Introduction
Single dose nevirapine (sdNVP) has been widely used in low resource settings for prevention of mother-to-child HIV transmission (pMTCT) and nevirapine continues to be used as part of more complex prophylactic and therapeutic regimens [1]. It is now well established that nevirapine used for pMTCT selects viral mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors (NNRTI) among a large proportion of exposed women [2-7]. Infants who fail prophylaxis and who acquire infection despite NVP exposure also develop resistance. Because transmission rates are low with antiretroviral prophylaxis, the number of children included in cohorts is small and infants have been mostly studied only 6-8 weeks after exposure [8-11].
The proportion of HIV-infected infants who have NNRTI mutations 6–8 weeks after sdNVP is usually higher than that observed among women at comparable times after exposure [2, 8, 11]. A meta-analysis estimated that the prevalence of NNRTI mutations 6 weeks after exposure was 56% [12] although higher rates (87%) have been observed in some sub-groups [13]. In contrast to adults where K103N predominates, the predominant mutation among infants is Y181C [2, 8, 11, 12]. Persistence of mutations has only been studied in small numbers of infants, but, like in adults, the prevalence of mutations declines with increasing time after exposure [9, 14]. Persistence is important as it is the mutations still present at the time of treatment initiation which predict virologic response to NNRTI-based treatment [15, 16]. For clinical and public health purposes, it is the prevalence of NNRTI mutations in infants and young children at the time of treatment initiation that needs to be accurately quantified.
It is also well established that standard methods of bulk population sequencing miss drug resistance mutations when they are present at low levels. Several more sensitive assays to detect minority variants have been developed, including real-time allele-specific PCR (AS-PCR), point mutation assays (LigAmp), oligonucleotide ligation assays (OLA) and pyrosequencing [7, 8, 11, 14, 17-20]. When these methodologies are used a larger proportion of children are found to harbor mutations [7, 8, 11, 14, 17-20]. However, the small numbers of children tested and the limited examination of later time points precludes a confident estimate of the proportion of children whose treatment may be compromised by these selected variants.
Here we examined the prevalence of drug resistance at the time of initiation of triple antiretroviral therapy in a large cohort of HIV-infected infants and young children in South Africa who had previously received sdNVP as part of pMTCT. We ascertained resistance using both standard genotyping and more sensitive AS-PCR methods for the Y181C and K103N mutations. We further investigated whether age and clinical characteristics would be associated with the detection of resistance mutations measured using both of these methods.
Methods
Samples from HIV-1 infected children
Samples for this study were collected at baseline of a randomized clinical trial designed to evaluate a novel strategy for preserving NVP as a component of treatment regimens for sdNVP-exposed children [21, 22]. Pre-treatment plasma samples were obtained from 255 HIV-1 infected children less than 24 months of age, who were exposed to sdNVP for pMTCT, and who met criteria for antiretroviral therapy at the time of recruitment. Children were enrolled at the Rahima Moosa Mother and Child Hospital in Johannesburg, South Africa between April 2005 and July 2007. Eligibility criteria for treatment included World Health Organization (WHO) stage III or IV disease, CD4% <25 if younger than 12 months or <20 if older than 12 months, or recurrent (> 2/year) or prolonged (>4 weeks) hospitalization for HIV-related complications. Prior to treatment, children were staged and samples were tested for HIV-1 RNA quantity (Roche version 1.5) and CD4 count and percent. Detailed histories were obtained and neither mothers nor children were reported to have been exposed to antiretroviral drugs other than sdNVP. Overall in the cohort, 28% of children were ever breastfed and the median duration of breastfeeding in the sub-group who initiated any breastfeeding was 60 days. Signed informed consent was obtained from the children's caregivers and the study was approved by the Institutional Review Boards of the University of the Witwatersrand and Columbia University.
Genotyping of the polymerase gene
Sequencing of the pol gene was done using an in-house assay certified by the Virology Quality Assessment Program (VQA) [23]. Briefly, viral RNA was isolated from plasma using a MagNa Pure LC Total Nucleic Acid Isolation kit on the MagNa Pure Automated System (Roche Diagnostics, Indianapolis, IN). A nested PCR was performed as previously described to generate a 1.7 kb amplicon spanning both the protease and reverse transcriptase genes. In cases where amplification of the pol gene was not obtained, the protease and reverse transcriptase regions were amplified separately. The first PCR was performed as previously described to generate a 985 bp amplicon of the reverse transcriptase gene [24], whereas the second nested PCR amplified 490 bp of the protease gene. The protease PCR used the same conditions as the reverse transcriptase PCR, but differed in that primers polCF (5’ GAAGGACACCAAATGAAAGACTGTAC) and polIRS (5’ ACTCTGGAATATTGCTGGTGATCC) were used for the initial reaction, and protease F (5’CTTCAGAACAGACCAGAGC 3’) and protease R (5’ CTCTTCTGTTAACGGCCATTG ) for the nested reaction. The PCR products were sequenced using BigDye Terminators v3.1 on an ABI3100 Genetic Analyzer (Applied Biosystems, Foster City, CA). Consensus sequences were aligned and manually edited using the Sequencher v4.5 software (GeneCodes, Ann Arbor, MI). Genotypic resistance was defined as the presence of resistance mutations associated with impaired drug susceptibility, using the Stanford Genotypic Resistance Interpretation Algorithm (http://hivdb.stanford.edu/) and the December 2009 International AIDS Society drug resistance mutation list [25]. Phylogenetic analysis of nucleic sequences was performed using MEGA 3.1 for internal quality purposes, and using reference sequences downloaded from Los Alamos (www.lanl.gov).
