Abstract
Docosahexanoic acid (DHA) is the most abundant omega-3 fatty acid in brain, and although considered essential, deficiency has not been linked to disease1,2. Despite the large mass of DHA in phospholipids, the brain does not synthesize it. DHA is imported across the blood-brain barrier (BBB) through the Major Facilitator Superfamily Domain 2a (Mfsd2a)3. Mfsd2a transports DHA as well as other fatty acids in the form of lysophosphatidylcholine (LPC). We identify two families displaying MFSD2A mutations in conserved residues. Patients exhibited a lethal microcephaly syndrome linked to inadequate uptake of LPC lipids. The MFSD2A mutations impaired transport activity in a cell-based assay. Moreover, when expressed in mfsd2aa zebrafish morphants, mutants failed to rescue microcephaly, BBB breakdown and lethality. Our results establish a link between transport of DHA and LPCs by MFSD2A and human brain growth and function, presenting the first evidence of monogenic disease related to transport of DHA in humans.
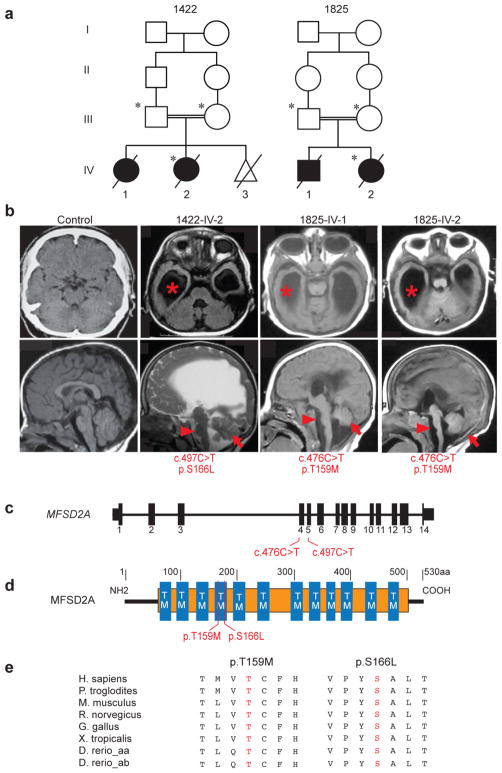
The two families, one from Libya and the other from Egypt, presented with microcephaly, developmental delay and intellectual disability, among other clinical characteristics, including hypotonia, hyperreflexia, spastic quadriparesis, seizures and ultimately death within the first few years (Supplementary Table 1). Consistent with a recessive mode of inheritance, both families demonstrated first-cousin parental consanguinity, each with two affected members (Fig. 1a), and no other non-genetic risk factors that would account for symptoms4. Brain imaging studies showed gross hydrocephalus, with hugely dilated lateral ventricles, effacement of the cortical surface, and cerebellar and brainstem hypoplasia/atrophy (Fig. 1b). We performed exome sequencing on both families and identified two rare protein-altering homozygous variants in the MFSD2A gene (NM_032793.3) (Supplementary Figs. 1 and 2). Family 1825 presented a chr1:40431005C>T variant, leading to a c.476C>T nucleotide and corresponding p.Thr159Met protein change. Family 1422 harbored a chr1:40431162C>T variant, leading to a c.497C>T nucleotide and corresponding p.Ser166Leu protein change (Fig. 1c,d). Both variants showed high damage prediction using standard programs (Supplementary Tables 2 and 3) and altered amino acid residues conserved throughout vertebrate evolution (Fig. 1e). Both mutations were in constitutively spliced exons and segregated in the respective family according to a fully penetrant recessive mode of inheritance (Supplementary Fig. 3). Moreover, exome sequencing results from both patients showed no rare deleterious homozygous variants in any gene listed in OMIM database to cause clinically relevant disease (Supplementary Tables 4 and 5).
Figure 1. MFSD2A mutations cause severe microcephaly and ventriculomegaly.
