Abstract
Latent transforming growth factor-β binding protein-1 (LTBP-1) is an extracellular protein that is structurally similar to fibrillin and has an important role in controlling transforming growth factor-β (TGF-β) signaling by storing the cytokine in the extracellular matrix and by being involved in the conversion of the latent growth factor to its active form. LTBP-1 is found as both a short (LTBP-1S) and long (LTBP-1L) forms, which are derived though the use of separate promoters. There is controversy regarding the importance of LTBP-1L, as Ltbp1L knockout mice showed multiple cardiovascular defects but the complete null mice did not. Here, we describe a third line of Ltbp1 knockout mice generated utilizing a conditional knockout strategy that ablated expression of both L and S forms of LTBP-1. These mice show severe developmental cardiovascular abnormalities and die perinatally; thus these animals display a phenotype similar to previously reported Ltbp1L knockout mice. We reinvestigated the other “complete” knockout line, and found that these mice express a splice variant of LTBP-1L and, therefore, are not complete Ltbp1 knockouts. Our results clarify the phenotypes of Ltbp1 null mice and re-emphasize the importance of LTBP-1 in vivo.
Keywords: Latent TGF-β binding protein, transforming growth factor β, persistent truncus arteriosus, conditional knockout mice
1. Introduction
Transforming growth factor-β (TGF-β), the prototype member of the TGF-β family, exerts numerous essential functions in vivo. TGF-β is involved in physiological processes, such as embryogenesis, organ development, and the maintenance of tissue homeostasis [1]. TGF-β also participates in pathological conditions, such as inflammation, tissue fibrosis, suppression and development of tumors, and the pathology of several connective tissue diseases [2]. There are three different TGF-β isoforms (TGF-β1, 2, and 3), all of which have been studied extensively and seem to have unique functions in vivo, as there is little overlap in the phenotypes of knockout mice for individual TGF-β isoforms [3–6].
The exquisite control of the production of active TGF-β cytokine, which is essential in maintaining physiological functions in vivo, is exerted at three levels; expression, storage, and activation. Unlike other members of the TGF-β family, such as the bone morphogenetic proteins, TGF-β is secreted from cells complexed non-covalently with its propeptide, referred to as the latency associated peptide (LAP). This complex of mature cytokine and LAP is called the small latent complex (SLC), and in this complex TGF-β is blocked from interacting with its receptor [7]. SLC can be secreted by itself, but LAP is usually covalently bound to a carrier protein by disulfide bonds [8]. This large carrier protein is named the latent TGF-β binding protein (LTBP), whereas the complex of SLC and LTBP is referred to as the large latent complex (LLC) [8–10]. LLC is incorporated into extracellular matrices (ECM) via interaction between LTBP and ECM components, such as fibrillin and fibronectin [11–13]. This allows latent TGF-β to be stored in the ECM in a ready-to-use state [14]. Mature TGF-β needs to be released from LLC and specifically from LAP via a process called activation, in order to bind to its high affinity surface receptor. Several mechanisms have been proposed to activate latent TGF-β including, but not limited to, mechanical traction by integrins [15–17], degradation of LLC by proteases [18–21], and release of free cytokine by mechanical shear [22].
The formation of LLC is essential for the control of TGF-β function. This has been shown by a series of studies with knock-in mice in which the cysteine residue in LAP used for binding to LTBP was replaced by a serine residue. These mice (Tgfb1C33S) showed severe systemic inflammation, development of gastrointestinal tumors, and decreased TGF-β signaling [23–25]. These phenotypes are similar to those observed with Tgfb1 null mice and indicate the importance of LLC formation for proper TGF-β1 activity.
LTBP-1 is the most studied isoform in the LTBP family, which is comprised of 4 members, LTBP-1, 2, 3, and 4. LTBP1 contains two transcription initiation sites controlled by independent promoters, resulting in long and short isoforms (LTBP-1L and LTBP-1S, respectively) [26]. LTBP-1 has an important role in regulating TGF-β availability, as LTBP-1 and 3 readily bind to all three isoforms of latent TGF-β, whereas LTBP-4 inefficiently interacts only with latent TGF-β1 and LTBP-2 does not bind to any latent TGF-β isoform [27]. LTBP-1 is involved not only in the activation of latent TGF-β, but also in storing the cytokine in the ECM. The C-terminal region of LTBP-1 interacts with fibrillin microfibrils [11]. LTBP-1 also deposits in the ECM in the absence of fibrillin microfibrils via interaction with fibronectin [18, 28, 29]. LTBP-1 is essential for the activation of latent TGF-β1 via interaction with integrin β6 [15].
The importance of LTBP-1 in vivo has been underscored by a series of studies with Ltbp1L knock out mice. Targeted deletion of either exon 1 or 2 led to the same phenotype, developmental cardiovascular abnormalities, such as persistent truncus arteriosus (PTA), interrupted aortic arch (IAA), atrial septal defect (ASD), ventricular septal defect (VSD), and thickening of cardiac valves due to aberrant epithelial-mesenchymal transformation (EMT) [30, 31]. Ltbp1L null mice showed complete penetrance of PTA and partial penetrance of IAA, which are perinatally lethal after closure of ductus arteriosus. As the targeted exons in these mice are upstream of the first exon of LTBP-1S, it was predicted that the Ltbp1L null mice retained the expression of LTBP-1S, and this was verified by Northern blotting [30].
However, a contradiction that was difficult to explain occurred when the complete knockout mouse of Ltbp1 in which exon 5, the first exon shared by LTBP-1L and -1S, was targeted in order to abolish expression of both isoforms was reported [32]. These total knock out mice were viable, fertile, showed only mild craniofacial abnormalities, but there was no developmental cardiovascular defects including PTA or IAA, as would have been expected with loss of Ltbp1L. The reason for the observed discrepancy between the expected and observed phenotypes of Ltbp1L knockout mice and these mice, hereafter designated as Ltbp1 ΔEx5 mice, was unclear.
