Significance
Genome inheritance requires the complete resolution of all intertwines within parental DNA. This is facilitated by fork rotation and precatenation of the newly replicated DNA. However, the general importance and frequency of fork rotation in vivo are poorly understood. We find that the evolutionarily conserved Timeless and Tipin proteins actively inhibit fork rotation in budding yeast. In their presence, fork rotation appears restricted to hard-to-replicate fragile sites. In their absence, excessive fork rotation leads to damage accumulating in the replicated sister chromatids, especially at known yeast fragile sites. Therefore, fork rotation appears to be restricted to contexts where it is absolutely required for unwinding, and this restriction is required to prevent precatenation inducing excessive chromosomal fragility.
Keywords: DNA replication, topoisomerases, DNA topology, fork rotation, DNA catenation
Abstract
Faithful genome duplication and inheritance require the complete resolution of all intertwines within the parental DNA duplex. This is achieved by topoisomerase action ahead of the replication fork or by fork rotation and subsequent resolution of the DNA precatenation formed. Although fork rotation predominates at replication termination, in vitro studies have suggested that it also occurs frequently during elongation. However, the factors that influence fork rotation and how rotation and precatenation may influence other replication-associated processes are unknown. Here we analyze the causes and consequences of fork rotation in budding yeast. We find that fork rotation and precatenation preferentially occur in contexts that inhibit topoisomerase action ahead of the fork, including stable protein–DNA fragile sites and termination. However, generally, fork rotation and precatenation are actively inhibited by Timeless/Tof1 and Tipin/Csm3. In the absence of Tof1/Timeless, excessive fork rotation and precatenation cause extensive DNA damage following DNA replication. With Tof1, damage related to precatenation is focused on the fragile protein–DNA sites where fork rotation is induced. We conclude that although fork rotation and precatenation facilitate unwinding in hard-to-replicate contexts, they intrinsically disrupt normal chromosome duplication and are therefore restricted by Timeless/Tipin.
During DNA replication, it is essential to completely unwind and remove all of the intertwining between the two strands of the template DNA double helix. This is achieved by the combined action of replicative helicases and topoisomerases. During elongation, replicative helicases force the strands apart, generating compensatory topological overwinding stress in the unreplicated region ahead of the fork. If overwinding accumulates, it prevents further DNA replication (1, 2). Relaxation of the stress is achieved either by topoisomerase action ahead of the fork, directly on the overwound region, or by coupling helicase action with rotation of the whole fork relative to the unreplicated DNA (Fig. S1). This latter pathway relaxes topological stress ahead of the fork at the expense of generating double-stranded intertwines behind the fork, often referred to as DNA precatenanes (3, 4). These intertwines are subsequently resolved by the action of type II topoisomerases. If type II topoisomerases do not completely resolve either the precatenanes or the full DNA catenanes formed at the completion of replication, the unresolved intertwines will cause chromosome bridging, nondisjunction, and aneuploidy (5). Fork rotation and DNA precatenation appear to be the primary pathway of unlinking when forks come together at the termination of DNA replication (6, 7). In addition, fork rotation appears to be a frequent event during elongation in vitro; it can support ongoing replication, and extensive precatenation is observed behind elongating forks (8–10). Therefore, the prevailing view is that the topological stress caused by DNA unwinding is resolved stochastically during elongation by both topoisomerase action ahead of the fork and fork rotation and DNA decatenation behind the fork (3, 11). At termination, the diminishing distance between converging replisomes is thought to prevent topoisomerase action ahead of the fork, leaving fork rotation as the primary pathway for DNA unwinding in this context. However, unlike viral replisomes and replication complexes established in in vitro systems, the eukaryotic replisome holoenzyme is composed of a far greater number of proteins and activities. These factors are thought to facilitate replication through the highly variable eukaryotic genomic landscape and coordinate DNA synthesis with other chromatid maturation processes such as chromatin assembly and cohesion establishment (5). These latter processes act on the same newly replicated DNA potentially braided by precatenation. How these activities occurring in the wake of the fork may be impeded by precatenation is unexplored (5).
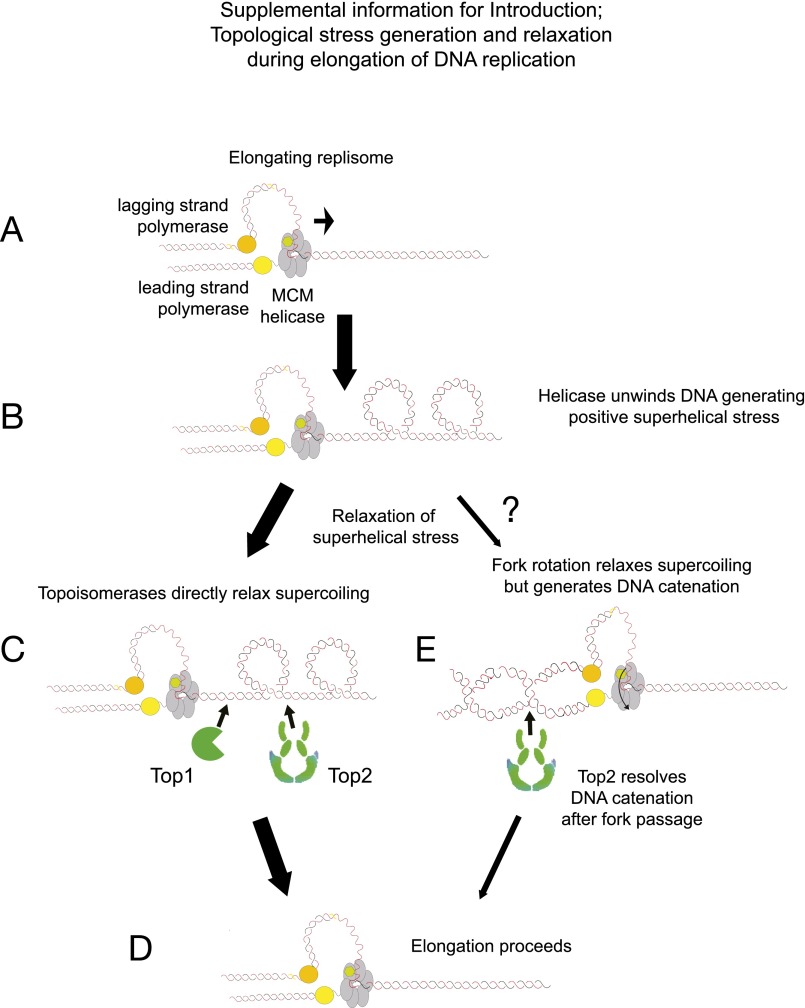
Fig. S1.
Model of topological stress generation and relaxation during elongation of DNA replication. (A) During elongation, unwinding of the parental template separates the parental strands but does not resolve the linkages that exist between the two strands. (B) The linkages between the strands are displaced into the region ahead of the fork, leading to this becoming overwound, that is, positively supercoiled. (C and D) This tension is normally resolved by the action of either topoisomerase I (Top1) or topoisomerase II (Top2) (C), which act effectively as “swivelases” ahead of the fork to generate a relaxed replication region (D). (E) However, Champoux and Been (4) proposed a second mode of topological stress unwinding where the helical tension is relaxed by rotation of the fork to generate DNA precatenation behind the fork. Although these linkages should not arrest forward elongation of replication, it is essential that the type II topoisomerase resolve all DNA catenation before the completion of cell division.
To study these events in vivo, we have directly examined fork rotation and precatenation in budding yeast. We show that fork rotation and DNA precatenation are not stochastic but rather are actively restricted to distinct contexts by the evolutionarily conserved homologs of Timeless/Tipin, Tof1/Csm3. Failure to regulate fork rotation leads to significantly elevated levels of DNA damage, particularly at known fragile sites. Therefore, the eukaryotic replisome appears to minimize rotation and precatenation, so that they are used to unwind DNA only when absolutely necessary to maintain genome stability.
Results
Fork Rotation During Replication Elongation Is Not Stochastic and Is Restricted to Distinct Genomic Contexts.
