Abstract
5-Aminolevulinate synthase is the first enzyme of the heme biosynthetic pathway in non-plant eukaryotes and some prokaryotes. The enzyme functions as a homodimer and requires pyridoxal 5′-phosphate as a co-factor. Although the roles of defined amino acids in the active site and catalytic mechanism have been recently explored using site-directed mutagenesis, much less is known about the role of the 5-aminolevulinate synthase polypeptide chain arrangement in folding, structure, and ultimately, function. To assess the importance of the continuity of the polypeptide chain, circularly permuted 5-aminolevulinate synthase variants were constructed through either rational design or screening of an engineered random library. One percent of the random library clones were active, and a total of 21 active variants had sequences different from that of the wild type 5-aminolevulinate synthase. Out of these 21 variants, 9 displayed unique circular permutations of the 5-aminolevulinate synthase polypeptide chain. The new termini of the active variants disrupted secondary structure elements and loop regions and fell in 100 amino acid regions from each terminus. This indicates that the natural continuity of the 5-aminolevulinate synthase polypeptide chain and the sequential arrangement of the secondary structure elements are not requirements for proper folding, binding of the cofactor, or assembly of the two subunits. Furthermore, the order of two identified functional elements (i.e. the catalytic and the glycine-binding domains) is apparently irrelevant for proper functioning of the enzyme. Although the wild type 5-aminolevulinate synthase and the circularly permuted variants appear to have similar, predicted overall tertiary structures, they exhibit differences in the arrangement of the secondary structure elements and in the cofactor-binding site environment. Taken together, the data lead us to propose that the 5-aminolevulinate synthase overall structure can be reached through multiple or alternative folding pathways.
5-Aminolevulinate synthase (ALAS)1 (EC 2.3.1.37) catalyzes the condensation of glycine and succinyl-CoA to form 5-aminolevulinic acid (ALA), CoA, and carbon dioxide. This is the first and the major regulatory reaction in the heme biosynthetic pathway in non-plant eukaryotes and the a-subclass of purple bacteria (1, 2). Mammals encode two distinct ALAS isoforms, housekeeping ALAS and erythroid-specific ALAS isoform; the latter is only expressed in developing erythrocytes (3, 4) and is responsible for ~90% of the total synthesized heme in the body. The gene encoding the human erythroid ALAS has been localized on the band Xp11.21 of the X chromosome (5), whereas the gene for the human housekeeping ALAS has been assigned to the band 3p21 of chromosome 3 (6). Mutations in the human erythroid ALAS have been associated with X-linked sideroblastic anemia (7–10), a disorder characterized by inadequate formation of heme and an overaccumulation of iron in erythroblast mitochondria (7).
Murine erythroid ALAS functions as a homodimer, with the active site residing at the subunit interface (11), and requires pyridoxal 5′-phosphate (PLP) as an essential cofactor (2). In murine erythroid ALAS, the PLP cofactor is covalently bound to the Lys-313 residue through a Schiff base linkage, forming the cofactor-protein complex termed internal aldimine (12). In addition, the conserved Lys-313 residue was reported to have a catalytic role (13, 14).
Steady-state kinetic analysis of the ALAS-catalyzed reaction indicated an Ordered Bi Bi mechanism, in which glycine binds before succinyl-CoA and ALA is dissociated from the enzyme last (15). The chemical mechanism of the ALAS reaction appears to be similar to that of other PLP-dependent enzymes catalyzing reactions involving amino acids (16, 17). Briefly, the binding of glycine leads to the formation of the PLP-glycine complex termed external aldimine, which upon removal of the pro-R proton of glycine yields a resonance-stabilized quinonoid intermediate. Subsequently, this intermediate reacts with the second substrate succinyl-CoA forming a putative α-amino-β-ketoadipate aldimine (14). The next steps involve the decarboxylation of the glycine-derived carboxyl group and formation of an aldimine to ALA. The release of ALA, or a protein conformation change associated with it, was suggested to be the rate-limiting step of the ALAS-catalyzed reaction (18).
Although the three-dimensional structure of ALAS has yet to be determined, the evolutionary proximity between ALAS and other members of the α-family of PLP-dependent enzymes of known three-dimensional structure made it possible to perform homology modeling studies of ALAS structure and function (17). The Arg-439 residue is involved in the binding of glycine by forming a salt bridge with its negatively charged α-carboxylate group (19). The Asp-279 residue appears to be positioned close to the pyridinium ring nitrogen of the PLP cofactor. Its negatively charged carboxylate group stabilizes the protonated form of the ring nitrogen, thus enhancing the electron sink capacity of the PLP cofactor (20). Tyr-121 has been shown to be involved in PLP cofactor binding by donating a hydrogen bond from its hydroxyl group to the phosphate oxygen of the PLP cofactor (21).
Whereas the roles of defined active site amino acids in the structure and catalytic mechanism of ALAS have been recently explored using site-directed mutagenesis (14, 19–21), much less is known about the role of the ALAS polypeptide chain arrangement in folding, final structure, and catalysis. Circular permutation, which disrupts the continuity of the polypeptide chain by placing the original N and C termini at new locations, has proved to be a valuable tool for studying the effects of polypeptide chain rearrangements on catalytic activity and folding of proteins (22, 23). With circular permutation of proteins, the natural N and C termini are covalently linked, and new termini are created upon cleavage of the circularized protein at a different sequence position (24, 25). The result is a change in the primary (i.e. order of the amino acid sequence), secondary, and possibly tertiary structures (24, 26). To date, the circular permutation approach has been successfully applied to more than 20 proteins. These include, for example, T4 lysozyme (22), aspartate transcarbamoylase (27), disulfide oxidoreductase DsbA (28), and dehydrofolate reductase (26). Circularly permuted variants have been constructed either by engineering recombinant variants with N and C termini at selected locations (23, 26, 29–31) or by randomly generating new termini (27, 28). In addition, circularly permuted chains have been generated with protein chemical modification methods involving the covalent linkage of the N and C termini, followed by hydrolysis of a single polypeptide bond at a different position from that of the linked original termini (24). The folding of the circularly permuted variants into functional proteins indicated that these variants can achieve a proper conformation for function and suggested that the amino acid sequence, and not the positioning of the N and C termini, determines the three-dimensional structure (28, 30, 31). Importantly, the identification of “functional elements,” which have been defined as the smallest continuous sequences required for catalytic activity (23), makes possible the exploration of the “architecture of enzyme function.” These functional elements, which are spread throughout the primary structure, define the functional active site in a properly folded enzyme (23).
Here, we analyze the role of the ALAS polypeptde chain in relation to folding and assembly of the holoenzyme. We report that the circular permutation of the ALAS polypeptide chain affects neither the folding/assembly of the ALAS subunits into the dimeric holoprotein nor the activity of the enzyme. The stable and active circularly permuted ALAS variants, however, appear to have different topologies.
EXPERIMENTAL PROCEDURES
Reagents
The following reagents were purchased as noted: Sigma, DEAE-Sephacel, β-mercaptoethanol, PLP, bovine serum albumin, α-ketoglutarate dehydrogenase, α-ketoglutarate, NAD+ thiamine pyrophosphate, succinyl-CoA, HEPES-free acid, aprotinin, pepstatin, leupeptin, phenylmethylsulfonyl fluoride, the bicinchoninic acid protein concentration determination kit and the gel filtration molecular weight markers kit (cytochrome c, carbonic anhydrase, bovine serum albumin, alcohol dehydrogenase, β-amylase); Fisher, glycerol, mono- and dibasic potassium phosphate, disodium EDTA dihydrate; IBF Biotechnics, Ultrogel AcA-44; Bio-Rad, sodium dodecyl sulfate-polyacrylamide gel electrophoresis reagents; New England Biolabs, Vent DNA polymerase, BamHI, SalI, Ecl136 II, T4 DNA ligase, T4 DNA polymerase Klenow fragment, T4 polynucleotide kinase, and alkaline phosphatase; Promega, RQ1 DNase; U. S. Biochemical Corp., T7 Sequenase version 2.0; Stratagene Inc., PfuTurbo DNA polymerase; Amersham Pharmacia Biotech, Superdex 200 gel filtration resin; Cybersyn Inc., DNA oligonucleotides. The Escherichia coli strain HU227 (32) was a gift from Dr. C. S. Russell (City University of New York), and the E. coli strain LC24 (33) was obtained from the American Type Culture Collection (ATCC).