AS-PCR for K103N and Y181C
PCR products were further tested for the K103N mutation using an allele-specific-PCR assay, as previously described [7, 24]. Analysis of synthetic plasmid mixtures and treatment-naïve samples showed a detection cut-off of 0.2% for the minor variant. All positive samples were re-tested twice and only those that were positive on all three repeats were considered true positives.
A second AS-PCR was designed to discriminate between the Y and C alleles at position 181 of the RT genes. The reaction was performed on the LightCycler® 480, using LightCycler® 480 SYBR Green 1 Master (Roche Applied Science, Mannheim GM) and primers 71F, 75R (internal control), 76R (TGT) and 77R (TAT) [18]. The primers were added to a final concentration of 300 nM, and the reaction performed in a total volume of 25 μl. Analysis of a dilution series of wild-type 181Y and mutant 181C plasmid mixtures and 80 treatment-naïve samples showed the lower limit of detection to be 0.4%.
Statistical analyses
We calculated the proportions of children with NNRTI mutations detected using each of these methods and compared proportions between groups using chi-squared tests. Stratified analyses were done where appropriate. Descriptive statistics, including medians and 25th to 75th percentile distributions to describe the inter-quartile range (IQR), were used. Data were analyzed using GraphPad InStat v3.06, San Diego, USA.
Results
Detection of HIV-1 drug resistance mutations using population sequencing
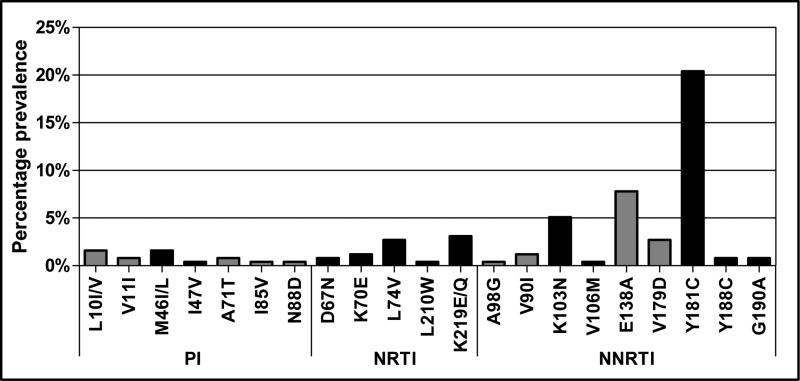
Population sequencing detected major antiretroviral drug resistance mutations from PI, NRTI and NNRTI drug classes in 31% (80/255) of all infants and children. As expected, NNRTI mutations were most prevalent and were present in 27% (69/255) of samples: 20% had Y181C and 5% K103N (Figure 1). The G190A and Y188C mutations were each detected in 2 samples and V106I/M in 1 sample. NNRTI minor mutations were also detected, specifically E138A (n=20), V179D (n=7), V90I (n=3) and A98G (n=1), as well as the NNRTI-associated polymorphisms K101Q/T (n=3), E138G/S (n=3), V179A (n=2), H221Y (n=2) and L234P (n=1). Nineteen samples had major NRTI mutations, specifically K219E/Q (n=8), L74V (n=7), K70E (n=3), D67N (n=2) and L210W (n=1). Other NRTI-associated mutations detected included T69A/N/S (n=21), V75L, V118I and T215A/I. The major PI mutations M46I/L and I47V were present in 4 samples, and minor PI mutations L10I/V, V11I, A71T, I85V, and N88D were also detected. The PI mutation T74S was found in 29 samples, and 1 sample had an insertion at position 35 in protease. All samples clustered with HIV-1 subtype C sequences except for one which was a subtype A.
Figure 1. Pattern of HIV-1 drug resistance mutations detected by population sequencing among 255 sdNVP-exposed children initiating antiretroviral therapy.
All major PI, NRTI and NNRTI mutations are shown (black bars). Minor mutations are shown in grey. Mutations are categorized as per the December 2009 IAS mutation list [25]
Detection of Y181C and K103N minority populations by AS-PCR
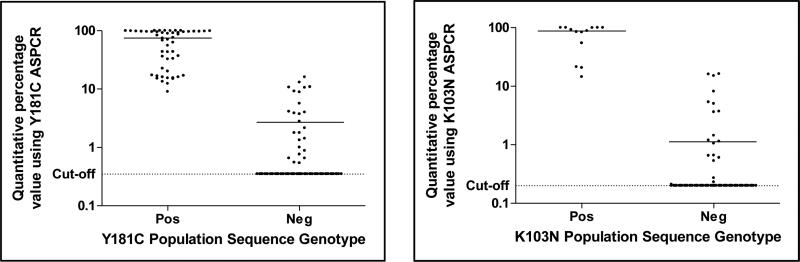
The Y181C mutation was detected by AS-PCR in all 52 samples that were positive by sequencing as well as an additional 10% (25/250) that were wild-type by sequencing. Two samples had the polymorphisms Y181F/S in the primer binding regions which interfered with the AS-PCR assay giving false positive results, and were excluded from analysis. An further three samples were all poorly amplified by AS-PCR, despite no apparent sequence differences in the primer binding regions, so were considered indeterminate and therefore also excluded. The median AS-PCR quantitative values (estimated % of viral population) for the 52 genotype positive samples was 74.9% (IQR 21.9 - 97.1), whereas for the 25 AS-PCR only positive samples it was 2.8% (IQR 1.0 - 8.9) (Figure 2).