To assess fork rotation in vivo, we adapted a previously described plasmid DNA catenation assay for use with yeast episomal plasmids (12, 13). The conditional loss of type II topoisomerase (Top2) activity during DNA replication leads to the accumulation of catenated replicated plasmids in yeast (14), due to the DNA precatenanes formed through fork rotation not being resolved by Top2. Assaying the extent of DNA catenation on these plasmids by agarose gel electrophoresis allows a direct assessment of the frequency of formation of precatenation and therefore fork rotation (the assay is fully outlined in Fig. S2). To conditionally remove topoisomerase II activity from cells specifically during DNA replication, we synchronized the top2-4 yeast strain (15) in G1, switched the cells to the restrictive temperature to ablate Top2 activity, and then released them into the cell cycle, allowing plasmid replication and formation of catenated sister chromatids. For analysis, we then purified DNA following replication without Top2, nicked the plasmids with a site-specific nuclease, and then resolved and quantified the entire distribution of relaxed and catenated plasmids (referred to as CatAn, where n = number of catenated linkages) by 2D gel electrophoresis, Southern blotting, and densitometry. Using this assay, we found that replication of the 5-kb yeast ARS/CEN episomal plasmid pRS316 produced a normal distribution of catenated states with a median of 13. Analysis of the tail of the distribution indicated that 14% of the population appeared highly catenated [i.e., with plasmids containing more than 20 catenations (CatAn >20)] (Fig. 1A). Bacterial Topo III can resolve precatenation in vitro by acting on gaps in the lagging strand (8). To eliminate the possibility that the activity of yeast Top3 may have led to an underestimation of fork rotation, we assessed DNA catenation of the plasmid in the absence of both Top2 and Top3. However, we did not observe any significant difference in DNA catenation of pRS316 in the absence of Top3 and Top2 compared with Top2 alone (Fig. S3A), indicating that Top3 does not resolve precatenanes in this context. Finally, to test whether residual Top2 activity in the ts allele could be modifying our results, we compared catenated plasmids generated immediately following DNA replication with catenated plasmids maintained for a further hour in a postreplicative block. No residual decatenation activity could be detected (Fig. S3B). Therefore, we conclude that our analysis of DNA catenation is providing a direct assessment of fork rotation and precatenation on these replicons.
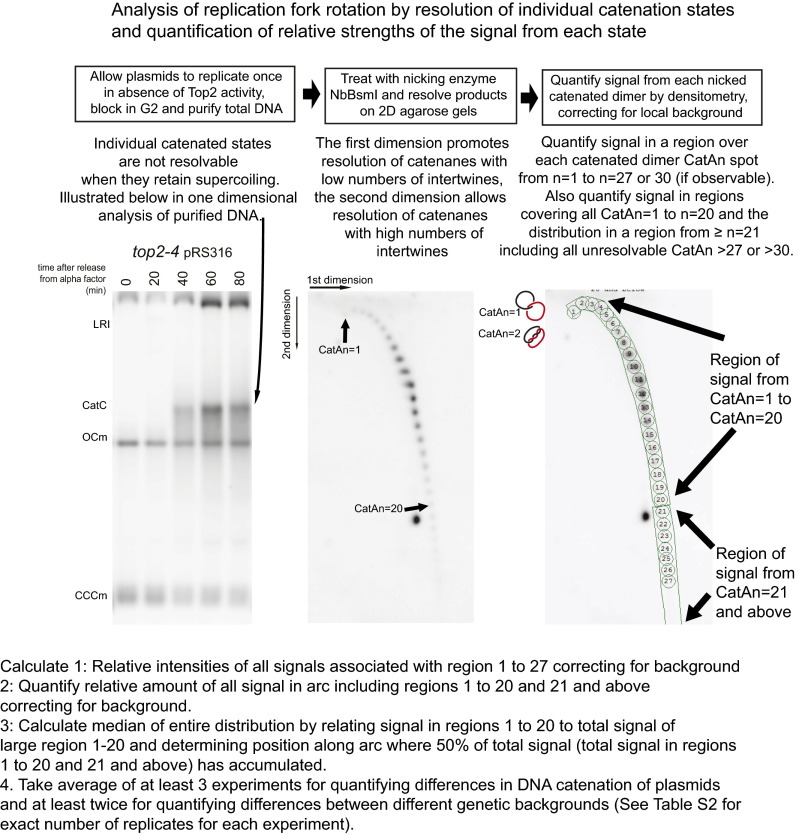
Fig. S2.
Quantification of fork rotation during DNA replication. In wild-type cells, Top2 resolves both precatenanes and catenanes formed during DNA replication. Therefore, to assay how often fork rotation and precatenation occur during replication on a plasmid replicon, we allowed one round of replication in the absence of Top2 activity and collected DNA from the cells blocked in G2 by nocodazole 80 min after release into the cell cycle (plasmid products are shown; Left). The catenated products of replication in this background are negatively supercoiled due to nucleosome deposition in the sister chromatids. This supercoiling of deproteinized plasmids normally compacts all catenated states into an unresolvable population. To resolve individual catenated states, the purified DNA is treated with a site-specific nicking enzyme to remove supercoiling but maintain the catenated nodes. The nicked DNA is resolved by agarose gel electrophoresis in two separate dimensions to be able to resolve both low- and high-catenated states before Southern blotting and probing for plasmid sequences (Middle). The relative intensities of each state were then calculated by densitometry in two ways. The first was by quantifying signal in equally sized regions centered on each catenated signal state on states 1–27 (or 1–30 for hypercatenated samples where the signal >27 was definable) and correcting for local background, and then expressing the relative intensity as a percentage of the total signal in all regions. The second measure was to compare the signal from the arc related to states 1–20 to states 21 and above. For this measure, regions were drawn around all states 1–20 and from the remainder of the arc relating to 21 and above. Then, each set of states is expressed as a percentage of the sum of both. This measure has the advantage of being able to quantify the signal from the individually unresolvable catenated states that produced detectable signal.
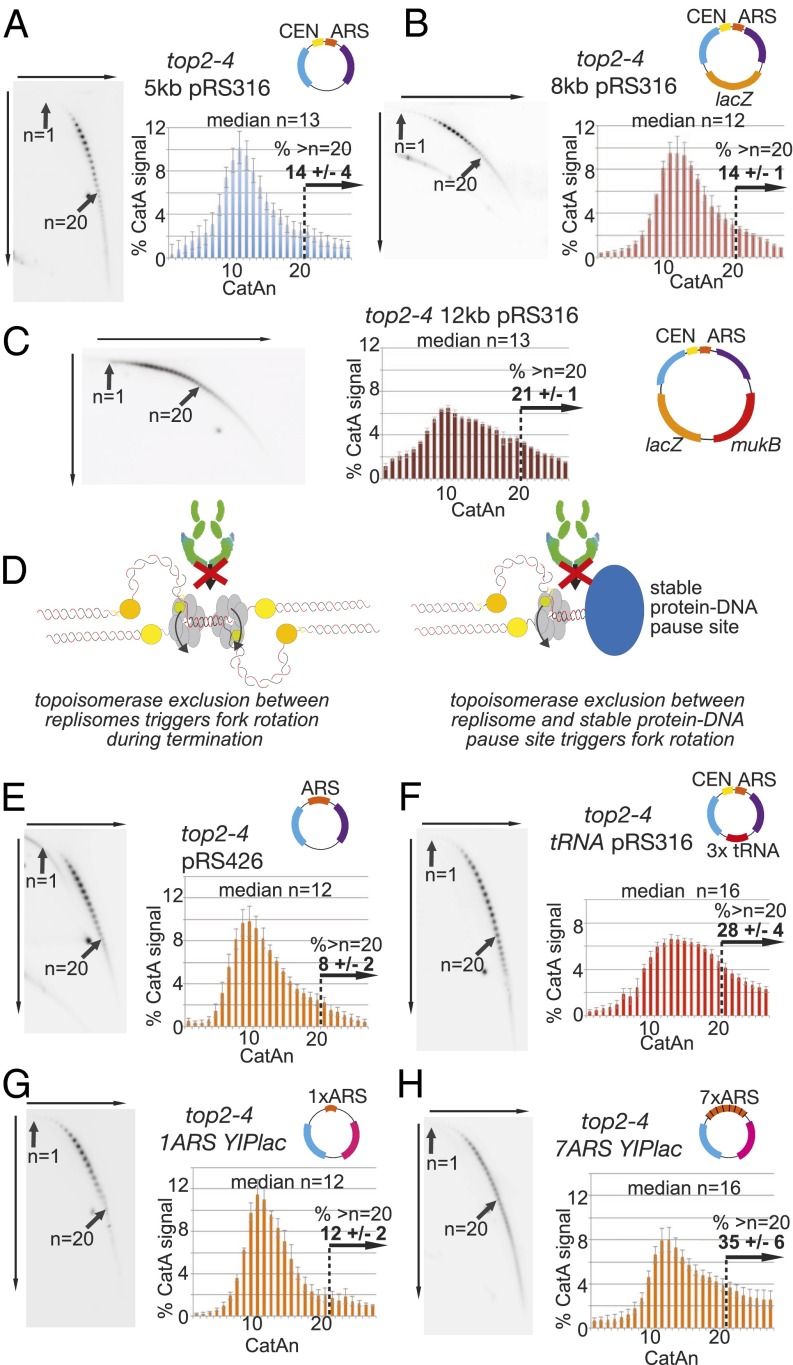
Fig. 1.