Construction of Plasmid pAC9
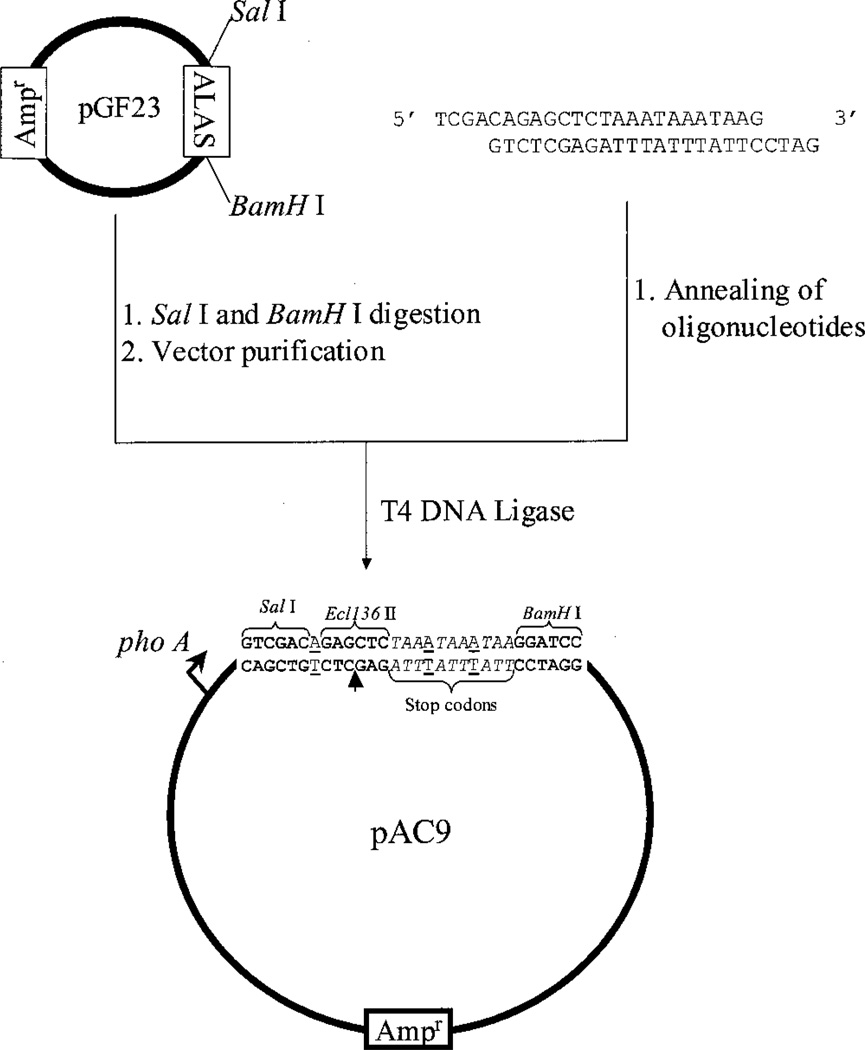
The pAC9 plasmid was used as the vector plasmid in the construction of the random library of circularly permuted ALAS variants. pAC9 contains the sequence for stop codons in the three possible reading frames and for three restriction enzyme sites (SalI, Ecl136 II, and BamHI). The Ecl136 II site was engineered so that the blunt 5′ and 3′ ends of the insert encoding the circularly permuted ALAS variants would be in frame with the ATG codon (located upstream of the SalI site (34)) and the first TAA of three stop codons sequence, respectively. Two additional nucleotides (downstream of the first TAA) were also included to generate the two other reading frames and, thus, to accommodate inserts that would not be in the same reading frame as the first TAA codon (Fig. 1). The cassette containing the sequences for the three stop codons and the three restriction enzyme sites was constructed by annealing two phosphorylated oligonucleotides (STOP1, 5′-TCGACAGAGCTCTAAATAAATAAG-3′, andrST-OP2, 5′-GATCCTTATTTATTTAGAGCTCTG-3′). Upon phosphorylation and annealing, the oligonucleotides were subcloned into pGF23 expression vector (34) previously digested with SalI and BamHI (Fig. 1). Competent E. coli DH5α cells were transformed with the ligated DNA by electroporation. The screening for the correct construct was performed by DNA sequencing according to dideoxy chain termination method (35).
Fig. 1. Design of the pAC9 expression plasmid.
Indicated are the steps performed to yield an expression plasmid containing a cassette with Ecl136 II as the cloning site and encoding stop codons in the three possible reading frames (see “Experimental Procedures” for details). pho A, alkaline phosphatase promoter; Ampr, ampicillin resistance gene; ALAS, ALAS encoding sequence. The underlined nucleotides in pAC9 indicate the inserted nucleotides to create the correct reading frames, and the arrow indicates the cloning site.
Construction of Plasmid pAC10
The pAC10 plasmid was used to produce sufficient amounts of ALAS cDNA fragment with engineered SalI sites at 5′ and 3′ ends. The SalI sites were engineered such that, upon circularization of the ALAS cDNA piece, the original ALAS reading frame would be maintained. The ALAS cDNA fragment with engineered SalI sites was obtained by PCR using pGF23 plasmid as a template (34) and subcloned into pGF23 vector, previously digested with SalI.
Construction of a Random Library of Circularly Permuted ALAS Variants
The experimental design entailed several modifications of the method developed by Graf and Schachman (27) (Fig. 2). Briefly, a cDNA fragment encoding the murine erythroid ALAS with engineered SalI sites at both the 5′ and 3′ ends was obtained by digestion of the pAC10 plasmid with SalI, followed by gel purification. The circularization was accomplished in a final volume reaction of 50 µl using T4 DNA ligase (8.0 units/µl) and 11 µg of DNA in ligase buffer (50 mM Tris-HCl, pH 7.5, 10 mM MgCl2,10 mM dithiothreitol, 1 mM ATP, 25 µg/ml bovine serum albumin). Efficiency of the ligation reaction was estimated by agarose gel electrophoresis. Upon heat inactivation of ligase, RQ1 DNase was added at a ratio of 0.002 units per µg of DNA, and the samples were incubated for 15 min at 16 °C. The samples were pooled, and the DNA was recovered by phenol extraction. The generated, linearized inserts were repaired, blunt-ended with Klenow DNA polymerase I fragment, and subcloned into pAC9 vector (Fig. 1) previously digested with Ecl136 II. Competent E. coli HU227 cells, which can only grow in a medium containing ALA or when harboring an ALAS expression plasmid, were transformed with the ligation reaction by electroporation. One-eighth of the transformation solution was plated onto permissive 2×YT-agar medium (1.6% bacto-tryptone, 1% bacto-yeast extract, 0.5% NaCl, 1.5% agar) containing 50 µg/ml ampicillin and 10 µg/ml ALA, to score the total number of colonies produced. The remaining transformation solution was plated onto selective 2 ×YT-agar medium containing 50 µg/ml ampicillin, to select only for active ALAS variants. DNA sequencing templates were prepared from colonies recovered on the selective medium, and the DNA sequences of the 5′ and 3′ ends of the fragments encoding the functional circularly permuted variants were determined by the dideoxy chain termination method (35).
Fig. 2. Experimental strategy for construction of circularly permuted ALAS variants-containing library.
