolutionary origin of peroxisomal proteins, Gabaldón et al. (60) found a substantial number of cases of unresolved ancestry that “imply the existence of homology to prokaryotic sequences without a tree that specifically supports a bacterial or archaeal origin.”
In their analysis of the yeast mitochondrial proteome (a set of 423 proteins), Karlberg et al. (48) found that about half had homologs in prokaryotes and, of this portion, 108 (25% of the total) were represented in bacteria and archaea as well as eukaryotes. The authors suggested that these 108 proteins might have been present in the last universal common ancestor (LUCA), and therefore conceivably some could already have been resident in the host cell before uptake of the α-proteobacterial endosymbiont. Other prokaryotic genes might have been contributed by a “slow-drip” process in which successions of LGT over a long period seeded the evolving eukaryotic genome with prokaryotic genes from diverse lineages (61). The results of an analysis of mitochondrial aminoacyl-tRNA synthetases (aaRSs), which indicated that “a majority of the mitochondrial aaRSs originate from within the bacterial domain, but none specifically from the α-Proteobacteria” (62), would seem consistent with this view. The point is that, within a phagotrophic model of mitochondrial origin, much of the mitochondrial NPC, at least that portion that is prokaryotic but not specifically α-proteobacterial, might well have been present in the protoeukaryotic host organism that assimilated the α-proteobacterial symbiont. As early as 1987, Cavalier-Smith (55) asserted that the host “was probably a facultative aerobe” and, as such, “it would probably already have most of the proteins now present in the mitochondrion, so they could be transferred into it and allow deletion of the homologous symbiont genes without any gene transfer being necessary”—an overreach, perhaps, in light of current knowledge about the composition of the mitochondrial proteome, but nevertheless an early explicit statement of the idea of a substantial host contribution to the mitochondrion of preendosymbiont proteins.
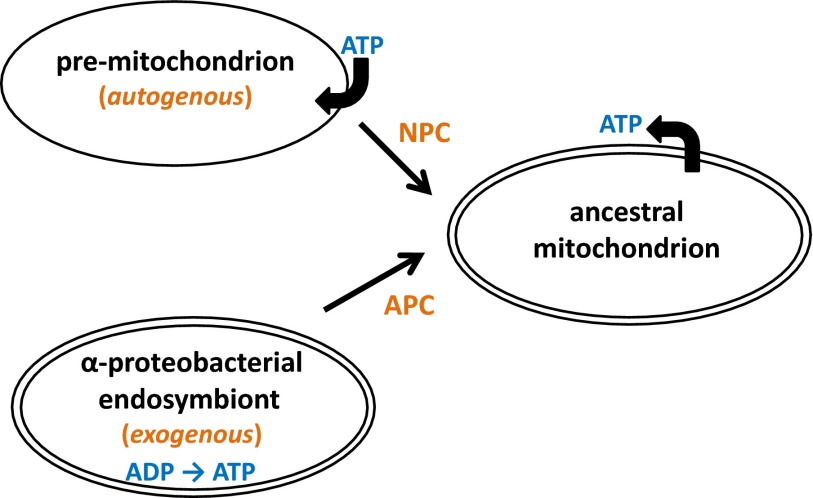
If we allow that certain proteins destined to be incorporated into the mitochondrion preexisted in the host, what might they have been doing there? Recently, in what I dubbed the “pre-endosymbiont hypothesis,” I raised the possibility that a membrane-bound organelle (“premitochondrion”) might have compartmentalized some of the metabolic activities that were eventually relocated to the mitochondrion (5). In effect, such an endogenously evolved host organelle could have provided an immediately available source of proteins for the endosymbiont-to-organelle transformation, with duplicate pathways being rationalized via preferential loss/transfer of endosymbiont genes and consequent marked reduction in the gene content of what would become the mitochondrial genome (Fig. 2).
Fig. 2.
