Abstract
Candida albicans cells are often detected with Streptococcus mutans in plaque biofilms from children affected with early childhood caries. The coadhesion between these 2 organisms appears to be largely mediated by the S. mutans–derived exoenzyme glucosyltransferase B (GtfB); GtfB readily binds to C. albicans cells in an active form, producing glucans locally that provide enhanced binding sites for S. mutans. However, knowledge is limited about the mechanisms by which the bacterial exoenzyme binds to and functions on the fungal surface to promote this unique cross-kingdom interaction. In this study, we use atomic force microscopy to understand the strength and binding dynamics modulating GtfB–C. albicans adhesive interactions in situ. Single-molecule force spectroscopy with GtfB-functionalized atomic force microscopy tips demonstrated that the enzyme binds with remarkable strength to the C. albicans cell surface (~2 nN) and showed a low dissociation rate, suggesting a highly stable bond. Strikingly, the binding strength of GtfB to the C. albicans surface was ~2.5-fold higher and the binding stability, ~20 times higher, as compared with the enzyme adhesion to S. mutans. Furthermore, adhesion force maps showed an intriguing pattern of GtfB binding. GtfB adhered heterogeneously on the surface of C. albicans, showing a higher frequency of adhesion failure but large sections of remarkably strong binding forces, suggesting the presence of GtfB binding domains unevenly distributed on the fungal surface. In contrast, GtfB bound uniformly across the S. mutans cell surface with less adhesion failure and a narrower range of binding forces (vs. the C. albicans surface). The data provide the first insights into the mechanisms underlying the adhesive and mechanical properties governing GtfB interactions with C. albicans. The strong and highly stable GtfB binding to C. albicans could explain, at least in part, why this bacterially derived exoenzyme effectively modulates this virulent cross-kingdom interaction.
Keywords: nanotechnology, bioengineering, caries, single-molecule force spectroscopy, dissociation rate, glucans
Introduction
Streptococcus mutans is often regarded as one of the main bacterial pathogens in dental caries, particularly in early childhood caries (ECC). ECC is characterized by heavy S. mutans infection (often >30% of the cultivable plaque flora), accompanied by protracted feeding of dietary sugars, such as sucrose (Gross et al. 2010; Parisotto et al. 2010; Hughes et al. 2012). This bacterial pathogen can rapidly orchestrate the assembly of cariogenic biofilms when frequently exposed to sucrose. Sucrose is utilized by S. mutans–derived exoenzymes (e.g., glucosyltransferases) to produce extracellular glucans, which enhance local accumulation of microbes and facilitate the buildup of cariogenic biofilms on the tooth surface (Paes Leme et al. 2006; Bowen and Koo 2011). However, S. mutans may not act alone in ECC, as other organisms contribute to caries pathogenesis (Takahashi and Nyvad 2011).
Results from previous clinical studies reveal that high numbers of the fungus Candida albicans are frequently detected with heavy infection by S. mutans in plaque biofilms from children with ECC (de Carvalho et al. 2006; Raja et al. 2010; Yang et al. 2012). These findings are intriguing because C. albicans usually does not bind well with S. mutans (Jenkinson et al. 1990; Gregoire et al. 2011). Rather, C. albicans adheres to oral mucosal and prosthetic surfaces and interacts with commensal streptococci to cause mucosal infections (Thein et al. 2009; Xu, Jenkinson, et al. 2014). C. albicans is well known to coadhere with Streptococcus gordonii and Streptococcus oralis (Jenkinson et al. 1990; Jenkinson and Douglas 2002; Diaz et al. 2012), enhancing fungal carriage and infectivity in vivo (Xu, Sobue, et al. 2014). Yet Candida was initially regarded as having little to no physical coadhesion with the pathogen S. mutans in the absence of sucrose. However, evidence from prior in vitro studies revealed that the adhesive interactions between C. albicans and S. mutans are greatly enhanced in the presence of sucrose (Branting et al. 1989; Gregoire et al. 2011) and that these conditions also promote biofilm formation (Pereira-Cenci et al. 2008; Metwalli et al. 2013; Falsetta et al. 2014).
Additional in vitro studies demonstrated that S. mutans–derived glucosyltransferase B (GtfB) binds avidly to C. albicans cell surfaces in an enzymatically active form (Gregoire et al. 2011). When sucrose is available, the surface-bound GtfB produces large amounts of glucans on the fungal surface. The glucans produced in situ provide enhanced binding sites for S. mutans, greatly promoting their adhesive interactions (Branting et al., 1989; Gregoire et al. 2011) and the development of highly virulent cospecies biofilms in vivo (Falsetta et al. 2014). Indeed, lack of gtfB expression by S. mutans impaired the ability of the bacterium to interact with C. albicans and form cospecies biofilms in the presence of sucrose (Falsetta et al. 2014). Clearly, GtfB plays a critical role mediating S. mutans–C. albicans coadherence. However, how the bacterial exoenzyme (GtfB) binds to and functions on the fungal surface to orchestrate this unique adhesive interaction between a bacterial oral pathogen and an opportunistic fungus remains to be elucidated.