Figure 2. Comparison of Y181C and K103N AS-PCR quantitative value relative to population genotype.
The actual quantitative values of the percent of the viral population found to be Y181C (left panel) and K103N (right panel) by AS-PCR stratified by genotype result are shown. Solid horizontal lines indicate the median quantitative value of AS-PCR positive samples in each group. 173 and 221 samples were negative for Y181C and K103N by population sequencing and AS-PCR, respectively.
The K103N mutation was detected in an additional 8% (21/255) of samples by AS-PCR. All of the samples positive for K103N by population sequencing were similarly positive by AS-PCR, with a median quantitative value of 87.8% (IQR, 54.3-99.5), whereas for the samples only positive on AS-PCR it was 1.1% (IQR, 0.6-5.0) (Figure 2). The polymorphism K103R was detected by population sequencing in 4 samples, and did not appear to affect the assay in that all 4 samples were negative by AS-PCR. An additional sample with an unusual K103T polymorphism had a quantitative AS-PCR value of 0.9 for K103N. Similar to K103R, this polymorphism falls out of the primer binding region and thus the low positive AS-PCR value was assumed to reflect the presence of K103N.
No samples had both K103N and Y181C detected by population sequence, and only 12 (4.7%) had both mutations detected by either method. Specifically 8 samples were positive for Y181C by population sequence and K103N by AS-PCR, 2 samples were positive for K103N by population sequence and Y181C by AS-PCR, and 2 samples were positive for both mutations by AS-PCR. In all cases where both mutations were detected, the infants were less than 6 months of age. One sample had both K103N and Y188C by population genotype, otherwise multiple major NNRTI mutations were not found.
Interestingly, the ratio of all Y181C-containing samples detected only by population sequencing relative to those detected by either method (52/77, 68%) was significantly higher than the ratio of all K103N-containing samples detected only by population sequencing (13/34, 38.2%), p=0.006. This indicates that K103N occurred mostly as a minority species and hence the K103N AS-PCR was particularly useful at revealing additional samples harboring this mutation.
Decline in NNRTI mutations with time since exposure
To assess the presence of NNRTI mutations over time since exposure to sdNVP, the 255 children were categorized into four age groups. Analysis of population genotypes showed a clear age trend with 45.7% of infants 0-6 months having major NNRTI mutations, compared to 23.7% at 6-12 months, 20% at 12-18 months and 0% at 18-24 months (p<0.0001) (Table 1) indicating that these mutations fade with age. The Y181C mutation was most common at all time-points and only Y181C and K103N persisted until 18 months. After 18 months all samples were wild-type by population sequencing. Other major NNRTI mutations V106M, Y188C and G190A were found in a small proportion of infants under 12 months but were absent in older children. Polymorphisms classified as minor mutations were generally present at low frequencies and did not decline over time.
Table 1.
NNRTI mutations detected by population sequencing and AS-PCR among 255 sdNVP-exposed children initiating antiretroviral therapy.
| 0-6 months | 6-12 months | 12-18 months | 18-24 months | Total | p-value** | |
|---|---|---|---|---|---|---|
| N=81 | N=97 | N=45 | N=32 | N=255 | ||
|
Mutation detection
| ||||||
| Total N (%) with detectable major NNRTI mutations: | ||||||
| By population sequencing | 37 (45.7%) | 23 (23.7%) | 9 (20.0%) | 0 | 69 (27.1%) | <0.0001 |
| By AS-PCR only | 13 (16.0%) | 14 (14.4%) | 1 (2.2%) | 5 (15.6%) | 34 (12.9%) | 0.13 |
| N (%) with detectable Y181C: | ||||||
| By population sequencing | 25 (30.9%) | 20 (20.6%) | 7 (15.6%) | 0 | 52 (20.4%) | 0.0025 |
| By AS-PCR only* | 10 (12.5%) | 11 (11.8%) | 1 (2.2%) | 3 (9.4%) | 25 (10.0%) | 0.27 |
| N (%) with detectable K103N: | ||||||
| By population sequencing | 10 (12.3%) | 1 (1.0%) | 2 (4.4%) | 0 | 13 (5.1%) | 0.0031 |
| By AS-PCR only | 16 (19.8%) | 3 (3.1%) | 0 | 2 (6.3%) | 21 (8.2%) | <0.0001 |
| N (%) with other major NNRTI mutations | ||||||
| V106IM | 0 | 1 (1.0%) | 0 | 0 | 1 (0.4%) | 0.65 |
| Y188C | 1 (1.2%) | 1 (1.0%) | 0 | 0 | 2 (0.8%) | 0.83 |
| G190A | 2 (2.5%) | 0 | 0 | 0 | 2 (0.8%) | 0.23 |
| N (%) with minor NNRTI mutations: | ||||||
| V90I | 2 (2.5%) | 0 | 1 (2.2%) | 0 | 3 (1.2%) | 0.37 |
| A98G | 0 | 1 (1.0%) | 0 | 0 | 1 (0.4%) | 0.65 |
| E138A | 8 (9.9%) | 5 (5.2%) | 6 (13.3%) | 1 (3.1%) | 20 (7.8%) | 0.23 |
| V179D | 1 (1.2%) | 4 (5.1%) | 2 (4.4%) | 0 | 7 (2.7%) | 0.43 |
One child aged 0-6 months and 4 children aged 6-12 month were excluded from analyses due to indeterminate Y181C AS-PCR results.