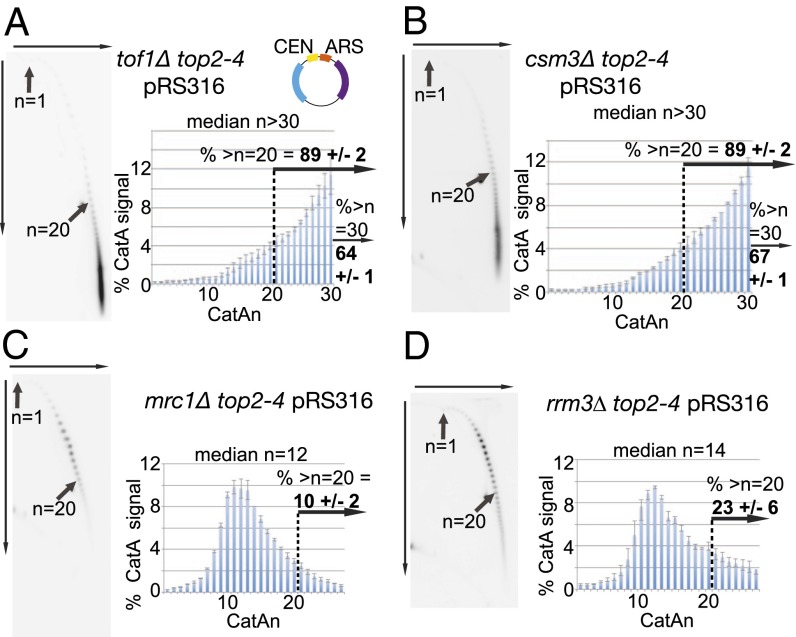
Fork rotation and DNA catenation occur upon replication through stable protein–DNA pause sites. (A–C and E–H) Cells containing the top2-4 allele and plasmid (A) pRS316, (B) pRS316 containing 3 kb or (C) 7 kb of additional bacterial sequence, (E) pRS426, (F) CEN and 3× tRNA gene plasmid tRNApRS316, (G) YIplac plasmid with 1×ARSH4, or (H) 7×ARSH4 origins in a YIplac plasmid (all plasmids are shown in cartoons) were assessed for DNA catenation following one round of DNA replication in the absence of Top2 activity (representative autoradiograms are shown; the top arrow indicates electrophoresis in the first dimension, and the side arrow indicates in the second dimension). Histograms show the relative intensity of catenanes containing 1–27 catenated links (CatAn), along with the median of the whole distribution and % of catenanes from plasmids with >20 catenanes. Arrows indicate the mobility of plasmids containing 1 (n = 1) and 20 catenanes (n = 20). Histograms and % of plasmids with >20 catenanes represent the average of ≥3 independent experiments. Error bars or values are average deviation. See Fig. S2 for a full explanation. Light blue, ampR gene; purple, URA3 gene; pink, TRP1 gene; others colors are as indicated. (D) Model for topoisomerase exclusion at both termination (Left) and stable protein–DNA structures (Right). As replisomes converge, topoisomerases are sterically inhibited from relaxing helical tension in the final few turns. Therefore, fork rotation is required for unwinding. Theoretically, a similar situation arises when a replisome approaches a stable protein–DNA complex.
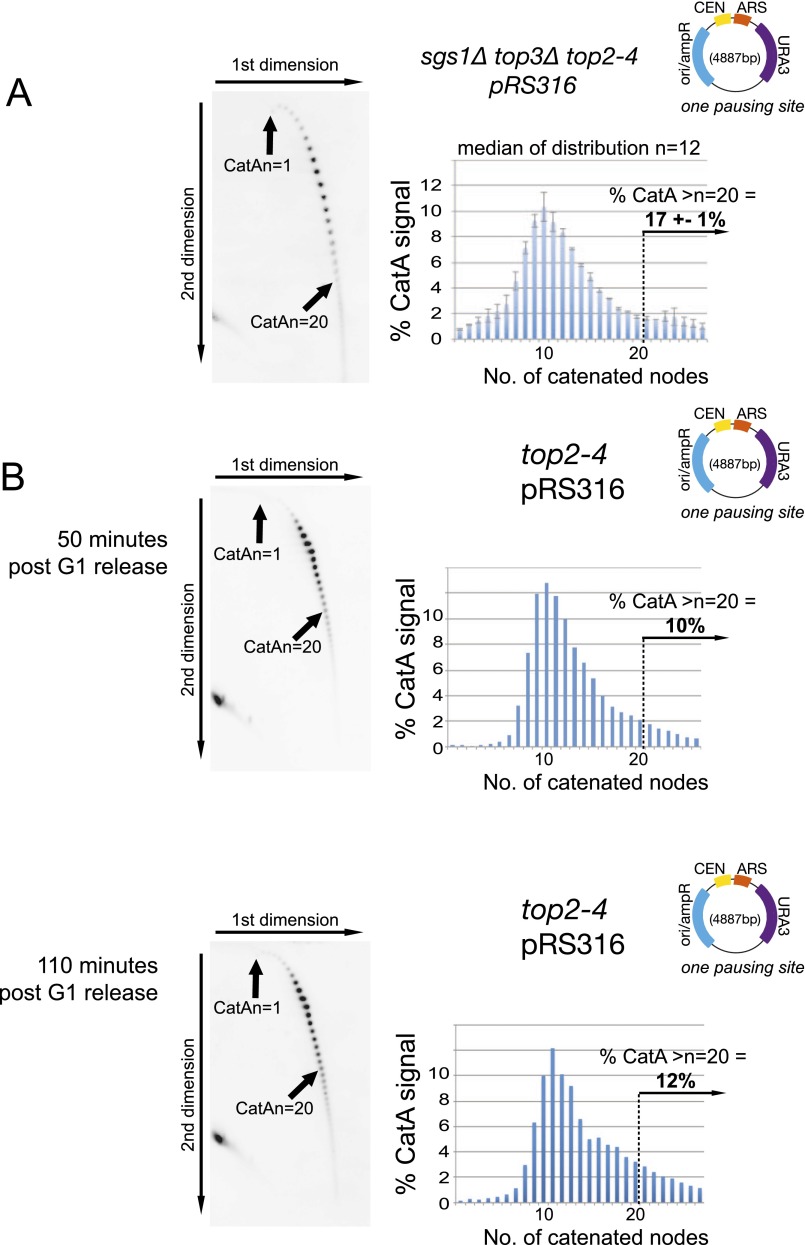
Fig. S3.
Quantification of DNA catenation is directly related to incidence of fork rotation on plasmids. (A) Deletion of TOP3 does not alter the level of DNA catenation generated in catenated plasmids. DNA catenation analysis of plasmid pRS316 in sgs1Δ top3Δ top2-4 cells was analyzed as in Fig. 1. SGS1 is deleted in this strain to allow relatively normal levels of proliferation of the cells. Histograms and % of plasmids with >20 catenanes represent the average of two independent experiments. Error bars or values are equal to the average deviation of the experiments. (B) No residual decatenation activity can be detected in top2-4 cells maintained under restrictive conditions. DNA catenation analysis of plasmid pRS316 in top2-4 cells isolated either 50 or 110 min after release from alpha factor arrest. DNA was analyzed as described in Fig. 1.
If fork rotation occurs stochastically during elongation, increasing the size of a replicon should increase the incidence of fork rotation during its replication. To assay whether increasing the size of the replicon leads to increased fork rotation, we first ligated 3 kb of DNA into pRS316, increasing the size of the plasmid by 60%. However, no difference in the distribution of DNA catenation was observed (median n = 12; 14% of plasmids had >20 catenations) (Fig. 1B). We then ligated a further 4 kb of DNA into the plasmid, increasing the size of the replicon by 140% (to 12 kb), and reanalyzed DNA catenation. On this plasmid, we observed a spreading of the distribution of catenated states, compared with the smaller plasmids, but did not see an increase of the median of the population (median n = 13; 21% of plasmids had >20 catenations) (Fig. 1C). Therefore, in vivo, increasing the size of the plasmid replicon does not significantly alter the overall extent of DNA catenation, indicating that elongation distance is not the primary determinant of the extent of fork rotation during DNA replication. Rather, our data suggest that the extent of fork rotation is regulated by distinct contexts within the replicon, such as the termination of DNA replication.
Fork rotation is predicted to be more frequent at termination, because the convergence of two replisomes spatially restricts topoisomerase access to the unreplicated DNA between them (11). In this model, the replisomes have to rotate to unwind the final few turns of DNA (Fig. 1D, Left). Projecting this model onto the different chromosomal contexts of DNA replication, we postulated that a similar situation would arise when the fork passes through stable protein–DNA complexes that are known to pause ongoing replication (16) (Fig. 1D, Right). When the fork approaches a stable protein–DNA complex that pauses replication, topoisomerases will be spatially inhibited from accessing the DNA between the encroaching replisome and the stable complex, potentially requiring fork rotation to allow ongoing replication. To test this hypothesis, we assessed whether pausing structures in plasmid replicons leads to increased fork rotation during replication. We compared similarly sized plasmids that contained either no known pause site (pRS426), a point centromere pause site (pRS316), or multiple pausing sites consisting of three tRNA genes and a centromere (tRNApRS316). Catenation analysis of pRS426 produced a slightly reduced distribution of catenated states compared with the one-pause site plasmid pRS316 (pRS426 median n = 12; 8% of plasmids with >20 catenations) (Fig. 1E). In contrast, a substantial increase in catenation was observed for the multiple-pause site plasmid tRNApRS316 (median n = 16; 28% of plasmids had >20 catenations) (Fig. 1F). Therefore, replication through tRNAs and potentially centromeres increases fork rotation and DNA catenation over and above the catenation that occurs at termination. We extended our analysis to inactive origins, where DNA-bound ORC and MCM proteins pause replication (16). We achieved this by comparing a seven-origin plasmid to a one-origin plasmid (17). On multiorigin plasmids, only one origin is activated and the remaining inactive origins are passively replicated (18). The seven-origin plasmid based on the YIplac plasmid became highly catenated during DNA replication (median n = 16; 35% of plasmids with >20 catenations) compared with a YIplac plasmid containing one copy of the origin (median n = 12; 12% of plasmids with >20 catenations) (Fig. 1 G and H) (a full summary of DNA catenation experiments is in Table S1). Together, these data indicate that replication through stable structures that pause DNA replication including tRNAs, inactive origins, and potentially centromeres (16) induces fork rotation and precatenation during replication elongation.