A murine erythroid ALAS cDNA obtained by digestion of pAC10 vector with SalI (see “Experimental Procedures”) was circularized, and the circularized DNA was subsequently treated with RQ1 DNase. The randomly linearized ALAS-encoding fragments were repaired and blunt-ended with T4 DNA polymerase Klenow fragment and subcloned into the pAC9 expression vector to yield a library of circularly permuted ALAS variants (see “Experimental Procedures” for details).
Rescue of Non-perfectly Permuted ALAS Variants
The repair of staggered ends created after DNase I treatment can lead to the deletion or insertion of nucleotides, which consequently can yield extra amino acids, and thus non-perfectly permuted polypeptide chains (27). To correct the 5′ and 3′ ends, the fragments encoding these variants were PCR-amplified using primers coding for the expected 5′ and 3′ sequences. The PfuTurbo DNA polymerase was used to minimize nonspecific incorporation of nucleotides at the 5′ and 3′ ends of the amplified inserts, and the PCR conditions were according to the manufacturer’s instructions. The obtained inserts were ligated into pAC9 vector, previously digested with Ecl136 II. Competent E. coli HU227 cells were transformed with the ligation, and the 5′ and 3′ end insert sequences of the functional rescued variants were verified by dideoxy DNA sequencing (35).
Construction of Rationally Designed Circularly Permuted ALAS Variants
Two ALAS cDNAs were ligated in tandem and in the original reading frame, using an ALAS expression plasmid, pGF23 (34), as the DNA template for two PCRs. In PCR1, the 5′ primer (NALAS-6, 5′-GCATATGTCGACTGACAGGAAGAGCAAGATTG-3′) contains the sequences for the first six amino acids of the mature ALAS and for a SalI site (sequence for SalI site is underlined) and the 3′ primer (RALAS-75, 5 ′-AGCTCAATTGAGCATAGGTGGTAAC-3′) contains the sequences for the terminal six amino acids and for an MfeI site (sequence for MfeI site is underlined). In PCR2, the 5′ primer (MALAS-76, 5′-TGCTCAATTGGACAGGAAGAGCAAG-3′) contains the sequences for the first six amino acids of the mature ALAS and for an MfeI site (sequence for MfeI site is underlined), and the 3′ primer (CALAS-7, 5′-GCAGCTGGATCCTTAAGCATAGGTGGTAACAT-3′) contains the sequences for the terminal six amino acids and for a BamHI site (sequence for BamHI site is underlined). The purified PCR1 product was digested with SalI and MfeI, whereas the purified PCR2 was digested with BamHI and MfeI. The digested PCR products were then ligated into pGF23 previously digested with SalI and BamHI. The generated plasmid, pAC1, encoding the two ALAS cDNAs in tandem was subsequently used as the PCR template in the construction of circularly permuted variants of designed N and C termini. The 5′ and 3′ primers were 5′-TATAGTCGACTGCT-GCATGCAACTTCTGTA-3′ and 5′-TATAGGATCCTTACACAGACAC-ATCTTGGAA-3′, respectively, for A475 variant; 5′-TATAGTCGACTA-TGGAAAACTTTGTGGAGA-3′ and 5′-TATAGGATCCTTACATCTGA-GGGCTGTGAAA-3′, respectively, for M451 variant; 5′-TATAGTCGA-CTTCTGGGGCTCTAGAATCTA-3′ and 5′-TATAGGATCCTTAGAGC-ATCATGGGAGGCAG-3′, respectively, for S351 variant; 5′-TATAGTC-GACTCAGTATGGAGCCCTGACCT-3′ and 5′-TATAGGATCCTTAGT-GGGCCACATCACACAA-3′, respectively, for Q271 variant; 5′-TATAG-TCGACTAAGTTTCATGTGGAGCTT-3′ and 5′-TATAGGATCCTTAG-CTGGTACCTGAGATATT-3′, respectively, for K156 variant; and 5′-G-CATATGTCGACTGGTTATGACCAATTTTTCA-3′ and 5′-TATAGGAT-CCTTAGAAAGCCTGGCTTCCAGT-3′, respectively, for Y167 variant.
Purification of Selected Circularly Permuted ALAS Variants
Recombinant wild type and circularly permuted ALAS variants were purified from E. coli overproducing cells containing the ALAS-encoding cDNAs under the control of the alkaline phosphatase (pho A) promoter (34). E. coli strain BL21(DE3) cells harboring the expression plasmids for Q69, N404, and N408 ALAS variants or E. coli strain LC24 cells (33) harboring the expression plasmid for L25 were grown in MOPS medium and harvested as described previously (34). Cell pellets were resuspended as in Ref. 34, with the exception of the pellet corresponding to the BL21(DE3) cells harboring the N404 variant, which was resuspended in 20 mM potassium phosphate buffer, pH 7.5, containing 5% glycerol, 1 mM EDTA, 20 µM PLP, 5 mM mercaptoethanol, and the following protease inhibitors: 1 µg/ml aprotinin, 1 µg/ml leupeptin, 1 µg/ml pepstatin, and 1 mg/ml phenylmethylsulfonyl fluoride. The steps following cell lysis and centrifugation were essentially as described in Ref. 34 with slight modifications. Specifically, the initial ammonium sulfate fractionation step was 20% for the Q69 variant, 25% for the N408 and L25 variants, and 28% for the N404 variant. After stirring for 10 min at 4 °C, the solution was centrifuged at 27,000 × g for 30 min at 4 °C, and the supernatant was further fractionated with ammonium sulfate to a final concentration of 30% (in the case of L25 and Q69 variants) or 38% (in the case of N408 and N404 variants). The chromatographic steps using an Ultrogel AcA and DEAE-Sephacel columns were as described previously (34), with the following modifications: the DEAE-Sephacel resin was washed with Buffer A until A280 was lower than 0.1 and the protein of interest (i.e. wild type or circularly permuted ALAS variants) was eluted with Buffer A (34) containing 60 mM KCl. Protein-containing fractions were pooled and concentrated in an Amicon 8050 stirred cell with an YM30 membrane. The purified and concentrated enzyme (wild type or variants) was stored under liquid nitrogen.
Protein Concentration Determination, Native PAGE, and SDS-PAGE
Protein concentration was determined by the bicinchoninic acid assay, according to the instructions supplied with the protein concentration determination kit, and using bovine serum albumin as standard. Protein purity was assessed by SDS-PAGE (36). The oligomeric state of the rationally designed ALAS variants was verified using native PAGE (37). The protein samples were prepared from E. coli cells, harboring the different expression plasmids, which were induced for protein expression and grown for 16 h at 37 °C (34). Cells were harvested and lysed using sonication, and the cell membranes were pelleted. Most of the supernatant protein was precipitated by addition of a saturated solution of ammonium sulfate to a final concentration of 50%. The precipitated protein was resuspended in 300 µl of 20 mM potassium phosphate buffer, pH 7.5, containing 15% glycerol. The resuspended protein was desalted using Bio-Rad Bio-Spin 6 gel filtration columns, and ~30 µg of the desalted protein samples were loaded onto the gel.
Molecular Mass Determination of ALAS Variants by Gel Filtration Chromatography
The native molecular masses of ALAS variants were determined by gel filtration chromatography on Superdex 200 column (1.0×50 cm). The Superdex 200 gel filtration column, which was connected to a PerkinElmer Life Sciences high pressure liquid chromatography system, was equilibrated with 20 mM potassium phosphate buffer, pH 7.5, containing 10% glycerol, 1 mM EDTA, 20 µM PLP, and 5 mM mercaptoethanol. The flow rate was set at 1 ml/min. The gel filtration molecular weight markers and the purified ALAS variants were dissolved in the same buffer and applied onto the column under the same conditions. The molecular masses of the ALAS variants were calculated using the linear equation obtained from the calibration curve, which was generated from the molecular weight markers.