P-values determined by comparing percentage of positive samples in each age category (Chi-squared test for trend)
In contrast to population sequencing there was no significant age trend for mutations detected by AS-PCR only (Table 1). However analysis of the K103N mutation showed that there was a significant decline with age that was particularly apparent in the first 6 months suggesting that minority K103N variants do not persist as long as Y181C in infants. Of the 5 samples that were positive for AS-PCR 18-24 months post-exposure to sdNVP but wild-type by sequencing, 3 were Y181C positive and 2 were K103N positive.
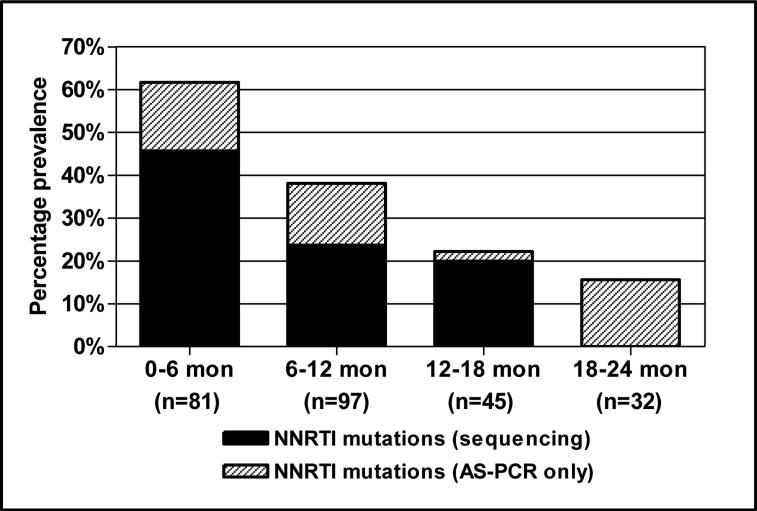
When population sequencing- and AS-PCR-derived data were combined, 61.9% of all infants 0-6 months of age harbored NNRTI resistance mutations prior to starting ART therapy (Table 1 and Figure 3). The frequency of resistance mutations declined significantly over time with 38.6% of 6-12 months and 22.2% of 12-18 month old children showing evidence of NNRTI resistance. Minority variants were still present in 15.6% of children 18-24 months after sdNVP.
Figure 3. Overall prevalence of NNRTI mutations among 255 sdNVP-exposed children initiating antiretroviral therapy.
Data show the prevalence of NNRTI mutations by genotyping (black bars) plus the additional prevalence when including samples that were only identified using the Y181C or the K103N AS-PCR (grey bars). Samples classified as indeterminate by Y181C AS-PCR were excluded.
Associations between resistance and clinical characteristics
In addition to age, a trend towards higher pre-treatment HIV-1 RNA quantities and greater likelihood of detecting NNRTI mutations was found (p=0.03). This association appeared to be at least partially explained by age. When stratified into age categories, there was no longer a consistent trend towards increasing resistance with higher pre-treatment viral load. Associations between increasing resistance and younger age were observed consistently across all viral load strata. There was no association between CD4 percentage, child gender, any breastfeeding and clinical stage and detection of NNRTI resistance by either population sequencing or AS-PCR.
Discussion
Most prior studies have described the frequency of drug resistance mutations among infants 6-8 weeks after sdNVP exposure [8-11, 13]. Here we quantified the frequency of resistance in a large cohort of HIV-infected infants and young children under 2 years of age at the time of initiating antiretroviral therapy. The most important finding is the clear age trend with 62% of infants 0-6 months of age harboring resistant variants compared to only 16% in the 18-24 month age group. After 18 months of age, mutations were only detected by AS-PCR. A recent trial confirms that virologic response is better if boosted PI-based therapy is initiated among sdNVP-exposed infants rather than NNRTI-based therapy [26]. We have examined the virologic responses of these children to their first line regimen of d4T, 3TC and LPV/r or RTV [27] and have examined whether pre-treatment resistance profile affects virological response [28]. As expected, there was no association between NNRTI mutations at treatment initiation and virological failure. Our data provide support for the World Health Organization (WHO) pediatric treatment guidelines that expanded recommendations for use of boosted PI-based regimen to all children under 2 years of age if drug-exposed as part of pMTCT interventions (previously only under one year).
Consistent with other pediatric studies, the Y181C mutation was most frequently detected [2, 9, 14, 17]. This is in contrast to studies in women where K103N predominates [2-6]. It is unclear why the virus transmitted to children develops different mutations to the ones present in their mothers. Viral loads are higher in infants and drug exposure may be greater due to the fact that children receive in utero exposure in addition to oral dosing after birth. In children, Y181C detected either by standard methods [26] or using ultra-sensitive methods [15, 16] have been associated with attenuated virologic response to nevirapine-based therapy. Intriguing new results in adults suggests that Y181C when present only at low frequencies may not have as marked an adverse effect on response to efavirenz-based therapy [29]. Unfortunately, for infants and young children treatment options are more limited and efavirenz cannot yet be used. Thus this option cannot be tested in this young age group.