Table S1.
Summary of DNA catenation quantification experiments
| Strain | Plasmid size, bp | Number of replicates | Median CatAn | % >20 |
| top2-4 pRS316 | 4,887 | 5 | 13 | 14 ± 4 |
| top2-4 8kb pRS316 | 7,936 | 3 | 12 | 14 ± 1 |
| top2-4 12kb pRS316 | 12,391 | 3 | 13 | 21 ± 1 |
| top2-4 pR426 | 5,726 | 5 | 12 | 8 ± 2 |
| top2-4 tRNApRS316 | 6,060 | 7 | 16 | 28 ± 4 |
| top2-4 1ori YIplac | 5,640 | 4 | 12 | 12 ± 2 |
| top2-4 7ori YIplac | 6,144 | 5 | 16 | 35 ± 6 |
| tof1Δ top2-4 pRS316 | 4,887 | 2 | >30 | 89 ± 2 |
| tof1Δ top2-4 pRS426 | 5,726 | 2 | >30 | 93 ± 1 |
| tof1Δ top2-4 tRNApRS316 | 6,060 | 2 | >30 | 95 ± 2 |
| csm3Δ top2-4 pRS316 | 4,887 | 2 | >30 | 89 ± 2 |
| mrc1Δ top2-4 pRS316 | 4,887 | 2 | 12 | 10 ± 2 |
| ctf4Δ top2-4 pRS316 | 4,887 | 2 | 12 | 17 ± 7 |
| rrm3Δ top2-4 pRS316 | 4,887 | 2 | 14 | 23 ± 6 |
| rrm3Δ top2-4 pRS426 | 5,726 | 2 | 12 | 10 ± 6 |
| rrm3Δ top2-4 tRNApRS316 | 6,060 | 2 | 18 | 38 ± 2 |
| sgs1Δ top3Δ top2-4 pRS316 | 4,887 | 2 | 12 | 17 ± 1 |
| top2-4 pRS316 + 200 mM HU | 4,887 | 2 | 11 | 11 ± 1 |
The median was calculated as explained in Fig. S2.
Timeless/Tof1 and Tipin/Csm3 Restrict Fork Rotation.
Because stable protein–DNA complexes both pause replication and cause fork rotation, we next examined whether factors that influence fork pausing also alter fork rotation. Deletion of the Timeless homolog TOF1 leads to reduced pausing at stable protein–DNA complexes (19). Therefore, we examined whether deletion of TOF1 alters fork rotation during replication of plasmid pRS316. In tof1Δ cells, we observed a radical increase in the number of DNA catenanes formed on the plasmid. The distribution of highly catenated states was dramatically shifted, with a median in excess of 30 (64% of the distribution containing >30 catenanes) (Fig. 2A). Therefore, Tof1 appears to restrict fork rotation during DNA replication on this replicon. We then examined how deletion of other replisome factors linked to Tof1/Timeless alters fork rotation on pRS316. Excessive fork rotation was also observed in cells deleted for the yeast homolog of Tipin, CSM3, the conserved partner of Tof1/Timeless in the replisome (20) (Fig. 2B). Claspin/Mrc1 and AND1/Ctf4 have also been reported to be linked to Tof1/Csm3 function (21, 22). However, deletion of neither MRC1 nor CTF4 increased fork rotation during replication of the plasmid (Fig. 2C and Fig. S4). We also examined the potential role of the protein displacement helicase RRM3 in fork rotation. Rrm3 is required in vivo to minimize fork pausing at stable protein–DNA structures, presumably by promoting their displacement to allow rapid replication passage (16). Deletion of RRM3 produced a significant increase in fork rotation on plasmid pRS316 (Fig. 2D); however, this increase appeared modest compared with tof1Δ or csm3Δ cells. We conclude that TOF1/CSM3 and RRM3 are involved in distinct pathways to inhibit fork rotation in vivo.
Fig. 2.
Yeast Timeless and Tipin homologs TOF1 and CSM3 and the displacement helicase RRM3 inhibit fork rotation and DNA catenation. DNA catenation analysis of plasmid pRS316 in top2-4 cells and the different deletion alleles (A) tof1Δ, (B) csm3Δ, (C) mrc1Δ, and (D) rrm3Δ were analyzed as in Fig. 1. Histograms and % of plasmids with >20 or >30 catenanes represent the average of ≥2 independent experiments. Error bars or values are average deviation. See Fig. S2 for a full explanation.
Fig. S4.
Deletion of CTF4 significantly increases DNA catenation on pRS316. DNA catenation analysis of plasmid pRS316 in ctf4Δ top2-4 cells, analyzed as in Fig. 1. Histograms and % of plasmids with >20 represent the average of two independent experiments. Error bars or values are equal to the average deviation of the experiments.
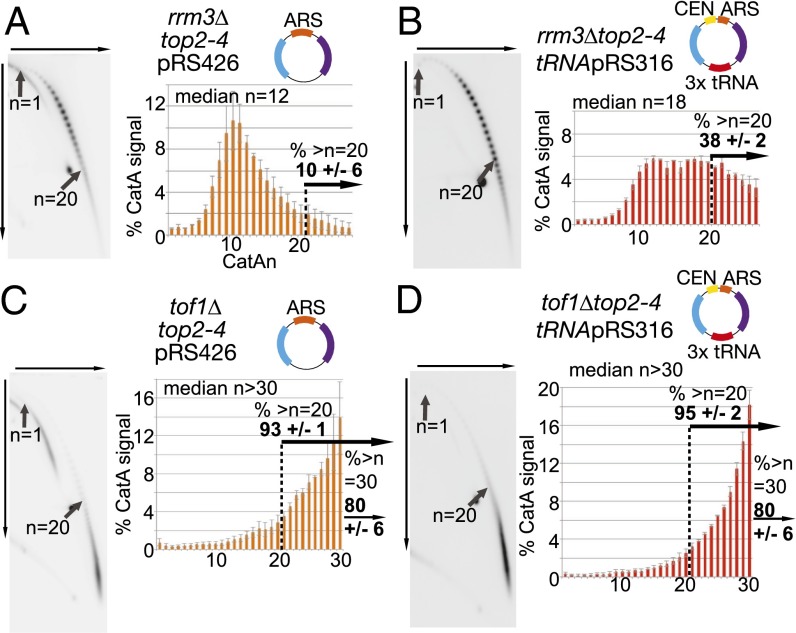
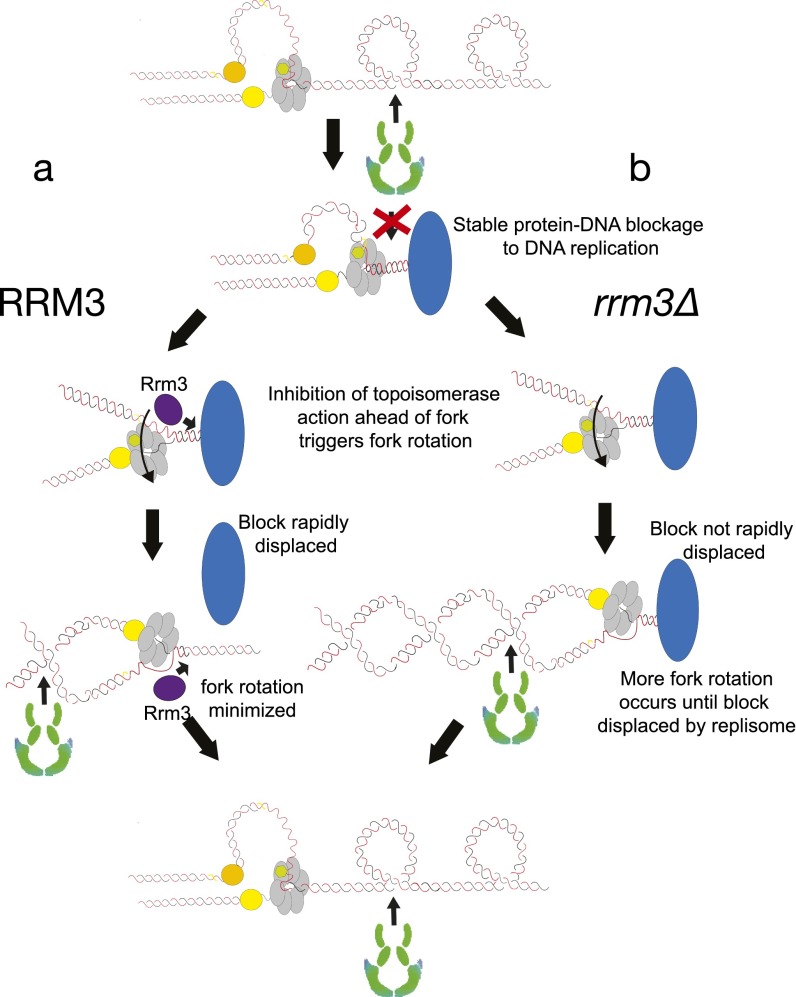
We next examined the effects on fork rotation of deleting either RRM3 or TOF1 on plasmids with and without pause sites. We observed that deletion of RRM3 increased fork rotation only on plasmids containing pause sites (Fig. 3 A and B compared with Fig. 1 E and F; Table S2). This suggests that Rrm3 is required to rapidly displace stable protein–DNA complexes and allows accelerated access of topoisomerases ahead of the fork, limiting the need for fork rotation at such sites (Fig. S5). In contrast, we observed that deletion of TOF1 caused hypercatenation of plasmids irrespective of the number of pause sites (Fig. 3 C and D). This suggests that Tof1/Csm3 more generally restricts fork rotation during DNA replication and not just at pause sites. Deletion of TOF1/TIMELESS is reported to slow replisome elongation rates (23). Potentially, excessive fork rotation in tof1/csm3 cells could be due to slower elongation, which could favor fork rotation. However, we did not detect any increase in DNA catenation occurring on plasmids replicated either in the presence of hydroxyurea, which would be predicted to arrest fork progression (Fig. S6A), or in cells deleted for the leading-strand polymerase elongation factor DPB3 (Fig. S6B), which causes a general slowing of DNA replication (Fig. S6C). Therefore, unrestricted fork rotation does not appear to be a consequence of general fork slowing but rather is directly related to Tof1/Csm3 function.