UV-visible Absorption and CD Spectrocopies
Shimadzu UV2100U UV-visible dual beam spectrophotometer was used to obtain all UV-visible absorption spectra. This spectrophotometer is equipped with thermostatically controlled cell holders and allows exporting data as ASCII files through the RS232 interface. CD spectra were obtained on a Jasco model 710 spectropolarimeter calibrated for both wavelength maxima and signal intensity using an aqueous solution of D-10-camphorsulfonic acid (38). CD spectra (6–2 µM enzyme) were recorded over the wavelength range 200–270 nm using a cylindrical cell of 0.1 cm path length and a total volume of 300 µl. The observed rotation degrees (θobs) were converted to molar ellipticity. All spectra were obtained at 25 °C and corrected for buffer contribution.
Determination of Glycine Dissociation Constant
The glycine dissociation constants for wild type ALAS and its variants were determined by spectrophotometrically titrating the proteins in 20 mM potassium phosphate, pH 7.5, containing 10% glycerol, 1 mM EDTA, and 5 mM mercaptoethanol with glycine at 20 °C. UV-visible spectra were recorded from 250 to 500 nm after each addition of glycine to the protein of interest. High concentration glycine stock solution (2 m) was used to ensure minimal dilution of the protein sample. The binding of glycine to the enzyme was determined by monitoring the increase of absorbance at 410 nm, which corresponds to the formation of external aldimine between PLP and glycine (19). The obtained absorbance change data were fit to the Equation 1,
| (Eq.1) |
where Y is the ratio between the absorbance increase at 410 nm (ΔA410) and the maximal absorbance increase at 410 nm (ΔAmax), and [Gly]tot is the total glycine concentration.
Steady-state Kinetic Characterization of ALAS Variants
The steady-state kinetic parameters , and k cat of ALAS and ALAS variants were determined at 20 °C using a continuous spectrophotometric assay as described previously (39). To determine the apparent Km and maximal velocities the data were analyzed in matrices of six glycine and six succinyl-CoA concentrations and fit to Equation 2.
| (Eq.2) |
To determine the Km and Vmvalues, the apparent and were fit to Equations 3 and 4,
| (Eq.3) |
| (Eq.4) |
where and are Michaelis constants for glycine and succinyl-CoA, respectively, and is the limiting value of when glycine concentration approaches zero.
Modeling of the Three-dimensional Structure of the Wild Type ALAS and Circularly Permuted ALAS Variants and Prediction of Secondary Structure of the Wild Type ALAS
The primary structure and the x-ray structure coordinates of the E. coli 8-amino-7-oxononanoate synthase (AONS) (40, 41) (Protein Data Bank accession code 1BS0) were used to model both the wild type ALAS and its circularly permuted variants. Molecular modeling was performed using the automated protein modeling server Swiss model (42–44). The First Approach Mode and the lower Blast limit of 0.0001 were used. Protein topology schematics (TOPS) (45, 46) were generated using the automated TOPS-generating server. The secondary structure of the ALAS was predicted using the Garnier (GOR IV) method (47).
RESULTS
Construction of a Random Library of Circularly Permuted ALAS Variants and Screening for Active ALAS Variants
To evaluate the role of the ALAS polypeptide chain in folding, assembly of the holoenzyme, and function, we constructed a library of randomly circularly permuted murine erythroid ALAS cDNAs using a modified version of the method described by Graf and Schachman (27). The overall scheme is depicted in Fig. 2. A circularized ALAS cDNA was synthesized such that the first and last residues of the natural ALAS were connected. The major modification of our method in relation to that described previously (27) was the omission of a sequence encoding a peptide linker between the natural N and C termini. In fact, as far as we are aware, this is the first time that circularly permuted variants have been constructed without a peptide linker between the natural termini of the protein. Typically, linkers have been introduced so that the two natural termini can be connected without steric strain (27, 28). The circular ALAS cDNA was randomly linearized upon digestion with RQ1 and any generated 5′- and 3′-protruding ends were repaired with DNA polymerase. The blunt-ended inserts encoding the library of circularly permuted ALAS variants were cloned into the pAC9 expression plasmid (Fig. 1), which was designed to contain an Ecl 136 II cloning site and stop codons in the three different frames, so that inserts that did not terminate in the same frame as that of ALAS could be accommodated (Fig. 1). To identify active circularly permuted ALAS variants, the hemA− E. coli strain HU227 was transformed with the library, and ~ 17,500 clones were screened for their ability to rescue the HU227 cells in media containing neither ALA nor heme. Of these, 180 transformants were identified as active, based on the ability to grow on ALAS selection medium (2× YT + ampicillin). Sequencing of the 5′ and 3′ ends of 172 active clones indicated that 21 corresponded to circularly permuted variants, whereas the remaining 151 clones encoded variants with the natural ALAS N and C termini. Thus, ~ 12% of the active ALAS variants were circularly permuted. However, out of the 21 active variants only 9 were identified as different circularly permuted ALAS variants.
Most of the active variants exhibited elongations at their 5′ end, which ranged 10–300 base pairs. In contrast, no variants with shortened termini were identified. The observed elongations most probably resulted from the RQ1 DNase treatment followed by the filling-in of the of staggered ends by the Klenow enzyme. DNase I produces both blunt and staggered ends. The latter, upon reaction with the Klenow enzyme, can yield 5′-terminal elongations and 3′-terminal deletions. Similar cDNA modifications were also encountered previously in the construction of random libraries (27, 28). Since the active enzymes with terminal elongations do not correspond to perfectly circularly permuted ALAS variants, these “defective” variants were “corrected” by PCR amplification using 5′ end primers encoding the new N termini and excluding the elongations corresponding to amino acids already present at the C termini. Eight of the nine different circularly permuted ALAS variants conferred heme prototrophy to the HU227 cells and thus, according to our selection criterion, were considered as active, perfectly circularly permuted ALAS variants (Fig. 3A). The removal of the ≈300-base pair long 5′ end extension from the ninth circular permuted ALAS variant yielded a protein incapable of rescuing HU227 cells grown on selective medium without ALA. It is possible that the N-terminal extension was required for folding and/or stability and enzymatic activity in this particular ALAS variant.
Fig. 3. Schematic diagram illustrating the amino acid sequence of the ALAS circularly permuted variants in relation to wild type ALAS.
Top, white bar represents the wild type (WT) ALAS sequence with highlighted amino acids previously reported to be essential for ALAS function (12, 19–21). The sequence numbering for the wild type erythroid ALAS is indicated at the top, and the amino acid numbering at the left of each variant indicates its N-terminal amino acid. Thus, for each variant the amino acid corresponding to the wild type ALAS N-terminal amino acid is represented by the junction between the black and white bars. A, active, perfectly circularly permuted ALAS variants obtained from the screening of a random library. B, rationally designed ALAS circularly permuted variants. Only A475 was found to be active, based on its ability to rescue HU227 cells on selective medium without ALA.
The new N termini of the eight active circularly permuted ALAS variants are mostly centered in protein regions spanning ~ 100 amino acids from either the N or the C terminus (Fig. 3A). However, if the protein regions C-terminal to Lys-313 are considered C-terminal domains, then the swapping of C-terminal domains in front of the natural ALAS N terminus appears to occur to a greater extent (up to 20.8%) than that of N-terminal domains after the natural ALAS C terminus (up to 13.4%) (Table I). The new termini of the active, circularly permuted ALAS variants fell both within predicted secondary structure elements and in predicted loop regions. That is, the new termini of E13, E16, L25, and E491 variants (Fig. 3A) are located in α-helices, whereas those of Q69, N404, and N408 variants (Fig. 3A) are at the border of a loop and α-helix regions. Finally, D472 had both termini in a loop region (Fig. 3A). No new termini of active ALAS variants were identified in central domains, which have been previously reported to entail active site residues (e.g. Lys-313 and Asp-279) (14, 20).