(a) Consanguineous families 1422 and 1825 designated by number in each generation. Circles: females, squares: males, slashes: deceased, triangle: spontaneous abortion, asterisk: sampled. (b) Upper: axial MRI, lower: parasagittal MRI. Images show enlarged lateral ventricles (asterisks), hypoplasia of the corpus callosum, brain stem (arrow heads) and cerebellum (arrows) in affected children. (c) Exonic structure of MFSD2A with location of the patient mutations. (d) Alignment of amino acid sequences of vertebrate MFSD2A showing the conservation of residues p.T159 and p.S166. (e) Location of mutations relative to predicted protein. TM: transmembrane domains, orange: Major facilitator superfamily, general substrate transporter domain.
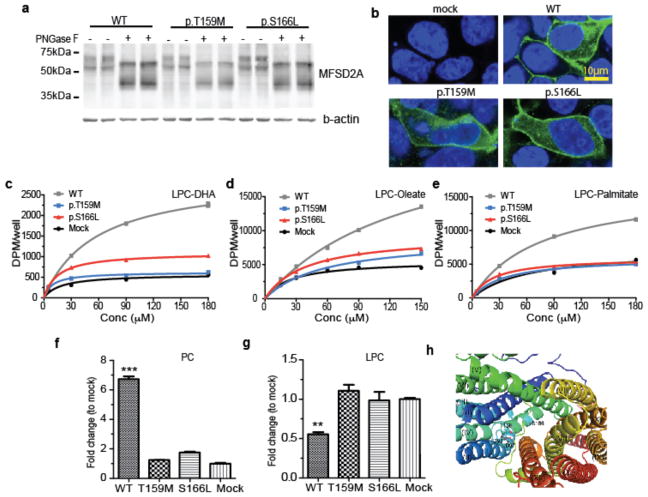
MFSD2A encodes a 12-pass transmembrane protein recently shown to be required for brain uptake of LPC containing DHA and other fatty acids in mice and implicated in the formation of the blood-brain barrier (BBB)3,5. MFSD2A is highly enriched in cerebral vasculature, where it is exclusively found in the endothelium constituting the BBB3,5,6, a localization pattern we documented, and which co-localized with GLUT1 in control human brain samples (Supplementary Fig. 4). Moreover, MFSD2A transcripts were expressed in a range of human tissues (Supplementary Fig. 5), but function in other organs is not described. The p.Thr159Met and p.Ser166Leu altering mutations were introduced into the MFSD2A human cDNA, expressed in HEK293 cells and examined by Western blot and immunofluorescence. Both were expressed at similar levels and showed reported post-translational modifications7 identical to WT (Fig. 2a). Furthermore, both mutant proteins were stably expressed and localized to the plasma membrane in a fashion similar to WT (Fig. 2b), suggesting that p.Thr159Met and p.Ser166Leu mutations did not destabilize the protein. To test for functional impairment, we used a cell-based assay with a range of concentrations of exogenous LPC-[14C]DHA, LPC-[14C]oleate and LPC-[3H]palmitate substrates following transfection into HEK293 cells (Fig. 2c–e). Both mutants exhibited transport activity similar to background in mock transfected cells for all LPC-lipids tested, indicating impaired LPC transporter activity. LPCs taken up by cells are esterified into phosphatidylcholine (PC) by cellular lysophosphatidylcholine acyltransferases (LPCATs) enzymes8. Thus, cells transfected with MFSD2A constructs were assessed for conversion of labeled LPC to PC (Fig. 2f,g and Supplementary Fig. 6). Cells expressing WT MFSD2A showed significantly greater conversion of exogenous LPC into membrane PC compared to cells expressing p.Thr159Met and p.Ser166Leu MFSD2A mutants, consistent with loss of transport function in mutants.
Figure 2. MFSD2A p.T159M and p.S166L mutations display impaired LPC transport.