Fig. 3.
Rrm3 limits the frequency of fork rotation and precatenation at stable protein–DNA sites, whereas Tof1 acts more generally. Cells deleted for RRM3 and containing the top2-4 allele and (A) plasmid pRS426 or (B) plasmid tRNApRS316 and cells deleted for TOF1 and containing the top2-4 allele and (C) plasmid pRS426 or (D) plasmid tRNApRS316 were cultured and collected, DNA was prepared, and DNA catenation analysis was carried out as described in Fig. 1. Error bars or values are average deviation.
Table S2.
Yeast strains
| Strains | Full genotype |
| top2-4 | top2-4 Mat a his4-539 lys2-801 ura3-52 (15) |
| top2-4 pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 |
| top2-4 pRS426 | top2-4 pRS426 Mat a his4-539 lys2-801 ura3-52 |
| top2-4 tRNA-pRS316 | top2-4 tRNA-pRS316 Mat a his4-539 lys2-801 ura3-52 |
| top2-4 tof1Δ pRS316 | top2-4 3tRNA-pRS316 Mat a his4-539 lys2-801 ura3-52 tof1::hphMX |
| top2-4 tof1Δ pRS426 | top2-4 pRS426 Mat a his4-539 lys2-801 ura3-52 tof1::hphMX |
| top2-4 tof1Δ tRNApRS316 | top2-4 tRNA-pRS316 Mat a his4-539 lys2-801 ura3-52 tof1::hphMX |
| top2-4 csm3Δ pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 csm3::natMX |
| top2-4 mrc1Δ pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 mrc1::natMX |
| top2-4 ctf4Δ pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 ctf4::natMX |
| top2-4 sgs1Δ top3Δ pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 sgs1::NATNT2 top3::hphMX |
| top2-4 dpb3Δ pRS316 | top2-4 pRS316 Mat a his4-539 lys2-801 ura3-52 dpb3::natMX |
| yST114 (wt) | MATa ade2-1 ura3-1 his3-11, trp1-1, can1-100 UBR1::pGAL-myc-UBR1 (HIS3), leu2-3 LEU2::pCM244 x3 |
| top2-td | yST114 + kanMX-tTA-tetO2-UB-DHFRts-myc-top2 |
| top2-td tof1Δ | yST114 + kanMX-tTA-tetO2-UB-DHFRts-myc-top2 tof1::hphMX |
| top2-td dpb3Δ | yST114 + kanMX-tTA-tetO2-UB-DHFRts-myc-top2 dpb3::natNT2 |
| dpb3Δ | yST114 + dpb3::natNT2 |
| top2-td mrc1Δ | top2-td + mrc1::hphMX |
| mrc1Δ | yST114 + mrc1::hphMX |
| tof1Δ | yST114 + tof1::hphMX |
Fig. S5.
Model for how Rrm3 displacement activity alters fork rotation at stable protein–DNA fragile sites. As the replisome approaches a stable protein–DNA complex, topoisomerases are inhibited from acting between the complex and the converging replisome. (A) If the Rrm3 helicase is active at the site then the protein complex will be rapidly displaced, allowing access of topoisomerases ahead of the fork and the curtailing of replisome rotation and DNA catenation. (B) If RRM3 is deleted then rotation continues until the replisome itself physically displaces the stable protein complex, thereby allowing access for topoisomerases ahead of the fork. However, in this case, more rotation-coupled unwinding is required until the protein complex is displaced.
Fig. S6.
Artificial slowing of replication elongation does not increase fork rotation and DNA catenation. (A) DNA catenation analysis of plasmid pRS316 in top2-4 cells released from alpha factor arrest into 200 mM hydroxyurea (HU). The cells were collected 80 min after release, and extracted DNA was analyzed as described in Fig. 1. (B) DNA catenation analysis of plasmid pRS316 in dpb3Δ top2-4 cells. (C) FACS analysis of dpb3Δ and dpb3Δ top2-td cells following release from alpha factor in the restrictive condition shows that deletion of DPB3 appears to significantly slow down S phase, consistent with its known role as a polymerase epsilon processivity factor. wt, wild-type.
Unrestricted Fork Rotation and DNA Catenation Cause Aneuploidy and Trigger a G2/M Arrest in Cells.
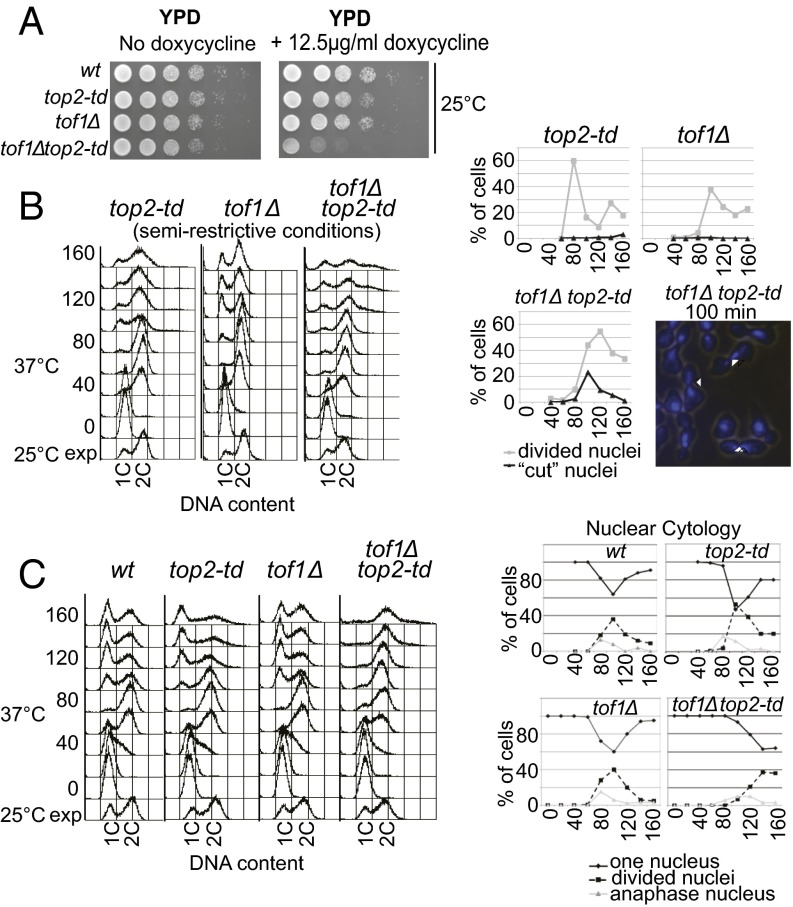
We next sought to confirm that loss of Tof1 function leads to excessive DNA catenation on endogenous chromosomes. Consistent with excessive fork rotation and DNA catenation occurring in the absence of Tof1, tof1Δ cells were acutely sensitive to a partial loss of Top2 activity, induced by transcriptional repression of the top2-td allele (Fig. 4A). We next examined the extent of chromosome missegregation in cells where TOF1 is deleted and Top2 is either partially depleted (Fig. 4B) or wholly depleted (Fig. 4C). It has previously been shown in budding yeast that unresolved DNA catenation does not inhibit cell-cycle progression but does cause aneuploidy following cell division (14) (Fig. 4C). If deletion of TOF1 causes excessive DNA catenation, then it would be predicted to cause aneuploidy at levels of Top2 activity that normally allow chromosome segregation. To test this prediction, we first used semirestrictive growth conditions that reduced Top2 dosage to a level just sufficient to allow normal segregation of endogenous chromosomes in a single cell cycle (Fig. 4B, Left) with very few “cut” cells [indicative of gross missegregation (14)], cytologically observed (Fig. 4B, Right and Fig. S7). However, in the absence of TOF1, this dosage of Top2 was not sufficient to prevent widespread missegregation of endogenous chromosomes and aneuploidy (Fig. 4B, Left), and cut cells were frequently observed in mitosis (Fig. 4B, Right). Therefore, deletion of TOF1 specifically sensitizes cells to partial loss of Top2, resulting in a phenotype that resembles a complete loss of Top2 (Fig. 4C). To control for the possibility that depleting Top2 in tof1Δ cells could also cause aneuploidy by general destabilization of the replisome, leading to frequent fork collapse, we repeated this set of experiments in mrc1Δ cells. Deletion of MRC1 destabilizes the replisome in response to replication stress in a similar manner to tof1Δ (21) but does not cause an increase in DNA catenation (Fig. 2C). We observed that mrc1Δ cells are not as sensitive to partial loss of Top2 as tof1Δ cells (Fig. S8A), and that deletion of MRC1 does not cause the marked increase in cut nuclei observed following depletion of Top2 in tof1Δ cells (Fig. S8B). These data indicate that the aneuploidy and missegregation observed in tof1Δ cells is not due to a general increase in fork arrest and collapse. Rather, our observations are consistent with loss of TOF1 leading to unrestricted fork rotation and formation of excessive DNA catenation on endogenous chromosomes.