A schematic view of the preendosymbiont hypothesis. The premitochondrion is seen as a membrane-bound entity endowed with a protein-import system and various ion/small-molecule transporters, compartmentalizing many of the metabolic functions of the mitochondrion. The premitochondrion is assumed to have evolved endogenously within the preeukaryote cell and to contain proteins that would later contribute to the NPC (non–α-proteobacterial component) of the contemporary mitochondrial proteome. An α-proteobacteria–like endosymbiont is converted to the ancestral mitochondrion, effectively “capturing” protein components and functions of the premitochondrion. The endosymbiont contributes the α-proteobacterial component (APC) of the mitochondrial proteome, which is largely directed toward specification of energy-generating capacity in the form of coupled electron transport/oxidative phosphorylation; as well, the mitochondrial inner and outer membranes are assumed to be endosymbiont-derived (57). The conversion of endosymbiont to ancestral mitochondrion is greatly facilitated by the existence of the reservoir of premitochondrial NPC proteins in the host cell. Endosymbiont-to-nucleus gene transfer (EGT), coupled with rationalization of redundant pathways, results in the formation of evolutionarily chimeric enzymatic pathways and protein complexes in the ancestral mitochondrion, as well as functional relocation of the products of some transferred α-proteobacterial genes to other cellular compartments. The premitochondrion is assumed to be a non–energy-generating organelle that imports ATP; a key innovation in the transition to the contemporary mitochondrion is the acquisition of an ADP/ATP transporter that reverses this flow, ultimately allowing the mitochondrion to become the primary site of ATP generation for cellular functions. Modified from ref. 5.
In some respects, in contemporary eukaryotic cells, the peroxisome encapsulates some of the features ascribed to the premitochondrion: an endogenously evolved (63), membrane-bound organelle compartmentalizing (in different eukaryotes) various activities involved in the oxidative metabolism of carbohydrates, lipids, amino acids, and purines, and the biosynthesis of certain lipids (56, 64, 65). Mitochondria and peroxisomes are known to be linked metabolically (sharing pathways such as β-oxidation of very long-chain fatty acids, with metabolites shuttling between the two organelles) as well as evolutionarily, with a significant fraction of peroxisomal proteins seemingly recruited from mitochondria (60). Peroxisomes have been implicated as the evolutionary source of the mitochondrial ADP/ATP exchanger (57), as well of the organellar division machinery (66). Thus, it is not inconceivable that the primitive peroxisome might have harbored a wider variety of metabolic processes, some of which were ultimately relocated to the evolving mitochondrion.
A Phagotrophic Perspective: 2015
Recent studies have reinforced an emerging view that eukaryotes arose from within (rather than as a sister group to) Archaea and are specifically affiliated with the TACK superphylum, comprising the phyla Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota (67–73). This inference, based largely on phylogenetic reconstructions, draws support from the finding of what were previously deemed to be eukaryotic signature proteins (ESPs) in TACK genomes, including archaeal homologs of actin, tubulin, ESCRT proteins, and several proteins involved in eukaryotic transcription and translation (69, 73). These new insights allow the possibility that the archaeal ancestor of the eukaryotic cell might have had more eukaryotic character than previously supposed, including a primitive ability to phagocytose. A recently proposed model of eukaryogenesis [“phagocytosing archaeon theory” (PhAT)] envisages an emerging amitochondriate host cell in which rampant LGT (a consequence of phagocytotic acquisition of genomic DNA from various sources) leads to “a highly mosaic host genome” (74). This theory is compatible with the view (5) that many of the proteins acquired later by the evolving mitochondrion (part of the NPC) could already have existed in the host cell (Fig. 2); it is also consistent with the idea that much of the evolution of eukaryotic cellular complexity occurred before the acquisition of an α-proteobacterium and its transformation into the mitochondrion (11, 57).
Concluding Remarks
Despite considerable progress over the past 15 y in elucidating the composition of the mitochondrial proteome in a variety of eukaryotes, we still cannot infer with any certainty the precise evolutionary origin of many of the mitochondrial proteins, particularly the prokaryotic portion of the NPC. Nor can we assert with confidence at what time point any individual protein was introduced into the evolving eukaryotic cell to become associated with the mitochondrion. At present, phagotrophic and synthrophic models of mitochondrial origin account in quite different ways for the NPC component of the mitochondrial proteome. What we still lack is compelling evidence, of the sort that convincingly demonstrated the α-proteobacterial nature of the mitochondrial genome, that will robustly turn the tide in favor of one or the other model.