Table I.
Occurrence and permutation extent of the active circularly permuted ALAS variants obtained from the random library
| Variant | Occurrence frequencya | Permutation extentb |
|---|---|---|
| % | ||
| E13 | 0.58 | 2.36 |
| E16 | 1.16 | 2.95 |
| L25 | 2.91 | 4.72 |
| Q69 | 1.74 | 13.36 |
| N404 | 0.58 | (–) 20.83 |
| N408 | 0.58 | (–) 20.04 |
| D472 | 1.16 | (–) 7.47 |
| E491 | 2.91 | (–) 3.73 |
Occurrence frequency represents the percentage of independent clones for each active variant obtained from screening the random library of circularly permuted ALAS variants.
Permutation extent was calculated relative to Lys-313, which was arbitrarily set as the reference residue, and thus negative values (–) represent the swapping of domains located C-terminal to Lys-313 in front of the wild type ALAS N terminus.
Rationally Designed Circularly Permuted ALAS Variants
To explore the possibility that the termini of the above inactive circularly permuted ALAS variants had disrupted essential secondary structural elements essential in folding, dimerization, and stability, six variants were designed to have their termini spanning the entire protein sequence and falling outside the sequence for secondary structural elements (Fig. 3B). Although these six circularly permuted ALAS variants exhibited similar levels of overproduction in HU227 cells and appeared to be dimers (data not shown), with exception of A475, they were inactive as defined by their inability to rescue the growth of HU227 cells in selective medium without ALA.
UV-CD, UV-visible Absorption Spectroscopic Characterization and Oligomeric State of Active ALAS Circularly Permuted Variants
To verify whether circular permutations in ALAS introduced substantial changes in secondary structure, CD spectra in the far-UV region (200–300 nm) were recorded for the wild type and four active circularly permuted enzymes. As shown in Fig. 4B, the wild type and circularly permuted ALAS variants displayed similar CD spectra, suggesting that no dramatic changes in the overall conformation and structural content were introduced.
Fig. 4.
A, the UV-visible absorption spectra of purified ALAS circularly permuted variants. The UV-visible absorption spectra were recorded in 20 mM potassium phosphate buffer at 20 °C and pH 7.5. Protein samples were 12.5 µm concentration. B, the normalized UV-CD spectra of purified ALAS circularly permuted variants. The UV-CD spectra were recorded in 20 mM potassium phosphate buffer at 20 °C and pH 7.5. Protein sample concentrations are as follows: wild type ALAS, 7.4 µm; L25, 6.3 µm; Q69, 6.9 µm; N404, 5.0 µm; N408, 8.0 µm. Curves are as follows: —, wild type ALAS; …, L25; –··–··, Q69;– – –, N404;–·–·–, N408.
Despite possessing the PLP cofactor, the absorption spectra of the purified, active, circularly permuted ALAS variants differed substantially from that of the wild type ALAS (Fig. 4A). The major spectral differences arise from the protein-bound PLP cofactor. The wild type ALAS-bound PLP has two distinct absorbance maxima at ≈330 and ≈420 nm. With the circularly permuted ALAS variants, these maxima are shifted toward shorter wavelengths (Fig. 4B), indicating that the local environment of the protein-bound PLP cofactor might be different in the circularly permuted variants and wild type ALAS.
To determine the oligomeric state of the native, active circularly permuted ALAS variants, their molecular masses were determined by gel filtration chromatography. All of the variants were found to have molecular masses close to that of the wild type ALAS (≈112 kDa (11)). The small differences probably reflect different Stoke radii between the circularly permuted variants and the wild type ALAS. Since the molecular mass of each subunit of the variants and wild type ALAS is ~56 kDa, as determined by SDS-PAGE (Fig. 5), the circularly permuted variants, as in wild type ALAS, are homodimers. The above findings, taken together, suggest that circular permutation in ALAS did not prevent folding, coupling of the PLP cofactor, or assembly of the two subunits into a functional homodimer.
Fig. 5. Determination of the molecular mass of the circularly permuted ALAS variants by gel filtration chromatography.
Each of the variants (≈6.0 µg) was independently applied to a Amersham Pharmacia Biotech Sephadex 200 filtration column and eluted with 20 mM potassium phosphate buffer containing 10% glycerol at 4 °C and pH 7.5 (flow rate 1.0 ml/min). The chromatogram depicted is that of L25 variant. Inset A, molecular mass calibration curve for the Sephadex 200 column. The standard proteins used were cytochrome c (12.4 kDa), carbonic anhydrase (29.0 kDa), bovine serum albumin (66.0 kDa), alcohol dehydrogenase (150.0 kDa), and β-amylase (200.0 kDa). Inset B, 15% SDS-PAGE of the purified wild type (WT) and circularly permuted ALAS variants. Approximately 7 µg of each protein sample were loaded per lane.
Glycine Dissociation Constants of Wild Type ALAS and ALAS Variants
The circularly permuted and wild type enzymes were titrated with glycine to determine the dissociation constants for formation of external aldimines with substrate. The titrations were performed at 20 °C, as some of the circularly permuted variants (e.g. Asn-408) tended to precipitate at 37 °C. The dissociation constants () of the circularly permuted variants ranged from values 2.65-fold smaller (for N404) to 2.25-fold greater (for Q69) than that of the wild type ALAS (Table II). Interestingly, the N404 variant exhibited the tightest binding for glycine, with an affinity increased almost 3-fold over that of wild type ALAS. Previously, we reported that the Arg-439 residue has an important role in the binding of the carboxylate group of the glycine substrate (19). It is tempting to speculate that the circular permutation of the ALAS polypeptide chain in the N404 variant made the glycine-binding domain and the Arg-439 residue more exposed than in the wild type ALAS and facilitated the binding of the glycine substrate. In contrast, the Arg-439 residue might be buried deeper in the protein structure and not readily accessible to glycine in the L25 and Q69 variants. However, the circular permutation in the N408 variant increased the Kd for glycine about 1.3-fold, and the protein appeared less stable upon glycine binding. It is possible that in this variant the integrity of the glycine-binding domain might have been disrupted despite the exposure of the Arg-439 residue. These results indicate that the C-terminal domain of the protein, which contains the Arg-439 residue, might mediate efficient glycine binding. The disruption of this domain and/or relocation of this domain into a more secluded environment inside the protein affect/s the glycine binding properties of the ALAS variant. However, at the primary structure level, this domain can be moved as a whole into the opposite terminus of the protein without affecting the glycine binding ability of the enzyme.
Table II.
Steady-state kinetic parameters of wild type (WT) ALAS and circularly permuted ALAS variants
| Protein |
|
kcat |
|
|
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| mM | s–1 | mM | µM | s–1 · mM–1 | s–1 · µM–1 | ||||||
| WT ALAS | 29.7 ± 0.8 | 0.016 ± 0.002 | 14.0 ± 2.0 | 11.0 ± 1.0 | 0.0011 | 0.0015 | |||||
| L25 | 32.0 ± 5.0 | 0.016 ± 0.003 | 10.7 ± 0.5 | 2.6 ± 0.4 | 0.0015 | 0.0062 | |||||
| Q69 | 67.0 ± 3.0 | ||||||||||
| N404 | 11.3 ± 0.2 | 0.057 ± 0.003 | 5.1 ± 0.2 | 21.0 ± 2.0 | 0.0112 | 0.0027 | |||||
| N408 | 40.0 ± 4.0 | 0.033 ± 0.005 | 19.0 ± 3.0 | 8.1 ± 0.3 | 0.0017 | 0.0041 | |||||
Steady-state Kinetic Characterization of Circularly Permuted ALAS Variants
To investigate the effects of circular permutation on the catalytic properties of ALAS, the steady-state kinetic parameters kcat, , and were determined (Table II). All of the circularly permuted variants exhibited enzymatic activities either comparable to, or even higher than, that of wild type ALAS (Table II). The circular permutation in the N404 variant increased the kcat value ~3.5-fold and increased the catalytic efficiency for glycine 10-fold. The catalytic efficiency for succinyl-CoA was increased ~2-fold. The L25 and N408 variants displayed similar kcat values (i.e. 1–2-fold) to that of the wild type enzyme, although the Km and the catalytic efficiency for succinyl-CoA were increased ~4-fold in the L25 variant. The increase in the overall enzymatic performance of some of the variants probably reflects improved catalytic efficiency toward the substrates (N404 variant for glycine, N408 variant for succinyl-CoA, and L25 variant for both substrates). The steady-state kinetic parameters for the Q69 variant were not determined, due to technical limitations associated with deviations of the kinetic mechanism from the ALAS Ordered Bi Bi kinetic mechanism (2). However, neither substrate nor product inhibitions were identified (data not shown).