Here, we explore the mechanical properties mediating GtfB binding to C. albicans and S. mutans surfaces by measuring the magnitude of adhesion force and stability using a single-molecule force spectroscopy (SMFS). The adhesion force (which indicates binding strength), dissociation rate (i.e., binding stability), and force map distribution (i.e., force localization) were measured between GtfB and microbial cell surfaces with atomic force microscopy (AFM) tips functionalized with GtfB. Our data reveal remarkable binding strength and binding stability of GtfB on C. albicans surfaces, both of which were several-fold higher than those observed for the same enzyme on S. mutans surfaces. Furthermore, we observed that GtfB, when bound to C. albicans, is highly active and produces more glucans (with distinct structure) than does GtfB bound to S. mutans surfaces. The results presented in this study provide the first insight into the biophysical underpinnings governing the adhesive interactions between a bacterially derived exoenzyme (GtfB) and the C. albicans cell surface, which could explain, at least in part, how GtfB effectively modulates this intriguing and virulent cross-kingdom association.
Materials and Methods
Microbial Strains and Culture Conditions
Candida albicans SC5314, a well-characterized strain whose genome has been sequenced, and Streptococcus mutans UA159, a proven virulent cariogenic pathogen, were used for single-molecule force measurements. C. albicans and S. mutans cultures were stored at −80 °C in tryptic soy broth and Sabouraud dextrose broth containing 20% glycerol. C. albicans and S. mutans cells were grown to midexponential phase (optical densities at 600 nm of 0.65 and 0.5, respectively) in ultrafiltered (10-kDa molecular-mass cutoff; Millipore, Billerica, MA, USA) yeast–tryptone extract broth (UFTYE; pH 5.5 and 7.0 for C. albicans and S. mutans, respectively) containing 1% (wt/vol) glucose and harvested by centrifugation (6,000 × g, 10 min, 4 °C) as described previously (Gregoire et al. 2011). The cells were then washed 3 times in phosphate-buffered saline (pH 7.0 to 7.2; HyClone Laboratories Inc., Logan, UT, USA) before cell immobilization.
Glucosyltransferase Enzymes
The GtfB (or GtfC) enzyme was prepared and purified to near homogeneity via hydroxylapatite column chromatography as detailed elsewhere (Koo et al. 2002). Glucosyltransferase activity was measured by the incorporation of [14C]-glucose from labeled sucrose (NEN Research Products, Boston, MA, USA) into glucans (Schilling and Bowen 1992; Koo et al. 2002). One unit of enzyme activity was defined as the amount of glucosyltransferase enzyme that incorporates 1 µmol of glucose into glucans over the 2-h reaction.
Immobilization of Microbial Cells and Functionalization of AFM Tips with GtfB
Prior to AFM analysis, microbial cells were immobilized on a poly-L-lysine-coated glass slide (Schaer-Zammaretti and Ubbink 2003). Briefly, a glass slide was cleaned by piranha solution (a mixture of sulfuric acid and hydrogen peroxide), followed by deposition of poly-L-lysine solution (0.1% w/v in H2O; Sigma-Aldrich, St. Louis, MO, USA) by overnight incubation. Washed cells were immobilized on a positively charged poly-L-lysine-coated glass slide for 1 h at room temperature. Then, the glass slide was gently washed with demineralized water to remove loosely adhered cells, and the slide was kept hydrated prior to AFM analysis. The immobilized cells were verified for viability using BacLight Live/Dead, as detailed in the Appendix.
To functionalize the AFM tips with GtfB, the cantilever tips were cleaned by immersing them in nitric acid for 5 min, followed by washing them in demineralized water 3 times. Then, AFM tips were exposed to saturating amounts of GtfB for 1 h at room temperature. The glucosyltransferase tips were dipped in 1% bovine serum albumin solution to block other active sites on the tips. GtfB functionalization was verified using our monoclonal antibody to GtfB and fluorescence imaging via Alexa Fluor 488–labeled goat anti-mouse IgG secondary antibody (488/519 nm; Life Technologies, Inc.). The enzymatic activity of the GtfB immobilized onto AFM tips was also verified by incubating GtfB tips with sucrose substrate in adsorption buffer containing Alexa Fluor 647–labeled dextran conjugate (647/668 nm; Molecular Probes Inc.) for 1 h at room temperature as described previously (Gregoire et al. 2011). Details of GtfB (and GtfC) functionalization of AFM tips and verification can be found in the Appendix.