The detection of NRTI and PI mutations in some children in this cohort was unexpected. To the best extent of our information, these infants and mothers had not been exposed to antiretroviral drugs either during pregnancy or breastfeeding, other than sdNVP. In addition, studies in South African have indicated that levels of transmitted resistance remain low [23], suggesting that in some cases these may be naturally occurring mutations. Unusual profiles of mutations, unrelated to the drugs used for prophylaxis, have also been noted among infants in the United States [30]. Given that there is a rapid expansion of antiretroviral therapy in South Africa, drug resistance patterns in both drug-naïve and drug-exposed children should continue to be evaluated to monitor the development of regimen-compromising polymorphisms and mutations that could be transmitted to or selected for in this population.
The samples in this study were genotypically analyzed using conventional population sequencing as well as allele-specific real-time PCR assays. Whilst population sequencing remains the gold-standard in genotypic analysis and provides a comprehensive description of all amino acids in the regions analyzed, it is comparatively insensitive, detecting resistant species in heterogeneous populations only when they are above 20% [31, 32]. Our data confirms that AS-PCR, with its detection levels of ~1% of minority species, provides a significantly more accurate representation of the extent and persistence of mutant species. Whist the assay is confined to analysis of one mutation at a time, selection of key mutations can provide a relevant indication of antiretroviral drug class resistance in patients. In our study, analysis of the Y181C and K103N mutations by AS-PCR was sufficient, as other NNRTI mutations were rare as shown by population sequencing. The AS-PCR identified mutant species correctly and quantified them accurately as all those with higher levels were also positive on the population sequence. In addition, 100% and 98% of the samples could be analyzed by the K103N and Y181C AS-PCR assays respectively. Only 5 samples (2%) were indeterminate on the Y181C assay, 2 of which were due to the presence of unusual amino acids within the primer binding region. However, in some cases polymorphisms can be tolerated as evidenced by the K103R genotype which did not interfere with the K103N assay. Overall, the high sensitivity and relative simplicity of the AS-PCR assay makes this methodology an attractive one for investigating drug resistance in samples where the mutational patterns can be anticipated. Further clarification of the clinical consequence of minority variant detection of Y181C and K103N in children is warranted.
Analysis of the samples using both conventional genotyping and the more sensitive AS-PCR allowed us to detect interesting differences between the dynamics of Y181C and K103N mutations in children. In more than two-thirds of cases, Y181C was detected by conventional genotyping with low level mutations detected by AS-PCR constituting the remainder. Whereas with K103N the reverse pattern was observed, with more than 60% of mutations detected only with the more sensitive AS-PCR. Although with both Y181C and K103N the prevalence of mutations declined with age, the steepness of the decline was more marked with K103N. Y181C therefore appears to be a more persisting mutation in children compared to adults. The presence of both Y181C and K103N in children may require longer periods before mutations fade sufficiently to allow for reintroducing NNRTI-based therapies.
In women, it was initially hypothesized that the clinical consequences of sdNVP in compromising virologic response to primary NNRTI therapy may be completely attenuated within 6 months of exposure [33]. Subsequent studies suggested that this wash-out period is more likely to be 12-18 months [34-36]. Studies in infants similarly support poor virologic responses to NNRTI-based therapy in sdNVP-exposed infants, especially when initiating therapy before the age of 12 months [15, 33] although this is not consistently observed [37]. Recent evidence indicates that a delay between exposure and treatment initiation should be avoided in infants. A landmark study demonstrated that infants fared better if treatment was initiated soon after diagnosis rather than waiting until clinical indicators were met [38]. Thus clinical programs should initiate therapy for infants as early as possible, which is also the time when the detection of resistance mutations is at its highest and most compromising.
Pre-treatment drug resistance screening is recommended in many resource-rich environments. It would be helpful to identify which children could be safety initiated on nevirapine-based therapy despite their history of exposure. The high costs and limited laboratory capacity, as well as the challenges of returning results to clinical sites rapidly so that treatment can be started almost immediately, preclude routine screening in any public service in sub-Saharan Africa to our knowledge. Our data also indicate that in children less than 6 months of age, over 60% have detectable resistance mutations. The costs of screening would need to be balanced against the money saved by initiating the less expensive regimen in the remaining 40%. Newer pMTCT regimens and more widespread use of maternal therapy may reduce the prevalence of NNRTI mutations among infected children making for a more favorable cost-benefit ratio of pre-treatment resistance screening.
A limitation of this study is its cross-sectional nature. We do not have data on what the prevalence of mutations was soon after sdNVP exposure to confirm in a longitudinal design that mutations faded over time. Nevertheless, we did not detect associations between markers of increased severity of disease and resistance once age was taken into account. Moreover, our sample is informative for its generalizability to clinical practice as it included a large cohort who had been exposed to sdNVP as used as part of routine public health programs. Thus it provides a useful estimate for policy purposes of the extent to which it is necessary to avoid primary NNRTI-based therapy in infants presenting at different ages who report a history of sdNVP exposure. These estimates may be useful for programs that do not have sufficient resources to provide boosted PI-based regimens to all exposed children who could ration use of this regimen to the youngest children only. Our estimates may also be useful for cost-effectiveness analyses of drug resistance testing in low resource settings.
Table 2.