To resolve this discrepancy, we disrupted the long and short forms of LTBP-1 by targeting exon 8, which is common for both forms and is never spliced out, by utilizing a conditional knockout Ltbp1 mouse generated by the International Knockout Mouse Consortium (IKMC) and the Cre-loxP system. The new Ltbp1 complete knockout mice, hereafter designated as Ltbp1 ΔEx8 mice, died perinatally, and showed PTA and IAA, similar to Ltbp1L knockout mice. We also reexamined Ltbp1 ΔEx5 mice and found that these mice express a splice variant of LTBP-1 by splicing out exon 5. These results indicate that LTBP-1 is essential in the developing cardiovascular system as was first reported.
2. Results
2.1 Generation of Ltbp1 ΔEx8 mice
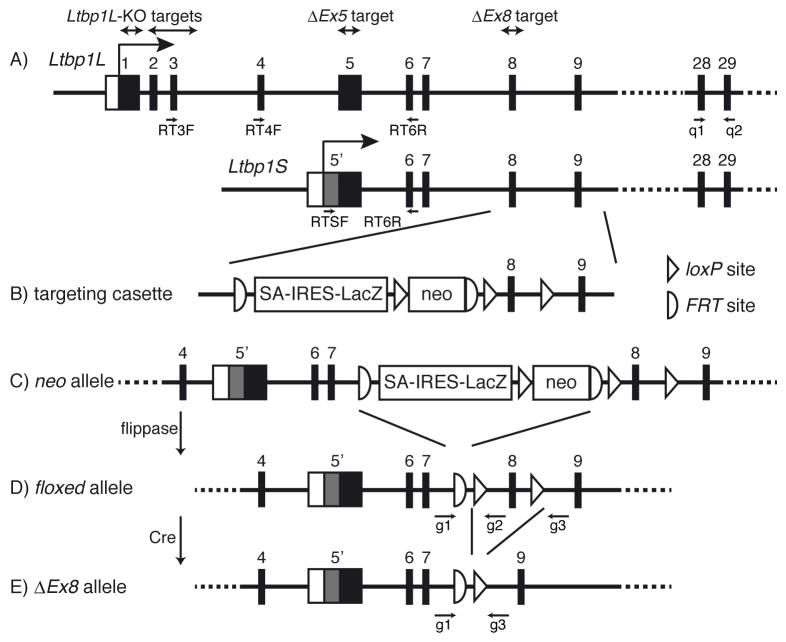
The European Conditional Mouse Mutagenesis Program (EUCOMM) Consortium, a member of IKMC, generated a gene-targeted mouse of Ltbp1 allele (Fig. 1A) by knocking in an L1L2_Bact_P cassette, which is a promoter-driven selection cassette (Fig. 1B), to produce a knockout-first allele (Fig. 1C). We obtained sperm from these mice, generated animals and crossed them with mice that ubiquitously express flippase to generate a conditional floxed allele, in which exon 8 of Ltbp1 is flanked by loxP sequences (Fig. 1D). These mice were crossed with mice that express Cre recombinase ubiquitously to generate the “ΔEx8” allele, which lacks the 103-base pair long exon 8 (Fig. 1E). The deletion should cause a frameshift mutation and result in the degradation of protein coding transcripts by nonsense-mediated decay (NMD). Heterozygous mice were bred to each other to produce Ltbp1 ΔEx8/ΔEx8 mice.
Figure 1. Generation of Ltbp1 total knockout allele (ΔEx8).
Schematic representation of the strategy to target the Ltbp1 allele is shown. (A) Wild type allele of Ltbp1L and Ltbp1S. Ltbp1S transcription and translation start upstream of the splice acceptor of the 5th exon of Ltbp1L. Gray box represents the Ltbp1S specific region. Targeted regions in each knockout mouse are shown above, and positions of primers used for PCR are shown below. (B) Structure of the targeting vector, L1L2_Bact_P. IRES, internal ribosome entry site; FRT, flippase recognition target; neo, neomycin resistance cassette; SA, splice acceptor. (C) Structure of the “neo allele” after homologous recombination. (D) Ltbp1 floxed allele. After excision of the LacZ-neo cassette by flippase, exon 8 is flanked by loxP sequences. (E) Ltbp1 ΔEx8 allele. Exon 8 is deleted after recombination by Cre recombinase.
2.2 Disruption of exon 8 abrogates expression of Ltbp1 mRNA and protein
We first performed experiments to verify if the targeting strategy effectively abrogated the expression of both long and short forms of LTBP-1. PCR genotyping of neonatal tissue showed bands of the expected size for the correct targeting (Fig. 2A). Quantitative PCR (qPCR) of cDNAs derived from mouse embryonic fibroblasts (MEFs) revealed minimum residual expression of Ltbp1 transcripts (Fig. 2B). As the targeting strategy was dependent on NMD, we assumed that the residual signal was detected from transcripts before they were subjected to NMD.