Fig. 4.
Deletion of TOF1 triggers excessive catenation of endogenous chromosomes and G2 cell-cycle arrest. (A) Viability assay of strains under the permissive condition of growth on YPD at 25 °C or with partial transcriptional repression of TOP2 (+doxycycline; DOX). (B) Deletion of TOF1 causes chromosome nondisjunction following partial depletion of Top2 activity. Cells were grown under semirestrictive conditions (YPD 37 °C + 12.5 µg/mL DOX) following synchronous release from G1 and analyzed for chromosome missegregation by FACS for DNA content (Left) or cytological analysis for cut and divided nuclei at the time points shown (min). Examples of cut cells (arrowheads) are shown (Right) [DAPI-stained DNA (blue) is shown over a light image of cells 100 min after release]. (C) The indicated strains were arrested in alpha factor, and Top2 was degraded (YP + 2% raffinose + 2% galactose; 37 °C + 25 µg/mL DOX). Samples were taken at midlog phase (25 °C exponential; exp) before alpha factor release (0) and at time points shown after release (min) for FACS analysis to assess DNA content (Left) or nuclear cytologies of cells (Right). The percentages of single (diamonds and solid line) and divided nuclei (squares and dashed line) are shown.
Fig. S7.
Western blot analysis of Top2 in exponential cells, grown under permissive and semirestrictive conditions in wild-type, top2-td, and tof1Δ top2-td cells. Pgk1 Western blot of the same lanes is shown for loading comparison.
Fig. S8.
(A) Viability assay of wild-type, top2-td, tof1Δ, tof1Δ top2-td, mrc1Δ, and mrc1Δ top2-td cells under the permissive condition of growth on YPD at 25 °C or conditions of partial transcriptional repression of TOP2 (+12.5 µg/mL doxycycline). (B) Partial depletion of Top2 activity does not lead to an increase in “cut” mitosis in mrc1Δ cells. top2-td, mrc1Δ, and mrc1Δ top2-td cells grown under semirestrictive conditions (YPD, 37 °C + doxycycline) following synchronous release from G1 were analyzed for cytological analysis for cut and divided nuclei and chromosomal aneuploidy by cellular DNA content using FACS analysis (Bottom).
Our plasmid analysis indicates that fork rotation is actively inhibited during DNA replication. This inhibition appears to be enforced through the cellular function of Tof1/Csm3. These data raise the question of why fork rotation is actively inhibited. Fork rotation provides an important alternative pathway for unwinding, because excessive fork rotation and the resultant DNA precatenation do not appear to be immediately lethal until Top2 becomes limiting. However, impaired Tof1/Timeless function leads to chronic genomic instability in cells. Increased levels of gross chromosomal rearrangements are observed in tof1Δ cells (24), whereas depletion of Timeless/Tipin orthologs in several systems causes constitutive DNA damage (20, 25). Potentially, the chronic genome-instability phenotypes and constitutive DNA damage in cells depleted of Tof1/Timeless could be a direct result of excessive fork rotation and precatenation occurring during DNA replication. We reasoned that if the excessive fork rotation was leading to chronic genomic instability, then such genome-instability phenotypes should be exacerbated when Top2 is depleted, because this would stabilize the precatenanes formed following fork rotation.
Consistent with the possibility that excessive DNA precatenation destabilizes replication-associated processes, we observed a slight delay in anaphase onset in tof1Δ cells partially depleted of Top2 (Fig. 4B) and a pronounced G2/M delay following full depletion of Top2 in tof1Δ cells (Fig. 4C). Cells deleted for TOF1 or depleted of Top2 alone did not arrest in G2 (Fig. 4 B and C) (14). These data suggest that a combination of tof1Δ and Top2 depletion generates defects in the newly replicated chromatids, triggering cell-cycle checkpoints and inhibiting mitosis.
Excessive Fork Rotation and DNA Precatenation Cause Premitotic DNA Damage at Yeast Protein–DNA Fragile Sites.
Deletion of Timeless causes constitutive DNA damage in human cells (25). Using H2A S129 phosphorylation (H2AS129P) as a marker for DNA damage, we also observed a significant increase in constitutive damage in tof1Δ cells in both S-phase and postreplicative whole-cell extracts (Fig. 5A). Depletion of Top2 alone led to less H2AS129P than in tof1Δ, but we observed the highest levels of H2AS129P in postreplicative cells with both tof1Δ and depleted Top2 (Fig. 5A). In these experiments, the cells were treated with nocodazole to avoid detection of damage generated during mitosis. Therefore, the combination of excessive formation and stabilization of DNA precatenation during DNA replication appeared to lead to the highest levels of DNA damage in these cells. Consistent with this, we also observed elevated activity of the DNA damage checkpoint effector kinase Rad53 in nocodazole-arrested tof1Δ top2-td cells using the in-blot kinase assay (26) but not in similarly arrested tof1Δ- or top2-depleted cells (Fig. 5B). In addition, examination of postreplicative protein extracts also showed that PCNA (proliferating cell nuclear antigen encoded by POL30) monoubiquitylation was increased in tof1Δ top2-td cells (Fig. 5C), indicating that postreplication repair (PRR) was generally active following excessive fork rotation and DNA precatenation. This would be consistent with either gaps being left in the newly replicated sister chromatids, triggering PRR, or with ongoing PRR being inhibited by an abnormal sister chromatid structure. Together, these data suggest that both excessive fork rotation and stabilization of precatenation lead to increased levels of constitutive DNA damage being detected on the newly replicated chromatids, with the combination of both sufficient to trigger widespread DNA repair and activation of the DNA damage checkpoint.
Fig. 5.
Unrestricted fork rotation and excessive DNA catenation cause DNA damage and extended postreplicative repair of replicated chromatids. Cells arrested in alpha factor were shifted to restrictive conditions (YP Raf Gal; 37 °C + 25 µg/mL DOX) and released into media containing nocodazole. (A) Samples from the time points indicated were analyzed by Western blot for phosphorylation of H2A S129 (Pgk1 was used for a loading control) and FACS for DNA content. (B) Samples were assayed for Rad53 activation using the Rad53 autophosphorylation assay. Control samples of MMS (methyl methanesulfonate)-treated exponential top2-td and rad53Δ top2-td cells are also shown. (C) PCNA ubiquitylation was monitored in postreplicative cells (80 min following release) by Western blotting. Blotting for PCNA in MMS-treated cells containing either His-tagged wild-type PCNA or His-tagged K164R confirmed the specificity of the PCNA antibody for monoubiquitylated PCNA (U1) or polyubiquitylated PCNA (U2). (D) Chromatin immunoprecipitation of H2A S129 phosphorylation at two tRNAs, tI(AAU)N1 and tA(UGC)L, and euchromatic loci in isogenic top2-td, tof1Δ, and tof1Δ top2-td strains 80 min after release. ChIP signal was normalized to amplification of input DNA before calculation of the relative change at each locus compared with wild-type cells. Data shown are the mean of three independent experiments ±1 SD.
