Significance
The antibacterial effectiveness of penicillins and cephalosporins is increasingly challenged by bacterial resistance, highlighting the increasing clinical importance of the carbapenems as the most potent, broad-spectrum class of β-lactam antibiotics. The carbapenems are produced commercially by total synthesis, rather than by lower-cost fermentation methods. The development of chemoenzymatic or semisynthetic means of manufacture requires elucidation of at least four central, undefined steps in the biosynthesis of thienamycin. The cobalamin-dependent radical S-adenosylmethionine (RS) superfamily enzyme ThnK performs two sequential methyl transfers in a stereodefined manner. This enzyme activity identifies critical intermediates in carbapenem biosynthesis, more clearly defining the enigmatic central steps of the pathway and exemplifying an unprecedented reaction sequence among RS enzymes.
Keywords: β-lactam antibiotics, carbapenem, radical SAM, cobalamin, methylase
Abstract
Despite their broad anti-infective utility, the biosynthesis of the paradigm carbapenem antibiotic, thienamycin, remains largely unknown. Apart from the first two steps shared with a simple carbapenem, the pathway sharply diverges to the more structurally complex members of this class of β-lactam antibiotics, such as thienamycin. Existing evidence points to three putative cobalamin-dependent radical S-adenosylmethionine (RS) enzymes, ThnK, ThnL, and ThnP, as potentially being responsible for assembly of the ethyl side chain at C6, bridgehead epimerization at C5, installation of the C2-thioether side chain, and C2/3 desaturation. The C2 substituent has been demonstrated to be derived by stepwise truncation of CoA, but the timing of these events with respect to C2–S bond formation is not known. We show that ThnK of the three apparent cobalamin-dependent RS enzymes performs sequential methylations to build out the C6-ethyl side chain in a stereocontrolled manner. This enzymatic reaction was found to produce expected RS methylase coproducts S-adenosylhomocysteine and 5′-deoxyadenosine, and to require cobalamin. For double methylation to occur, the carbapenam substrate must bear a CoA-derived C2-thioether side chain, implying the activity of a previous sulfur insertion by an as-yet unidentified enzyme. These insights allow refinement of the central steps in complex carbapenem biosynthesis.
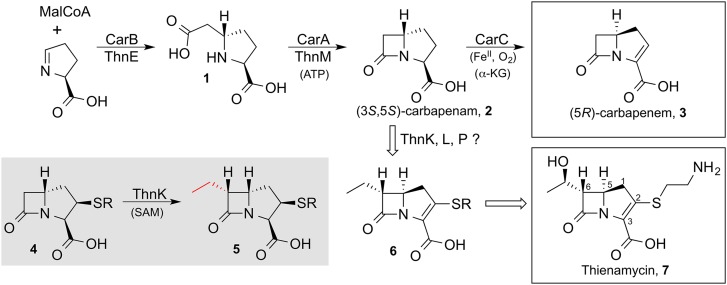
Bacterial resistance to penicillin and cephalosporin has placed increased clinical reliance on the carbapenem class of β-lactam antibiotics, notably synthetic variants of the naturally occurring thienamycin (Fig. 1, thienamycin 7) (1). Approximately 50 “complex” carbapenems are known, distinguished by a vinylthio substituent at C2 and a short alkyl chain at C6. These natural products have been isolated from actinomyces, whereas the lone “simple” carbapenem, carbapenem-3-carboxylic acid 3, is known from an enterobacterium, Pectobacterium carotovorum (1). Despite the evolutionary distance between the producers, the first two biosynthetic steps to each are shared (2). Thus, CarB encoded in the latter and ThnE from Streptomyces cattleya, and, correspondingly, CarA and ThnM give (3S,5S)-carbapenam 2 (Fig. 1). At this point, the two pathways diverge in a striking manner, with the simple carbapenem 3 produced by both configurational inversion at the bridgehead and C2/3 desaturation mediated by CarC (3). In contrast, the path to thienamycin 7, and likely to all complex carbapenems, involves an entirely separate sequence of transformations.
Fig. 1.
Biosynthesis of thienamycin and activity of ThnK. Two enzymes, ThnE and ThnM, form the (3S,5S)-carbapenam 2, which is then acted on by other enzymes, likely including ThnK, ThnL, and ThnP, to form thienamycin. Correspondingly, CarB and CarA can also form 2, which CarC can convert to the simple carbapenem 3. The demonstrated activity of ThnK is highlighted in gray.
Apart from the identification of ThnR, ThnH, and ThnT in the stepwise truncation of CoA to the C2-cysteamine side chain of thienamycin (4), the timing of these tailoring events with respect to the introduction of the C2 substituent onto the carbapenam(em) nucleus, alkylation at C6, ring inversion, and desaturation remain shrouded in mystery, although compound 6 is hypothesized to be an intermediate. Unique to the complex carbapenems are three apparent cobalamin-dependent radical S-adenosylmethionine (RS) enzymes, ThnK, ThnL, and ThnP (all annotated as class B RS methylases) (5), which likely play critical roles in the central undefined steps of thienamycin biosynthesis.
RS enzymes are commonly identified by a conserved Cx3Cx2C motif, typically responsible for coordinating a [4Fe-4S] cluster (6), which enables this protein family to perform more than 40 biochemical transformations identified to date, including sulfur insertion and methylation, which are relevant to the thienamycin biosynthesis (7). The three thienamycin RS enzymes also contain N-terminal extensions thought to be cobalamin (B12)-binding domains, apparently in keeping with early observations that CoII and cobalamin increase carbapenem titers in cultures of producing strains (8). Indeed, the low turnover of RS enzymes may account for the fact that thienamycin cannot be produced by cost-effective fermentation methods, and thus all carbapenems for clinical use are manufactured synthetically (1).