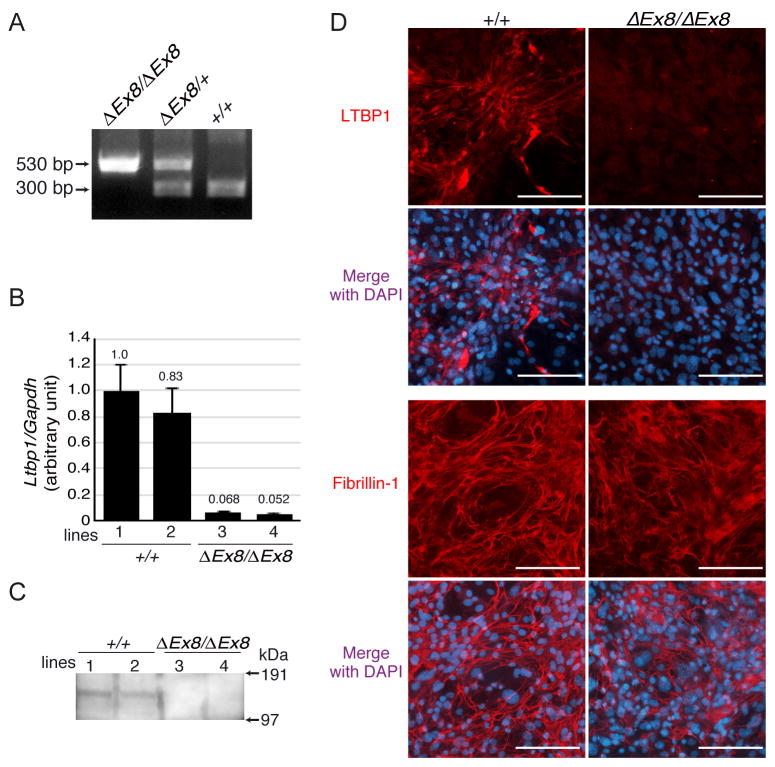
Figure 2. Deletion of Ltbp1 exon 8 effectively abrogates expression of LTBP-1.
(A) Genotyping by PCR. The PCR genotyping with genomic DNA shows bands with predicted sizes for wild type and ΔEx8 alleles. (B) qPCR of cDNA from 2 independent lines of MEFs per genotype. Each sample was measured in triplicate and normalized to Gapdh. Relative expression level of Ltbp1 in line 1 was defined as 1.0, and the mean ± standard deviation is shown. Significantly lower levels of signal were detected in Ltbp1ΔEx8/ΔEx8 MEFs compared to wild type MEFs. (C) LTBP-1 protein analysis. Concentrated conditioned media of MEF cultures were blotted with anti-LTBP-1 antibody. LTBP-1 was not detected in Ltbp1ΔEx8/ΔEx8 MEFs. (D) MEFs immunostaining. LTBP-1 was detected in wild type MEFs in fibrillar structures, but staining was abolished in Ltbp1ΔEx8/ΔEx8 MEF cultures (upper panels). Staining with anti-fibrillin-1 antibody showed that microfibril formation was not affected (lower panels). Bars 100 μm.
We next tried to determine if the residual mRNA produced a measurable amount of immunoreactive protein. Conditioned medium from MEF cultures was collected, concentrated, and analyzed by Western blotting with an anti LTBP-1 antibody. The result verified that cells from Ltbp1ΔEx8/ΔEx8 embryos do not produce detectible protein with the antibody under these conditions (Fig. 2C). In addition, we immunostained ECM produced by MEFs, which were cultured for 7 days, with an anti LTBP-1 antibody. The immunocytochemistry also showed the lack of LTBP-1 deposition on fibrillin-1 microfibrils in cells from Ltbp1ΔEx8/ΔEx8 mice, whereas wild type MEFs developed abundant fibrillar LTBP-1 meshwork (Fig. 2D).
These data confirmed that deletion of exon 8 causes a frameshift mutation followed by NMD of the transcripts and the absence of protein production as measured by both immunoblotting of medium from cultured cells and immunostaining of cells consistent with the lack of RNA for mature LTBP-1.
2.3 Ltbp1 ΔEx8 mice die perinatally of defective great vessel development and resemble Ltbp1L knockout mice
Genotyping of postnatal pups from crosses of Ltbp1 ΔEx8 heterozygous parents revealed that no homozygous pups survived longer than 1 day, whereas the genotype frequency of embryos did not deviate from the expected Mendelian distribution (Table 1).
Table 1.
Genotype frequency of offspring from Ltbp1ΔEx8/+ intercrosses
| Number of mice (%) of genotype | ||||
|---|---|---|---|---|
|
| ||||
| stage | +/+ | ΔEx8/+ | ΔEx8/ΔEx8 | Total No. |
| Pre-natal | 15 (27%) | 29 (52%) | 12 (21%) | 56 |
| Post-natal | 10 (27%) | 27 (73%) | 0 (0%) | 37 |
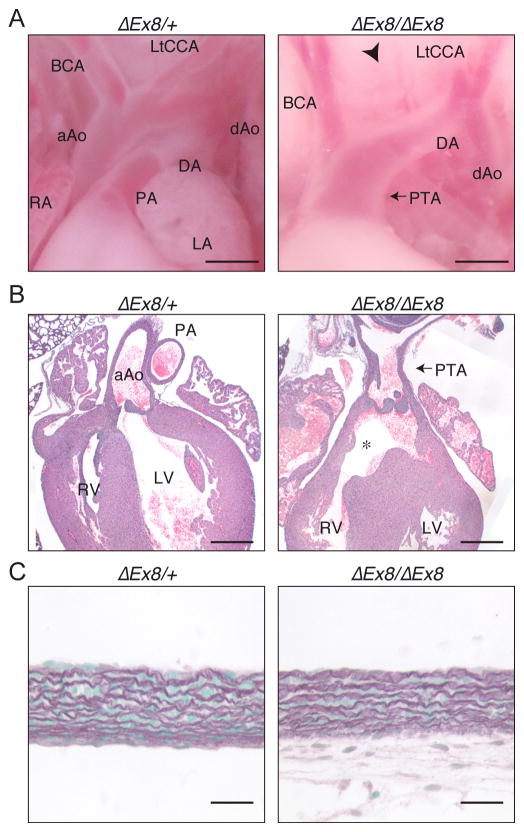
Careful analysis of newborns revealed that homozygote animals displayed multiple cardiovascular abnormalities including PTA and IAA (Fig. 3A). Histological analysis showed that the homozygote pups also had VSD and thickening of truncal semilunar valves (Fig. 3B). We investigated if the development of aortic structure was affected, by staining the elastic fibers. The layered structure of the elastic lamellae in the aorta was not affected by the loss of LTBP-1 (Fig. 3C).