The apparent infliction of DNA damage following excessive DNA precatenation would predict that sites of constitutive fork rotation should be especially vulnerable to this pathway, potentially leading these sites to have elevated levels of DNA damage even in unchallenged wild-type cells. Others have identified all of the yeast genomic loci that exhibit constitutively elevated levels of DNA damage in wild-type yeast cells and have defined such sites as the yeast equivalent of mammalian fragile sites (27). Interestingly, this set includes tRNAs and inactive origins where elevated levels of DNA precatenation are likely to occur. To test whether DNA damage detected at these fragile sites is linked to increased precatenation during DNA replication, we examined how loss of Top2 and/or Tof1 altered H2A S129 phosphorylation at candidate fragile sites. ChIP analysis for H2AS129P demonstrated that the absence of Top2 and/or Tof1 during DNA replication led to significantly greater increases of DNA damage at two distinct genomic tRNA pausing sites compared with two tested euchromatic sites (Fig. 5D). Increased DNA damage was not due to passage through mitosis, because these cells were arrested in nocodazole following passage through one S phase. We conclude that the elevated frequencies of fork rotation and DNA precatenation that occur at stable protein–DNA fragile sites are linked to DNA damage and fragile-site instability.
Discussion
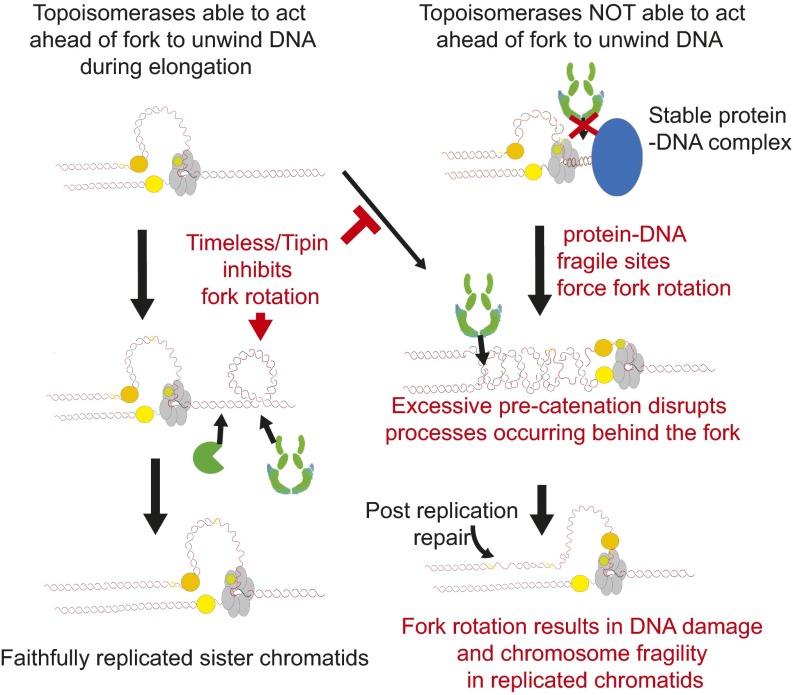
Previous studies of DNA precatenation have demonstrated the potential for fork rotation and precatenation to facilitate ongoing DNA replication in the face of topological stress in vitro (8–10). Here we show that this capability is actively restricted during DNA replication in budding yeast through the activity of the evolutionarily conserved Timeless/Tipin factors Tof1/Csm3. When this complex is present, fork rotation and precatenation are not determined by replicon size. Rather, it appears that the presence of distinctive genomic features within the replicon alter how often fork rotation occurs during replication elongation. Outside of termination (6), we show that the presence of tRNA genes, inactive origins, and possibly centromeres in plasmid replicons also increases the incidence of fork rotation during DNA replication. All these genomic contexts pause ongoing replication, presumably due to the stability of the protein–DNA complexes that they generate (16). We speculate that these large stable complexes inhibit topoisomerase access to the underlying DNA, preventing relaxation of topological stress ahead of the fork, which results in fork rotation at these known hard-to-replicate and fragile loci (Fig. S9).
Fig. S9.
Model of causes and consequences of fork rotation and precatenation. During normal DNA replication the Tof1/Csm3 structured replisome inhibits fork rotation, and topoisomerases act ahead of the fork (Left). In certain genomic contexts, such as stable protein–DNA structures, topoisomerases are impeded from acting ahead of the fork (Right Top). Here fork rotation aids unwinding, resulting in elevated precatenation. Excessive precatenation could impede several processes, including Okazaki fragment maturation, resulting in gaps in the newly replicated chromosomes that require repair and leave chromatids fragile.
Our data suggest that the yeast replisome rarely rotates unless topoisomerase action ahead of the fork proves insufficient for ongoing replication. Potentially, topoisomerase action could be similarly inhibited in other genomic contexts, for example at heterochromatin. Regions of transcriptionally induced overwinding, such as converging genes (28) or nuclear membrane-attached genes (29), could also induce such high levels of topological stress that topoisomerase action ahead of the fork would be insufficient for ongoing replication. Therefore, in all these contexts, fork rotation could facilitate replication, resulting in local elevated DNA precatenation.
This restriction of fork rotation requires both Tof1 and Csm3, the yeast homologs of Timeless/Tipin. Deletion of TOF1/CSM3 leads to diminished pausing at stable protein–DNA sites (19). Our data suggest that in wild-type cells, Tof1/Csm3 inhibit fork rotation and therefore DNA unwinding at such sites, contributing to pausing. In the absence of Tof1/Csm3 this inhibition is lifted, leading to a more rapid transit of the pause site, using fork rotation to unwind the DNA. Mechanistically, we speculate that Tof1/Csm3 could inhibit fork rotation in either of two ways. Loss of Timeless/Tipin orthologs could disrupt replisome configuration (21, 22, 30). Potentially, the mutant replisome could have a lower resistance to fork rotation than wild-type. Alternatively, Tof1 has been reported to interact with eukaryotic topoisomerase I (31). This interaction could maximize the local concentration of topoisomerase activity at the fork and minimize the need for fork rotation.
Whatever the exact mechanism of rotation inhibition, our data suggest that fork rotation and precatenation are minimized to prevent the generation of DNA damage in the newly replicated sister chromatids. Because yeast fragile sites accumulate both DNA catenation and DNA damage in pathways regulated by Tof1 and Top2 activity, our observations argue that excessive fork rotation and DNA precatenation are closely linked with endogenous replicative damage and chromosome fragility. We hypothesize that this is through one of two pathways. First, the incidence of fork rotation and DNA catenation could actually be a comarker for other aberrant DNA transactions. High levels of topological stress ahead of a fork can lead to fork reversal (32) as well as fork rotation (9). Consistent with this possibility, fork reversal is observed following trapping of Top1 complexes ahead of the fork (33). Potentially, in a population of cells, both reversal and rotation could be taking place within topologically stressed regions, and inappropriate processing of reversed forks would lead to damage. Alternatively, high local levels of fork rotation and DNA precatenation could lead to braiding of the newly replicated sister chromatids (Fig. S9).
Such braiding would inhibit several processes that occur in the immediate wake of the fork. For example, Okazaki fragment maturation could be inhibited by excessive precatenation, consistent with the high levels of PCNA ubiquitylation we observe (34). This model also predicts that intra-S checkpoint signaling and cohesion establishment would be inhibited. Because loss of Tof1/Timeless leads to gaps in the replicated chromosomes, loss of checkpoint signaling, and mitotic cohesion defects (20), we currently favor this possibility. However, further investigation is required to determine the exact nature of the connection between fork rotation, DNA precatenation, and fragile-site instability.
Materials and Methods
Yeast Strains.
Full genotypes are listed in Table S2.
Plasmids.
Plasmids pRS426 and pRS316 have been described previously (13). To construct tRNApRS316, a fragment containing tA(AGC)F, tY(GUA)F1, and tF(GAA)F was produced by gene synthesis and SmaI-cloned into pRS316. 7ARS-YIplac204 and 1ARS-YIplac204 were a gift from J. Diffley (Cancer Research UK, Hertfordshire, United Kingdom) (35). To allow resolution on the same gel, 1ARS-YIplac204 was extended by the AatII-EcoRI fragment of pRS316 (containing the URA3 marker) cloned into AatII-MfeI. For larger plasmids, a 3,067-bp BamHI/EcoRI fragment of pGT60 was cloned into pRS316. An additional 4,400 bp was added by cloning full-length bacterial mukB into NotI/SpeI sites of the 8-kb plasmid.
Media and Cell-Cycle Synchronization.
Spot test and alpha factor release experiments were carried out as described (14). For top2-4 strains, YPD [yeast peptone media + 2% (wt/vol) glucose] was used. Nocodazole at 10 μg/mL was used where indicated.
Assessing Plasmid DNA Catenation.
DNA was purified for resolution and nonradioactive Southern blotting, and detection was carried out as described (13). For catenation 2D gels, the DNA was nicked with either Nb.BsmI or Nb.BsrDI (NEB). DNA was first resolved in a 0.4% agarose (MegaSieve; Flowgen) gel in 1× TBE (Tris/borate/EDTA) at 1.2 V/cm for 13–24 h before being excised and embedded into a 0.8–1.2% (depending on plasmid size) agarose gel and resolved at 2–4.8 V/cm in 1× TBE. Probes were generated from pRS316 including URA3. Images were acquired by ImageQuant LAS 4000 (GE Healthcare) and analyzed using ImageQuant TL software.