Modeling of Wild Type ALAS and Its Circularly Permuted Variants
To gain insight into how the circular permutation of the ALAS polypeptide chain could possibly affect its three-dimensional structure, homology modeling for the structures of the wild type ALAS and its variants was performed. The high degree of the amino acid sequence similarity between wild type ALAS and AONS allowed us to predict the three-dimensional structures of ALAS and its circularly permuted variants (Fig. 6). The sequences used in the models for the polypeptide chains of ALAS and its variants were at least 36% homologous to the template AONS sequence. The major fitness parameters of the models, estimated by the WHAT IF program (48), indicated that the predicted three-dimensional structures were of good quality (data not shown).
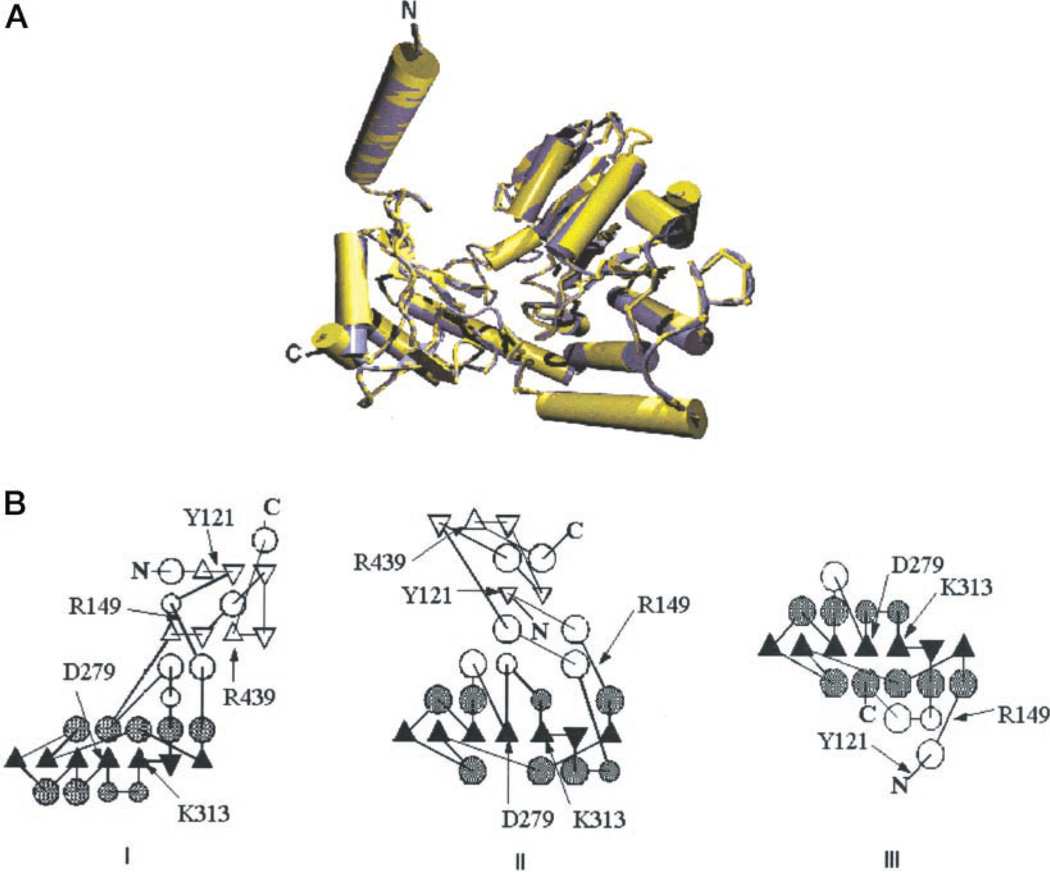
Fig. 6.
A, the three-dimensional structure model of the wild type ALAS (monomeric). The superimposed three-dimensional structures of the wild type ALAS (yellow) and 7-amino-8-oxononanoate synthase (39) (Protein Data Base code 1BS0) (blue). B, the protein topology schematics (TOPS) of the predicted three-dimensional structures of wild type ALAS and circularly permuted ALAS variants. I, wild type ALAS, E16, L25, D472, and E491 variants. The two possible structural domains are shown. II, E13 and Q69 variants. III, N404 and N408 variants. The approximate positions of functional amino acid residues are indicated by arrows. The pointing triangles represent the β-strands “out of” or “into” the plane of the diagram, respectively; the circles represent α-helices (45). The /3-strands and the α-helices of the α/β structure central core are represented in solid black and patterned black, respectively.
To simplify the comparison of the structural features of the models, two-dimensional structure topology schematics (TOPS) were generated based on the predicted three-dimensional structures (Fig. 6B). Essentially, TOPS are two-dimensional schematic representations of protein structures. They represent the structure as a sequence of secondary structure elements and illustrate the relative spatial position and direction of these elements (46). The unifying feature among wild type and ALAS variants is, as with AONS, the presence of a typical α/β structure with a seven-stranded β-sheet as the central core in which six strands are parallel (Fig. 6B). The wild type ALAS Asp-279 and Lys-313 residues are located on neighboring (β-strands of the central core (Fig. 6B). The structural differences in the predicted three-dimensional structure models are subtle and appear to reflect a slightly different arrangement of the secondary structure elements (but not of their content, as expected from the similar CD spectra). Specifically, in wild type ALAS and half of the ALAS circularly permuted variants, the β-sheet central core is surrounded by 9 α-helices, with five of the α-helices located on the side of the sheet facing the active site of the enzyme and the other four on the side of the β-sheet facing the surface of the protein (Fig. 6B,I). The ALAS circularly permuted variants L25 and Q69 have α-helices equally distributed, albeit less organized, on both sides of the β-sheet central core (Fig. 6B, II), and the N404 and N408 ALAS variants display a third α-helical arrangement around the β-sheet central core (Fig. 6B, III). It should be also noted that the predicted three-dimensional structures of the N404 and N408 variants do not contain the Arg-439 residue (Fig. 6B, III), as this protein region was not included in the modeling of the two variants, given the low BLAST score.
DISCUSSION
ALAS is the first PLP-dependent enzyme and the first enzyme of the tetrapyrrole biosynthetic pathway to be investigated using circular permutation of the polypeptide chain. The goal of this work was to use circular permutation of the ALAS amino acid sequence to probe the roles of the natural N and C termini and of the order of the secondary structure elements in folding and assembly of an active, dimeric enzyme. Circular permutation of proteins, such as trypsin inhibitor (24), dihydrofolate reductase (26), aspartate transcarbamoylase (27, 31), E. coli DsbA (28), glucose transporter (30), Bacillus β-glucanase (49), and spectrin (50), has revealed that circularly permuted polypeptide chains can fold into stable and active proteins, albeit with, possibly, different topologies from those of the natural proteins. Most of the proteins so far circularly permuted (slightly over 20) have been small monomers, which have required linkers connecting the naturally close N and C termini in order to yield properly folded and active proteins (23, 28, 29, 50, 51). The characterization of active, circularly permuted ALAS variants, obtained through functional screening of a library of randomly circularized ALAS variants without linkers between their termini, has permitted us to conclude that circularly permuted ALAS variants are capable of folding into functional, native-like conformations, and thus that the natural location of the termini and sequential arrangement of the secondary structure elements are not critical determinants of the final protein fold, PLP cofactor attachment, or assembly of the two subunits into an active, dimeric enzyme.
