Significance
Bacterial pathogens evolve rapidly in the face of clinical interventions and therapeutics; one mechanism that can promote this evolution is their ability to acquire novel DNA sequences, known as horizontal gene transfer (HGT). Here, we studied HGT in clinical isolates of Vibrio cholerae, the causative agent of cholera, and found that a horizontally transferred element inhibits another mechanism of HGT—natural transformation. The element that inhibits natural transformation is globally distributed among V. cholerae isolates. We show, however, that there has been a rise in the prevalence of strains that lack this inhibitory element. Thus, our results suggest that in the future there may be an increase in the role of natural transformation on the evolution of this pathogen.
Keywords: DNase, integrating conjugative element, horizontal gene transfer, evolution
Abstract
Natural transformation is one mechanism of horizontal gene transfer (HGT) in Vibrio cholerae, the causative agent of cholera. Recently, it was found that V. cholerae isolates from the Haiti outbreak were poorly transformed by this mechanism. Here, we show that an integrating conjugative element (ICE)-encoded DNase, which we name IdeA, is necessary and sufficient for inhibiting natural transformation of Haiti outbreak strains. We demonstrate that IdeA inhibits this mechanism of HGT in cis via DNA endonuclease activity that is localized to the periplasm. Furthermore, we show that natural transformation between cholera strains in a relevant environmental context is inhibited by IdeA. The ICE encoding IdeA is globally distributed. Therefore, we analyzed the prevalence and role for this ICE in limiting natural transformation of isolates from Bangladesh collected between 2001 and 2011. We found that IdeA+ ICEs were nearly ubiquitous in isolates from 2001 to 2005; however, their prevalence decreased to ∼40% from 2006 to 2011. Thus, IdeA+ ICEs may have limited the role of natural transformation in V. cholerae. However, the rise in prevalence of strains lacking IdeA may now increase the role of this conserved mechanism of HGT in the evolution of this pathogen.
The causative agent of the diarrheal disease cholera, Vibrio cholerae, is annually responsible for 3.5 million infections worldwide (1). This facultative pathogen naturally resides in temperate aquatic environments and causes disease when ingested in contaminated food or water. A critical nutrient for Vibrio species in the aquatic environment is the chitinous exoskeleton of crustacean zooplankton (2–4). Chitin is an insoluble polysaccharide composed of β-1,4-linked GlcNAc. In addition to serving as a carbon and nitrogen source, chitin also induces a physiological state in V. cholerae known as natural competence (5). In this state, bacteria can take up DNA from the extracellular environment and integrate this DNA into their chromosomes by homologous recombination. This cumulative process of DNA uptake and integration is known as natural transformation and is one mechanism for horizontal gene transfer (HGT) in V. cholerae. HGT by natural transformation is used by pathogenic microbes to evolve in the face of clinical intervention and immune pressure. Indeed, in V. cholerae, this mechanism of HGT is hypothesized to have generated an antigenic variant, the O139 outbreak strain, through homologous recombination and replacement of the locus responsible for O-antigen biosynthesis (6–9).
Another mechanism of HGT in V. cholerae is integrating conjugative elements (ICEs) of the SXT/R391 family. These elements can range from ∼80 to 110 Kb in size and contain all of the genes required for conjugative transfer into naive hosts (10, 11); they integrate in a site-specific manner into the 5′ end of the highly conserved prfC (peptide-chain-release factor C) gene (10–12). The first natural transfer of an ICE into V. cholerae likely occurred between 1980 and 1985 (10, 13) and, by the 1990s, virtually all clinical isolates of V. cholerae contained an ICE (13). These elements confer resistance to multiple antibiotics, and it is likely that widespread use of antibiotics has rapidly selected for strains containing ICEs. There are at least 10 genetically distinct ICEs circulating in the V. cholerae population (11). These ICEs share a core set of genes, but have varied gene content at distinct sites. The most common ICE in V. cholerae is VchInd5, which is present in ∼77% of currently sequenced clinical isolates (10, 11). It is hypothesized that the current (seventh) pandemic of cholera originated in the Bay of Bengal, and strains have spread globally from this region in three overlapping waves of transmission (13, 14). Strains containing VchInd5 are globally distributed, indicating that the original transfer of VchInd5 into V. cholerae may have occurred in this region.
In 2010, cholera spread to Haiti, a region that previously lacked this disease (15, 16). Phylogenetic and Bayesian analyses indicate that all strains in Haiti share a common ancestor, which was introduced into the region at the outset of the epidemic (16, 17). Consistent with this finding, strains from Haiti ubiquitously harbor a VchInd5 ICE. Throughout the epidemic, strains have acquired mutations that are likely generated intrinsically, and there is no evidence of horizontal gene transfer among these isolates (16). Consistent with this finding, strains from the Haiti outbreak were found to be poorly transformed by chitin-induced natural competence (16).
In this study, we identify and characterize an ICE-encoded DNase present on VchInd5 that inhibits HGT by natural transformation in V. cholerae. We also assess the role and prevalence of this DNase in limiting transformation among clinical isolates from Haiti and Bangladesh.
Results and Discussion
An ICE-Encoded DNase, IdeA, Is Necessary and Sufficient to Inhibit Natural Transformation of Haiti Outbreak Isolates.
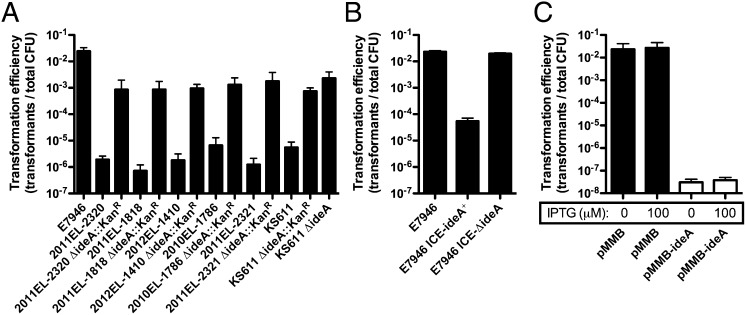
Previously, it was shown that V. cholerae isolates from the Haiti outbreak were poorly transformable (16). To confirm this finding, we performed natural transformation assays with exogenously added PCR products as a source of transforming DNA (tDNA), and used chitin beads as a source of chitin on which to grow V. cholerae and induce natural competence. As expected, all Haiti isolates tested displayed transformation efficiencies that were 1,000–10,000-fold lower than E7946, a clinical isolate collected in 1978 in Bahrain (18) (Fig. 1A). Isolates from the Haiti outbreak have been fully sequenced, and it was previously shown that these strains contain all of the genes required for natural transformation (15, 16); this is further supported by our data, which demonstrates that though reduced, all isolates still had measurable transformation efficiencies (Fig. 1A). For this reason, we hypothesized that Haiti strains may contain a gene that inhibits natural transformation, which is not present in the ancestral E7946 strain. One genetic island that differs between these strains is an ICE that is present in the Haiti strains but absent in E7946. This ICE is 97.7 Kb in length and is most similar to the globally disseminated VchInd5 ICE. Analyzing the genes on this ICE in silico, we found that one gene, Vch1786_I0128, encoded a protein that was highly homologous to the well-characterized DNA endonucleases EndA from Escherichia coli (56% identity and 66% similarity) and Dns from V. cholerae (52% identity and 64% similarity). Therefore, we have named this gene ICE-encoded DNA endonuclease A (ideA). It was previously shown that the DNA endonuclease Dns inhibits natural transformation in V. cholerae, supporting a potential role for IdeA in inhibiting transformation among Haiti isolates (19); to test this, we generated isogenic ideA mutants and found that this resulted in a 100–1,000-fold enhancement in transformation efficiency among Haiti outbreak strains (Fig. 1A). Deletion of ideA in these strains was performed by allelic replacement with a drug resistance marker, which can have pleiotropic effects. To confirm that the phenotype observed was due specifically to deletion of ideA, we generated an unmarked in-frame deletion of this gene in one Haiti isolate, KS611. As expected, the in-frame ΔideA mutant had a phenotype similar to the strain where ideA was replaced with an antibiotic resistance marker (Fig. 1A). Cumulatively, these results indicate that IdeA is necessary for limiting natural transformation among strains from the Haiti outbreak.