We should not, of course, exclude postendosymbiont LGT, either before or after the emergence of the LECA, as an additional source of NPC mitochondrial proteins. An important example is the early replacement of the original mtDNA-encoded bacterial-type RNA polymerase (today found only in jakobid flagellates) with a nucleus-encoded, single-subunit, phage T3/T7-like RNA polymerase that now functions as the mitochondrial transcriptase in all other eukaryotes (75). Convincing cases of LGT events of this sort might be recognized through comparative studies of particular mitochondrial metabolic pathways, but such studies will likely require direct biochemical confirmation in addition to sequence comparisons. Post-LECA lineage-specific LGT from particular bacterial or archaeal groups, as well as lineage-specific evolution of novel mitochondrial proteins, must certainly be invoked to account for the substantial differences in overall composition: e.g., between the yeast and human mitochondrial proteomes (32).
Accordingly, there is a continuing place for comparative mitochondrial proteomics in better defining the conserved functional core of the mitochondrion: that cohort of proteins that was present in the LMCA and has been preserved throughout eukaryotes, despite evident lineage-specific reworking of the mitochondrial proteome. Particular efforts should be made to more solidly identify and define prokaryote-derived NPC proteins that might be part of this core. To date, few studies have examined in depth the mitochondrial proteome in early diverging eukaryotes; such studies may hold important additional clues to the origin and evolution of the mitochondrial proteome, as they have in the case of the mitochondrial genome.
Acknowledgments
I am grateful to Tom Cavalier-Smith for comments that were most helpful in the preparation of this review, in particular for pointing out that the peroxisome might have had the preconditioning function that I have ascribed “to an evanescent separate premitochondrial compartment.”
Footnotes
The author declares no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Symbioses Becoming Permanent: The Origins and Evolutionary Trajectories of Organelles,” held October 15–17, 2014, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Symbioses.
This article is a PNAS Direct Submission. P.J.K. is a Guest Editor invited by the Editorial Board.
References
- 1.Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14(3):255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 2.Margulis L. Origin of Eukaryotic Cells. Yale Univ Press; New Haven, CT: 1970. [Google Scholar]
- 3.O’Malley MA. The first eukaryote cell: An unfinished history of contestation. Stud Hist Philos Biol Biomed Sci. 2010;41(3):212–224. doi: 10.1016/j.shpsc.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Poole AM, Gribaldo S. Eukaryotic origins: How and when was the mitochondrion acquired? Cold Spring Harb Perspect Biol. 2014;6(12):a015990. doi: 10.1101/cshperspect.a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray MW. The pre-endosymbiont hypothesis: A new perspective on the origin and evolution of mitochondria. Cold Spring Harb Perspect Biol. 2014;6(3):a016097. doi: 10.1101/cshperspect.a016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kannan S, Rogozin IB, Koonin EV. MitoCOGs: Clusters of orthologous genes from mitochondria and implications for the evolution of eukaryotes. BMC Evol Biol. 2014;14(1):237. doi: 10.1186/s12862-014-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degli Esposti M, et al. Evolution of mitochondria reconstructed from the energy metabolism of living bacteria. PLoS ONE. 2014;9(5):e96566. doi: 10.1371/journal.pone.0096566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Wu M. Phylogenomic reconstruction indicates mitochondrial ancestor was an energy parasite. PLoS ONE. 2014;9(10):e110685. doi: 10.1371/journal.pone.0110685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lane N. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 2014;6(5):a015982. doi: 10.1101/cshperspect.a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Wu M. An integrated phylogenomic approach toward pinpointing the origin of mitochondria. Sci Rep. 2015;5:7949. doi: 10.1038/srep07949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalier-Smith T. The neomuran revolution and phagotrophic origin of eukaryotes and cilia in the light of intracellular coevolution and a revised tree of life. Cold Spring Harb Perspect Biol. 2014;6(9):a016006. doi: 10.1101/cshperspect.a016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McInerney JO, O’Connell MJ, Pisani D. The hybrid nature of the Eukaryota and a consilient view of life on Earth. Nat Rev Microbiol. 2014;12(6):449–455. doi: 10.1038/nrmicro3271. [DOI] [PubMed] [Google Scholar]