The new termini of the active, circularly permuted ALAS variants interrupted both predicted secondary structure elements and loop regions, although most of the new termini were centered in 100 amino acid regions from each of the natural ALAS termini (Fig. 3), indicating that specific regions of the protein are more tolerant to disruption (27, 28). The UV CD spectra of the four purified, circularly permuted ALAS variants are not significantly different from that of the wild type ALAS, confirming that all of the characterized ALAS variants fold into an almost identical wild type conformation regardless of the position of new N and C termini and absence of a polypeptide linker between the original termini. However, the circularly permuted ALAS variants exhibited differences in the PLP-binding sites; namely the UV-visible spectra maxima of the purified variants shifted toward shorter wavelengths. As with AONS (41), the ≈425 nm maximum can be attributed to a planar configuration, in which the electrons of the double bond between the NZ of the Lys-313 residue and the C4A of the PLP are conjugated with the delocalized electrons of the pyridine ring. The 330 nm maximum might reflect the non-coplanar form of the internal aldimine. The distortion of the PLP-protein complex brought about by the local environment of the polypeptide chain should interfere with the conjugation and should produce changes in the absorbance spectrum of the protein. The differences in the absorbance spectra of circularly permuted ALAS variants suggest that the local environments within the active sites of the variants, created by different arrangements of their polypeptide chains, differ significantly from that of the wild type ALAS. Significantly, these findings validate the plasticity of the PLP binding and active site and indicate that as long as ALAS and the polypeptide chain folds in place for the binding of the cofactor and for the catalytic residues to be in the correct proximity, then a PLP fold and an active ALAS are attainable.
The use of the E. coli hemA− HU227 strain (32) as the initial biological screen, although powerful, had some limitations. Specifically, the inability of providing heme prototrophy to this strain could be due to inappropriate folding, proteolytic degradation, malfunction in PLP binding, or assembly of the two subunits or could just emulate ALAS variants, which had enzymatic activities too low to compensate the ALA and heme requirements of the hemA− strain (32). By studying the active, circularly permuted ALAS variants (Fig. 3A), as defined through the biological screen of the random library, we selected for variants that were biologically functional, with a wild type like fold and resistant to proteolytic degradation. In contrast, the generation of six rationally designed circularly permuted ALAS variants (Fig. 3B )permitted us to establish that five of the variants were dimers, but the lack or the low enzymatic activity (as defined by the inability to rescue the growth of the hemA− strain) cannot be ascribed, at present, to misfolding, malfunction in PLP binding, or lowered enzymatic activities.
The comparable specific activities of the purified circularly permuted variants and wild type ALAS (Table II) suggest tertiary structural homology of their active sites. However, despite the overall tertiary structural homology, the arrangement of the secondary structural elements seems to differ (Fig. 6B). These results led us to hypothesize that the same ALAS overall structure can be reached through multiple folding pathways in which adjacent secondary structural elements interact to form folding units. In fact, unfolding studies of circularly permuted proteins have demonstrated that, regardless of their structural similarity to the wild type protein counterpart, their mechanisms of folding may strongly differ from that of the wild type (50). Nevertheless, even if multiple protein folding pathways exist, “there may be essential nucleation sites that are common to all pathways,” as postulated by Hennecke et al. (28). The identification of such nucleation sites or “modules,” which are compact structural units essential for correct folding and/or catalysis, can be achieved through circular permutation (23). Significantly, the identification of the “folding modules” should reveal the essential features to reach a PLP-binding fold.
The functional mapping of the ALAS polypeptide chain, using circular permutation clearly, indicates that there are at least two continuous regions or functional elements (23), which have defined functions in ALAS catalysis. The first functional element spans from the Phe-70 residue to the Gly-403 residue of the ALAS polypeptide chain. The integrity of this element is essential for ALAS enzymatic activity, but it can be moved around the polypeptide chain, without apparently affecting seriously ALAS function. This functional element contains residues involved in binding of the PLP cofactor and in enhancing the properties of the PLP cofactor in catalysis (i.e. Lys-313, Tyr-121, and Asp-279) (12, 20, 21). We designated this functional element as the catalytic domain of the ALAS. Curiously, this functional element is a sub-domain of a previously defined 49-kDa, “core catalytic domain of erythroid ALAS” (52). The second functional element appears to span from the Asn-404 residue to the C terminus of the protein (i.e. Ala-509) and entails the Arg-439 residue, which was reported to be involved in binding of the glycine substrate (19). This functional element also can be moved around the polypeptide chain and disrupted at specific locations without abolishing enzymatic activity. The relocation of this intact functional element (or glycine binding domain) to the natural N terminus of ALAS yielded a circularly permuted variant (i.e. N404 variant) with enhanced glycine binding, enzymatic activity, and catalytic efficiency (Table II). Thus, the change in the order of the functional elements actually improved the function of ALAS. This finding represents a novel view of the enzyme architecture.
The idea that protein structure can be explained in terms of building blocks (folding and functional elements or modules) and that the integrity of these building blocks is essential for folding and enzyme function has been put forward previously (23). Circular permutation is a particularly suitable approach for the identification of folding and functional elements, as the cleavage of the peptide bond within (but not outside) these elements will prevent folding/function (23). The identification of two functional elements in ALAS not only supports the above proposal of functional elements as the “building blocks” of protein structure but also reveals that the order of these building blocks is not critical for protein function, as long as the proper folding of the protein brings them together for catalysis to occur. Indeed, modeling of the circularly permuted variants indicated a different arrangement of the secondary structure elements (Fig. 6B), suggesting that a wild type-like tertiary structure can be achieved through alternative arrangement of the secondary structure elements of the polypeptide chain. Current research in our laboratory is aimed at defining whether the ALAS overall structure can be reached through alternative or multiple folding pathways.