Flow Cytometry, Western Blotting, In-Blot Kinase Assay, and Chromatin Immunoprecipitation.
Western blotting and Rad53 kinase assay were carried out as described (14). Antibodies used were anti-H2AP (Abcam; ab15083), anti-PGK1 (Invitrogen; 459250), and anti-PCNA [5E6/2] (Abcam; ab70472). Detection of PCNA ubiquitylation was as described (34), and chromatin immunoprecipitation was carried out as described (36).
An extended description of all methods used in this study is provided in SI Materials and Methods.
SI Materials and Methods
Yeast Strains.
Yeast containing top2-td was derived from W303-1a (MATa ade2-1 ura3-1 his3-11, trp1-1 leu2-3, can1-100) (14); top2-4 cells were derived from ref. 15. Full genotypes are listed in Table S1.
Media and Cell-Cycle Synchronization.
Top2-td cell cultures for alpha factor release experiments were prepared as described previously (2). For plasmid experiments with top2-4 strains, yeast cells were grown in minimal media, 2% glucose selecting for the plasmid (−ura or −trp) to log phase at 25 °C before transferring to YP, +ade, 2% glucose and grown to midlog phase. Cells were then arrested in G1 with 10 μg/mL alpha factor until 90% of cells were unbudded (120 min). The culture was incubated at 37 °C for 1 h and cells were released from the block into YP, +ade, 2% glucose. Time 0 was designated as the time of the addition of the first wash. Nocodazole was added to cultures at 10 μg/mL. Samples were taken at the indicated time points, pelleted, and frozen on dry ice.
Flow Cytometry Analysis.
For FACS analysis, cells were prepared as described previously (14).
DNA Preparation.
DNA was extracted as described in Baxter et al. (13).
Gel Electrophoresis for Detection of Plasmid Catenation.
For catenation 2D gels, the DNA was nicked with either Nb.BsmI or Nb.BsrDI (NEB) according to the manufacturer’s instructions.
Nicked catenanes were separated in the first dimension on a 0.4% agarose (MegaSieve; Flowgen) gel in 1× TBE at 1.2 V/cm for 13–17 h at room temperature. The respective lanes were excised and embedded into a 0.8–1.2% (depending on plasmid size) agarose (MegaSieve; Flowgen) gel and run at 2–4.8 V/cm in 1× TBE (in the cold room if more than 2 V/cm was used). Nonradioactive Southern blotting and detection were carried out as described (3). Blots were probed with either DNA amplified from sequences of pRS316 including the URA3 sequences or from pRS314 including the TRP1 gene as appropriate. Nonsaturating exposures of the blot were acquired by ImageQuant LAS 4000 (GE Healthcare), and densitometry analysis was carried out using ImageQuant TL software. Overexposed images were taken to clearly identify the CatAn = 1 signal, which was often weak in nonsaturating exposures.
Protein Extraction and Analysis.
Whole-protein extracts were prepared by alkaline lysis followed by TCA precipitation. Cells were resuspended in lysis buffer (1.85 M NaOH, 7.5% β-mercaptoethanol) and incubated on ice for 15 min. Proteins were precipitated with TCA (6.4% final concentration) on ice for 10 min. Precipitates were resuspended in HU buffer (8 M urea, 5% SDS, 200 mM Tris⋅HCl, pH 6.8, 1 mM EDTA, 1.5% DTT, 0.1% bromophenol blue) and separated by 15% SDS/PAGE. Phosphorylation of S129 of H2A and PGK1 expression were detected using an antibody against H2AP (Abcam; ab15083) and anti-PGK1 (Invitrogen; 459250). Detection of PCNA ubiquitylation was as described in Karras and Jentsch (34) using an anti-PCNA antibody [5E6/2] (Abcam; ab70472).
Rad53 Kinase Assay.
In situ autophosphorylation assay was carried out as described (26).
Chromatin Immunoprecipitation of H2A S129 Phosphorylation.
Synchronized cultures were fixed with 1% formaldehyde for 10 min at room temperature before immunoprecipitation with an anti–phospho-H2A S129 antibody (Abcam; ab15083). ChIP assays were performed essentially as in ref. 36. Immunoprecipitated and input DNA was quantified by quantitative PCR. Enrichment was analyzed at tRNA loci tI(AAU)N1 and tA(UGC)L and within the euchromatic loci ASI1 and HXT7 (primer sequences are available upon request). ChIP signal was normalized to amplification from input DNA and expressed relative to wild-type. Data shown are the mean enrichment ±1 SD.
Acknowledgments
We thank members of the Genome Damage and Stability Centre for discussions and critical reading of the manuscript. This work was funded by the Royal Society (J.B.), Biotechnology and Biological Sciences Research Council United Kingdom (S.A.S.), and Cancer Research UK (A.L.C. and J.A.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1505356112/-/DCSupplemental.
References
- 1.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 2.Bermejo R, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21(15):1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell. 2012;45(6):710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Champoux JJ, Been MD. Topoisomerases and the swivel problem. In: Alberts B, editor. Mechanistic Studies of DNA Replication and Genetic Recombination. Academic; New York: 1980. pp. 809–815. [Google Scholar]
- 5.Baxter J. “Breaking up is hard to do”: The formation and resolution of sister chromatid intertwines. J Mol Biol. 2015;427(3):590–607. doi: 10.1016/j.jmb.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21(1):103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 7.Sundin O, Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: Dissection of the final stages of SV40 DNA replication. Cell. 1981;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- 8.Hiasa H, Marians KJ. Topoisomerase III, but not topoisomerase I, can support nascent chain elongation during theta-type DNA replication. J Biol Chem. 1994;269(51):32655–32659. [PubMed] [Google Scholar]
- 9.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94(6):819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 10.Lucas I, Germe T, Chevrier-Miller M, Hyrien O. Topoisomerase II can unlink replicating DNA by precatenane removal. EMBO J. 2001;20(22):6509–6519. doi: 10.1093/emboj/20.22.6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proc Natl Acad Sci USA. 2001;98(15):8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez-Robles ML, et al. Interplay of DNA supercoiling and catenation during the segregation of sister duplexes. Nucleic Acids Res. 2009;37(15):5126–5137. doi: 10.1093/nar/gkp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxter J, et al. Positive supercoiling of mitotic DNA drives decatenation by topoisomerase II in eukaryotes. Science. 2011;331(6022):1328–1332. doi: 10.1126/science.1201538. [DOI] [PubMed] [Google Scholar]
- 14.Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30(6):790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 15.Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41(2):553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 16.Ivessa AS, et al. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell. 2003;12(6):1525–1536. doi: 10.1016/s1097-2765(03)00456-8. [DOI] [PubMed] [Google Scholar]
- 17.Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89(7):3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brewer BJ, Fangman WL. Initiation preference at a yeast origin of replication. Proc Natl Acad Sci USA. 1994;91(8):3418–3422. doi: 10.1073/pnas.91.8.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18(10):3894–3902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leman AR, Noguchi E. Local and global functions of Timeless and Tipin in replication fork protection. Cell Cycle. 2012;11(21):3945–3955. doi: 10.4161/cc.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424(6952):1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 22.Errico A, et al. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO J. 2009;28(23):3681–3692. doi: 10.1038/emboj.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19(5):699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 24.Putnam CD, Hayes TK, Kolodner RD. Specific pathways prevent duplication-mediated genome rearrangements. Nature. 2009;460(7258):984–989. doi: 10.1038/nature08217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci USA. 2006;103(48):18143–18147. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellicioli A, et al. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18(22):6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nat Struct Mol Biol. 2010;17(3):299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeppsson K, et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 2014;10(10):e1004680. doi: 10.1371/journal.pgen.1004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bermejo R, et al. The replication checkpoint protects fork stability by releasing transcribed genes from nuclear pores. Cell. 2011;146(2):233–246. doi: 10.1016/j.cell.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho W-H, et al. Human Tim-Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci USA. 2013;110(7):2523–2527. doi: 10.1073/pnas.1222494110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park H, Sternglanz R. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast. 1999;15(1):35–41. doi: 10.1002/(SICI)1097-0061(19990115)15:1<35::AID-YEA340>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 32.Postow L, et al. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem. 2001;276(4):2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 33.Ray Chaudhuri A, et al. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol. 2012;19(4):417–423. doi: 10.1038/nsmb.2258. [DOI] [PubMed] [Google Scholar]
- 34.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141(2):255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka S, Diffley JF. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 2002;16(20):2639–2649. doi: 10.1101/gad.1011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers AL, et al. The INO80 chromatin remodeling complex prevents polyploidy and maintains normal chromatin structure at centromeres. Genes Dev. 2012;26(23):2590–2603. doi: 10.1101/gad.199976.112. [DOI] [PMC free article] [PubMed] [Google Scholar]