AFM Methodology
All force measurements were conducted in fluid-phase at room temperature under phosphate-buffered saline (HyClone Labora-tories Inc.) using an MFP-3D AFM (Asylum Research, Santa Barbara, CA, USA). Silicon nitride probes (TR400PSA, Olympus, Tokyo, Japan) with a resonance frequency of ~11 kHz and spring constant of ~0.02 N/m and a nominal tip radius of 20 nm were calibrated via the thermal tune method (Hutter and Bechhoefer 1993) and examined optically before every experiment was conducted. To locate microbial cells, cells immobilized on glass surfaces were imaged in contact mode at randomly selected locations at 1 Hz. When isolated C. albicans or S. mutans was located in the image, a region of interest was defined over the cell surface, and an adhesion map was obtained by recording 50 × 50 force-distance curves within the region (5 × 5 µm and 0.5 × 0.5 µm for C. albicans and S. mutans, respectively). Cantilever deflection data upon retraction from cell surfaces were acquired and converted to force data. Force measurements obtained from loosely immobilized microbes (detached/moved during AFM probing) were discarded. The force-distance curves were obtained from at least 20 individual microbial cells from at least 3 distinct culture preparations per strain.
Statistical Analyses
The data were analyzed by pairwise comparisons of multiple groups with regression models based on the ranked values. Kruskal-Wallis tests, which are nonparametric and based on ranks, were used for 2-group comparisons. The significance level was set at 5%.
Results
Functionalization of AFM Tips and Immobilization of Microbial Cells
We used AFM tips functionalized with GtfB to investigate the physical interactions between the enzyme and single cells of C. albicans or S. mutans. Stable functionalization of AFM tips and preparation of immobilized microbial cells are key requirements for reliable detection of intermolecular forces (Hinterdorfer and Dufrêne 2006). To determine whether AFM tips were successfully functionalized with GtfB, we used 2 complementary methods. We initially verified whether GtfB molecules were immobilized onto AFM tips using fluorescently labeled anti-GtfB monoclonal antibody (Alexa 488). Fluorescent imaging showed that GtfB was attached and located on the tip of the AFM cantilever probe (in green; Fig. 1B). Then, we incubated GtfB-functionalized AFM tips with sucrose containing Alexa 647–labeled dextran conjugate to assess enzymatic activity of the immobilized GtfB. As shown in Figure 1C, glucans (in red) were produced on the enzyme-functionalized AFM tip, indicating GtfB bound to the probe without affecting its biological activity. Fluorescence was not detected in “no sucrose” control (Appendix Fig. 1), indicating that the fluorescence from the probe (incubated with sucrose) is due to synthesis of glucans in situ. Immobilization of microbial cells on the glass slide was also verified before and after force measurements to confirm that the cells remained anchored to the glass surfaces during force measurement (Fig. 1D, E). Furthermore, microbial cells were viable after immobilization, as determined via BacLight Live/Dead.
Figure 1.

Single-molecule atomic force microscopy (AFM) analysis of the adhesive interactions between glucosyltransferase B (GtfB) and microbial cell surfaces (Candida albicans or Streptococcus mutans). (A) AFM tips were functionalized with GtfB, and cells were immobilized on the glass surface. (B) Fluorescent image of GtfB-functionalized AFM tip. Alexa 488–labeled monoclonal antibody (goat anti-mouse IgG [H+L]–HRP) was bound to GtfB on the AFM tips to verify the local functionalization. (C) Fluorescent image of glucans formed on the GtfB-functionalized AFM tip. In situ glucan production was determined by incubating GtfB-functionalized AFM tips with sucrose substrate containing Alexa Fluor 647–labeled dextran conjugate to verify the enzymatic activity of GtfB on the AFM tips. (D) AFM scanning image of immobilized C. albicans on poly-L-lysine-coated glass slide. (E) AFM scanning image of immobilized S. mutans on poly-L-lysine-coated glass slide.
Single-molecule Force Measurements
GtfB–C. albicans interactions
Although GtfB has been shown to bind avidly to C. albicans, the biophysical properties mediating GtfB–C. albicans adhesive interactions are unknown. Thus, we used SMFS with GtfB-attached AFM tips to measure the magnitude of force, force distribution/localization, and dynamics of GtfB binding to the surface of single microbial cells. First, we determined the adhesion force and rupture length (binding strength measurements), and the data are shown in Figure 2A and B. We observed a large portion of highly adhesive interactions ranging from 1 to 2 nN and rupture lengths between 600 and 700 nm. Interestingly, the frequency of nonadhesive GtfB–C. albicans interaction was rather high (>25%), suggesting heterogeneous binding of GtfB on the C. albicans surface. To further detail the location of GtfB-Candida interactions, we used the GtfB tips to map the distribution of GtfB binding strength on microbial cell surface. Adhesion force maps revealed that large portions of the C. albicans surface have remarkably high binding affinities to GtfB, while some areas were devoid of detectable binding events (Fig. 3A).
Figure 2.