- 13.Baum DA, Baum B. An inside-out origin for the eukaryotic cell. BMC Biol. 2014;12(1):76. doi: 10.1186/s12915-014-0076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lang BF. Mitochondria and the origin of eukaryotes. In: Löffelhardt W, editor. Endosymbiosis. Springer; Vienna: 2014. pp. 3–18. [Google Scholar]
- 15.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283(5407):1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 16.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4(9):a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gray MW, Doolittle WF. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray MW. The endosymbiont hypothesis revisited. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 19.Raff RA, Mahler HR. The non symbiotic origin of mitochondria. Science. 1972;177(4049):575–582. doi: 10.1126/science.177.4049.575. [DOI] [PubMed] [Google Scholar]
- 20.Mahler HR, Raff RA. The evolutionary origin of the mitochondrion: A nonsymbiotic model. Int Rev Cytol. 1975;43:1–124. doi: 10.1016/s0074-7696(08)60067-4. [DOI] [PubMed] [Google Scholar]
- 21.Uzzell T, Spolsky C. Mitochondria and plastids as endosymbionts: A revival of special creation? Am Sci. 1974;62(3):334–343. [PubMed] [Google Scholar]
- 22.Lang BF, et al. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387(6632):493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 23.Burger G, Gray MW, Forget L, Lang BF. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol. 2013;5(2):418–438. doi: 10.1093/gbe/evt008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW. Compositional complexity of the mitochondrial proteome of a unicellular eukaryote (Acanthamoeba castellanii, supergroup Amoebozoa) rivals that of animals, fungi, and plants. J Proteomics. 2014;109(0):400–416. doi: 10.1016/j.jprot.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Taylor SW, Fahy E, Ghosh SS. Global organellar proteomics. Trends Biotechnol. 2003;21(2):82–88. doi: 10.1016/S0167-7799(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 26.Warnock DE, Fahy E, Taylor SW. Identification of protein associations in organelles, using mass spectrometry-based proteomics. Mass Spectrom Rev. 2004;23(4):259–280. doi: 10.1002/mas.10077. [DOI] [PubMed] [Google Scholar]
- 27.Richly E, Chinnery PF, Leister D. Evolutionary diversification of mitochondrial proteomes: Implications for human disease. Trends Genet. 2003;19(7):356–362. doi: 10.1016/S0168-9525(03)00137-9. [DOI] [PubMed] [Google Scholar]
- 28.Smith DGS, et al. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: Direct analysis by tandem mass spectrometry. J Mol Biol. 2007;374(3):837–863. doi: 10.1016/j.jmb.2007.09.051. [DOI] [PubMed] [Google Scholar]
- 29.Atteia A, et al. A proteomic survey of Chlamydomonas reinhardtii mitochondria sheds new light on the metabolic plasticity of the organelle and on the nature of the α-proteobacterial mitochondrial ancestor. Mol Biol Evol. 2009;26(7):1533–1548. doi: 10.1093/molbev/msp068. [DOI] [PubMed] [Google Scholar]
- 30.Panigrahi AK, et al. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9(2):434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabaldón T, Huynen MA. Shaping the mitochondrial proteome. Biochim Biophys Acta. 2004;1659(2-3):212–220. doi: 10.1016/j.bbabio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 32.Gabaldón T, Huynen MA. From endosymbiont to host-controlled organelle: The hijacking of mitochondrial protein synthesis and metabolism. PLOS Comput Biol. 2007;3(11):e219. doi: 10.1371/journal.pcbi.0030219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yip CY, Harbour ME, Jayawardena K, Fearnley IM, Sazanov LA. Evolution of respiratory complex I: “Supernumerary” subunits are present in the α-proteobacterial enzyme. J Biol Chem. 2011;286(7):5023–5033. doi: 10.1074/jbc.M110.194993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brandt U. Energy converting NADH:quinone oxidoreductase (complex I) Annu Rev Biochem. 2006;75:69–92. doi: 10.1146/annurev.biochem.75.103004.142539. [DOI] [PubMed] [Google Scholar]