Associations between pre-treatment NNRTI resistance mutations detected by population sequencing and AS-PCR and clinical characteristics.
| Clinical characteristics | Y181C by population genotype | K103N by population genotype | Y181C by AS-PCR only* | K103N by AS-PCR only | Other major NNRTI mutations | No NNRTI mutations detected | p-value** |
|---|---|---|---|---|---|---|---|
| Age of child | |||||||
| 0-12 months | 45/178 (25.3%) | 11/178 (6.2%) | 18/173 (10.4%) | 9/178 (5.1%) | 4/178 (2.2%) | 86/178 (48.3%) | |
| 12-24 months | 7/77 (9.1%) | 2/77 (2.6%) | 4/77 (5.2%) | 2/77 (2.6%) | 0 | 62/77 (80.5%) | <0.0001 |
| Gender | |||||||
| Male | 26/127 (20.5%) | 3/127 (2.4%) | 10/124 (8.1%) | 5/127 (3.9%) | 2/127 (1.6%) | 78/127 (61.4%) | |
| Female | 26/128 (20.3%) | 10/128 (7.8%) | 12/126 (9.5%) | 6/128 (4.7%) | 2/128 (1.6%) | 70/128 (54.7%) | 0.29 |
| Pre-treatment CD4% | |||||||
| 0-20 | 33/141 (23.4%) | 6/141 (4.3%) | 14/138 (10.1%) | 5/141 (3.5%) | 3/141 (2.1%) | 77/141 (54.6%) | |
| >=20 | 19/114 (16.7%) | 7/114 (6.1%) | 8/112 (7.1%) | 6/114 (5.3%) | 1/114 (0.9%) | 71/114 (62.3%) | 0.28 |
| Pre-Rx viral load*** | |||||||
| <100,000 copies/ml | 0 | 1/17 (5.9%) | 1/16 (6.3%) | 0 | 0 | 14/17 (82.4%) | |
| 100,000-750,000 | 10/60 (16.7%) | 3/60 (5.0%) | 4/59 (6.8%) | 5/60 (8.3%) | 1/60 (1.7%) | 36/60 (60.0%) | |
| ≥750,000 copies/ml | 38/160 (23.8%) | 9/160 (5.6%) | 16/157 (10.2%) | 4/160 (2.5%) | 2/160 (1.3%) | 88/160 (55.0%) | 0.0314 |
| Clinical stage**** | |||||||
| I | 5/39 (12.8%) | 3/39 (7.7%) | 2/37 (5.4%) | 3/39 (7.7%) | 0 | 24/39 (61.5%) | |
| II | 1/11 (9.1%) | 0 | 2/11 (18.2%) | 1/11 (9.1%) | 0 | 7/11 (63.6%) | |
| III | 27/126 (21.4%) | 5/126 (4.0%) | 12/123 (9.8%) | 3/126 (2.4%) | 2/126 (1.6%) | 74/126 (58.7%) | |
| IV | 18/76 (23.7%) | 5/76 (6.6%) | 6/76 (7.9%) | 4/76 (5.3%) | 2/76 (2.6%) | 41/76 (53.9%) | 0.25 |
Five Y181C AS-PCR indeterminate samples were excluded from analysis
P-values determined by comparing all subjects with NNRTI mutations to those without (Chi-squared tests)
Eighteen samples had no corresponding viral load data
Three samples had no corresponding clinical staging data
Acknowledgments
We would like to thank the participants and their care-givers and the clinical and administrative study team.
Sources of funding: The clinical study was supported in part by grants from the National Institutes of Child Health and Human Development (NICHD) HD 47177 and Secure the Future Foundation. The laboratory work was funded by a grant from the National Department of Health in South Africa as part of the Operational Plan for the Comprehensive HIV and AIDS Care, Management and Treatment of South Africa.
Footnotes
Author Contributions: AC, EJA, LM and LK participated in the study design. GH performed the genotyping and sequence analysis. AC, GS, TM participated in the clinical management of the participants and data collection. GH and LM conducted the data analysis and wrote the first draft. All authors participated in reviewing and commenting on the manuscript.
Part of this work was presented at the 16th Conference on Retroviruses and Opportunistic Infections (CROI), Montreal, Canada, 7-11 February 2009.