Fig. 1.
A putative ICE-encoded DNase, IdeA, inhibits natural transformation of Haiti outbreak strains. (A) Natural transformation of the indicated strains on chitin beads with 2 μg of PCR product that confers resistance to spectinomycin. (B) Effect of ICE from KS611 and KS611 ΔideA on natural transformation when conjugatively transferred to E7946. All strains in B contain a ΔlacZ::SpecR mutation. Strains were transformed with 2 μg of PCR product that confers resistance to kanamycin. (C) Natural transformation of E7946 harboring empty vector (pMMB) or an IdeA expression vector (pMMB-ideA) in the presence of the indicated concentration of IPTG with 2 μg of a PCR product that confers resistance to spectinomycin. White bars signify that no transformants were obtained and indicate the limit of detection in the assay. All data are shown as the mean ± SD and the result of at least two independent experiments.
Next, we sought to determine if IdeA was sufficient to inhibit natural transformation. To that end, we conjugatively transferred the ICE from KS611 into a marked (ΔlacZ::SpecR) derivative of E7946 to generate what we refer to as E7946 ICE-ideA+. When we tested this isolate, we found that the KS611-ICE was indeed sufficient to inhibit natural transformation of E7946 (Fig. 1B). To confirm that this inhibitory activity was dependent on IdeA, we conjugatively transferred the ICE from KS611 ΔideA into E7946 to generate E7946 ICE-ΔideA, and found that this strain had a transformation efficiency similar to the parent strain (Fig. 1B). Furthermore, we cloned ideA onto pMMB67EH (abbreviated pMMB henceforth), an IPTG-inducible expression vector to yield pMMB-ideA and showed that ectopic expression of IdeA in E7946 inhibits transformation efficiency independently from the ICE (Fig. 1C). This vector inhibited natural transformation of E7946 even without any inducer added, which is likely due to leaky expression of IdeA. These results indicate that IdeA is both necessary and sufficient to inhibit natural transformation of V. cholerae. These results are also consistent with previous work, which showed that constitutive expression of Dns completely inhibited natural transformation (19).
IdeA Acts in cis to Inhibit Natural Transformation of V. cholerae and Is Periplasmically Localized.
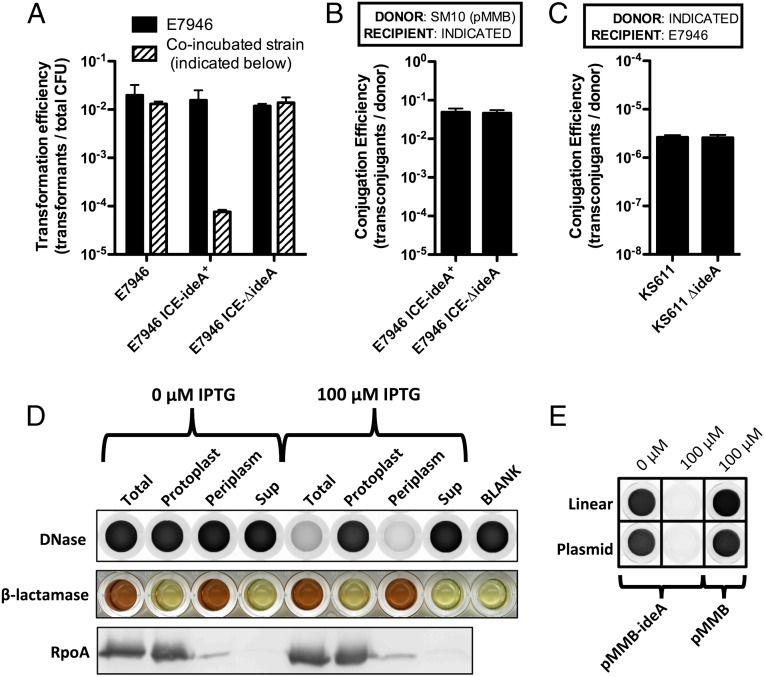
Like E. coli EndA, IdeA has a predicted signal sequence, which indicates that this DNase may be secreted into the periplasm and/or extracellularly. If IdeA were secreted into the extracellular space, we would predict that this DNase could act in trans to inhibit natural transformation. Alternatively, if IdeA were only secreted into the periplasmic space, we would predict that it would only inhibit natural transformation in cis (e.g., only inhibit transformation of those cells expressing ideA). To test this hypothesis, we coincubated E7946 lacking ideA, with strains ±ideA to determine if this DNase could act in trans to inhibit natural transformation using exogenously added PCR products as a source of tDNA. We found that the transformation efficiency of E7946 was unchanged regardless of the IdeA expression status of the coincubated strain, suggesting that IdeA only acts in cis to inhibit natural transformation (Fig. 2A, black bar).
Fig. 2.
IdeA is a periplasmically localized DNA endonuclease that acts in cis to inhibit natural transformation of V. cholerae. (A) Natural transformation assays where E7946 was coincubated with the indicated strain on chitin. Cells were transformed with 0.5 μg of a PCR product that confers resistance to kanamycin. Transformation efficiency of E7946 is shown in black bars and that of the coincubating strain in stippled bars. (B) Conjugation of pMMB from an E. coli SM10 donor into the indicated V. cholerae strains. (C) Conjugation of the ICE from the indicated donor V. cholerae strain into a naive E7946 recipient. (D) Enzyme activity assays and Western blot analysis of cellular fractions of E7946 containing pMMB-ideA grown in the presence of the indicated concentration of IPTG. DNase activity, indicated by a loss of fluorescent signal in this assay (i.e., clearing from black to white) was measured in each fraction using 2 μg of linear DNA as a substrate for cleavage. β-lactamase activity, a periplasmic marker, was measured in cellular fractions using nitrocefin, a colorimetric substrate that changes color from yellow to red in the presence of β-lactamase activity. RpoA, a cytoplasmic marker, was also assessed by Western blot analysis in each cellular fraction. (E) DNase activity assay of periplasmic fractions of E7946 containing the indicated plasmid grown in the presence of the indicated concentration of IPTG using linear DNA (salmon sperm DNA) or supercoiled plasmid DNA (pGhost9) as a substrate for measuring DNase activity. Data in A–C are shown as the mean ± SD and the result of at least two independent experiments. Data in D and E are representative of at least two independent experiments.