- 35.Gabaldón T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J Mol Biol. 2005;348(4):857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 36.Cardol P. Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: A highly conserved subunit composition highlighted by mining of protein databases. Biochim Biophys Acta. 2011;1807(11):1390–1397. doi: 10.1016/j.bbabio.2011.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Gawryluk RMR, Chisholm KA, Pinto DM, Gray MW. Composition of the mitochondrial electron transport chain in acanthamoeba castellanii: Structural and evolutionary insights. Biochim Biophys Acta. 2012;1817(11):2027–2037. doi: 10.1016/j.bbabio.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Zíková A, Schnaufer A, Dalley RA, Panigrahi AK, Stuart KD. The F(0)F(1)-ATP synthase complex contains novel subunits and is essential for procyclic Trypanosoma brucei. PLoS Pathog. 2009;5(5):e1000436. doi: 10.1371/journal.ppat.1000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balabaskaran Nina P, et al. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol. 2010;8(7):e1000418. doi: 10.1371/journal.pbio.1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lapaille M, et al. Atypical subunit composition of the chlorophycean mitochondrial F1FO-ATP synthase and role of Asa7 protein in stability and oligomycin resistance of the enzyme. Mol Biol Evol. 2010;27(7):1630–1644. doi: 10.1093/molbev/msq049. [DOI] [PubMed] [Google Scholar]
- 41.Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35(14):4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desmond E, Brochier-Armanet C, Forterre P, Gribaldo S. On the last common ancestor and early evolution of eukaryotes: Reconstructing the history of mitochondrial ribosomes. Res Microbiol. 2011;162(1):53–70. doi: 10.1016/j.resmic.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 43.Zíková A, et al. Trypanosoma brucei mitochondrial ribosomes: Affinity purification and component identification by mass spectrometry. Mol Cell Proteomics. 2008;7(7):1286–1296. doi: 10.1074/mcp.M700490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK. Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci USA. 2009;106(24):9637–9642. doi: 10.1073/pnas.0901631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dolezal P, Likic V, Tachezy J, Lithgow T. Evolution of the molecular machines for protein import into mitochondria. Science. 2006;313(5785):314–318. doi: 10.1126/science.1127895. [DOI] [PubMed] [Google Scholar]
- 46.Koumandou VL, et al. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48(4):373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gabaldón T, Huynen MA. Reconstruction of the proto-mitochondrial metabolism. Science. 2003;301(5633):609. doi: 10.1126/science.1085463. [DOI] [PubMed] [Google Scholar]
- 48.Karlberg O, Canbäck B, Kurland CG, Andersson SGE. The dual origin of the yeast mitochondrial proteome. Yeast. 2000;17(3):170–187. doi: 10.1002/1097-0061(20000930)17:3<170::AID-YEA25>3.0.CO;2-V. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurland CG, Andersson SGE. Origin and evolution of the mitochondrial proteome. Microbiol Mol Biol Rev. 2000;64(4):786–820. doi: 10.1128/mmbr.64.4.786-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin W, Hoffmeister M, Rotte C, Henze K. An overview of endosymbiotic models for the origins of eukaryotes, their ATP-producing organelles (mitochondria and hydrogenosomes), and their heterotrophic lifestyle. Biol Chem. 2001;382(11):1521–1539. doi: 10.1515/BC.2001.187. [DOI] [PubMed] [Google Scholar]
- 51.Sommer MS, Schleiff E. Protein targeting and transport as a necessary consequence of increased cellular complexity. Cold Spring Harb Perspect Biol. 2014;6(8):a016055. doi: 10.1101/cshperspect.a016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lang BF, Burger G. Mitochondrial and eukaryotic origins: A critical review. In: Maréchal-Drouard L, editor. Mitochondrial Genome Evolution, Advances in Botanical Research. Vol 63. Elsevier; Amsterdam: 2012. pp. 1–20. [Google Scholar]