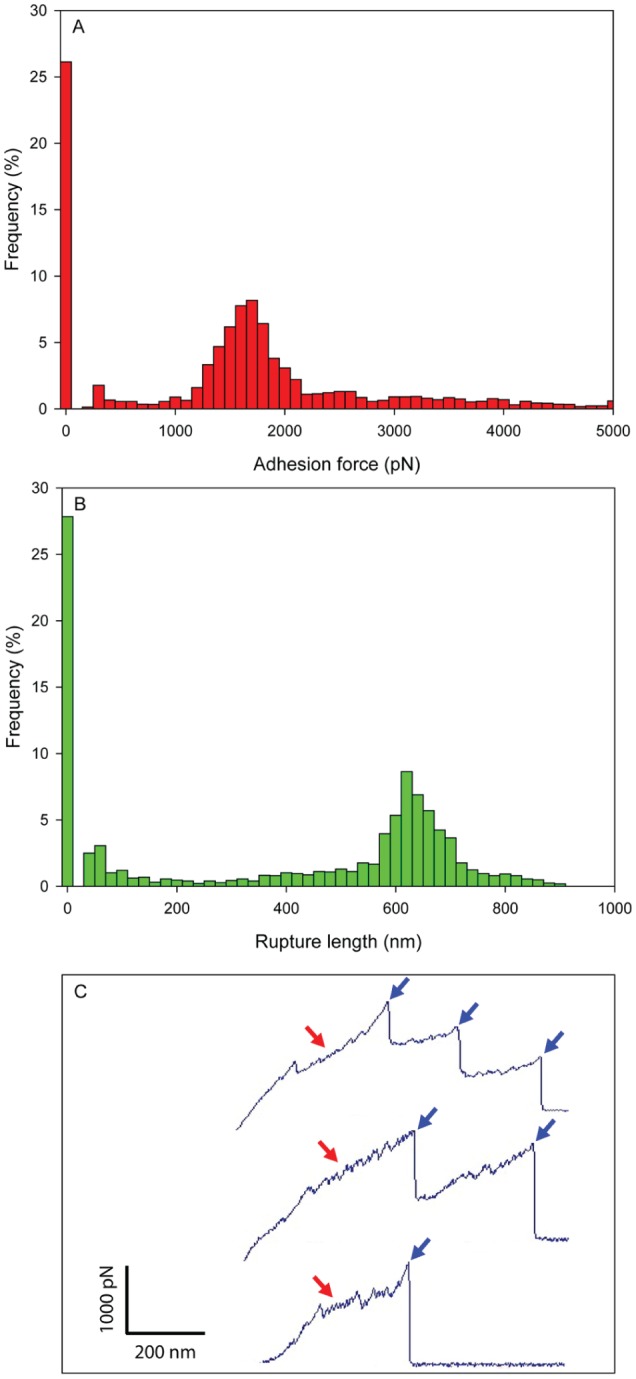
Binding force and rupture length of glucosyltransferase B on Candida albicans surfaces. (A) Adhesion force histogram, (B) rupture length histogram, and (C) representative retraction force curves. Blue arrows indicate force plateau, while red arrows indicate sawtooth pattern of force curves. The maximum adhesion force and the rupture length from last peak were used to generate the histograms. This figure is available in color online at http://jdr.sagepub.com.
Figure 3.
Adhesion force maps of Candida albicans and Streptococcus mutans surfaces. (A) Adhesion force map of C. albicans, (C) adhesion force map of S. mutans, and (B, D) adhesion force maps of C. albicans and S. mutans under same range.
Force-distance curves can display distinct adhesion profiles that indicate the types of interactions taking place and provide clues to the underlying physics (Cappella and Dietler 1999). Representative retraction force-distance curves of GtfB–C. albicans revealed striking sawtooth-like patterns (red arrows) and force plateaus (blue arrows; Fig. 2C). Sawtooth patterns reflect the sequential unfolding of multiple tandem-repeat domains, while force plateaus correspond with the mechanical unzipping of β-sheet interactions (Dufrêne 2014). It implies that the bindings of GtfB–C. albicans are dissociated through initial unfolding of the bonds, followed by sequential unzipping, thereby suggesting a stable complex between GtfB and C. albicans. Furthermore, the observed multiple force plateaus suggest the sequential or simultaneous breakages of multiple bonds during the retraction of the GtfB tip.
GtfB–S. mutans interactions
Following binding measurements with C. albicans, we determined the adhesion strength and dynamics between GtfB and S. mutans. Interestingly, the adhesion strengths of GtfB–S. mutans interactions were substantially lower than those of GtfB–C. albicans. Binding forces ranged from 0.3 to 1.0 nN and rupture lengths within 200 nm (Fig. 4A, B). However, GtfB adhesive interactions on S. mutans cell surfaces were distributed more evenly over the cell surface (Fig. 3C), despite the lower magnitude of the binding force (vs. GtfB–C. albicans interactions). Indeed, the frequency of nonadhesive interactions between GtfB and S. mutans was <10%. Force map images show that the distribution of adhesion forces of GtfB–S. mutans is more homogeneous than GtfB–C. albicans interactions (Fig. 3A, C); furthermore, when adjusted to the same force range, the higher strength of the GtfB–C. albicans interactions is also evident (Fig. 3B, D). In addition, the shape of the force-distance curves for GtfB–S. mutans adhesions were mostly single peak with force plateaus (blue arrows), indicating that the bond is broken mainly via unzipping without unfolding.