It was recently shown that in Gram-negative bacteria, like V. cholerae, tDNA taken up during natural competence accumulates in the periplasmic space (20–22). This DNA then serves as a reservoir for tDNA that can be taken into the cytoplasm and integrated into the host chromosome (20, 22, 23). Because IdeA acts in cis to inhibit natural transformation, we hypothesized that IdeA is periplasmically localized and acts at this site to deplete the tDNA reservoir of the cell. As mentioned previously, another mechanism of HGT is conjugative transfer of DNA. During conjugation, DNA is transferred through a pilus that spans the periplasmic space. As a result, we would expect that a periplasmically localized DNase would not affect conjugative transfer. To formally test this, we assessed whether expression of IdeA affects conjugation in V. cholerae. First, we tested conjugation of pMMB, an RP4 family plasmid, from an E. coli donor into a V. cholerae recipient ±ideA. As expected, we found that IdeA had no effect on recipients during conjugation (Fig. 2B). Next, we tested whether IdeA affected donors during conjugation. To that end, we assessed conjugative transfer of the ICE using donor strains ±ideA into a naïve V. cholerae recipient. Again, as expected, we found that IdeA did not affect conjugation in donor cells (Fig. 2C). These data are consistent with periplasmic localization of IdeA.
To test this hypothesis further, we determined the subcellular localization of IdeA DNase activity in cells. To measure DNase activity, we incubated DNA with SYBR Safe, a DNA dye that fluoresces when intercalated into dsDNA. If the DNA is digested to nucleotides via the action of a DNase, then the dye no longer intercalates, and fluorescence is lost. Thus, the reduction in fluorescence in this assay is a direct result of DNase activity. We found that the endogenous level of IdeA expression was below the limit of detection in this assay. Therefore, we overexpressed IdeA in E7946 via pMMB-ideA, and under these conditions, we observed IPTG-inducible DNase activity in total cell lysates (Fig. 2D). Of note, E7946 containing pMMB-ideA is inhibited for natural transformation without inducer (Fig. 1C). This result is consistent with leaky expression being sufficient to inhibit transformation, but is below the limit of detection in our DNase activity assay (Fig. 2D). Next, we determined where IdeA localizes in cellular fractions. These fractions were also assessed for β-lactamase activity, a periplasmic marker, and RpoA levels, a cytoplasmic marker. We found that IdeA-dependent DNase activity was predominantly localized to the periplasmic fraction (Fig. 2D). This result is consistent with our phenotypic analyses and supports periplasmic localization of IdeA in V. cholerae.
IdeA Is a DNA Endonuclease.
The IdeA-dependent DNase activity observed above could represent DNA exonuclease or endonuclease activity; to define its mode of action, we performed the fluorescence-based DNase activity assay using supercoiled plasmid DNA as a substrate for cleavage. A DNA endonuclease would remain capable of acting on this substrate, whereas a DNA exonuclease would not, due to the absence of a terminal end to act on. When we performed this experiment, we find that plasmid DNA is efficiently degraded by IdeA, indicating that, as predicted by homology, this enzyme is a DNA endonuclease (Fig. 2E).
A VchInd5 ICE Inhibits Natural Transformation of V. cholerae Isolates from Bangladesh.
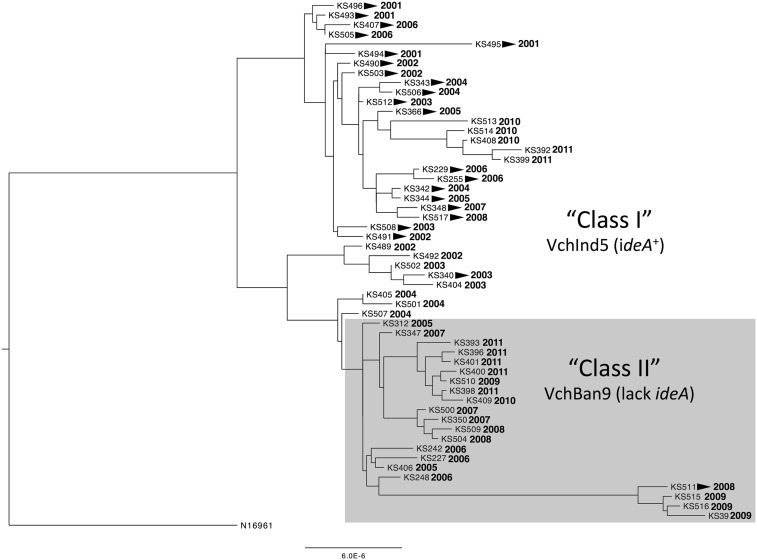
The prototypic ICE containing ideA is VchInd5, which is globally distributed among clinical isolates. Also, the ideA gene is in a variable region within the ICE known as hotspot 4 (HS4) (11). So, we next wanted to assess the prevalence of ideA among clinical isolates from a site located near the Bay of Bengal, the region where the current (seventh) pandemic originated and has spread from. To that end, we studied 54 clinical isolates from the International Center for Diarrheal Disease Research, Bangladesh (ICDDR, B) that were collected between 2001 and 2011. Draft genomes of all 54 isolates were determined as described (24), and a phylogenetic tree was constructed by mapping to the ancestral clinical isolate N16961 isolated in 1975 in Bangladesh (25) (Fig. 3). Importantly, N16961 lacks an ICE as well as other recently characterized mobile genetic elements. Thus, the phylogenetic analysis performed here demonstrates the relatedness of the core V. cholerae genome, and is unaffected by the presence or absence of large genomic islands like ICEs.
Fig. 3.
Phylogenetic tree of 54 V. cholerae isolates obtained from the ICDDR, B. Phylogeny is based solely on SNPs. Strain designations and the year of isolation are indicated at the tips of branches. All strains are of two serotypes, Inaba or Ogawa, and strains of the Inaba serotype are indicated by arrowheads next to strain designations. The scale bar indicates the nucleotide substitutions per site.
Using the annotated draft genomes of our 54 clinical isolates from Bangladesh, we determined that all strains contained an ICE; however, there were two distinct ICEs among strains from this region. One ICE contained ideA, and was most similar to VchInd5, whereas the other ICE lacked ideA and was most similar to VchBan9 (11). When we analyzed the phylogenetic distribution of the core genome of strains harboring each of these ICEs, we found that these groups clustered independently (Fig. 3), which indicates that there has been little to no conjugative transfer of ICEs between strains during the decade-long sampling period.
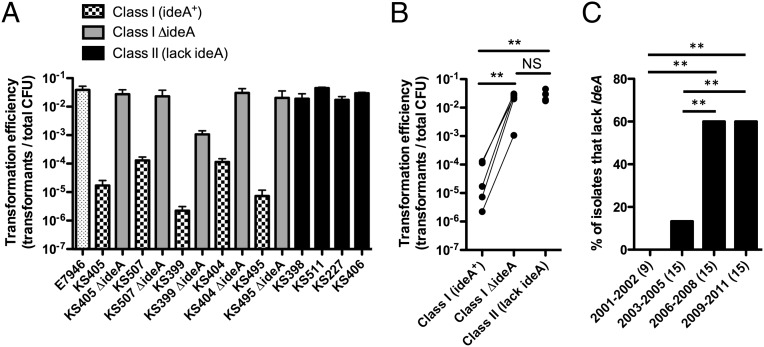
Next, we sought to determine if IdeA inhibits natural transformation among clinical isolates from Bangladesh. Strains harboring VchInd5 were denoted class I isolates, and those harboring VchBan9 were denoted class II isolates. The transformation efficiency of class I isolates was 100–10,000-fold lower than class II isolates (Fig. 4A). We generated isogenic ΔideA mutants of the class I strains and found that transformation efficiencies were, in most cases, increased to the level of the class II strains (Fig. 4 A and B). Thus, these data are consistent with what was observed among Haiti outbreak strains, and indicate that ICE-encoded IdeA also inhibits natural transformation among Bangladeshi isolates.