References
- 1.Fowler MG, Lampe MA, Jamieson DJ, Kourtis AP, Rogers MF. Reducing the risk of mother-to-child human immunodeficiency virus transmission: past successes, current progress and challenges, and future directions. Am J Obstet Gynecol. 2007;197:S3–9. doi: 10.1016/j.ajog.2007.06.048. [DOI] [PubMed] [Google Scholar]
- 2.Eshleman SH, Mracna M, Guay LA, Deseyve M, Cunningham S, Mirochnick M, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). Aids. 2001;15:1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JA, Li JF, Morris L, Martinson N, Gray G, McIntyre J, Heneine W. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192:16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn L, Sinkala M, Kankasa MP, Kasonde P, Thea DM, Aldrovandi GM. Nevirapine resistance viral mutations after repeat use of nevirapine for prevention of perinatal HIV transmission. J Acquir Immune Defic Syndr. 2006;42:260–262. doi: 10.1097/01.qai.0000214820.26281.df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flys TS, Mwatha A, Guay LA, Nakabiito C, Donnell D, Musoke P, et al. Detection of K103N in Ugandan women after repeated exposure to single dose nevirapine. Aids. 2007;21:2077–2082. doi: 10.1097/QAD.0b013e3282703847. [DOI] [PubMed] [Google Scholar]
- 6.Kassaye S, Lee E, Kantor R, Johnston E, Winters M, Zijenah L, et al. Drug resistance in plasma and breast milk after single-dose nevirapine in subtype C HIV type 1: population and clonal sequence analysis. AIDS Res Hum Retroviruses. 2007;23:1055–1061. doi: 10.1089/aid.2007.0045. [DOI] [PubMed] [Google Scholar]
- 7.Loubser S, Balfe P, Sherman G, Hammer S, Kuhn L, Morris L. Decay of K103N mutants in cellular DNA and plasma RNA after single-dose nevirapine to reduce mother-to-child HIV transmission. Aids. 2006;20:995–1002. doi: 10.1097/01.aids.0000222071.60620.1d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flys TS, McConnell MS, Matovu F, Church JD, Bagenda D, Khaki L, et al. Nevirapine resistance in women and infants after first versus repeated use of single-dose nevirapine for prevention of HIV-1 vertical transmission. J Infect Dis. 2008;198:465–469. doi: 10.1086/590160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinson NA, Morris L, Gray G, Moodley D, Pillay V, Cohen S, et al. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr. 2007;44:148–153. doi: 10.1097/QAI.0b013e31802b920e. [DOI] [PubMed] [Google Scholar]
- 10.Kurle SN, Gangakhedkar RR, Sen S, Hayatnagarkar SS, Tripathy SP, Paranjape RS. Emergence of NNRTI drug resistance mutations after single-dose nevirapine exposure in HIV type 1 subtype C-infected infants in India. AIDS Res Hum Retroviruses. 2007;23:682–685. doi: 10.1089/aid.2006.0167. [DOI] [PubMed] [Google Scholar]
- 11.Church JD, Omer SB, Guay LA, Huang W, Lidstrom J, Musoke P, et al. Analysis of nevirapine (NVP) resistance in Ugandan infants who were HIV infected despite receiving single-Dose (SD) NVP versus SD NVP plus daily NVP up to 6 weeks of age to prevent HIV vertical transmission. J Infect Dis. 2008;198:1075–1082. doi: 10.1086/591503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arrive E, Newell ML, Ekouevi DK, Chaix ML, Thiebaut R, Masquelier B, et al. Prevalence of resistance to nevirapine in mothers and children after single-dose exposure to prevent vertical transmission of HIV-1: a meta-analysis. Int J Epidemiol. 2007;36:1009–1021. doi: 10.1093/ije/dym104. [DOI] [PubMed] [Google Scholar]
- 13.Eshleman SH, Hoover DR, Chen S, Hudelson SE, Guay LA, Mwatha A, et al. Resistance after single-dose nevirapine prophylaxis emerges in a high proportion of Malawian newborns. Aids. 2005;19:2167–2169. doi: 10.1097/01.aids.0000194800.43799.94. [DOI] [PubMed] [Google Scholar]
- 14.Micek MA, Blanco AJ, Beck IA, Dross S, Matunha L, Montoya P, et al. Nevirapine resistance by timing of HIV type 1 infection in infants treated with single-dose nevirapine. Clin Infect Dis. 2010;50:1405–1414. doi: 10.1086/652151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLeod IJ, Rowley CF, Thior I, Wester C, Makhema J, Essex M, Lockman S. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J Clin Virol. 2010;48:162–167. doi: 10.1016/j.jcv.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowley CF, Boutwell CL, Lee EJ, MacLeod IJ, Ribaudo HJ, Essex M, Lockman S. Ultrasensitive detection of minor drug-resistant variants for HIV after nevirapine exposure using allele-specific PCR: clinical significance. AIDS Res Hum Retroviruses. 2010;26:293–300. doi: 10.1089/aid.2009.0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troyer RM, Lalonde MS, Fraundorf E, Demers KR, Kyeyune F, Mugyenyi P, et al. A radiolabeled oligonucleotide ligation assay demonstrates the high frequency of nevirapine resistance mutations in HIV type 1 quasispecies of NVP-treated and untreated mother-infant pairs from Uganda. AIDS Res Hum Retroviruses. 2008;24:235–250. doi: 10.1089/aid.2007.0138. [DOI] [PubMed] [Google Scholar]
- 18.Palmer S, Boltz V, Martinson N, Maldarelli F, Gray G, McIntyre J, et al. Persistence of nevirapine-resistant HIV-1 in women after single-dose nevirapine therapy for prevention of maternal-to-fetal HIV-1 transmission. Proc Natl Acad Sci U S A. 2006;103:7094–7099. doi: 10.1073/pnas.0602033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flys TS, Donnell D, Mwatha A, Nakabiito C, Musoke P, Mmiro F, et al. Persistence of K103N-containing HIV-1 variants after single-dose nevirapine for prevention of HIV-1 mother-to-child transmission. J Infect Dis. 2007;195:711–715. doi: 10.1086/511433. [DOI] [PubMed] [Google Scholar]