(a, b) Western blot and immunolocalization of MFSD2A (WT), mutant p.T159M, and p.S166L proteins expressed in HEK293 cells. (c–e) Concentration-dependent transport of LPC-[14C]DHA, LPC-[14C]oleate, and LPC [14C]palmitate after 30 min of MFSD2A (WT), mutant p.T159M, and p.S166L proteins expressed in HEK293 cells. (f) Comparison of biological incorporation of radiolabeled LPC-[14C]oleate into phosphatidylcholine (PC). (g, f) Quantification of radiolabeled PC and LPC bands from TLC plates shown in Supplementary Fig 6. (h) View of the internal cavity of human MFSD2A Experiments were performed twice with triplicates. Data are expressed as mean ± SEM. ***p<0.001, ** p <0.01. Specific p values from left to right: p<0.001, p=0.0019.
A molecular explanation for loss of function of mutations was inferred from the recently solved atomic resolution structure of MelB9,10, the E. coli sodium-melibiose transporter, which is ~ 54 % similar to MFSD2A, making it a suitable candidate for modeling patient mutations. T159 is homologous to T121 in MelB, which was shown to hydrogen bond with aspartate residues D93 and D97, essential for sodium binding, and critical for transport activity (Fig. 2h and Supplementary Note). In contrast, S166 was not conserved in E. coli, but was located at the putative substrate-binding site as predicted by the MelB structure. Therefore, both mutations are predicted to interfere with transport activity (Fig. 2h and Supplementary Note).
To validate the functional impact of the human mutations in vivo, we developed a zebrafish model of MFSD2A deficiency. MFSD2A has two paralogs in zebrafish, mfsd2aa and mfsd2ab, sharing 58 % and 62 % identity to the human amino acid sequence, respectively. Whole-mount in situ hybridization showed expression of mfsd2aa and mfsd2ab mRNA throughout the developing nervous system (Supplementary Fig. 7). As expected based on high amino acid sequence identity, both mfsd2aa and mfsd2ab exhibit similar transport activity of LPC ligands, and to a similar level as human MFSD2A (Supplementary Fig. 8). We thus tested genetic causality of the human mutations by knocking down endogenous mfsd2aa and mfsd2ab using morpholinos (MO). Lethality was observed for each mfsd2aa and mfsd2ab morpholino, indicating they play non-redundant roles and are required for survival (Fig. 3a, Supplementary Fig 9). Lethality in morphants was also consistent with postnatal lethality in human and mouse with MFSD2A deficiency (40 % of Mfsd2a KO die postnatally7). However, since the majority of mfsd2aa and mfsd2ab morphants died before neural maturation (i.e. BBB forms at 3 dpf 21 in zebrafish) it is unlikely that lethality is caused solely through disruption of neural function.
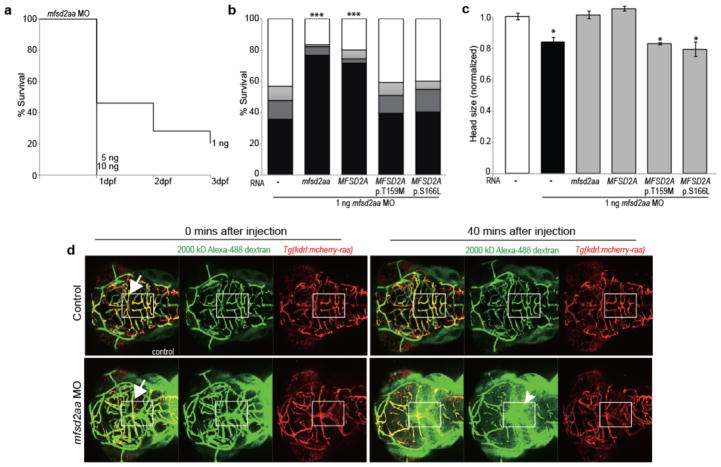
Figure 3. Zebrafish mfsd2aa morphants present disrupted BBB integrity, lethality and microcephaly.