Figure 3. Phenotype of Ltbp1 ΔEx8 mice resembles that of Ltbp1L knockout mice.
(A) Macroscopic view of the origin of great arteries of neonates. Heterozygotes showed a normal structure (left), but homozygous pups showed impaired separation of ascending aorta and pulmonary artery (PTA) and interrupted aortic arch between left common carotid artery (LtCCA) and brachiocephalic artery (BCA) (arrowhead). Bars 0.5 mm. (B) HE staining of heart sections of neonates. PTA and VSD (asterisk) are visible. Cardiac valves are hypercellular compared to control mice. Bars 400 μm. (C) Elastic staining of the aorta. The lamellar structure of the aorta did not reveal any differences between homozygous and control mice. The absence of cellular material below the elastic lamellae in the left panel is the result of the loss of the adventitia during sample preparation. Bars 20 μm. aAo, ascending aorta; dAo, descending aorta; DA, ductus arteriosus; LA, left atrium; LV, left ventricle; PA, pulmonary artery; RA, right atrium; RV, right ventricle.
These results clearly show that deletion of exon 8 and abrogation of the expression of LTBP-1 is perinatally lethal, which is consistent with our previous analysis of Ltbp1L knockout mice. We were then prompted to investigate Ltbp1 ΔEx5 mice to elucidate the reason why these mice are not severely affected in the absence of LTBP-1.
2.4 Expression of LTBP-1 in Ltbp1 ΔEx5 mice
As Ltbp1 ΔEx5 mice were shown by Western blotting to lack LTBP-1 in skin fibroblasts, we investigated other tissues that strongly express LTBP-1, namely lungs and heart. By Northern blot analysis of RNA purified from wild type lungs, we visualized two bands corresponding to Ltbp1L and 1S, whereas we observed only the Ltbp1L band with RNA from the heart. Surprisingly, RNA from Ltbp1 ΔEx5 mice still displayed the higher band corresponding to Ltbp1L, although the lower band was absent (Fig. 4A).
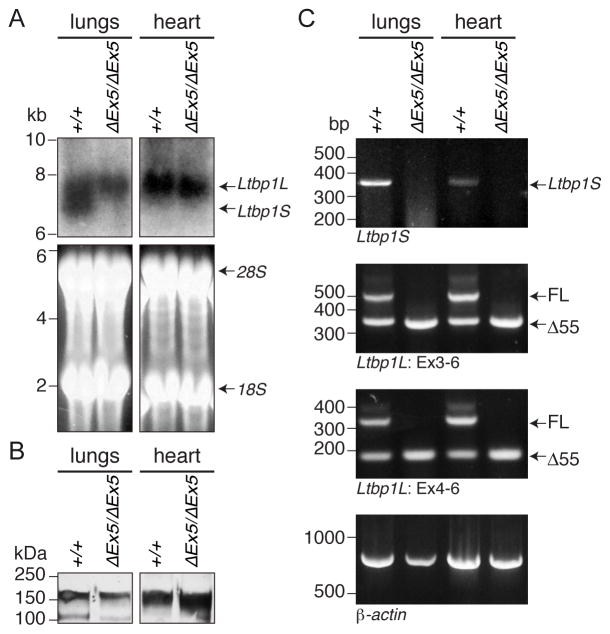
Figure 4. Ltbp1 ΔEx5 mice lack LTBP-1S but express Δ55 form of LTBP-1L.
(A) Northern blotting. Total RNA was extracted from newborn lungs and hearts of wild type and Ltbp1 ΔEx5 mice and utilized for Northern Blotting analysis. Ltbp1S transcripts were detected only in lungs of wild type samples, whereas transcripts of Ltbp1L were detected in both tissues of wild type and Ltbp1 ΔEx5 mice. (B) LTBP-1 protein analysis. Total protein extracts from newborn lungs and hearts of wild type and Ltbp1 ΔEx5 mice were subjected to plasmin digestion to liberate matrix-bound proteins. The released, truncated LTBP-1 was visualized by Western blotting with a rabbit polyclonal anti-LTBP-1 antibody (Ab 39). Bands around the predicted mass of 150-kDa were detected in all samples. (C) RT PCR. RT PCR was performed with lung and heart cDNAs from wild type and Ltbp1 ΔEx5 mice. The primer set specific for Ltbp1S amplified only wild type templates (top panel). Primers flanking exon 5 (Ex3-6 or Ex4-6) revealed expression of a full length (FL) and an Ltbp1L splice variant lacking exon 5 (Δ55) in wild type samples, whereas only Δ55 was amplified in Ltbp1 ΔEx5 samples (second and third panel, respectively).
We identified a protein form corresponding to LTBP-1 in samples prepared from both wild type and Ltbp1 ΔEx5 mice by Western blot analysis of the insoluble ECM fraction of lungs and heart (Fig. 4B). The two major LTBP-1 isoforms cannot be distinguished by Western blotting of these preparations, as tissues were digested with plasmin during the extraction to release bound LTBP-1 and this yields a truncated polypeptide contained in both LTBP-1S and LTBP-1L forms.