- 20.Moorthy A, Gupta A, Bhosale R, Tripathy S, Sastry J, Kulkarni S, et al. Nevirapine resistance and breast-milk HIV transmission: effects of single and extended-dose nevirapine prophylaxis in subtype C HIV-infected infants. PLoS One. 2009;4:e4096. doi: 10.1371/journal.pone.0004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coovadia A, Abrams EJ, Stehlau R, Meyers T, Martens L, Sherman G, et al. Reuse of nevirapine in exposed HIV-infected children after protease inhibitor-based viral suppression: a randomized controlled trial. Jama. 2010;304:1082–1090. doi: 10.1001/jama.2010.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coovadia A, Abrams EJ, Strehlau R, Martens L, Sherman G, Meyers T, et al. Randomized clinical trial of switching to nevirapine-based therapy for infected children exposed to nevirapine prophylaxis. 5th IAS Conference on HIV Pathogenesis, Treatment and Prevention; Cape Town, South Africa. 19-22 July 2009. [Google Scholar]
- 23.Pillay V, Ledwaba J, Hunt G, Rakgotho M, Singh B, Makubalo L, et al. Antiretroviral drug resistance surveillance among drug-naive HIV-1- infected individuals in Gauteng Province, South Africa in 2002 and 2004. Antivir Ther 2008. 13(Suppl 2):101–107. [PubMed] [Google Scholar]
- 24.Coovadia A, Hunt G, Abrams EJ, Sherman G, Meyers T, Barry G, et al. Persistent Minority K103N Mutations among Women Exposed to Single-Dose Nevirapine and Virologic Response to Nonnucleoside Reverse-Transcriptase Inhibitor-Based Therapy. Clin Infect Dis. 2009;48:462–472. doi: 10.1086/596486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: December 2009. Top HIV Med. 2009;17:138–145. [PubMed] [Google Scholar]
- 26.Palumbo P, Lindsey JC, Hughes MD, Cotton MF, Bobat R, Meyers T, et al. Antiretroviral treatment for children with peripartum nevirapine exposure. N Engl J Med. 2010;363:1510–1520. doi: 10.1056/NEJMoa1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reitz C, Coovadia A, Ko S, Meyers T, Strehlau R, Sherman G, et al. Initial response to protease-inhibitor-based antiretroviral therapy among children less than 2 years of age in South Africa: effect of cotreatment for tuberculosis. J Infect Dis. 2010;201:1121–1131. doi: 10.1086/651454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunt G, Taylor BS, Coovadia A, Abrams E, Sherman G, Meyers T, et al. Development of Drug Resistance among a Cohort of HIV-infected Infants Exposed to Nevirapine for PMTCT Initiating PI-based ART in South Africa.. 16th Conference on Retroviruses and Opportunistic Infections; Montreal, Canada. 8-11 February 2009. [Google Scholar]
- 29.Halvas EK, Aldrovandi GM, Balfe P, Beck IA, Boltz VF, Coffin JM, et al. Blinded, multicenter comparison of methods to detect a drug-resistant mutant of human immunodeficiency virus type 1 at low frequency. J Clin Microbiol. 2006;44:2612–2614. doi: 10.1128/JCM.00449-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karchava M, Pulver W, Smith L, Philpott S, Sullivan TJ, Wethers J, Parker MM. Prevalence of drug-resistance mutations and non-subtype B strains among HIV-infected infants from New York State. J Acquir Immune Defic Syndr. 2006;42:614–619. doi: 10.1097/01.qai.0000225871.87456.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Church JD, Jones D, Flys T, Hoover D, Marlowe N, Chen S, et al. Sensitivity of the ViroSeq HIV-1 genotyping system for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagn. 2006;8:430–432. doi: 10.2353/jmoldx.2006.050148. quiz 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson JA, Li JF, Wei X, Lipscomb J, Bennett D, Brant A, et al. Simple PCR assays improve the sensitivity of HIV-1 subtype B drug resistance testing and allow linking of resistance mutations. PLoS ONE. 2007;2:e638. doi: 10.1371/journal.pone.0000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 34.Kuhn L, Semrau K, Ramachandran S, Sinkala M, Scott N, Kasonde P, et al. Mortality and virologic outcomes after access to antiretroviral therapy among a cohort of HIV-infected women who received single-dose nevirapine in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2009;52:132–136. doi: 10.1097/QAI.0b013e3181ab6d5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lockman S, Team AOS. Lopinavir/ritonavir+Tenofovir/Emtricitabine is superior to Nevirapine+Tenofovir/Emtracitabine for women with prior exposure to single-dose Nevirapine.. 16th Conference on Retroviruses and Opportunistic Infections; Montreal, Canada. Feb 8-11 2009. [Google Scholar]
- 36.Stringer JS, McConnell MS, Kiarie J, Bolu O, Anekthananon T, Jariyasethpong T, et al. Effectiveness of non-nucleoside reverse-transcriptase inhibitor-based antiretroviral therapy in women previously exposed to a single intrapartum dose of nevirapine: a multi-country, prospective cohort study. PLoS Med. 2010;7:e1000233. doi: 10.1371/journal.pmed.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musoke PM, Barlow-Mosha L, Bagenda D, Mudiope P, Mubiru M, Ajuna P, et al. Response to antiretroviral therapy in HIV-infected Ugandan children exposed and not exposed to single-dose nevirapine at birth. J Acquir Immune Defic Syndr. 2009;52:560–568. doi: 10.1097/qai.0b013e3181b93a5a. [DOI] [PubMed] [Google Scholar]
- 38.Violari A, Cotton MF, Gibb DM, Babiker AG, Steyn J, Madhi SA, et al. Early antiretroviral therapy and mortality among HIV-infected infants. N Engl J Med. 2008;359:2233–2244. doi: 10.1056/NEJMoa0800971. [DOI] [PMC free article] [PubMed] [Google Scholar]