SYSTEMS BIOLOGY Correction for “miRNA proxy approach reveals hidden functions of glycosylation,” by Tomasz Kurcon, Zhongyin Liu, Anika V. Paradkar, Christopher A. Vaiana, Sujeethraj Koppolu, Praveen Agrawal, and Lara K. Mahal, which appeared in issue 23, June 9, 2015, of Proc Natl Acad Sci USA (112:7327–7332; first published May 26, 2015; 10.1073/pnas.1502076112).
The authors note that Fig. 1 appeared incorrectly. The corrected figure and its legend appear below.
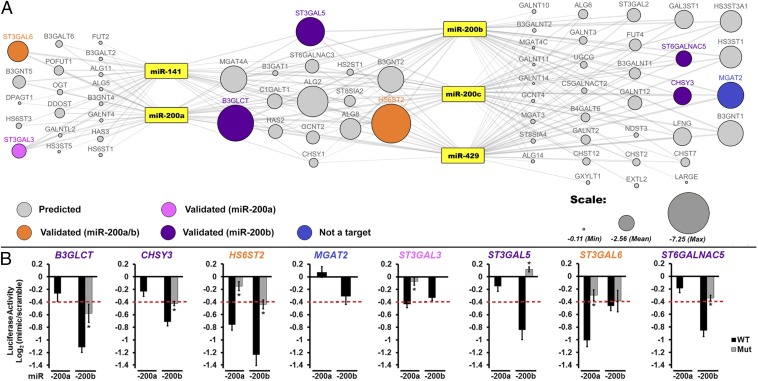
Fig. 1.
A broad network of glycogenes is targeted by miR-200f. (A) Network of predicted glycogene targets for miR-200f. Gray lines connect miRNA to predicted glycogene targets (circles). The size of the circle reflects combined miRSVR scores for all miR-200f predictions. Targets tested by luciferase assay are colored-coded as shown in the key in the figure. (B) Graphical representation of luciferase data. Luciferase–3′-UTR reporter constructs were cotransfected with miRNA or scramble mimics (60 nM) in HEK 293T/17 cells. Luciferase signal was assayed 24 h posttransfection. The graphs display average log2 (mimic/scramble). n = 3 biological replicates; *P < 0.005, t test. For wild-type constructs (black bars) displaying a ≥25% decrease (red line), mutants of the appropriate binding region were tested (gray bars; see Table S2). Fig. S1 shows the effect of all five miR-200f members on select constructs. Error bars represent the SD.