2.5 Ltbp1 ΔEx5 mice express the Δ55 variant of LTBP-1L by alternative splicing
We performed several reverse transcription-PCR (RT PCR) amplifications using template cDNA derived from lungs and heart. With primers designed to be specific for Ltbp1S, PCR products were detected with wild type mice but were absent in Ltbp1 ΔEx5 mice (Fig. 4C top panel). The band intensity was stronger in lungs than in heart, which is consistent with the absence of Ltbp1S observed by Northern blot analysis (Fig. 4A). Next we designed forward primers in either exon 3 or 4 and a reverse primer in exon 6 to detect Ltbp1L. PCR products of the expected size for full-length Ltbp1L transcripts (532 and 340 base pairs long for 3F–6R and 4F–6R templates, respectively) were detected only with samples isolated from wild type mice and not from Ltbp1 ΔEx5 mice. However, these reactions produced shorter bands in both wild type and Ltbp1 ΔEx5 samples (Fig. 4C second and third panels). These bands were approximately 170 base pairs shorter than the intact bands. Sequencing of these PCR products revealed that these bands are products of the previously reported Δ55 variant of Ltbp1L, which lacks 168 bases of exon 5 [33].
Together with the Northern and Western blotting results, these data showed that Ltbp1 ΔEx5 mice lack Ltbp1S but retain expression of the Δ55 variant of Ltbp1L.
3. Discussion
In this study, we show that the complete knockout of both isoforms of LTBP-1 leads to PTA and IAA and subsequent perinatal lethality. This result strongly supports the idea that LTBP-1, and probably the TGF-β signaling mediated by it, is essential in the development of the great arteries and cardiac cushions, as we previously reported [30]. The present results clarify the controversy as to whether Ltbp1 complete null mice are viable or not and whether LTBP-1 is indispensable for murine development.
Phenotypes of Ltbp1 ΔEx8 mice reproduced those described for Ltbp1L knockout mice, implying that the major role of LTBP-1 in the pre-natal development of cardiovascular system is conducted by LTBP-1L and that LTBP-1S does not play an important role. As Ltbp1L knockout and Ltbp1 ΔEx8 mice are perinatally lethal, it is impossible to investigate specific postnatal functions of LTBP-1L with these mutant mice.
The observed discrepancies between the previously reported general Ltbp1 knockout (i.e. Ltbp1 ΔEx5) and Ltbp1L knockout mice can be attributed to the expression of an alternatively spliced form of LTBP-1L in the former mice. Drews et al. showed successful targeting of Ltbp1 ΔEx5 with Southern blotting and genomic DNA PCR, and concluded in their paper that the expression of LTBP-1 protein was absent in Ltbp1 ΔEx5 mice based on the result of Western blotting of skin fibroblast extracts [32]. We speculate that these fibroblasts normally produce predominantly LTBP-1S, and low levels of LTBP-1L and/or the Δ55 variant. Therefore, it is possible that the presence of the LTBP-1L form was too low to be detected in the experiments of Drews et al.
Ltbp1 ΔEx5 mice have a shorter snout, and show less fibrosis with reduced phosphorylation of Smad2 in hepatic tissue after the ligation of the bile duct. These phenotypes imply that LTBP-1S may be involved specifically in hepatic fibrosis and craniofacial development. Another explanation for the phenotypes is that the 56 amino acids missing in the Δ55 variant are essential for LTBP-1 function in the liver and skull and their absence results in the hepatic fibrosis and craniofacial abnormalities. It should be mentioned that this region contains 2 putative N-glycosylation sites, which may be critical for the proper function of LTBP-1 [33].
Finally, we have shown that the deletion of exon 8 is sufficient to abrogate expression of both long and short form of LTBP-1. Three splice variants, Δ41, Δ53 and Δ55 have been reported for the Ltbp1 locus [33, 34]. From the gene sequence, it was predicted that deletion of exon 8 would abrogate all protein products including these splice variants by NMD. This underscores the reliability and usefulness of the Ltbp1 conditional knockout line we used in the present study. This line can be a powerful tool for the investigation of spatially and temporally specific functions of LTBP-1, especially in postnatal life, by using appropriate Cre expressing mice.
4. Materials and Methods
4.1 Generation of Ltbp1 targeted mice
The Ltbp1 conditional knockout mouse (Ltbp1tm1a(EUCOMM)Wtsi) was generated at Wellcome Trust Sanger Institute under EUCOMM Consortium, which is a member of IKMC, and was stored and distributed by MRC Harwell on behalf of The European Mouse Mutant Archive (EMMA). Targeting strategy is outlined in Fig. 1 [35]. Briefly, the targeting vector was designed to introduce a promoter-driven selection cassette named “L1L2_Bact_P cassette” at position 75248411 of the chromosome 17 upstream of exon 8, which is 103 bases long. Another loxP site was inserted at position 75249265 in intron 8 (Fig. 1B). The targeting vector was introduced into ES cells JM8.N4 derived from C57BL/6N and cells were selected for the homologous recombination by short range PCR. ES cells with resultant “neo” allele (Fig. 1C) were injected into blastocysts of C57BL/6Brd-Tyrc-Brd; C57BL/6Brd-Tyrc-Brd mice to produce chimeric mice and heterozygous offspring. Mice were tested with series of short range PCR and qPCR. Details of targeting strategy and the quality control of ES cells and engineered mice can be found on the website of International Mouse Phenotype Consortium that followed IKMC [36].