Although the carbapenem RS enzymes likely perform several intermediate biosynthetic steps and potentially use unprecedented chemistry, the challenge of deciphering their activities is exacerbated by their intrinsic instability, unknown substrates, and multiple possible reactions. We have taken a first step in illuminating the obscure center of the thienamycin biosynthetic pathway by identifying substrates for ThnK and establishing that this enzyme performs two consecutive methylations to generate the C6-ethyl side chain stereospecifically (Fig. 1). Although other RS cobalamin-dependent enzymes that act on carbon have been investigated previously (9–12), to our knowledge this is the first in vitro characterization of a RS methylase acting sequentially to form a side chain.
Results and Discussion
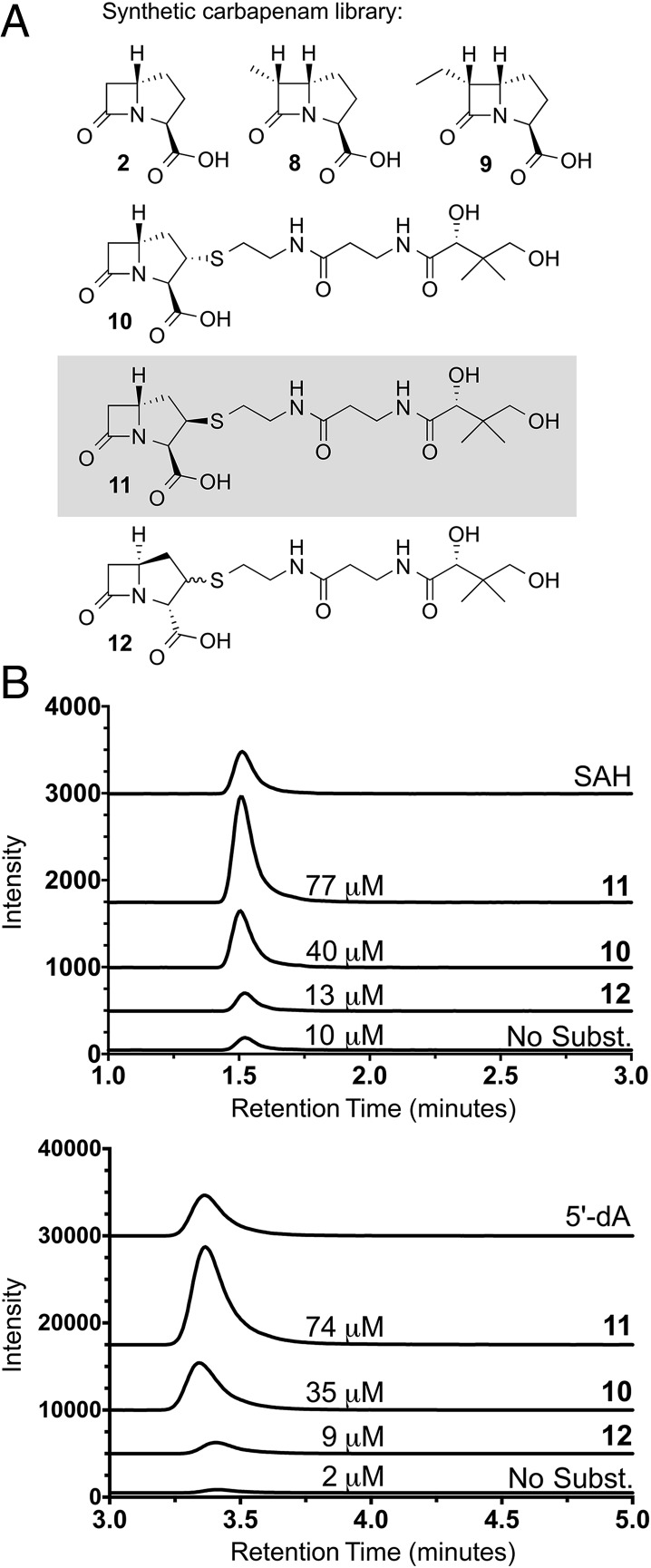
A library of candidate substrates and product standards was prepared, including the simple (3S,5S)-carbapenam 2 and related C2- and C6-substituted derivatives 8–12. The library was biased toward structures with the (3S,5S) stereochemistry, matching that of the ThnM product 2, and those bearing the 6R configuration (8, 9), matching that of thienamycin (Fig. 2A). A pantetheinyl moiety was selected for the side chain at C2 (10–12), in keeping with the OA-6129 series of metabolites that has been isolated from carbapenem producers (13).
Fig. 2.
Carbapenam synthetic library and detection and quantification of SAM-derived products of ThnK. (A) Compounds synthesized to probe ThnK activity. (B) LC-MS/MS detection of enzymatic SAM-derived coproducts SAH (385.4→136 m/z) and 5′-dA (252.1→136 m/z). The EICs for the transitions of SAH and 5′-dA are shown for each reaction. The reaction with compound 11 yielded the highest levels of these SAM-derived coproducts. Quantified levels of SAH and 5′-dA are shown for each reaction.