(a) Intracardiac injection of 2000-kD dextran into mfsd2aa morpholino (MO)-injected and control embryos. Arrows: colocalization of dextran (green) and cranial blood vessels (red). Arrowhead: dextran extravasation into the brain parenchyma. (b) Kaplan–Meier plot of mfsd2aa morphants survival (n=457 embryos). (c) mfsd2aa MO (1ng) was co-injected with zebrafish wild-type mfsd2aa mRNA (50ng; n=118), human wild-type MFSD2A mRNA (50ng; n=135), or mutated p.T159M and p.S166L human MFSD2A mRNA (50ng; n=120 and 107 respectively). Bars represent the cumulative % of survival after 1 (dark grey), 2 (light grey) and 3 (black) days post fertilization (dpf). (d) Comparison of the head size of control and mfsd2aa morphants (n=20). *** p <0.001, * p <0.05. Specific p values from left to right: p<0.001, p<0.001, p=0.0143, p=0.0302, p=0.0310.
Remarkably, even at the 1 ng dose, about 50 % of mfsd2aa morphants died by 1 dpf, with only ~ 20 % remaining by 3 dpf (Fig. 3a, Supplementary Fig. 9). Because the observed lethal phenotype in fish at 24 hpf is more severe than in human and mouse, other species-specific effects of mfsd2aa and mfsd2ab depletion that are unrelated to lipid transport cannot be excluded. Given that mfsd2aa and mfsd2ab morphants both show lethality, we focused our genetic tests arbitrarily on mfsd2aa. To corroborate pathogenicity of the human mutations, we co-injected human MFSD2A WT or mutated mRNA or zebrafish mfsd2aa mRNA with mfsd2aa MO in zebrafish zygotes. We observed that either zebrafish or human WT mRNA largely rescued the lethal phenotype whereas co-injection of the human p.Thr159Met or p.Ser166Leu mutant MFSD2A mRNA failed to reverse the lethality (Fig. 3b). Moreover, mfsd2aa morphants present with microcephaly similar to patients with MFSD2A mutations and while WT zebrafish or human mRNA rescued the phenotype, human p.Thr159Met or p.Ser166Leu mutant MFSD2A did not (Fig. 3c), providing further support for genetic causality of the human mutations. However, morphant phenotypes may include off-target effects22, so additional studies of germline mutants will be needed to validate the observed phenotypes.
Mfsd2a has been implicated in the formation of the BBB as indicated by increased uptake of fluorescently labeled dextran beads through vesicular transport into brain parenchyma of Mfsd2a KO mice5. Given these findings, we tested whether mfsd2aa morphants present disruption of the BBB. The vascular-labeled Tg(kdrl:mcherry-ras)s896 zebrafish line was utilized in order to visualize blood vessels11. Intracardiac injection of 10 kD and 2,000 kD Alexa 488-conjugated dextran at 3 dpf was followed by live imaging. As expected, fluorescent dextran was confined within vessels of control-injected embryos (Fig. 3d, Supplementary Figs. 10 and 11, and Supplementary videos 1, 2 and 13–16). In contrast, dextran extravasated extensively from vessels as evidenced by fluorescence within the brain parenchyma of mfsd2aa morphant brains (36/83 embryos tested) (Fig. 3d, Supplementary Figs. 10 and 11, and Supplementary videos 3, 4 and 17–20). Finally, co-injection with either zebrafish and human mRNA largely rescued dextran extravasation, whereas co-injection with the human p.Thr159Met or p.Ser166Leu mutant MFSD2A mRNA failed to reverse extravasation (Fig. 3d, Supplementary Figs. 10 and 11, and Supplementary videos 5–12 and 21–36). Moreover, 10 % of mfsd2aa, but not mfsd2ab, morphant brains exhibit internal hemorrhage (Supplementary Fig. 12), consistent with breakdown of the BBB. The similar dextran extravasation phenotype in mouse and zebrafish suggests conserved function in mediating BBB integrity, and although we did not observe evince of blood-derived products in patients T2 MRI images (Supplementary Fig. 13), the resulting hydrocephalus may have its root in defective BBB function.