Figure 4.
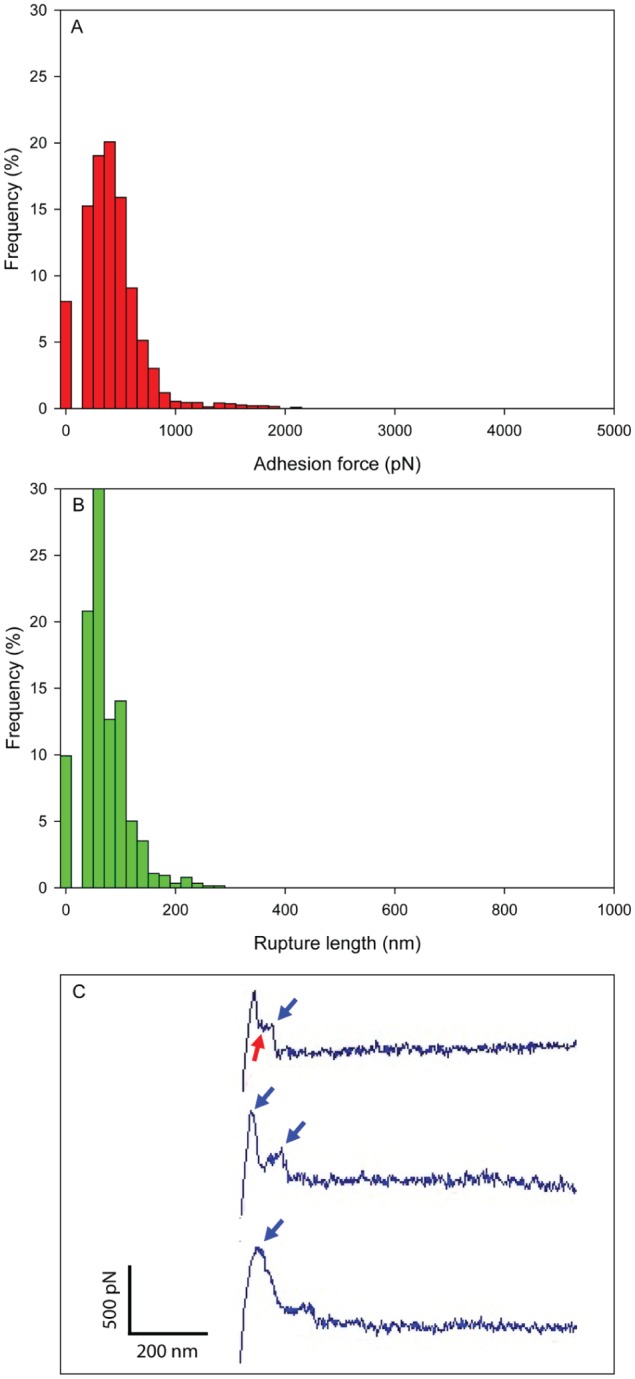
Binding force and rupture length of glucosyltransferase B (GtfB) on Streptococcus mutans surfaces. (A) Adhesion force histogram, (B) rupture length histogram, and (C) representative retraction force curves. Blue arrows indicate force plateau, while red arrows indicate sawtooth pattern of force curves. The maximum adhesion force and the rupture length from last peak were used to generate the histograms. This figure is available in color online at http://jdr.sagepub.com.
To confirm the specificity of the observed force distribution peaks and rupture events between GtfB and microbial surfaces, 2 control force experiments using albumin- or GtfC-coated AFM tips were carried out. Both albumin and GtfC show very low binding strength with minimal adhesion events to either S. mutans or C. albicans (vs. GtfB-functionalized tips; Appendix Fig. 2), which agrees well with limited GtfC binding to microbial cell surfaces observed previously (as reviewed; Bowen and Koo 2011). This figure is available in color online at http://jdr.sagepub.com.
Dissociation Rates of GtfB Bindings to C. albicans and S. mutans Surfaces
In addition to adhesion strength, binding stability is an equally important factor contributing to protein interaction and function on microbial surfaces (Dufrêne 2014). Binding stability can be determined from the kinetic off-rate by exploring the dependence of the binding strength with the force loading rate (Evans and Ritchie 1997; Lee et al. 2007). Thus, we determined the mean adhesion forces of GtfB–C. albicans (and GtfB–S. mutans) at various loading rates. Then, the characteristic dissociation rate—kinetic off rate at zero force, koff(0)—was estimated by equation 1:
where kB is the Boltzmann constant, T is the absolute temperature, and r is the loading rate. From the plot of F vs. ln r (Fig. 5), we can obtain the slope kBT/xβ from equation 1. Extrapolated to F = 0, equation 1 gives a value for koff = rxβ/kBT.