Work began with the best behaved of these three RS enzymes. ThnK was cloned from S. cattleya under experimental conditions that yielded moderate levels of soluble protein when expressed with a C-terminal His6-tag in Escherichia coli Rosetta 2(DE3). Because ThnK was expected to use both an iron-sulfur (Fe/S) cluster and cobalamin, protein production was conducted in ethanolamine-M9 medium, which facilitates uptake of externally supplied hydroxocobalamin (HOCbl) into E. coli, and with concurrent expression of the Azotobacter vinelandii isc operon, encoded on plasmid pDB1282. ThnK was purified under strictly anaerobic conditions. Its UV-visible spectrum revealed a 420-nm shoulder (SI Appendix, Fig. S1) typical of a bound Fe/S cluster. The relative stoichiometry of iron and sulfide bound by the as-isolated protein was determined to be 7.4 ± 1.4 and 3.7 ± 0.8 per polypeptide, respectively, consistent with the presence of a [4Fe-4S] cluster. Apparent excess iron has been observed with other RS enzymes (11, 14), and was similarly seen in an alanine variant control.
To determine whether any of our library compounds was a substrate for ThnK, we developed an LC-MS analytical screen whereby enzymatic reactions were monitored for appearance of the expected RS methylase coproducts, S-adenosylhomocysteine (SAH) and 5′-deoxyadenosine (5′-dA) (15, 16). Initial screens were conducted with ThnK, S-adenosylmethionine (SAM), and potential substrates in the presence of methyl viologen and NADPH as the Fe/S cluster reductants (10). The (3S,5S)-carbapenam and related C6-methyl and ethyl derivatives (2, 8, and 9) gave nearly background (no substrate control) levels of SAH and 5′-dA (SI Appendix, Fig. S2). In contrast, however, these SAM-derived coproducts accumulated when ThnK was incubated with C2-substituted carbapenams, suggesting that the full methyl transfer reaction(s) had occurred with these substrates (Fig. 2B). We also examined alternative Fe/S cluster reductants, including dithionite and the flavodoxin/flavodoxin reductase/NADPH reducing system, but these did not yield improved activity (SI Appendix, Fig. S3).
To assess enzymatic turnover, we quantified the amounts of SAH and 5′-dA by LC-MS/MS (Fig. 2B). Compounds bearing the now (3R,5R) stereochemistry with a pantetheine side chain at C2 gave the greatest levels of turnover. Therefore, we tested each C2 diastereomer (compound 10 or 11) individually. The (2R,3R,5R)-carbapenam 11 yielded approximately twofold greater amounts of SAM coproducts. These stereochemical preferences were found to be reproducible (SI Appendix, Fig. S4), and SAH and 5′dA were present in approximately equimolar amounts in each reaction.
To test the role of the [4Fe-4S] cluster in catalysis, we generated a variant of ThnK in which the conserved cluster binding motif Cx3Cx2C was changed from three cysteine residues to three alanine residues. The variant protein was prepared as described above for wild-type (WT) ThnK and incubated with substrate 11. The inactivated enzyme did produce low levels of SAH, but no detectable 5′-dA (SI Appendix, Fig. S5), consistent with reliance of the WT enzyme on a functional Fe/S cluster. ThnK was also expressed in M9 medium lacking HOCbl. The purified protein with substrate 11 did not produce SAH or 5′dA; however, enzymatic activity could be rescued if cobalamin was added to the reaction mixture (SI Appendix, Fig. S6).