Fig. 4.
IdeA+ ICEs inhibit natural transformation among isolates from the ICDDR, B, and the prevalence of strains lacking IdeA increases in this region from 2001 to 2011. (A) Natural transformation of the indicated strains on chitin with 2 μg of a PCR product that confers resistance to the antibiotic kanamycin. Data are shown as the mean ± SD. (B) The mean transformation efficiencies obtained in A are compared by grouping strains based on the ICE they harbor. For class I strains, a line connects data from each parental isolate with its isogenic ideA mutant. Statistical significance was determined by two-tailed Student's t test. (C) The prevalence of strains that lack ideA is indicated within the binned years, and the number of isolates in each bin is indicated in parentheses. Statistical comparisons were made by two-tailed Z-tests. Data from A and B are the result of at least two independent experiments. **P < 0.01.
Strains Lacking IdeA Increase in Prevalence in Bangladesh from 2001 to 2011.
Next, we assessed the temporal prevalence of ideA+ strains isolated from the ICDDR, B. Based on the phylogeny of these strains, we did not find clustering based on the date of isolation, indicating that there may be multiple strains endemic to the area sampled by the ICDDR, B over this time period (Fig. 3). However, we found that there was a statistically significant increase in the prevalence of strains lacking ideA over the sampled period (Fig. 4C) due to a relative reduction in the number of isolates containing VchInd5 and an increase in the prevalence of strains containing VchBan9.
Natural Transformation Is Inhibited by IdeA in a Physiologically Relevant Microcosm Model of Chitin-Induced Competence.
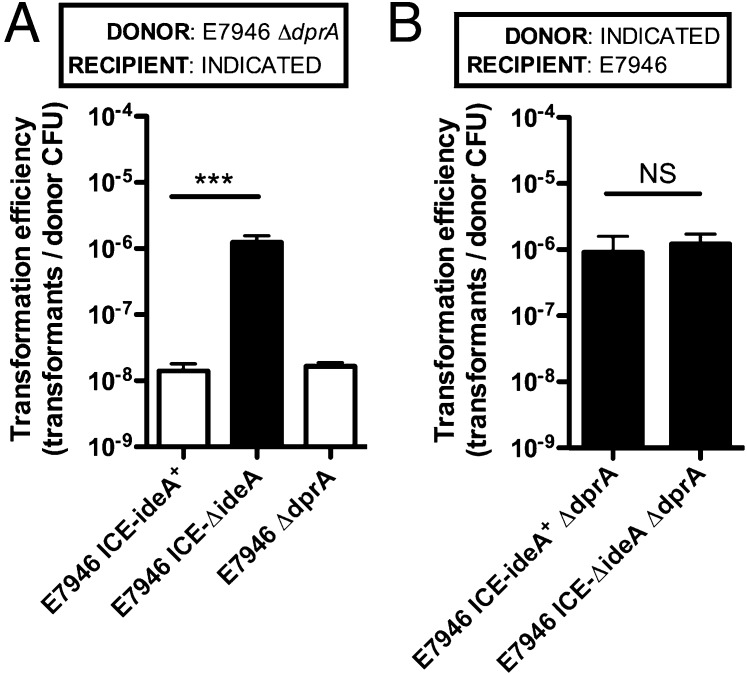
Thus far, we have assessed chitin-induced natural transformation of V. cholerae on purified chitin beads using PCR products as a source of tDNA. Though these studies have been informative in dissecting the mechanism of IdeA inhibition of natural transformation, the conditions used were not physiologically relevant for HGT in this pathogen. In the environment, V. cholerae strains likely obtain DNA from neighboring strains and closely related species that cocolonize chitinous surfaces. To assess if IdeA inhibits natural transformation under physiologically relevant conditions, we performed microcosm experiments to assess HGT between two V. cholerae strains coincubated on shrimp shells as a source of chitin. In these assays, the donor and recipient strains were differentially marked with distinct antibiotic resistance genes, and HGT by natural transformation was assessed by determining the number of double-resistant transformants following coincubation. Because we wanted to assess HGT in only one direction, donor strains in these experiments were ΔdprA mutant strains. DprA mutants are still capable of taking up DNA; however, these cells cannot integrate the DNA into their genome by homologous recombination (26, 27). To confirm that dprA mutants are not transformable, we coincubated two ΔdprA strains in these assays; however, as expected, no double-resistant transformants were obtained (Fig. 5A). This indicates that double-resistant transformants can only form as a result of DNA transfer from ΔdprA donors to dprA+ recipients in these assays. Furthermore, because DprA is specifically required only for HGT by natural transformation, the lack of transformants in this experiment indicates that double-resistant transformants obtained in this assay are the result of HGT by natural transformation.
Fig. 5.
IdeA inhibits natural transformation of V. cholerae in a physiologically relevant microcosm model of HGT. (A and B) HGT between two V. cholerae strains coincubated on chitin from shrimp shells without exogenously added DNA. In A, the donor strain is ΔVC1807::KanR, whereas recipients are ΔlacZ::SpecR. In B, all donor strains are lacZ::KanR, and the recipient is ΔVC1807::SpecR. Additional information about the genotypes of donor and recipient strains are indicated in each panel. White bars signify that no transformants were obtained and indicate the limit of detection in the assay. All data are shown as mean ± SD and the result of three independent experiments. Statistical differences were assessed by two-tailed Student’s t test. ***P < 0.001; NS, not significant.
When we coincubated an E7946 ΔdprA donor with recipient strains ±ideA, we find that IdeA inhibits natural transformation at least 100-fold to the limit of detection (Fig. 5A). These results are consistent with those obtained when tDNA was added exogenously (Fig. 1 A and B), indicating that even under physiologically relevant conditions, IdeA strongly inhibits HGT by natural transformation. Because DNase activity may be liberated when cells are lysing/providing DNA for transformation in these microcosm assays, we also wanted to assess if IdeA inhibits the ability of strains to release genomic DNA as a source of tDNA. To test this hypothesis, we incubated ΔdprA donors ±ideA with E7946 as the recipient strain, and find that IdeA does not affect the ability of V. cholerae to release tDNA (Fig. 5B). This result suggests that even if IdeA is liberated during coincubation of strains on chitin, the DNase activity present is not high enough to limit transformation in trans.
Concluding Remarks
HGT via natural transformation is a multistep process that requires first binding extracellular DNA via a type IV pilus structure (28). The retraction of this pilus along with a periplasmic DNA binding protein, ComEA, aids in bringing this DNA into the periplasmic space in Gram-negative microorganisms like V. cholerae (22). Next, a single strand of this DNA is brought into the cytoplasm via the action of the inner membrane translocase ComEC (23). During this process, DNA accumulates in the periplasmic space and is a reservoir of tDNA (20, 22, 23). Here, we show that IdeA, an ICE-encoded DNA endonuclease, inhibits natural transformation by several orders of magnitude in V. cholerae by depleting this reservoir, which is consistent with prior work showing that the DNA endonuclease Dns also depletes tDNA from the periplasm (22, 23).