Figure 5.

Dynamic adhesion force spectra depending on the loading rate. (A) Candida albicans and (B) Streptococcus mutans. The mean adhesion forces vs. the logarithm of the loading rate were straight lines for both C. albicans and S. mutans. The kinetic off rate constant of dissociation at zero force of C. albicans was significantly lower (~20-fold) than the one of S. mutans, indicating that bindings of glucosyltransferase B on C. albicans are substantially more stable than they are on S. mutans surfaces.
As shown in Figure 5, the most probable forces for bond breakage (F) of GtfB to both cell surfaces increased approximately linearly with the logarithm of the loading rate (r), in line with previous observations from receptor-ligand systems (Evans and Ritchie 1997; Evans and Ludwig 2000; Lee et al. 2007). However, loading rate at zero force estimated from the plot of GtfB–C. albicans (0.1 pNs-1) was ~11 times lower than that of GtfB–S. mutans (1.08 pNs-1), resulting in substantially (~20 times) lower dissociation rate of GtfB–C. albicans (4·10-4s-1; vs. GtfB–S. mutans, 7·10-3s-1). These findings suggest that the GtfB–C. albicans complex dissociates much slower than GtfB–S. mutans, implying that the binding stability of GtfB–C. albicans is substantially higher than that of GtfB–S. mutans.
The distinct pattern of GtfB binding to C. albicans and S. mutans surfaces also affected the enzymatic function on the microbial surfaces. GtfB adsorbed on C. albicans surfaces produced more glucans containing a higher proportion of α-1,6 glucosyl linkages than those formed on S. mutans surfaces (Appendix Figs. 3, 4), confirming previous findings (Gregoire et al. 2011). It is possible that the differences in the binding properties of GtfB observed on each microbial cell surface cause distinctive conformational changes of GtfB structure, which could affect the catalytic functions of the surface-adsorbed enzyme. Collectively, our data clearly show that GtfB has a remarkably strong and stable binding to C. albicans surfaces, while showing very distinctive biophysical properties compared with GtfB–S. mutans adhesive interactions.
Discussion
AFM is a powerful tool for measuring the binding properties between 2 interacting surfaces (Busscher et al. 2008; van der Mei et al. 2008; Wessel et al. 2014). Importantly, the AFM cantilever can also be functionalized with biomolecules (e.g., proteins) to determine adhesive interactions between specific proteins and host or bacterial surfaces at the single molecule level (Dupres et al. 2007; Hinterdorfer and Dufrêne 2006; Sullan et al. 2015). However, SMFS has not yet been extensively used to study the binding properties/mechanics between extracellular proteins and oral microbial surfaces.
We previously identified a fascinating interaction between S. mutans and C. albicans that is mediated primarily by a bacterially derived exoenzyme (GtfB) that binds to the fungal surface (Gregoire et al. 2011; Falsetta et al. 2014). However, direct measurements of forces and dynamics of GtfB binding to the fungal cell surfaces have not been reported. Thus, we have optimized and applied SMFS-AFM to understand the biophysical properties mediating the observed GtfB–C. albicans interaction.
Our data reveal that bonds between GtfB and C. albicans have characteristic forces of separation on the order of nanonewtons and low dissociation rates, indicating that the binding is strong and stable, roughly equivalent to the strength of a covalent bond (Grandbois et al. 1999). Accordingly, the GtfB–C. albicans complex is strongly stabilized possibly via a “dock, lock, and latch” mechanism (Ponnuraj et al. 2003) between GtfB and cell wall components of C. albicans. Surprisingly, GtfB binding force and stability are greater on C. albicans surfaces than on S. mutans. Interestingly, the spatial distribution of GtfB adhesion strength on the Candida surface reveals a highly heterogeneous pattern, indicating that the exoenzyme may bind with more affinity to specific areas on the C. albicans cell wall surface. Indeed, fluorescence imaging of glucans produced in situ by GtfB bound to the C. albicans surface showed localized areas of glucan accumulation rather than homogeneous coating by the exopolysaccharides (Gregoire et al. 2011). This agrees well with the GtfB binding pattern observed here.