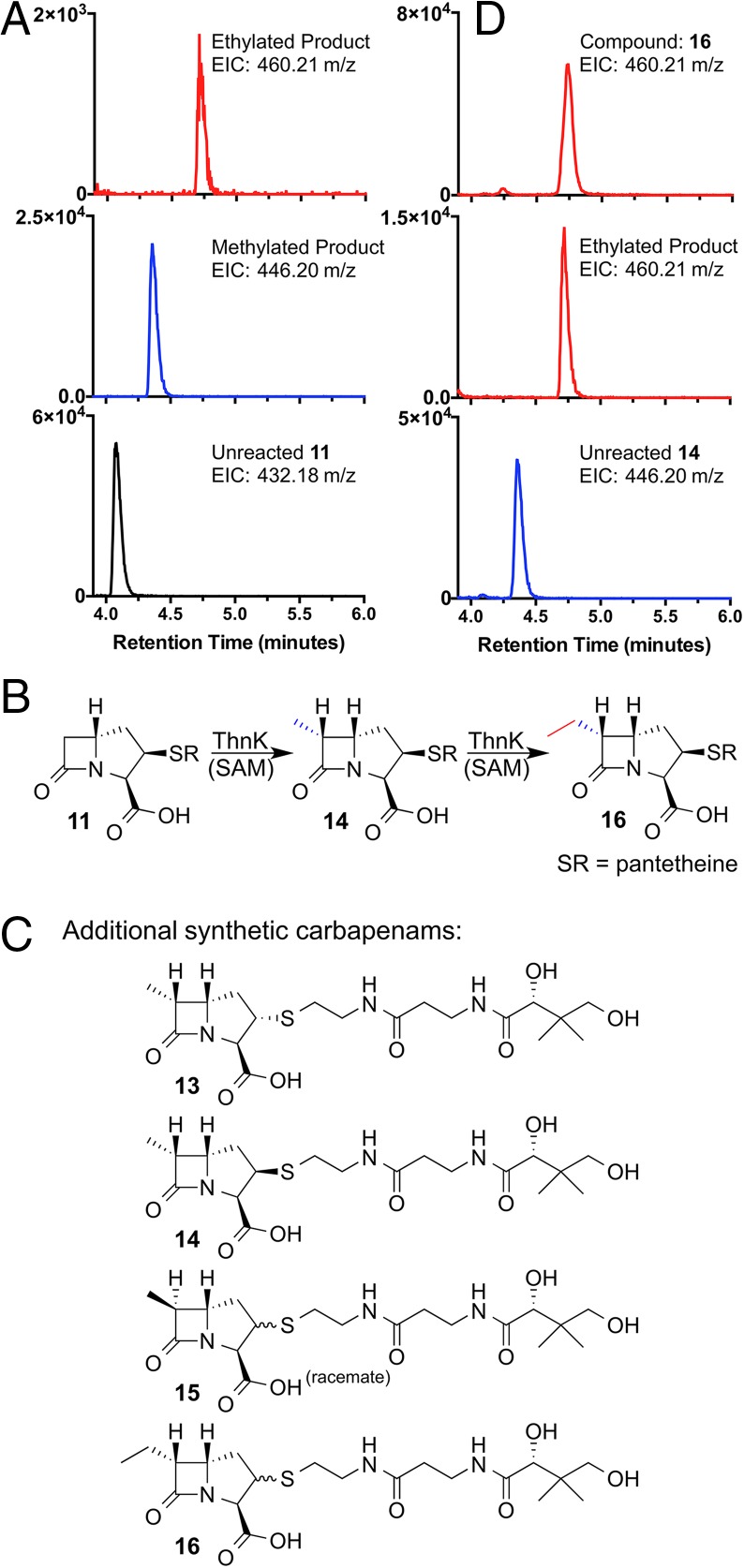
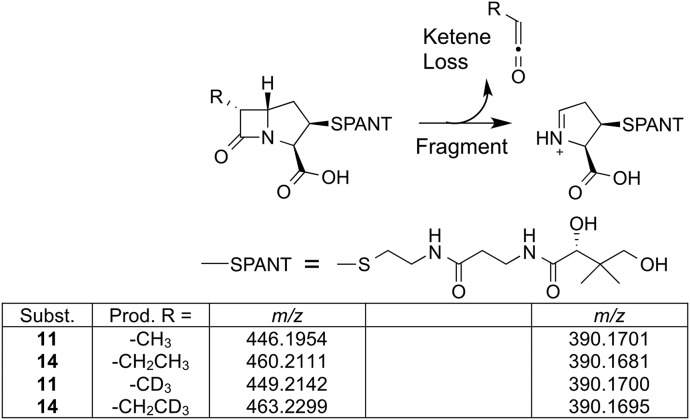
We next sought to determine how the substrate was transformed during the reaction with ThnK. Assays with 11 were analyzed by ultraperformance liquid chromatography-high resolution mass spectrometry (UPLC-HRMS). Exact masses were obtained matching methylated (calc: m/z 446.1961; found: m/z 446.1954) and twice-methylated (calc: m/z 460.2117; found: m/z 460.2104) modifications of the substrate. The extracted-ion chromatograms (EICs) for these masses showed retention times that were consistent with increasing hydrophobicity relative to the substrate (Fig. 3A). A highly diagnostic β-lactam mass fragment (calc: m/z 390.1699) corresponding to the loss of a ketene from cleavage of the β-lactam ring (17) was observed (found: m/z 390.1701) (Fig. 4), which was further consistent with methylation at C6.
Fig. 3.
ThnK product detection and additional synthetic compounds. (A) UPLC-HRMS detection of carbapenams from the ThnK reaction with compound 11. The bottom, middle, and top traces show the EICs (m/z ± 0.05) for unreacted substrate (432.18 m/z), methylated product (446.20 m/z), and ethylated product (460.21 m/z), respectively. (B) Summary of ThnK activity. (C) Additional compounds synthesized to analyze the ThnK reaction. Note: Compound 15 is a racemate and diastereomeric at C2; compounds 13 and 14 are diastereomeric at C2, but enriched for the diastereomer shown (approximately 3:1 and 3:2, respectively); and compound 16 is diastereomeric at C2. (D) UPLC-HRMS detection of carbapenams from the ThnK reaction with compound 14 compared with synthetic standard 16. The bottom, middle, and top traces show the EICs (m/z ± 0.05) for unreacted substrate 14 (446.20 m/z), ethylated product (460.21 m/z), and C6-ethyl synthetic standard (460.21 m/z), respectively. The ethylated product of 11 (A) and the ethlyated product of 14 match the synthetic standard 16 (D).
Fig. 4.
Detection of β-lactam parent and fragment ions. UPLC-HRMS was used to detect carbapenam parent and fragment ions from ThnK reactions with 11 or 14. Characteristic MS fragmentation of β-lactam–containing compounds results in loss of a substituted (R) ketene as depicted. Using d3-SAM or natural abundance SAM in the enzymatic reaction affects the observed m/z of the parent ion, but not the observed m/z of the fragmentation ion, consistent with methylation occurring at C6.
To establish SAM as the methyl donor, we performed additional ThnK reactions with S-adenosyl-l-[methyl-d3]methionine (d3-SAM). The corresponding M+3 m/z shift (Fig. 4 and SI Appendix, Fig. S7) was observed for the methylated product, and an M+5 m/z shift was seen for the twice-methylated product (SI Appendix, Fig. S7), fully in accordance with formation of the ethyl side chain at C6 in thienamycin by successive methyl transfers.