Our data suggest that strains lacking ideA have recently increased in prevalence in Bangladesh, which indicates that there may be an increase in the rates of HGT observed in V. cholerae; this could increase rates of transfer of virulence determinants, including recently described variant alleles of the cholera toxin B subunit gene, ctxB (29). Though cholera toxin is encoded by a lysogenic filamentous phage that is independently capable of being mobilized, this does not preclude a role for natural transformation in accelerating the spread of this virulence determinant. Similarly, ICEs are mosaic with specific hotspots and variable regions, which contain varied gene content (10, 11). One mechanism that can explain how these mosaic ICEs have been generated is via tandem ICE integration followed by inter-ICE recombination (30, 31). These mosaic ICEs, however, could also have been generated by natural transformation. Furthermore, if natural transformation contributes to ICE mosaicism, we would predict that the presence or absence of ideA on host ICEs would impact ICE recombination. A defined study assessing the relative contribution of conjugative transfer, phage lysogeny, and natural transformation on bacterial evolution under physiologically relevant conditions, however, has not been performed, and will be the focus of future studies.
Our results demonstrate that in V. cholerae, one type of horizontally transferred element, an ICE, can inhibit natural transformation, another mechanism of HGT. The first known natural occurrence of an ICE in V. cholerae likely occurred in the 1980s and was most similar to a VchInd5 or SXT element (10, 11, 13). These ICEs confer resistance to sulfamethoxazole, chloramphenicol, trimethoprim, and streptomycin. During the time that these ICEs were first observed in V. cholerae, antibiotic use in the treatment of diarrheal diseases became prevalent. Thus, it is likely that the selective advantage afforded by these ICEs were these antibiotic resistance determinants. Therefore, it is possible that ideA is a “stowaway” that was present on the ICE that was first transferred into V. cholerae and does not confer a specific selective advantage to the organism. Though its acquisition may have been serendipitous, this DNase may have had a profound impact on limiting the evolution and adaptability of V. cholerae strains harboring this element.
Materials and Methods
Bacterial Strains and Culture Conditions.
Strains used in this study are listed in Table S1. All clinical isolates were obtained as previously described (32). Strains were routinely grown in LB and plated on LB agar. When appropriate, media was supplemented with 200 μg/mL spectinomycin, 50 μg/mL kanamycin, and/or 50 μg/mL ampicillin.
Table S1.
Strains used in this study
| Strain name in text | Description | SRA accession no. | Source |
| N16961 | V. cholerae clinical isolate obtained in Bangladesh in 1975 | (25) | |
| E7946 | Streptomycin resistant derivative of a V. cholerae clinical isolate obtained in Bahrain in 1978 | (18) | |
| 2011EL-2320 | V. cholerae clinical Isolate from the Haiti outbreak | (16) | |
| 2011EL-1810 | V. cholerae clinical Isolate from the Haiti outbreak | (16) | |
| 2012EL-1410 | V. cholerae clinical Isolate from the Haiti outbreak | (16) | |
| 2010EL-1786 | V. cholerae clinical Isolate from the Haiti outbreak | (16) | |
| 2011EL-2321 | V. cholerae clinical Isolate from the Haiti outbreak | (16) | |
| KS611 | V. cholerae clinical Isolate from the Haiti outbreak | (37) | |
| 2011EL-2320 ΔideA::KanR | 2011EL-2320 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| 2011EL-1810 ΔideA::KanR | 2011EL-1810 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| 2012EL-1410 ΔideA::KanR | 2012EL-1410 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| 2010EL-1786 ΔideA::KanR | 2010EL-1786 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| 2011EL-2321 ΔideA::KanR | 2011EL-2321 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| KS611 ΔideA::KanR | V. cholerae KS611 containing an insertion/deletion mutation where the ideA gene was replaced with a KanR marker | This study | |
| KS611 ΔideA | V. cholerae KS611 containing an in-frame deletion of ideA as well as a lacZ::KanR mutation (the lacZ mutation was used as the selected marker during cotransformation) | This study | |
| E7946 ΔlacZ | E7946 containing a deletion/insertion mutation where the lacZ gene was replaced with a SpecR marker | This study | |
| E7946 ICE-ideA+ | A strain where the ICE from KS611 was conjugatively transferred into E7946 ΔlacZ | This study | |
| E7946 ICE-ΔideA | A strain where the ICE from KS611 ΔideA was conjugatively transferred into E7946 ΔlacZ | This study | |
| SM10 (pMMB) | E. coli SM10 strain harboring the pMMB empty vector | (39) | |
| E7946 (pMMB-ideA) | E7946 harboring the pMMB-IdeA plasmid (generated by conjugating plasmid from an Sm10 donor into E7946) | This study | |
| E7946 (pMMB) | E7946 harboring the pMMB empty vector | This study | |
| BL21 (pMMB-ideA) | E. coli BL21 harboring the pMMB-IdeA plasmid | This study | |
| BL21 (pMMB) | E. coli BL21 harboring the pMMB empty vector | This study | |
| KS493 | V. cholerae clinical Isolate from the ICDDR, B | SRS883111 | This study |
| KS494 | V. cholerae clinical Isolate from the ICDDR, B | SRS883428 | This study |
| KS495 | V. cholerae clinical Isolate from the ICDDR, B | SRS883429 | This study |
| KS496 | V. cholerae clinical Isolate from the ICDDR, B | SRS883491 | This study |
| KS490 | V. cholerae clinical Isolate from the ICDDR, B | SRS884567 | This study |
| KS489 | V. cholerae clinical Isolate from the ICDDR, B | SRS884568 | This study |
| KS492 | V. cholerae clinical Isolate from the ICDDR, B | SRS884569 | This study |
| KS491 | V. cholerae clinical Isolate from the ICDDR, B | SRS884570 | This study |
| KS503 | V. cholerae clinical Isolate from the ICDDR, B | SRS884571 | This study |
| KS502 | V. cholerae clinical Isolate from the ICDDR, B | SRS884573 | This study |
| KS512 | V. cholerae clinical Isolate from the ICDDR, B | SRS884574 | This study |
| KS404 | V. cholerae clinical Isolate from the ICDDR, B | SRS884575 | This study |
| KS508 | V. cholerae clinical Isolate from the ICDDR, B | SRS884576 | This study |
| KS507 | V. cholerae clinical Isolate from the ICDDR, B | SRS884577 | This study |
| KS340 | V. cholerae clinical Isolate from the ICDDR, B | SRS884578 | This study |
| KS506 | V. cholerae clinical Isolate from the ICDDR, B | SRS884968 | This study |
| KS342 | V. cholerae clinical Isolate from the ICDDR, B | SRS884969 | This study |
| KS343 | V. cholerae clinical Isolate from the ICDDR, B | SRS884970 | This study |
| KS405 | V. cholerae clinical Isolate from the ICDDR, B | SRS884971 | This study |
| KS312 | V. cholerae clinical Isolate from the ICDDR, B | SRS884972 | This study |