Footnotes
This work was supported in part by National Institutes of Health Grant DK52053 (to G. C. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: ALAS, 5-aminolevulinate synthase; ALA, 5-aminolevulinate; AONS, 7-amino-8-oxononanoate synthase; PLP, pyridoxal 5′-phosphate; PAGE, polyacrylamide gel electrophoresis; PCR, polymerase chain reaction; MES, 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Ferreira GC, Gong J. J. Bioenerg. Biomembr. 1995;27:151–159. doi: 10.1007/BF02110030. [DOI] [PubMed] [Google Scholar]
- 2.Ferreira GC. In: Inorganic Biochemistry and Regulatory Mechanisms of Iron Metabolism. Ferreira GC, Moura JJG, Franco R, editors. Wiley-VCH: Weinheim, Germany; 1999. pp. 15–34. [Google Scholar]
- 3.Sadlon TJ, Dell’Oso T, Surinya KH, May BK. Int. J. Biochem. Cell Biol. 1999;31:1153–1167. doi: 10.1016/s1357-2725(99)00073-4. [DOI] [PubMed] [Google Scholar]
- 4.Nakajima O, Takahashi S, Harigae H, Furuyama K, Hayashi N, Sassa S, Yamamoto M. EMBO J. 1999;18:6282–6289. doi: 10.1093/emboj/18.22.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter PD, Willard HF, Gorski JL, Bishop DF. Genomics. 1992;13:211–213. doi: 10.1016/0888-7543(92)90223-f. [DOI] [PubMed] [Google Scholar]
- 6.Bishop DF, Henderson AS, Astrin KH. Genomics. 1990;7:207–214. doi: 10.1016/0888-7543(90)90542-3. [DOI] [PubMed] [Google Scholar]
- 7.Bottomley SS. In: Wintrobe’s Clinical Hematology. Lee GR, editor. Baltimore: Lippincott Williams & Wilkins; 1999. pp. 1022–1045. [Google Scholar]
- 8.Bottomley SS, May BK, Cox TC, Cotter PD, Bishop DF. J. Bioenerg. Biomembr. 1995;27:161–168. doi: 10.1007/BF02110031. [DOI] [PubMed] [Google Scholar]
- 9.Cotter PD, May A, Li L, Al-Sabah AI, Fitzsimons EJ, Cazzola M, Bishop DF. Blood. 1999;93:1757–1769. [PubMed] [Google Scholar]
- 10.Harigae H, Furuyama K, Kimura A, Neriishi K, Tahara N, Kondo M, Hayashi N, Yamamoto M, Sassa S, Sasaki T. Br. J. Haematol. 1999;106:175–177. doi: 10.1046/j.1365-2141.1999.01479.x. [DOI] [PubMed] [Google Scholar]
- 11.Tan D, Ferreira GC. Biochemistry. 1996;35:8934–8941. doi: 10.1021/bi952918m. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira GC, Neame PJ, Dailey HA. Protein Sci. 1993;2:1959–1965. doi: 10.1002/pro.5560021117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira GC, Vajapey U, Hafez O, Hunter GA, Barber MJ. Protein Sci. 1995;4:1001–1006. doi: 10.1002/pro.5560040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter GA, Ferreira GC. Biochemistry. 1999;38:3711–3718. doi: 10.1021/bi982390w. [DOI] [PubMed] [Google Scholar]
- 15.Fanica-Gaignier M, Clement-Metral J. Eur. J. Biochem. 1973;40:19–24. doi: 10.1111/j.1432-1033.1973.tb03164.x. [DOI] [PubMed] [Google Scholar]
- 16.John RA. Biochim. Biophys. Acta. 1995;1248:81–96. doi: 10.1016/0167-4838(95)00025-p. [DOI] [PubMed] [Google Scholar]
- 17.Ferreira GC. In: Biochemistry and Molecular Biology of Vitamin B6 and PQQ-dependent Proteins. Iriarte A, Kagan HM, Martinez- Carrion MM, editors. Basel: Birkhauser Verlag; 2000. pp. 257–263. [Google Scholar]
- 18.Hunter GA, Ferreira GC. J. Biol. Chem. 1999;274:12222–12228. doi: 10.1074/jbc.274.18.12222. [DOI] [PubMed] [Google Scholar]
- 19.Tan D, Harrison T, Hunter GA, Ferreira GC. Biochemistry. 1998;37:1478–1484. doi: 10.1021/bi971928f. [DOI] [PubMed] [Google Scholar]
- 20.Gong J, Hunter GA, Ferreira GC. Biochemistry. 1998;37:3509–3517. doi: 10.1021/bi9719298. [DOI] [PubMed] [Google Scholar]
- 21.Tan D, Barber MJ, Ferreira GC. Protein Sci. 1998;7:1208–1213. doi: 10.1002/pro.5560070516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T, Bertelsen E, Benvegnu D, Alber T. Biochemistry. 1993;32:12311–12318. doi: 10.1021/bi00097a006. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura T, Iwakura M. J. Biol. Chem. 1999;274:19041–19047. doi: 10.1074/jbc.274.27.19041. [DOI] [PubMed] [Google Scholar]
- 24.Goldenberg DP, Creighton TE. J. Mol. Biol. 1983;165:407–413. doi: 10.1016/s0022-2836(83)80265-4. [DOI] [PubMed] [Google Scholar]
- 25.Rote KV, Hough R, Goldenberg D, Rechsteiner MC. J. Biol. Chem. 1989;264:1156–1162. [PubMed] [Google Scholar]
- 26.Uversky VN, Kutyshenko VP, Protasova N, Rogov VV, Vassilenko KS, Gudkov AT. Protein Sci. 1996;5:1844–1851. doi: 10.1002/pro.5560050910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graf R, Schachman HK. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hennecke J, Sebbel P, Glockshuber R. J. Mol. Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 29.Hennecke J, Glockshuber R. Biochemistry. 1998;37:17590–17597. doi: 10.1021/bi981888v. [DOI] [PubMed] [Google Scholar]
- 30.Gutknecht R, Manni M, Mao Q, Erni B. J. Biol. Chem. 1998;273:25745–25750. doi: 10.1074/jbc.273.40.25745. [DOI] [PubMed] [Google Scholar]
- 31.Zhang P, Schachman HK. Protein Sci. 1996;5:1290–1300. doi: 10.1002/pro.5560050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JM, Brathwaite O, Cosloy SD, Russell CS. J. Bacteriol. 1989;171:2547–2552. doi: 10.1128/jb.171.5.2547-2552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huala E, Moon AL, Ausubel FM. J. Bacteriol. 1993;173:382–390. doi: 10.1128/jb.173.1.382-390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira GC, Dailey HA. J. Biol. Chem. 1993;268:584–590. [PubMed] [Google Scholar]
- 35.Sanger F, Nicklen S, Coulson AR. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 37.Gabriel D. Methods Enzymol. 1971;22:568–578. [Google Scholar]
- 38.Chen GC, Yang JT. Anal. Lett. 1977;10:1195–1207. [Google Scholar]
- 39.Hunter GA, Ferreira GC. Anal. Biochem. 1995;226:221–224. doi: 10.1006/abio.1995.1217. [DOI] [PubMed] [Google Scholar]
- 40.Alexeev D, Alexeeva M, Baxter RL, Campopiano DJ, Webster SP, Sawyer L. J. Mol. Biol. 1998;284:401–419. doi: 10.1006/jmbi.1998.2086. [DOI] [PubMed] [Google Scholar]
- 41.Webster SP, Alexeev D, Campopiano DJ, Watt RM, Alexeeva M, Sawyer L, Baxter RL. Biochemistry. 2000;39:516–528. doi: 10.1021/bi991620j. [DOI] [PubMed] [Google Scholar]
- 42.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 43.Peitsch MC. Biochem. Soc. Trans. 1996;24:274–279. doi: 10.1042/bst0240274. [DOI] [PubMed] [Google Scholar]
- 44.Peitsch MC. Bio/Technology. 1995;13:658–660. [Google Scholar]
- 45.Sternberg MJ, Thornton JM. J. Mol. Biol. 1977;110:269–283. doi: 10.1016/s0022-2836(77)80072-7. [DOI] [PubMed] [Google Scholar]
- 46.Flores TP, Moss DS, Thornton JM. Protein Eng. 1994;7:31–37. doi: 10.1093/protein/7.1.31. [DOI] [PubMed] [Google Scholar]
- 47.Garnier J, Gibrat JF, Robson B. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 48.Vriend G. J. Mol. Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 49.Hahn M, Piotukh K, Borriss R, Heinemann U. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10417–10421. doi: 10.1073/pnas.91.22.10417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Viguera AR, Blanco FJ, Serrano L. J. Mol. Biol. 1995;247:670–681. doi: 10.1006/jmbi.1994.0171. [DOI] [PubMed] [Google Scholar]
- 51.Buchwalder A, Szadkowski H, Kischner K. Biochemistry. 1992;31:1621–1630. doi: 10.1021/bi00121a006. [DOI] [PubMed] [Google Scholar]
- 52.Munakata H, Yamagami T, Nagai T, Yamamoto M, Hayashi N. J. Biochem. (Tokyo) 1993;114:103–111. doi: 10.1093/oxfordjournals.jbchem.a124123. [DOI] [PubMed] [Google Scholar]