We obtained sperm of Ltbp1neo/+ mice and performed in vitro fertilization at New York University Langone Medical Center. Ltbp1neo/+ mice were crossed with B6.Cg-Tg(ACTFLPe)9205Dym/J mice (The Jackson Laboratory), which express FLP1 recombinase under human ACTB promoter, to produce conditional “floxed” allele (Fig. 1D). These mice were crossed with C57BL/6NTac-Gt(ROSA)26Sortm16(cre)Arte (Taconic) which express Cre recombinase under ROSA26 promoter to generate Ltbp1 ΔEx8 mice (Fig. 1E). Genotyping PCR was performed using Taq DNA polymerase (Roche) with primers g1, 2, 3 shown in Fig. 1E (Table 2).
Table 2.
Primer sequences
| primers | sequence |
|---|---|
| mLtbp1 g1 | cacttgtctacctcatagccgattctacag |
| mLtbp1 g2 | gaaaatccgaaccaatggcgcaggtccatg |
| mLtbp1 g3 | cgcagatctgtaagcaatcaattccccatctg |
| mLtbp1 q1 | ctcttacgacactggtggcgagaactac |
| mLtbp1 q2 | acccaggttgagtgttcaaacagtaacc |
| mGapdh S | ccatcaccatcttccaggag |
| mGapdh AS | cacacccatcacaaacatgg |
| mLtbp1 RT3F | ccctcaagccaaagccatca |
| mLtbp1 RT4F | aggagcaaggcacggcaccc |
| mLtbp1 RT6R | tgctgctgggcgtgctggta |
| mLtbp1 RTSF | caagttcatggatactaagc |
| mActb RT F | atctggcaccacaccttctacaatgagctgcg |
| mActb RT R | cgtcatactcctgcttgctgatccacatctgc |
Generation and phenotypes of Ltbp1L knockout mice and Ltbp1 ΔEx5 mice have been previously reported [30, 32]. All procedures were performed in accordance with the Regulation on Animal Experimentation at New York University Langone Medical Center and were approved by the Institutional Animal Care and Use Committee.
4.2 Mouse Embryonic Fibroblasts (MEFs)
MEFs were isolated from wild type and Ltbp1 ΔEx8 embryos at 13.5 days post-coitum as previously reported [28]. Briefly, embryos were digested with 0.125% trypsin-EDTA after decapitation and removal of visceral tissues. Cell suspensions were filtrated with cell strainer and cultured in DMEM (Corning) supplemented with 2 mM glutamine, 100 units/100 μg/mL penicillin/streptomycin (Corning), and 10% FBS at 37 °C in 5% CO2.
For LTBP-1 protein analysis, MEFs were cultured in serum free DMEM/Ham’s F12 50/50 Mix (Corning) supplemented with 2 mM glutamine, 100 units/100 μg/mL penicillin/streptomycin (Corning). Collected conditioned media were concentrated 50 fold with Ultracel-30K (EMD Millipore) and subjected for Western blotting.
4.3 Quantitative PCR (qPCR)
Cultured MEFs were homogenized and total RNA was extracted with TRIzol reagent (Life Technologies) and RNeasy Mini Kit (Qiagen), followed by reverse transcription with SuperScript® III Reverse Transcriptase (Life Technologies) with random hexamers. qPCR analyses were performed in triplicate with QuantiFAST SYBR Green PCR Kit (Qiagen) using StepOnePlus™ Real-Time PCR Systems (Life Technologies). Quantification was performed with the relative standard curve method and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) using StepOne™ Software. Relative expression of Ltbp1 in the wild type sample was defined as 1.0 and data are presented as means ± SD. Positions of primers are illustrated in Fig. 1A, and the sequences are provided in Table 2.
4.4 Isolation and plasmin digestion of ECM-associated protein from tissue
Neonatal lung and heart were homogenized in RIPA buffer (50 mM Tris-HCl (pH 7.2), 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate) and the insoluble fraction was collected by centrifugation, followed by digestion with plasmin (0.1 U/ml; Roche) in PBS at 37°C for 1 hour. Supernatants were collected after centrifugation and concentrated using Ultracel-30K (EMD Millipore) and subjected for Western blotting.
4.5 Western blotting
Protein samples from MEFs or neonatal tissues were separated with SDS-PAGE and transferred to PVDF or nitrocellulose membranes, respectively. Membranes were blocked with 5% skim milk, and blotted with anti-LTBP-1 polyclonal antibody (Ab39, a generous gift from Drs. K Miyazono and C–H Heldin [37], or ab78294 (Abcam)) as a primary antibody and horseradish peroxidase-conjugated anti-Rabbit IgG antibody (GE Healthcare Life Science) as a secondary antibody. Chemiluminescent reations were performed with either ECL Western Blotting Substrate (Thermo Scientific) or Western Lightning Chemiluminescence Plus-ECL (Perkin Elmer), and signals were detected with X-ray films or ImageQuant LAS 4000 System (GE Healthcare Life Science).
4.6 Immunocytochemistry
MEFs were cultured on gelatin-coated coverslips in DMEM (Corning) supplemented with 2 mM glutamine, 100 units/100 μg/mL penicillin/streptomycin (Corning) and 10% FBS at 37 °C in 5% CO2 for 7 days. After washing with PBS, cells were fixed with 100% ethanol at room temperature for 10 minutes and blocked with 2% bovine serum albumin. The primary antibodies used were anti-LTBP-1 polyclonal (Ab39) and anti-fibrillin-1 polyclonal (9543, a generous gift from Dr. L. Sakai)[38], and the secondary antibody used was AlexaFluor 594-conjugated anti-Rabbit IgG antibody (Life Technologies). Cells were mounted with ProLong® Gold Antifade Mountant with DAPI (Life Technologies), and were observed with a Nikon Eclipse Ni fluorescence compound microscope.