| KS406 | V. cholerae clinical Isolate from the ICDDR, B | SRS884973 | This study |
| KS501 | V. cholerae clinical Isolate from the ICDDR, B | SRS884974 | This study |
| KS344 | V. cholerae clinical Isolate from the ICDDR, B | SRS884977 | This study |
| KS366 | V. cholerae clinical Isolate from the ICDDR, B | SRS884979 | This study |
| KS248 | V. cholerae clinical Isolate from the ICDDR, B | SRS884980 | This study |
| KS229 | V. cholerae clinical Isolate from the ICDDR, B | SRS884981 | This study |
| KS407 | V. cholerae clinical Isolate from the ICDDR, B | SRS884982 | This study |
| KS505 | V. cholerae clinical Isolate from the ICDDR, B | SRS884983 | This study |
| KS242 | V. cholerae clinical Isolate from the ICDDR, B | SRS884984 | This study |
| KS255 | V. cholerae clinical Isolate from the ICDDR, B | SRS884985 | This study |
| KS227 | V. cholerae clinical Isolate from the ICDDR, B | SRS884986 | This study |
| KS347 | V. cholerae clinical Isolate from the ICDDR, B | SRS884987 | This study |
| KS348 | V. cholerae clinical Isolate from the ICDDR, B | SRS884988 | This study |
| KS500 | V. cholerae clinical Isolate from the ICDDR, B | SRS884990 | This study |
| KS350 | V. cholerae clinical Isolate from the ICDDR, B | SRS884991 | This study |
| KS504 | V. cholerae clinical Isolate from the ICDDR, B | SRS884993 | This study |
| KS509 | V. cholerae clinical Isolate from the ICDDR, B | SRS884994 | This study |
| KS511 | V. cholerae clinical Isolate from the ICDDR, B | SRS884995 | This study |
| KS517 | V. cholerae clinical Isolate from the ICDDR, B | SRS884996 | This study |
| KS516 | V. cholerae clinical Isolate from the ICDDR, B | SRS884997 | This study |
| KS510 | V. cholerae clinical Isolate from the ICDDR, B | SRS885000 | This study |
| KS515 | V. cholerae clinical Isolate from the ICDDR, B | SRS885001 | This study |
| KS408 | V. cholerae clinical Isolate from the ICDDR, B | SRS885002 | This study |
| KS39 | V. cholerae clinical Isolate from the ICDDR, B | SRS885003 | This study |
| KS409 | V. cholerae clinical Isolate from the ICDDR, B | SRS885004 | This study |
| KS513 | V. cholerae clinical Isolate from the ICDDR, B | SRS885005 | This study |
| KS514 | V. cholerae clinical Isolate from the ICDDR, B | SRS885006 | This study |
| KS393 | V. cholerae clinical Isolate from the ICDDR, B | SRS885007 | This study |
| KS392 | V. cholerae clinical Isolate from the ICDDR, B | SRS885008 | This study |
| KS396 | V. cholerae clinical Isolate from the ICDDR, B | SRS885010 | This study |
| KS399 | V. cholerae clinical Isolate from the ICDDR, B | SRS885012 | This study |
| KS398 | V. cholerae clinical Isolate from the ICDDR, B | SRS885013 | This study |
| KS401 | V. cholerae clinical Isolate from the ICDDR, B | SRS885014 | This study |
| KS405 ΔideA | KS405 containing an in-frame deletion of ideA as well as a lacZ::SpecR (the lacZ mutation was the selected marker for cotransformation) | This study | |
| KS507 ΔideA | KS507 containing an in-frame deletion of ideA as well as a lacZ::SpecR (the lacZ mutation was the selected marker for cotransformation) | This study | |
| KS399 ΔideA | KS399 containing an in-frame deletion of ideA as well as a lacZ::SpecR (the lacZ mutation was the selected marker for cotransformation) | This study | |
| KS404 ΔideA | KS404 containing an in-frame deletion of ideA as well as a lacZ::SpecR (the lacZ mutation was the selected marker for cotransformation) | This study | |
| KS495 ΔideA | KS495 containing an in-frame deletion of ideA as well as a lacZ::SpecR (the lacZ mutation was the selected marker for cotransformation) | This study | |
| E7946 ΔdprA | E7946 containing an in-frame deletion in dprA as well as a KanR cassette in place of VC1807, a frame-shifted transposase (the VC1807 mutation was the selected marker during cotransformation) | This study | |
| E7946 ΔdprA | E7946 containing an in-frame deletion in dprA as well as ΔlacZ::SpecR (the lacZ mutation was the selected marker during cotransformation) | This study | |
| E7946 ICE-ideA+ ΔdprA | E7946 ICE-ideA+ containing an in-frame deletion of dprA and a lacZ::KanR mutation (this mutation in lacZ replaces the SpecR cassette present in the parent isolate and was the selected marker for cotransformation) | This study | |
| E7946 ICE-ΔideA ΔdprA | E7946 ICE-ΔideA containing an in-frame deletion of dprA and a lacZ::KanR mutation (this mutation in lacZ replaces the SpecR cassette present in the parent isolate and was the selected marker for cotransformation) | This study |
Generating Mutant Strains and Plasmids.
Mutant constructs were generated by splicing-by-overlap extension PCR as previously described (33). For details, see SI Materials and Methods. All primers used to generate new mutant constructs are listed in Table S2.
Table S2.
Oligonucleotides used in this study
| Name | Sequence (5′→3′) | Description |
| Primers for making mutant constructs | ||
| BBC125 | CCATAATCGCCAAGGTGTTTTG | ΔideA or ΔIdeA::KanR F1 (up arm) |
| BBC131 | gtcgacggatccccggaatATTCATGTAAATTGGCCAAAAATGAC | ΔideA::KanR deletion R1 (up arm) |
| BBC132 | gaagcagctccagcctacaTGCTAGCATGTATCCACAACCTAAC | ΔideA::KanR deletion F2 (down arm) |
| BBC129 | GTTAGGTTGTGGATACATGCTAGCAATTCATGTAAATTGGCCAAAAATGAC | ΔideA R1 (up arm) |
| BBC130 | GTCATTTTTGGCCAATTTACATGAATTGCTAGCATGTATCCACAACCTAAC | ΔideA F2 (down arm) |
| BBC126 | GGCTTTGAGCAAGATATCCAGTATG | ΔideA or ΔideA::KanR R2 (down arm) |
| ABD123 | ATTCCGGGGATCCGTCGAC | KanR cassette for ΔIdeA::KanR F (middle) |
| ABD124 | TGTAGGCTGGAGCTGCTTC | KanR cassette for ΔIdeA::KanR R (middle) |
| ABD820 | CGCTCTTATCTGCTTGGATAATGG | ΔdprA F1 (up arm) |
| ABD998 | gtcgacggatccccggaatCATTAACTGGCATCATCAACC | ΔdprA R1 (up arm) |
| ABD999 | gaagcagctccagcctacaTAGCTATGATGATGGATATTTTGATG | ΔdprA F2 (down arm) |
| ABD823 | TGAAGTACAAGGCCAGTTACTGG | ΔdprA R2 (down arm) |
| prm361 | attccggggatccgtcgacCTGCAGTTCagaagcagctccagcctaca | ΔdprA mini-FRT F (middle) |
| Prm362 | tgtaggctggagctgcttctGAACTGCAGgtcgacggatccccggaat | ΔdprA mini-FRT R (middle) |
| Primers for confirming mutant strains | ||
| BBC122 | ATGGTAAGCCGCTTGACTGG | ICE status F |
| BBC124 | CAGTAGGAACAAGGCCTACAGC | ICE integrated (used with BBC122 = 702 bp product if ICE is integrated in prfC) R |
| BBC123 | GCACGTCCGAAAAGCAGGAC | ICE absent (used with BBC122 = 523 bp product if ICE is not integrated in prfC) R |
| BBC120 | CGAAGGGGCTCTTCATCTTGC | ideA status F |
| BBC121 | TCTGGGCGAGACTCAATACATC | ideA status R (used with BBC120 = 862 bp product if IdeA is present and 190 bp if strain has an in-frame deletion in IdeA) |
| ABD725 | GAAGCAGCTCCAGCCTACA | dprA status F |
| BBC190 | AACAGCAGGAAACCACGCG | dprA status (used with ABD725 = 280 bp product if the strain has an in-frame deletion in dprA) R |
Bacterial Conjugations and Natural Transformation Assays.