To secure the stereochemistry of the first methylation, we carried out the synthesis of a set of C6-methylcarbapenams (compounds 13–15; Fig. 3C) and an assay with ThnK. Whereas compounds 13 and 15 displayed trace activity and no measurable activity, respectively, compound 14 afforded unequivocal production of SAH and 5′-dA (SI Appendix, Fig. S8). This result underscores the importance of the stereochemical relationship between the C3-carboxylate and the pantetheinyl side chain, and confirms that the C6-methyl is attached in the R-configuration, as occurs in thienamycin. An ultraperformance liquid chromatography-electrospray mass spectrometry analysis showed that the ethylated product of 14 (resulting from a single methyl transfer) matched the exact mass and retention time of the ethylated product of compound 11 (Fig. 3 A and D). In further support of the twice-methylated compound, the (6R)-ethyl standard 16 was synthesized and determined to match the aforementioned products in terms of retention time, exact mass, and fragmentation pattern, as expected (Fig. 3 A and D). The observed activity of ThnK is summarized in Fig. 3B.
Mechanistically, ThnK resembles GenK, a cobalamin-dependent RS C-methylase in gentamicin biosynthesis (10), with both producing near-equimolar amounts of SAH and 5′-dA during catalysis. Early labeling experiments established that the C6-ethyl side chain of thienamycin is derived from methionine (18). A labeled carbon is also found in the C6-hydroxymethyl side chain of northienamycin, a minor cometabolite of thienamycin, suggesting a singly-methylated intermediate in the formation of the ethyl side chain (18). The present in vitro analysis of ThnK agrees with these early experiments. An experiment incorporating chiral [1H, 2H, 3H-methyl]-l-methionine demonstrated that the absolute configuration of the terminal methyl in the C6 side chain is largely retained, consistent with an overall methyl transfer process involving two inversions and a methyl-cobalamin intermediate (19). The B12-dependence of ThnK reactions established above supports the role of a methylcobalamin (MeCbl) intermediate, as has been implicated in other methyltransferases, including TsrM (9) and GenK (10). Although the tryptophan methylase TsrM may use a similar intermediate, the enzyme likely is mechanistically distinct from ThnK and GenK, because it does not produce detectable levels of 5′-dA (9).
The catalysis of consecutive methylations observed for ThnK likely occurs elsewhere in nature. Within the carbapenem family, the carpetimycins have three carbons in their C6 side chain (8), suggesting that a ThnK ortholog may be responsible for three methyl transfers. The natural product pactamycin has traditionally been compared with thienamycin, because both possess a hydroxyethyl side chain (18); however, a recent genetic knockout study has suggested that more than one enzyme takes part in attaching the ethyl group (20). Although sequential methylations appear not to occur by a single enzyme in pactamycin biosynthesis, other RS enzymes, including Swb9 (21) and PoyB or PoyC (22), in quinomycin and polytheonamide biosynthesis, respectively, have been implicated in multiple methylations of a single substrate chain.
These seemingly related biochemical examples notwithstanding, the data reported here establish notable catalytic roles for ThnK. It methylates in two distinct chemical environments (α to the β-lactam ring and on a methyl group). The second methyl transfer presumably involves a primary radical intermediate, a particularly high-energy species. By identifying favorable substrates for ThnK and elucidating its activity, we have gained additional clues about the unknown central steps in thienamycin biosynthesis as well. With formation of the ethyl side chain accounted for, ThnL and ThnP can provisionally be assigned nonmethyltransferase functions, potentially distinct chemistry for the RS-cobalamin family, given that both proteins are essential for carbapenem production (23, 24). Because ThnK activity was observed only when a C2 side chain was present (compounds 10–12 vs. 2, 8, and 9), we deduce that C2-thioether formation, likely mediated by ThnL or ThnP, precedes C6-methylation in the pathway. The stereochemical preference for (2R)-pantetheinyl carbapenams 11 and 14 indicates that the thiol is likely introduced in the 2-exo configuration. Furthermore, the observation of activity with substrates matching the (3S,5S)-carbapenam 2 at C5 (compounds 10, 11, and 14) suggests that bridgehead epimerization occurs after C6-methylation (ethylation). Consequently, C2-thiol attachment, C6-alkylation, and C5-epimerization likely occur in that order. The orchestration of these biosynthetic events with C2/3-desaturation and maturation of the C2-cysteamine will be addressed in due course.
Materials and Methods
General methods are presented in SI Appendix.
Cloning and Expression of S. cattleya ThnK.
The thnK gene was PCR amplified from genomic DNA of S. cattleya (NRRL8057) using the following primers: 5′-CAAGACATATGACCGTCCCCGCCGCGC-3′, 5′-TATCTCGAGCCGCTGCTCGGTCAGGACGGG-3′. The thnK PCR product was digested using NdeI and XhoI and ligated into the vector pET29b (Novagen) to obtain the C-terminal 6-His construct of thnK. The plasmid was sequence-verified. The pET29b:thnK was transformed into E. coli Rosetta 2(DE3) for expression of ThnK as a C-terminal His6 construct. The thnK Rosetta strain was also transformed with the vector pDB1282, containing the ISC operon with arabinose induction (25). The resulting strain was grown in 4 × 3.5 L of ethanolamine-M9 medium (26), which included HOCbl and supplemented with FeCl3 (∼0.14 mM), at 37 °C and shaken at 180 rpm. When the optical density of the cultures reached 0.3 at 600 nm, 25 μM FeCl3, 150 μM cysteine, and 0.1% arabinose were added. When the optical density of the fermentation reached 0.4–0.6 (8–10 h), the cultures were chilled on ice for ∼45 min, after which an additional 25 μM FeCl3, 150 μM cysteine, and 0.1% arabinose were added. ThnK expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside, and the cultures were shaken at 180 rpm overnight at 19 °C. Cells were collected by centrifugation at 4,000 × g for 12 min, then frozen on liquid nitrogen and stored for later use.