Importantly, the C. albicans–adsorbed GtfB not only retains its enzymatic activity but also affects glucan structure that the enzyme synthesizes on the fungal surface (Gregoire et al. 2011). The tight and spatially heterogeneous binding of GtfB and changes in the glucan products (elevated α-1,6-linked glucose) suggest highly specific interactions with cell wall components of C. albicans. The exact identities of the GtfB binding sites on the fungal surface are unknown. It was previously found that GtfB binds in an active form to purified mannans and β-1,3 glucans (Falsetta et al. 2014), which are found in the C. albicans cell wall (Gow et al. 2012). Among them, mannans could be one of the potential surface ligands because these polysaccharides are particularly abundant at the outermost layer (Gow et al. 2012). However, the composition and molecular architecture of the cell wall of C. albicans are quite complex (Klotz et al. 2007; Li and Palecek 2008), and other surface-associated glycoproteins and adhesins (e.g., Als and Eap1) potentially interact with GtfB. Clearly, further studies are needed to identify the binding partners as well as their spatial localization on the fungal surface that mediates GtfB–C. albicans adhesion.
The data presented here provide new details about the biophysical properties of GtfB-microbe interactions and the first insights about the mechanisms by which GtfB binds to C. albicans surfaces. The exact reasons why S. mutans releases GtfB to bind to their own and other microbial surfaces remain to be elucidated. One possibility is that S. mutans may export GtfB to facilitate biofilm formation as the exoenzyme binds to other microbes and to the saliva-coated apatitic surface in active form (Bowen and Koo 2011). When sucrose is available, the surface-bound GtfB produces glucans locally that mediate bacterial binding to teeth and coadherence with C. albicans and other bacteria, promoting the initiation of cariogenic biofilm (the preferred environment of S. mutans, where it can thrive by creating highly acidic niches).
The fascinating observation of stronger and highly stable GtfB binding to C. albicans than to the surface of its own bacterial producer provides a mechanistic rationale for why GtfB binds so effectively and functions onto the C. albicans surface (Gregoire et al. 2011), which plays a key role in modulating the exopolysaccharide synthesis in situ and the development of virulent cospecies biofilms (Falsetta et al. 2014). We are currently identifying the molecular ligands of GtfB on the C. albicans cell wall using our developed SMFS-AFM combined with genetic approaches (e.g., mutant strains defective of cell wall components). At the same time, further understanding is worthy of detailed exploration regarding how the conformation of the GtfB structure changes upon binding to the fungal surface and what effects it has on the catalytic activity of the enzyme.
Author Contributions
G. Hwang, H. Koo, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; G. Marsh, contributed to data acquisition and analysis, critically revised the manuscript; L. Gao, contributed to data acquisition, drafted the manuscript; R. Waugh, contributed to data analysis and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We are thankful to Dr. William H. Bowen (Center for Oral Biology, University of Rochester) for providing the anti-GtfB monoclonal antibody and mutanase and dextranase enzymes.
Footnotes
This work was supported in part by the National Institutes of Health (P01-HL018208), the National Science Foundation (EFRI-1137186), and the Nano/Bio Interface Center through the National Science Foundation (NSEC DMR08-32802).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Bowen WH, Koo H. 2011. Biology of Streptococcus mutans–derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45(1):69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branting C, Sund ML, Linder LE. 1989. The influence of Streptococcus mutans on adhesion of Candida albicans to acrylic surfaces in vitro. Arch Oral Biol. 34(5):347–353. [DOI] [PubMed] [Google Scholar]
- Busscher HJ, Norde W, van der Mei HC. 2008. Specific molecular recognition and nonspecific contributions to bacterial interaction forces. Appl Environ Microbiol. 74(9):2559–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappella B, Dietler G. 1999. Force-distance curves by atomic force microscopy. Surf Sci Rep. 34(1):1–104. [Google Scholar]
- de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DM. 2006. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol. 51(11):1024–1028. [DOI] [PubMed] [Google Scholar]
- Diaz PI, Xie Z, Sobue T, Thompson A, Biyikoglu B, Ricker A, Ikonomou L, Dongari-Bagtzoglou A. 2012. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 80(2):620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne YF. 2014. Atomic force microscopy in microbiology: New structural and functional insights into the microbial cell surface. MBio. 5(4):e01363-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupres V, Verbelen C, Dufrêne YF. 2007. Probing molecular recognition sites on biosurfaces using AFM. Biomaterials. 28(15):2393–2402. [DOI] [PubMed] [Google Scholar]
- Evans E, Ludwig F. 2000. Dynamic strengths of molecular anchoring and material cohesion in fluid biomembranes. J Phys Condens Matter. 12(8A):A315. [Google Scholar]