4.7 Histological analysis and Elastic Staining
Whole P0 neonates were fixed with 3.7% formaldehyde and embedded in paraffin block. Tissues were sectioned, and subjected to hematoxylin-eosin staining and Weigert’s Resorcin-Fuchsin staining with standard protocol.
4.8 Northern Blotting
Total RNA was isolated from wild type and Ltbp1 ΔEx5 neonatal lungs and hearts, using Trizol Reagent (Life Technologies) according to the manufacturer’s protocol. Northern blotting was performed as previously described [30]. Briefly, RNA samples were separated by denaturing gel electrophoresis on 0.8% agarose gels, and the gel was stained with ethidium bromide to observe the amount of 18S and 28S RNAs. RNAs were blotted on a nylon membrane by capillary transfer and hybridized in ExpressHyb Hybridization Solution (Clontech) with [α-32P]-dCTP-labeled probe synthesized using Ready-To-Go DNA Labeling Beads (-dCTP) (GE Healthcare Life Science) and mouse Ltbp1L cDNA [39].
4.9 Reverse transcription-PCR (RT PCR)
Total RNA isolated from wild type and Ltbp1 ΔEx5 neonatal lung and heart was transcribed using Reverse Transcriptase AMV (Roche). The resulting cDNA was used for RT PCR analyses. Reactions for Ltbp1S-specific transcripts were performed with primers RTSF and RT6R, and those for Ltbp1L transcripts were performed with either primer sets RT3F/RT6R or RT4F/RT6R. PCR products of the latter reaction were sequenced with ABI PRISM® 377 XL DNA analyzer (Life Technologies). Positions of primers used are illustrated in Fig. 1A, and the sequences are provided in Table 2.
Acknowledgments
We thank Ms. M Vassallo, Mr. J Ambrogio, Mr. A Chubak and Mr. J Ponessa for excellent technical assistance. We also acknowledge EUCOMM Consortium for providing us with Ltbp1 conditional knockout mice, and K Miyazono and C-H Heldin for Ab39. The authors wish to acknowledge the use of the New York University Langone Medical Center (NYUMC) Histopathology and Transgenic Mouse Cores supported by the NYUMC Cancer Center grant P30CA016087. This work was supported by NIH grants R01 CA034282 and P01 AR49698 to DB Rifkin. M Horiguchi was supported by fellowships from Banyu Life Science Foundation International and the Uehara Memorial Foundation.
Abbreviations
- aAo
ascending aorta
- ASD
atrial septal defect
- BCA
brachiocephalic artery
- CCA
common carotid artery
- DA
ductus arteriosus
- dAo
descending aorta
- ECM
extracellular matrix
- EMMA
The European Mouse Mutant Archive
- EMT
epithelial-mesenchymal transformation
- EUCOMM
The European Conditional Mouse Mutagenesis Program
- FRT
flippase recognition target
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- IAA
interrupted aortic arch
- IKMC
International Knockout Mouse Consortium
- IRES
internal ribosome entry site
- LA
left atrium
- LAP
latency associated peptide
- LLC
large latent complex
- LTBP
latent TGF-β binding protein
- LV
left ventricle
- MEF
mouse embryonic fibroblast
- NMD
nonsense-mediated decay
- PA
pulmonary artery
- PTA
persistent truncus arteriosus
- RA
right atrium
- RT PCR
reverse transcription-PCR
- RV
right ventricle
- SA
splice acceptor
- SLC
small latent complex
- TGF-β
transforming growth factor-β
- VSD
ventricular septal defect
Footnotes
6. Author Contributions
MH, VT and DBR designed the study, RW provided materials, MH, VT, and KH performed experiments, MH, VT, KH, and DBR analyzed the data, MH and DBR wrote the paper.
References
- 1.Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–91. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 2.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 3.Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, et al. Abnormal lung development and cleft palate in mice lacking TGF-beta 3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–21. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 4.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–70. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kulkarni AB, Huh CG, Becker D, Geiser A, Lyght M, Flanders KC, et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci U S A. 1993;90:770–4. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in mulitfocal inflammatory disease. Nature. 1992;359:693–9. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rifkin DB. Latent transforming growth factor-beta (TGF-beta) binding proteins: orchestrators of TGF-beta availability. J Biol Chem. 2005;280:7409–12. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- 8.Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–53. [PMC free article] [PubMed] [Google Scholar]
- 9.Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-beta 1-binding protein in the assembly and secretion of TGF-beta 1. EMBO J. 1991;10:1091–101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gleizes PE, Beavis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-beta binding protein-1 that mediates bonding to the latent transforming growth factor-beta1. J Biol Chem. 1996;271:29891–6. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- 11.Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, et al. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem. 2003;278:2750–7. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 12.Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-beta 1 associates to fibroblast extracellular matrix via latent TGF-beta binding protein. J Cell Biol. 1994;124:171–81. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana L, Chen Y, Prijatelj P, Sakai T, Fässler R, Sakai LY, et al. Fibronectin is required for integrin alphavbeta6-mediated activation of latent TGF-beta complexes containing LTBP-1. FASEB J. 2005;19:1798–808. doi: 10.1096/fj.05-4134com. [DOI] [PubMed] [Google Scholar]