Anaerobic Purification of ThnK.
Enzyme purification was performed in an anaerobic chamber (Coy). Frozen cells were resuspended in lysis buffer (10% glycerol, 300 mM KCl, 50 mM Hepes pH 8.0, 10 mM β-mercaptoethanol, and 2–5 mM imidazole) along with lysozyme (1 mg/mL final volume), PMSF (1 mM), DNase (∼0.1 mg/mL final volume), and optional HOCbl (∼0.4 mg/g cell paste). Cells were lysed by sonication, and insoluble material was removed by centrifugation at 50,000 × g for 1 h at 4 °C. Soluble ThnK was allowed to bind to preequilibrated TALON CoII resin (Clontech) and was then purified by immobilized metal-affinity chromatography. The resin was washed with 5 mM and 10 mM imidazole in lysis buffer. The protein was eluted using 250 mM imidazole in the lysis buffer. The enzyme was concentrated using an Amicon centrifugal filter device (Millipore) with a 10-kDA molecular weight limit. Using a PD-10 column (GE Biosciences), the protein was transferred into assay buffer (10% glycerol, 300 mM KCl, 50 mM Hepes pH 7.5, and 10 mM β-mercaptoethanol).
ThnK Protein Analysis.
The concentration of ThnK was determined using the Bradford assay (27) with a Varian Cary 50 Bio UV/visible spectrophotometer using Coomassie Protein Assay Reagent (Thermo Scientific). A concentration correction factor was obtained by amino acid analysis (Molecular Structure Facility, University of California Davis). The amount of iron bound by ThnK was determined by established methods (28, 29), and the amount of sulfide was determined as reported previously (30).
ThnK Activity Assays.
ThnK was mixed with 50–100 mM Hepes pH 7.5, 200 mM KCl, 1 mM SAM or d3-SAM, 1 mM methyl viologen, 4 mM NADPH, and 1 mM substrate. Initial activity screens also included 0.5 mM HOCbl and 0.5 mM MeCbl, although the addition of cobalamin for activity was later deemed unnecessary. Assays of potential substrates lacking a C2 substituent also included 1 mM cysteine and 1 mM CoA. When used, dithionite (3 mM) or flavodoxin (90 μM), flavodoxin reductase (9 μM), and NADPH (1 mM) were supplemented at the indicated concentrations in place of methyl viologen and NADPH (default reductant unless noted otherwise). Assays were conducted anaerobically at room temperature and were typically run for 1–2 h.
ThnK SAH and 5′-dA Quantification Assays.
ThnK (50 μM final concentration) was mixed with 100 mM Hepes pH 7.5, 200 mM KCl, 1 mM SAM or d3-SAM, 1 mM methyl viologen, 4 mM NADPH, and 1 mM substrate. Assays were run anaerobically for 2 h and 10 min at room temperature.
No-Cobalamin ThnK Expression, Purification, and Activity.
The ThnK WT Rosetta strain was grown in standard M9 medium lacking cobalamin but supplemented with FeCl3 (∼0.14 mM) and thiamine (0.074 mM). Expression and purification of the no-cobalamin ThnK was conducted as described for the WT protein, but no HOCbl was added during purification. Assays were conducted with 50 μM ThnK and 1 mM HOCbl or 1 mM MeCbl when used, and were otherwise as described for ThnK from ethanolamine medium.
Knockout of Cx3Cx2C Motif in ThnK and Variant Protein Expression, Purification, and Activity.
The pET29b:thnK plasmid served as a template for a two-step PCR method that used the following primers: 5′-CGC GGC GCC CCC TAC TCG GCT GCC TTC GCC GAC TGG-3′ and 5′-CCA GTC GGC GAA GGC AGC CGA GTA GGG GGC GCC GCG-3′. The C206A C210A C213A PCR product was digested with NdeI and XhoI and ligated into the pET29b vector. The resulting plasmid was sequenced and found to be without error. The mutant thnK vector was transformed into an E. coli Rosetta 2(DE3) strain containing the pDB1282 plasmid. Expression, purification, and assays of the mutant ThnK (85 μM final concentration) were conducted as described for the WT protein.
UPLC-MS Analysis of Carbapenams.
UPLC-MS detection of carbapenams was conducted on a Waters Acquity H-Class UPLC system equipped with a Waters Acquity BEH UPLC column packed with an ethylene-bridged hybrid C18 stationary phase (2.1 mm × 50 mm; 1.7 μm), followed by a multiwavelength UV-Vis diode array detector, coupled with accurate mass analysis by a Waters Xevo-G2 Q-ToF ESI mass spectrometer.
Mobile phase: 0–1 min 100% water + 0.1% formic acid, 1–7.5 min up to 80% acetonitrile + 0.1% formic acid, 7.5–8.5 min isocratic 80% acetonitrile + 0.1% formic acid, 8.5–10 min 100% water + formic acid. The flow rate was 0.3 mL/min. The first minute of eluant was discarded before MS analysis.
LC-MS Analysis of SAH and 5′-dA.