Conjugation and natural transformation assays were performed essentially as previously described (34, 35). For details, see SI Materials and Methods.
Fractionation of V. cholerae.
Cells were grown in M9+glucose because LB medium is autofluorescent in the same channel used to visualize DNase activity. Cells were grown to midexponential phase, then 109 cells were fractionated as previously described (36). For details, see SI Materials and Methods.
DNase Activity Assay.
DNase activity was measured using the DNA intercalating dye SYBR Safe (Invitrogen). For details, see SI Materials and Methods.
β-Lactamase Activity Assay.
Assays were performed using nitrocefin (Thermo Scientific) according to the manufacturer’s instructions. For details, see SI Materials and Methods.
Western Blot Analysis.
Six microliters of each cellular fraction was electrophoretically separated on a 10% polyacrylamide gel. Then proteins were transferred to a nitrocellulose membrane. Blots were then probed for RpoA using a mouse monoclonal antibody (Santa Cruz Biotechnology), and visualized using a Cy5-labeled α-mouse IgG secondary antibody (Invitrogen). Blots were scanned on an FLA-9000IR instrument.
Phylogenetic Tree Construction.
All 54 clinical isolates obtained from the ICDDR, B were sequenced and de novo assembled as previously described (37). Reads from whole-genome sequencing experiments are publicly accessible in the Sequence Read Archive (SRA) under BioProject ID PRJNA279349. SRA accession numbers for each isolate are provided in Table S1. Contig sequences for each strain were submitted to the REALPHY online tool along with the published N16961 genome as a reference genome for phylogenetic analysis (38). The tree is shown rooted to N16961.
SI Materials and Methods
Generating Mutant Strains and Plasmids.
To generate mutants where a gene was replaced/disrupted with an antibiotic resistance marker (e.g., ΔideA::KanR, VC1807::KanR, and lacZ::KanR), we used chitin-induced natural transformation as described below. For unmarked mutants (e.g., ΔideA and ΔdprA), we used cotransformation as previously described (33). All mutants were confirmed by PCR and/or sequencing.
To generate pMMB-ideA, we amplified IdeA from KS611 genomic DNA using primers BBC180 (5′-tatatagaattcATGAATAGAACTAATTTTCTCG-3′) and BBC181 (5′- tatataggatccCTAGCAGCTGCCTGATACATAGC-3′) and cloned this insert into the EcoRI and BamHI sites of pMMB67EH. The insert in pMMB-ideA was confirmed by sequencing with primers ABD598 (5′-ATTAATCATCGGCTCGTATAATGTG-3′) and BBC185 (5′-GCGTTCTGATTTAATCTGTATCAGG-3′).
Bacterial Conjugations and Natural Transformation Assays.
For transfer of RP4 containing plasmids from E. coli into V. cholerae, 500 μL of overnight cultures of donor and recipient strains were mixed 1:1 and concentrated 10-fold before spreading onto a sterile mixed cellulose esters filter (0.8 μM size cutoff; Millipore) placed on an LB plate. Reactions were incubated at 37 °C for 4 h. Then, the membrane was transferred to 1 mL of LB and vortexed vigorously to dislodge cells. Reactions were then plated onto media selective for donors, recipients, or transconjugates, and conjugation efficiency was reported as transconjugates per donor.
Conjugative transfer of an ICE element from one V. cholerae strain to another was performed as previously described (34). Briefly, overnight cultures of donor and recipients were subcultured in LB broth 1:100 and grown at 37 °C shaking for 2 h. Then, mitomycin C was added to donor strains to a final concentration of 20 ng/mL Cells were grown for an additional 1 h shaking at 37 °C. Next, 500 μL of donors and recipients were mixed, washed, and concentrated 10-fold before spreading onto a filter placed on an LB plate. Reactions were then incubated at 37 °C for 16 h before plating as described above. The ICE from KS611 confers resistance to trimethoprim, and recipient strains were resistant to spectinomycin. Therefore, transconjugates were selected by plating on trimethoprim and spectinomycin.
For natural transformation assays, ∼108 cfus of midexponential growth phase V. cholerae were placed in 1 mL of “instant ocean” (7 g/L; aquarium systems) containing 100 μL of chitin beads (New England Biolabs). Chitin beads were used because they promoted higher and more reproducible transformation efficiencies compared with chitin powder. Cells were incubated statically for 16–24 h at 30 °C to induce natural competence. Then, tDNA was added directly to each reaction and incubated statically for an additional 16–24 h at 30 °C. Next, reactions were vortexed vigorously and 500 μL of each reaction was transferred to 1 mL of LB and outgrown with shaking at 37 °C for 1 h. Assays were then plated on selective media to quantify transformants, as well as media lacking antibiotics to quantify total cfus. Transformation efficiency was defined as transformants per total cfus. For most transformation assays, the tDNA conferred resistance to kanamycin or spectinomycin (as indicated) by replacing VC1807, a frame-shifted transposase gene with an antibiotic resistance gene.
For microcosm transformation assays, the conditions used were essentially as described above, except the source of chitin used in assays was chitin from shrimp cells (Sigma-Aldrich). In these assays, the indicated V. cholerae strains were coincubated on these shrimp shells and induced for competence as outlined above. No exogenous DNA was added in these microcosm transformation assays; instead, cells were coincubated statically for 48 h at 30 °C. Strains were differentially marked as indicated in the text and transformation was assessed by plating as described above.
DNase Activity Assay.
For DNase assays, 5 μL of the indicated cellular fraction was added to 100 μL of DNase reaction buffer [10 mM Tris HCl (pH 8.0), 100 mM NaCl, 2 mM KCl, 2 mM CaCl2, and 10× SYBR Safe (Invitrogen)] containing 2 μg of the indicated DNA substrate. Because Mg2+ is essential for the activity of DNA endonucleases, assays were started by the addition of MgSO4 to a final concentration of 30 mM. Reactions were then incubated statically at room temperature for 5 min before assessing fluorescence of intercalated SYBR Safe on an FLA-9000IR instrument (GE Healthcare Life Sciences). Loss of signal is indicative of DNase activity in these assays. Supercoiled plasmid preps of pGhost9 were confirmed to be intact and unnicked by gel electrophoresis as well as resistance to cleavage with T7 exonuclease (New England Biolabs).
For assessing the activity of cellular fractions, the genomic DNA of cells was accounted for in total and protoplast fractions and additional linear DNA substrate (salmon sperm DNA) was added to obtain 2 μg total DNA in each reaction. This was done so that DNase activity could be compared across all cellular fractions.
Fractionation of V. cholerae.