- 53.Martin W, Müller M. The hydrogen hypothesis for the first eukaryote. Nature. 1998;392(6671):37–41. doi: 10.1038/32096. [DOI] [PubMed] [Google Scholar]
- 54.Cavalier-Smith T. Endosymbiotic origin of the mitochondrial envelope. In: Schwemmler W, Schenk HEA, editors. Endocytobiology II. de Gruyter; Berlin: 1983. pp. 265–279. [Google Scholar]
- 55.Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci. 1987;503(1):55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- 56.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52(Pt 2):297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 57.Cavalier-Smith T. Origin of mitochondria by intracellular enslavement of a photosynthetic purple bacterium. Proc Biol Sci. 2006;273(1596):1943–1952. doi: 10.1098/rspb.2006.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Esser C, Martin W, Dagan T. The origin of mitochondria in light of a fluid prokaryotic chromosome model. Biol Lett. 2007;3(2):180–184. doi: 10.1098/rsbl.2006.0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thiergart T, Landan G, Schenk M, Dagan T, Martin WF. An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biol Evol. 2012;4(4):466–485. doi: 10.1093/gbe/evs018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gabaldón T, et al. Origin and evolution of the peroxisomal proteome. Biol Direct. 2006;1(1):8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lester L, Meade A, Pagel M. The slow road to the eukaryotic genome. BioEssays. 2006;28(1):57–64. doi: 10.1002/bies.20344. [DOI] [PubMed] [Google Scholar]
- 62.Brindefalk B, Viklund J, Larsson D, Thollesson M, Andersson SGE. Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol Biol Evol. 2007;24(3):743–756. doi: 10.1093/molbev/msl202. [DOI] [PubMed] [Google Scholar]
- 63.Gabaldón T. Evolutionary considerations on the origin of peroxisomes from the endoplasmic reticulum, and their relationships with mitochondria. Cell Mol Life Sci. 2014;71(13):2379–2382. doi: 10.1007/s00018-014-1640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Duve C. The origin of eukaryotes: A reappraisal. Nat Rev Genet. 2007;8(5):395–403. doi: 10.1038/nrg2071. [DOI] [PubMed] [Google Scholar]
- 65.Titorenko VI, Rachubinski RA. The life cycle of the peroxisome. Nat Rev Mol Cell Biol. 2001;2(5):357–368. doi: 10.1038/35073063. [DOI] [PubMed] [Google Scholar]
- 66.Pan R, Hu J. The conserved fission complex on peroxisomes and mitochondria. Plant Signal Behav. 2011;6(6):870–872. doi: 10.4161/psb.6.6.15241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105(51):20356–20361. doi: 10.1073/pnas.0810647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foster PG, Cox CJ, Embley TM. The primary divisions of life: A phylogenomic approach employing composition-heterogeneous methods. Philos Trans R Soc Lond B Biol Sci. 2009;364(1527):2197–2207. doi: 10.1098/rstb.2009.0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guy L, Ettema TJG. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19(12):580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 70.Kelly S, Wickstead B, Gull K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc Biol Sci. 2011;278(1708):1009–1018. doi: 10.1098/rspb.2010.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Soc. 2012;279(1749):4870–4879. doi: 10.1098/rspb.2012.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams TA, Embley TM. Archaeal “dark matter” and the origin of eukaryotes. Genome Biol Evol. 2014;6(3):474–481. doi: 10.1093/gbe/evu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guy L, Saw JH, Ettema TJG. The archaeal legacy of eukaryotes: A phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6(10):a016022. doi: 10.1101/cshperspect.a016022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martijn J, Ettema TJ. From archaeon to eukaryote: The evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41(1):451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 75.Shutt TE, Gray MW. Bacteriophage origins of mitochondrial replication and transcription proteins. Trends Genet. 2006;22(2):90–95. doi: 10.1016/j.tig.2005.11.007. [DOI] [PubMed] [Google Scholar]