LC-MS detection of SAH and 5′-dA was performed with an Agilent 1200 HPLC system equipped with an autosampler and an Agilent ZORBAX Rapid Resolution Extend-C18 column (4.6 mm × 50 mm; 1.8 μm particle size) and coupled to an Agilent 6410 QQQ mass spectrometer. Assay mixtures were quenched with H2SO4, and insoluble material was pelleted before analysis.
Method 1 (compounds 2, 8, and 9): The methodology was similar to that reported previously (31). Mobile phase: The initial conditions included 95% solvent A (40 mM aqueous ammonium acetate, pH 6.0, with glacial acetic acid and 5% methanol) and 5% solvent B (methanol), 0.5–5 min up to 13% B, 5–6.5 min up to 27% B, 6.5–7.0 min up to 53% B, 7.0–7.5 min isocratic 53% B, 7.5–8 min down to 5% B, and 8.0–10.0 min reequilibration at 5% B.
Method 2 (compounds 10–15): Mobile phase: The initial conditions included 90.5% solvent A (0.1% formic acid, pH 2.6) and 9.5% solvent B (methanol), 0–0.2 min 9.5% B, 0.2–3.3 min up to 9.8% B, 3.3–4.1 min up to 30% B, 4.1–4.7 min up to 60% B, 4.7–5.1 min isocratic 60% B, 5.1–5.5 min down to 9.5% B, and 5.5–8.7 min isocratic 9.5% B.
Data collection and analysis were conducted with the associated MassHunter software package (Agilent). Detection of SAH and 5′-dA was performed using electrospray ionization in the positive mode (ESI+) with multiple reaction monitoring. The masses used for the detection of SAH and 5′-dA are shown in SI Appendix, Table S1. Quantification of SAH and 5′d-A levels was conducted using MassHunter software with appropriate standard curves and a tyrosine internal standard. The mass transition from the parent ion to product ion 1 (SI Appendix, Table S1) was used for quantification.
Synthesis of Substrates.
The (3S,5S)-carbapenam (2) was synthesized as described previously (32). Compounds 8, 9, 13, 14, 16 (33), 10–12 (34, 35), and 15 (36, 37) were derived from previous synthetic strategies as indicated. Details are presented in SI Appendix.
Supplementary Material
Acknowledgments
We thank R. F. Li and M. Freeman for initial thnK cloning and C. Hastings for preparation of the (3S,5S)-carbapenam. We are also grateful to I. P. Mortimer for high-resolution MS data and J. Tang for assistance with high-temperature NMR experiments. This work was supported by National Institutes of Health Grants AI01437 and GM103268.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1508615112/-/DCSupplemental.
References
- 1.Coulthurst SJ, Barnard AML, Salmond GPC. Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Microbiol. 2005;3(4):295–306. doi: 10.1038/nrmicro1128. [DOI] [PubMed] [Google Scholar]
- 2.Bodner MJ, et al. Definition of the common and divergent steps in carbapenem β-lactam antibiotic biosynthesis. ChemBioChem. 2011;12(14):2159–2165. doi: 10.1002/cbic.201100366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Stapon A, Blanchfield JT, Townsend CA. Three unusual reactions mediate carbapenem and carbapenam biosynthesis. J Am Chem Soc. 2000;122(1483):9296–9297. [Google Scholar]
- 4.Freeman MF, Moshos KA, Bodner MJ, Li R, Townsend CA. Four enzymes define the incorporation of coenzyme A in thienamycin biosynthesis. Proc Natl Acad Sci USA. 2008;105(32):11128–11133. doi: 10.1073/pnas.0804500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, van der Donk WA, Liu W. Radical-mediated enzymatic methylation: A tale of two SAMS. Acc Chem Res. 2012;45(4):555–564. doi: 10.1021/ar200202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broderick JB, Duffus BR, Duschene KS, Shepard EM. Radical S-adenosylmethionine enzymes. Chem Rev. 2014;114(8):4229–4317. doi: 10.1021/cr4004709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frey PA, Hegeman AD, Ruzicka FJ. The radical SAM superfamily. Crit Rev Biochem Mol Biol. 2008;43(1):63–88. doi: 10.1080/10409230701829169. [DOI] [PubMed] [Google Scholar]
- 8.Williamson J. The biosynthesis of thienamycin and related carbapenems. Crit Rev Biotechnol. 1986;4(1):111–131. [Google Scholar]
- 9.Pierre S, et al. Thiostrepton tryptophan methyltransferase expands the chemistry of radical SAM enzymes. Nat Chem Biol. 2012;8(12):957–959. doi: 10.1038/nchembio.1091. [DOI] [PubMed] [Google Scholar]
- 10.Kim HJ, et al. GenK-catalyzed C-6′ methylation in the biosynthesis of gentamicin: Isolation and characterization of a cobalamin-dependent radical SAM enzyme. J Am Chem Soc. 2013;135(22):8093–8096. doi: 10.1021/ja312641f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen KD, Wang SC. Initial characterization of Fom3 from Streptomyces wedmorensis: The methyltransferase in fosfomycin biosynthesis. Arch Biochem Biophys. 2014;543:67–73. doi: 10.1016/j.abb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C, et al. Delineating the biosynthesis of gentamicin x2, the common precursor of the gentamicin C antibiotic complex. Chem Biol. 2015;22(2):251–261. doi: 10.1016/j.chembiol.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okabe M, et al. Studies on the OA-6129 group of antibiotics, new carbapenem compounds, I: Taxonomy, isolation and physical properties. J Antibiot (Tokyo) 1982;35(10):1255–1263. doi: 10.7164/antibiotics.35.1255. [DOI] [PubMed] [Google Scholar]