- 14.Horiguchi M, Ota M, Rifkin DB. Matrix control of transforming growth factor-beta function. J Biochem. 2012;152:321–9. doi: 10.1093/jb/mvs089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–28. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 16.Wipff PJ, Hinz B. Integrins and the activation of latent transforming growth factor beta1 - an intimate relationship. Eur J Cell Biol. 2008;87:601–15. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Munger JS, Sheppard D. Cross talk among TGF-beta signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3:a005017. doi: 10.1101/cshperspect.a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taipale J, Saharinen J, Hedman K, Keski-Oja J. Latent transforming growth factor-beta 1 and its binding protein are components of extracellular matrix microfibrils. J Histochem Cytochem. 1996;44:875–89. doi: 10.1177/44.8.8756760. [DOI] [PubMed] [Google Scholar]
- 19.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–76. [PMC free article] [PubMed] [Google Scholar]
- 20.Ge G, Greenspan DS. BMP1 controls TGFbeta1 activation via cleavage of latent TGFbeta-binding protein. J Cell Biol. 2006;175:111–20. doi: 10.1083/jcb.200606058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sato Y, Rifkin DB. Inhibition of endothelial cell movement by pericytes and smooth muscle cells: activation of a latent transforming growth factor-beta 1-like molecule by plasmin during co-culture. J Cell Biol. 1989;109:309–15. doi: 10.1083/jcb.109.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahamed J, Burg N, Yoshinaga K, Janczak CA, Rifkin DB, Coller BS. In vitro and in vivo evidence for shear-induced activation of latent transforming growth factor-beta1. Blood. 2008;112:3650–60. doi: 10.1182/blood-2008-04-151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshinaga K, Obata H, Jurukovski V, Mazzieri R, Chen Y, Zilberberg L, et al. Perturbation of transforming growth factor (TGF)-beta1 association with latent TGF-beta binding protein yields inflammation and tumors. Proc Natl Acad Sci U S A. 2008;105:18758–63. doi: 10.1073/pnas.0805411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shibahara K, Ota M, Horiguchi M, Yoshinaga K, Melamed J, Rifkin DB. Production of gastrointestinal tumors in mice by modulating latent TGF-beta1 activation. Cancer Res. 2013;73:459–68. doi: 10.1158/0008-5472.CAN-12-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ota M, Horiguchi M, Fang V, Shibahara K, Kadota K, Loomis C, et al. Genetic suppression of inflammation blocks the tumor-promoting effects of TGF-beta in gastric tissue. Cancer Res. 2014;74:2642–51. doi: 10.1158/0008-5472.CAN-13-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koski C, Saharinen J, Keski-Oja J. Independent promoters regulate the expression of two amino terminally distinct forms of latent transforming growth factor-beta binding protein-1 (LTBP-1) in a cell type-specific manner. J Biol Chem. 1999;274:32619–30. doi: 10.1074/jbc.274.46.32619. [DOI] [PubMed] [Google Scholar]
- 27.Saharinen J, Keski-Oja J. Specific sequence motif of 8-Cys repeats of TGF-beta binding proteins, LTBPs, creates a hydrophobic interaction surface for binding of small latent TGF-beta. Mol Biol Cell. 2000;11:2691–704. doi: 10.1091/mbc.11.8.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, et al. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: Role of fibrillins and fibronectin. J Cell Physiol. 2012;227:3828–36. doi: 10.1002/jcp.24094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallas SL, Sivakumar P, Jones CJ, Chen Q, Peters DM, Mosher DF, et al. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J Biol Chem. 2005;280:18871–80. doi: 10.1074/jbc.M410762200. [DOI] [PubMed] [Google Scholar]
- 30.Todorovic V, Frendewey D, Gutstein DE, Chen Y, Freyer L, Finnegan E, et al. Long form of latent TGF-beta binding protein 1 (Ltbp1L) is essential for cardiac outflow tract septation and remodeling. Development. 2007;134:3723–32. doi: 10.1242/dev.008599. [DOI] [PubMed] [Google Scholar]
- 31.Todorovic V, Finnegan E, Freyer L, Zilberberg L, Ota M, Rifkin DB. Long form of latent TGF-beta binding protein 1 (Ltbp1L) regulates cardiac valve development. Dev Dyn. 2011;240:176–87. doi: 10.1002/dvdy.22521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drews F, Knobel S, Moser M, Muhlack KG, Mohren S, Stoll C, et al. Disruption of the latent transforming growth factor-beta binding protein-1 gene causes alteration in facial structure and influences TGF-beta bioavailability. Biochim Biophys Acta. 2008;1783:34–48. doi: 10.1016/j.bbamcr.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Oklu R, Hesketh TR, Metcalfe JC, Kemp PR. Expression of alternatively spliced human latent transforming growth factor beta binding protein-1. FEBS Lett. 1998;435:143–8. doi: 10.1016/s0014-5793(98)01054-0. [DOI] [PubMed] [Google Scholar]
- 34.Oklu R, Metcalfe JC, Hesketh TR, Kemp PR. Loss of a consensus heparin binding site by alternative splicing of latent transforming growth factor-beta binding protein-1. FEBS Lett. 1998;425:281–5. doi: 10.1016/s0014-5793(98)00257-9. [DOI] [PubMed] [Google Scholar]
- 35.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–42. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Mouse Phenotype Consortium. Ltbp1 - IKMC Project: 40539. 2014 [Google Scholar]
- 37.Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, et al. TGF-beta 1 binding protein: a component of the large latent complex of TGF-beta 1 with multiple repeat sequences. Cell. 1990;61:1051–61. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- 38.Charbonneau NL, Dzamba BJ, Ono RN, Keene DR, Corson GM, Reinhardt DP, et al. Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J Biol Chem. 2003;278:2740–9. doi: 10.1074/jbc.M209201200. [DOI] [PubMed] [Google Scholar]
- 39.Noguera I, Obata H, Gualandris A, Cowin P, Rifkin DB. Molecular cloning of the mouse Ltbp-1 gene reveals tissue specific expression of alternatively spliced forms. Gene. 2003;308:31–41. doi: 10.1016/s0378-1119(03)00463-3. [DOI] [PubMed] [Google Scholar]