- Evans E, Ritchie K. 1997. Dynamic strength of molecular adhesion bonds. Biophys J. 72(4):1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes the virulence of plaque-biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 10(2):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbois M, Beyer M, Rief M, Clausen-Schaumann H, Gaub HE. 1999. How strong is a covalent bond? Science. 283(5408):1727–1730. [DOI] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 77(18):6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, Asnani K, Griffen AL. 2010. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 48(11):4121–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinterdorfer P, Dufrêne YF. 2006. Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods. 3(5):347–55. [DOI] [PubMed] [Google Scholar]
- Hughes CV, Dahlan M, Papadopolou E, Loo CY, Pradhan NS, Lu SC, Mathney JM, Bravoco A, Kent RL, Jr, Tanner AC. 2012. Aciduric microbiota and mutans streptococci in severe and recurrent severe early childhood caries. Pediatr Dent. 34(2):e16–e23. [PMC free article] [PubMed] [Google Scholar]
- Hutter JL, Bechhoefer J. 1993. Calibration of atomic-force microscope tips. Rev Sci Instrum. 64(7):1868–1873. [Google Scholar]
- Jenkinson HF, Douglas LJ. 2002. Candida interactions with bacterial biofilms. In: Brogden KA, Guthmiller JM, editors. Polymicrobial diseases. Washington (DC): ASM Press; p. 357–373. [PubMed] [Google Scholar]
- Jenkinson HF, Lala HC, Shepherd MG. 1990. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infect Immun. 58(5):1429–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE, Jr, Lipke PN, El-Azizi M. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol. 45(4):363–370. [DOI] [PubMed] [Google Scholar]
- Koo H, Pearson SK, Scott-Anne K, Abranches J, Cury JA, Rosalen PL, Park YK, Marquis RE, Bowen WH. 2002. Effects of apigenin and tt-farnesol on glucosyltransferase activity, biofilm viability and caries development in rats. Oral Microbiol Immunol. 17(6):337–343. [DOI] [PubMed] [Google Scholar]
- Lee CK, Wang YM, Huang LS, Lin S. 2007. Atomic force microscopy: determination of unbinding force, off rate and energy barrier for protein–ligand interaction. Micron. 38(5):446–461. [DOI] [PubMed] [Google Scholar]
- Li F, Palecek SP. 2008. Distinct domains of the Candida albicans adhesin Eap1p mediate cell-cell and cell-substrate interactions. Microbiology. 154(4):1193–1203. [DOI] [PubMed] [Google Scholar]
- Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. 2013. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9(10):e1003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA. 2006. The role of sucrose in cariogenic dental biofilm formation: new insight. J Dent Res. 85(10):878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisotto TM, Steiner-Oliveira C, Silva CM, Rodrigues LK, Nobre-dos-Santos M. 2010. Early childhood caries and mutans streptococci: a systematic review. Oral Health Prev Dent. 8(1):59–70. [PubMed] [Google Scholar]
- Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EM, Del Bel Cury AA, ten Cate JM, Crielaard W. 2008. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch Oral Biol. 53(8):755–764. [DOI] [PubMed] [Google Scholar]
- Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Hook M, Narayana SV. 2003. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 115(2):217–228. [DOI] [PubMed] [Google Scholar]
- Raja M, Hannan A, Ali K. 2010. Association of oral candidal carriage with dental caries in children. Caries Res. 44(3):272–276. [DOI] [PubMed] [Google Scholar]
- Schaer-Zammaretti P, Ubbink J. 2003. Imaging of lactic acid bacteria with AFM-elasticity and adhesion maps and their relationship to biological and structural data. Ultramicroscopy. 97(1):199–208. [DOI] [PubMed] [Google Scholar]
- Schilling KM, Bowen WH. 1992. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infec Immun. 60(1):284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullan RM, Li JK, Crowley PJ, Brady LJ, Dufrêne YF. 2015. Binding forces of Streptococcus mutans P1 adhesin. ACS Nano. 9(2):1448–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. 2011. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 90(3):294–303. [DOI] [PubMed] [Google Scholar]
- Thein ZM, Seneviratne CJ, Samaranayake YH, Samaranayake LP. 2009. Community lifestyle of Candida in mixed biofilms: a mini review. Mycoses. 52(6):467–475. [DOI] [PubMed] [Google Scholar]
- van der Mei HC, Rustema-Abbing M, de Vries J, Busscher HJ. 2008. Bond strengthening in oral bacterial adhesion to salivary conditioning films. Appl Environ Microbiol. 74(17):5511–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel SW, Chen Y, Maitra A, van den Heuvel ER, Slomp AM, Busscher HJ, van der Mei HC. 2014. Adhesion forces and composition of planktonic and adhering oral microbiomes. J Dent Res. 93(1):84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jenkinson HF, Dongari-Bagtzoglou A. 2014. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 29(3):99–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sobue T, Thompson A, Xie Z, Poon K, Ricker A, Cervantes J, Diaz PI, Dongari-Bagtzoglou A. 2014. Streptococcal co-infection augments Candida pathogenicity by amplifying the mucosal inflammatory response. Cell Microbiol. 16(2):214–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XQ, Zhang Q, Lu LY, Yang R, Liu Y, Zou J. 2012. Genotypic distribution of Candida albicans in dental biofilm of Chinese children associated with severe early childhood caries. Arch Oral Biol. 57(8):1048–1053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