- 14.Allen KD, Wang SC. Spectroscopic characterization and mechanistic investigation of P-methyl transfer by a radical SAM enzyme from the marine bacterium Shewanella denitrificans OS217. Biochim Biophys Acta. 2014;1844(12):1–10. doi: 10.1016/j.bbapap.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan F, et al. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132(11):3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bauerle MR, Schwalm EL, Booker SJ. Mechanistic diversity of radical S-adenosylmethionine (SAM)-dependent methylation. J Biol Chem. 2015;290(7):3995–4002. doi: 10.1074/jbc.R114.607044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitscher LA, Showalter HD, Shirahata K, Foltz RL. Chemical-ionization mass spectrometry of beta-lactam antibiotics. J Antibiot (Tokyo) 1975;28(9):668–675. doi: 10.7164/antibiotics.28.668. [DOI] [PubMed] [Google Scholar]
- 18.Williamson JM, et al. Biosynthesis of the β-lactam antibiotic, thienamycin, by Streptomyces cattleya. J Biol Chem. 1985;260(8):4637–4647. [PubMed] [Google Scholar]
- 19.Houck DR, Kobayashi K, Williamson JM, Floss HG. Stereochemistry of methylation in thienamycin biosynthesis: Example of a methyl transfer from methionine with retention of configuration. J Am Chem Soc. 1986;108:5365–5366. [Google Scholar]
- 20.Lu W, Roongsawang N, Mahmud T. Biosynthetic studies and genetic engineering of pactamycin analogs with improved selectivity toward malarial parasites. Chem Biol. 2011;18(4):425–431. doi: 10.1016/j.chembiol.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe K, et al. Escherichia coli allows efficient modular incorporation of newly isolated quinomycin biosynthetic enzyme into echinomycin biosynthetic pathway for rational design and synthesis of potent antibiotic unnatural natural product. J Am Chem Soc. 2009;131(26):9347–9353. doi: 10.1021/ja902261a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Freeman MF, et al. Metagenome mining reveals polytheonamides as posttranslationally modified ribosomal peptides. Science. 2012;338(6105):387–390. doi: 10.1126/science.1226121. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez M, et al. Mutational analysis of the thienamycin biosynthetic gene cluster from Streptomyces cattleya. Antimicrob Agents Chemother. 2011;55(4):1638–1649. doi: 10.1128/AAC.01366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Lloyd EP, Moshos KA, Townsend CA. Identification and characterization of the carbapenem MM 4550 and its gene cluster in Streptomyces argenteolus ATCC 11009. ChemBioChem. 2014;15(2):320–331. doi: 10.1002/cbic.201300319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanz ND, et al. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 2012;516:125–152. doi: 10.1016/B978-0-12-394291-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 26.Bandarian V, Matthews RG. Measurement of energetics of conformational change in cobalamin-dependent methionine synthase. Methods Enzymol. 2004;380:152–169. doi: 10.1016/S0076-6879(04)80007-7. [DOI] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Beinert H. Micro methods for the quantitative determination of iron and copper in biological material. Methods Enzymol. 1978;54:435–445. doi: 10.1016/s0076-6879(78)54027-5. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy MC, et al. Evidence for the formation of a linear [3Fe-4S] cluster in partially unfolded aconitase. J Biol Chem. 1984;259(23):14463–14471. [PubMed] [Google Scholar]
- 30.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal Biochem. 1983;131(2):373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 31.Grove TL, Radle MI, Krebs C, Booker SJ. Cfr and RlmN contain a single [4Fe-4S] cluster, which directs two distinct reactivities for S-adenosylmethionine: Methyl transfer by SN2 displacement and radical generation. J Am Chem Soc. 2011;133(49):19586–19589. doi: 10.1021/ja207327v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stapon A, Li R, Townsend CA. Synthesis of (3S,5R)-carbapenam-3-carboxylic acid and its role in carbapenem biosynthesis and the stereoinversion problem. J Am Chem Soc. 2003;125(51):15746–15747. doi: 10.1021/ja037665w. [DOI] [PubMed] [Google Scholar]
- 33.Ohta T, Sato N, Kimura T, Nozoe S, Izawa K. Chirospecific synthesis of (+)-PS-5 from l-glutamic acid. Tetrahedron Lett. 1988;29(34):4305–4308. [Google Scholar]
- 34.Ueda Y, Damas CE, Belleau B. Nuclear analogs of β-lactam antibiotics, XVIII: A short synthesis of 2-alkylthiocarbapen-2-em-3-carboxylate. Can J Chem. 1983;61(9):1996–2000. [Google Scholar]
- 35.Ueda Y, Damas CE, Vinet V. Nuclear analogs of β-lactam antibiotics, XIX: Syntheses of racemic and enantiomeric p-nitrobenzyl carbapen-2-em-3-carboxylate. Can J Chem. 1983;61:2257–2263. [Google Scholar]
- 36.Baxter AJG, Dickenson KH, Roberts PM, Smale TC, Southgate R. Synthesis of 7-oxo-1-axabicyclo[3.2.0]hept-2-ene-2-carboxylates: The olivanic acid ring system. J Chem Soc Chem Commun. 1979:236–237. [Google Scholar]
- 37.Bateson JH, Quinn AM, Smale TC, Southgate R. Olivanic acid analogues, part 2: Total synthesis of some C(6)-substituted 7-oxo-1-azabicyclo[3.2.0]hept-2-ene-2-carboxylates. J Chem Soc Perk T. 1985;1(11):2219–2234. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.