For protoplast and periplasmic fractions, 109 cells were resuspended in 100 μL of polymyxin B buffer [50 mM Tris HCl (pH 7.6), 10 mM EDTA, 150 mM NaCl, 500 mM sucrose, and 1 mg/mL polymyxin B] and incubated on ice for 30 min. Then, this suspension was centrifuged at 16,000 × RCF for 30 min at 4 °C. The supernatant following this treatment represented the periplasmic fraction, whereas the pellet represented the protoplast fraction. Whole cells and protoplast fractions were then sonicated in a horn cup sonicator to release their soluble contents.
For supernatant fractions, cells were pelleted and the supernatant was moved to a fresh tube. EDTA was added to 4 mM, and then this fraction was concentrated on a 10 kDa molecular weight cutoff spin column (Millipore) to achieve cell equivalents (i.e., if 109 cells in 1 mL was concentrated 10-fold to 100 µL for cellular fractionation, we concentrated supernatants 10-fold as well).
β-Lactamase Activity Assay.
Assays were performed by adding 3.0 μL of each cellular fraction to 100 μL of β-lactamase reaction buffer [50 mM NaPO4 (pH 7.0), 4 mM MgSO4, and 100 μg/mL nitrocefin] in 96-well plates. Reactions were incubated statically at room temperature for 30 min. A colorimetric change from yellow to red is indicative of β-lactamase activity in these assays, and A550 was measured to quantify β-lactamase activity. Plates were also scanned on a flatbed scanner for visualization of β-lactamase activity.
Acknowledgments
We thank the Tufts Core Facility for sequencing and computational support, and Brian K. Hammer and Eryn Bernardy for providing strains and helpful discussions. This work was supported by US National Institutes of Health Grants AI055058 (to A.C.) and AI058935 (to S.B.C.). A.C. is a Howard Hughes Medical Institute investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive database, www.ncbi.nlm.nih.gov/sra/. For a list of accession numbers, see Table S1.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1509097112/-/DCSupplemental.
References
- 1.Harris JB, LaRocque RC, Qadri F, Ryan ET, Calderwood SB. Cholera. Lancet. 2012;379(9835):2466–2476. doi: 10.1016/S0140-6736(12)60436-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruzzo C, Vezzulli L, Colwell RR. Global impact of Vibrio cholerae interactions with chitin. Environ Microbiol. 2008;10(6):1400–1410. doi: 10.1111/j.1462-2920.2007.01559.x. [DOI] [PubMed] [Google Scholar]
- 3.Huq A, et al. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45(1):275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colwell RR. Global climate and infectious disease: The cholera paradigm. Science. 1996;274(5295):2025–2031. doi: 10.1126/science.274.5295.2025. [DOI] [PubMed] [Google Scholar]
- 5.Meibom KL, Blokesch M, Dolganov NA, Wu CY, Schoolnik GK. Chitin induces natural competence in Vibrio cholerae. Science. 2005;310(5755):1824–1827. doi: 10.1126/science.1120096. [DOI] [PubMed] [Google Scholar]
- 6.Blokesch M, Schoolnik GK. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 2007;3(6):e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stroeher UH, et al. Genetic rearrangements in the rfb regions of Vibrio cholerae O1 and O139. Proc Natl Acad Sci USA. 1995;92(22):10374–10378. doi: 10.1073/pnas.92.22.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waldor MK, Colwell R, Mekalanos JJ. The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc Natl Acad Sci USA. 1994;91(24):11388–11392. doi: 10.1073/pnas.91.24.11388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldor MK, Mekalanos JJ. Vibrio cholerae O139 specific gene sequences. Lancet. 1994;343(8909):1366. doi: 10.1016/s0140-6736(94)92504-6. [DOI] [PubMed] [Google Scholar]
- 10.Spagnoletti M, et al. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. MBio. 2014;5(4):01356–14. doi: 10.1128/mBio.01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozniak RA, et al. Comparative ICE genomics: Insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5(12):e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32(1):99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 13.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477(7365):462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Didelot X, et al. The role of China in the global spread of the current cholera pandemic. PLoS Genet. 2015;11(3):e1005072. doi: 10.1371/journal.pgen.1005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chin CS, et al. The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364(1):33–42. doi: 10.1056/NEJMoa1012928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katz LS, et al. Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio. 2013;4(4):00398–13. doi: 10.1128/mBio.00398-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Azarian T, et al. Phylodynamic analysis of clinical and environmental Vibrio cholerae isolates from Haiti reveals diversification driven by positive selection. MBio. 2014;5(6):01824–14. doi: 10.1128/mBio.01824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller VL, DiRita VJ, Mekalanos JJ. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989;171(3):1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blokesch M, Schoolnik GK. The extracellular nuclease Dns and its role in natural transformation of Vibrio cholerae. J Bacteriol. 2008;190(21):7232–7240. doi: 10.1128/JB.00959-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gangel H, et al. Concerted spatio-temporal dynamics of imported DNA and ComE DNA uptake protein during gonococcal transformation. PLoS Pathog. 2014;10(4):e1004043. doi: 10.1371/journal.ppat.1004043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seitz P, Blokesch M. DNA-uptake machinery of naturally competent Vibrio cholerae. Proc Natl Acad Sci USA. 2013;110(44):17987–17992. doi: 10.1073/pnas.1315647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seitz P, et al. ComEA is essential for the transfer of external DNA into the periplasm in naturally transformable Vibrio cholerae cells. PLoS Genet. 2014;10(1):e1004066. doi: 10.1371/journal.pgen.1004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seitz P, Blokesch M. DNA transport across the outer and inner membranes of naturally transformable Vibrio cholerae is spatially but not temporally coupled. MBio. 2014;5(4):01409–01414. doi: 10.1128/mBio.01409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazinski DW, Camilli A. Homopolymer tail-mediated ligation PCR: A streamlined and highly efficient method for DNA cloning and library construction. Biotechniques. 2013;54(1):25–34. doi: 10.2144/000113981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406(6795):477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergé M, Mortier-Barrière I, Martin B, Claverys JP. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol Microbiol. 2003;50(2):527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 27.Karudapuram S, Zhao X, Barcak GJ. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J Bacteriol. 1995;177(11):3235–3240. doi: 10.1128/jb.177.11.3235-3240.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat Rev Microbiol. 2004;2(3):241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 29.Grim CJ, et al. Genome sequence of hybrid Vibrio cholerae O1 MJ-1236, B-33, and CIRS101 and comparative genomics with V. cholerae. J Bacteriol. 2010;192(13):3524–3533. doi: 10.1128/JB.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burrus V, Waldor MK. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol. 2004;186(9):2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009;5(12):e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weil AA, et al. Memory T-cell responses to Vibrio cholerae O1 infection. Infect Immun. 2009;77(11):5090–5096. doi: 10.1128/IAI.00793-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc Natl Acad Sci USA. 2014;111(24):8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 35.Dalia AB, Lazinski DW, Camilli A. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio. 2014;5(1):e01028–e13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerny G, Teuber M. Differential release of periplasmic versus cytoplasmic enzymes from Escherichia coli B by polymixin B. Arch Mikrobiol. 1971;78(2):166–179. doi: 10.1007/BF00424873. [DOI] [PubMed] [Google Scholar]
- 37.Seed KD, et al. Evolutionary consequences of intra-patient phage predation on microbial populations. eLife. 2014;3:e03497. doi: 10.7554/eLife.03497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol Biol Evol. 2014;31(5):1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fürste JP, et al. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]