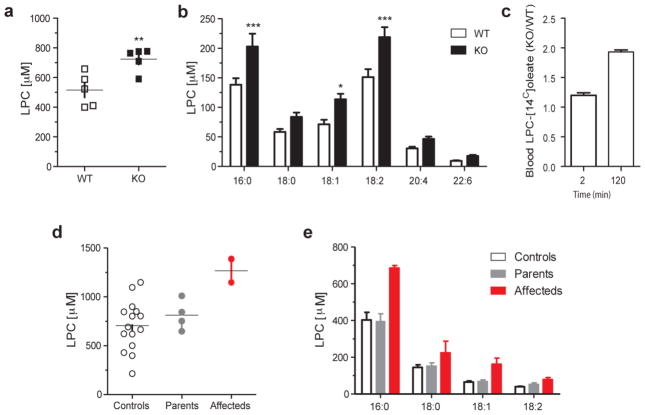
LPCs are synthesized by the liver and circulate via albumin at levels equivalent to unesterified fatty acids in plasma, and serve as the chemical carrier for DHA uptake via Mfsd2a. The specific roles for LPCs in brain growth remain only partially characterized, but their abundance in blood and efficient conversion to PCs in cellular membranes suggest a key role in cellular homeostasis and growth, likely playing roles in neural progenitor proliferation and neurite outgrowth12,13. Brain tissue from MFSD2A mutated patients was not available for our studies, precluding quantification of brain DHA concentrations. Since Mfsd2a KO mice exhibit reduced transport of plasma LPCs into brain3, we hypothesized that plasma LPC uptake by Mfsd2a at the BBB affects plasma LPC levels, and should result in increased plasma LPC levels, to serve as a surrogate marker for defective LPC transport. We thus quantified plasma LPC levels in Mfsd2a KO mice, which we found increased by 40 % compared with controls (p = 0.0094) (Fig. 4a). Increased plasma LPCs in Mfsd2a KO mice were found in the major LPC species compared with controls (Fig. 4b). Consistent with increased plasma LPC steady-state levels in Mfsd2a KO mice, and with the finding that brain uptake of LPCs in Mfsd2a KO mice were reduced between 85–90 % depending on the LPC species3, tracer studies of intravenous-injected LPC-[14C]oleate showed increased levels of plasma LPC-[14C]oleate in Mfsd2a KO mice at 2 h post-injection (Fig. 4c). Given these results, we tested whether MFSD2A mutated patients also have increased plasma levels of LPCs relative to heterozygous parents and healthy age-matched controls. Lipidomic analysis indicated that total plasma LPCs were increased in the probands relative to their heterozygous parents and controls (Fig. 4d). Similar to the findings in Mfsd2a KO mice, the common plasma LPC species containing 16:0, 18:0, 18:1 and 18:2 length fatty acids were increased in the sera from MFSD2A mutated patients, consistent with a defect in LPC uptake at the BBB (Fig. 4e). In summary, lipid analysis pointed to elevated serum LPC levels, likely as a result of failed cellular uptake due to the lack of MFSD2A activity. Because MFSD2A can transport both DHA as well as other long chain fatty acids linked to LPC3, it remains unknown the degree to which defects in transport of specific LPCs contribute to the human phenotype.
Figure 4. Total plasma LPC and individual LPC species by lipidomic mass spectrometry.
(a,b) Concentration of total plasma LPC and common C16-22 chain lenght LPC species from WT (n=5) and Mfsd2a KO (n=5) mice, with 3 technical replicates. (c) Quantification of injected LPC [14C]oleate over time in the plasma of Mfsd2a KO mice (n=4) realtive to WT (n=3) littermates. (d, e) Total plasma LPC and common LPC species concentrations from age matched controls, unaffected parents and affected individuals from families 1422 and 1825. Analysis was performed once with 3 technical replicates from two independent plasma samples collected on different days. * p <0.05, ** p <0.01, *** p <0.001. Specific p values from left to right: p=0.0094, p=0.0004, p=0.0372, p=0.0002.
It has been reported that Mfsd2a KO mice have leaky BBB due to increased transcytosis of pinocytotic vesicles at the luminal side of the BBB endothelium5. It is questionable whether leaky BBB due to transcytosis is causative for defective brain growth. Indeed, pericyte deficiency mouse models due to mutations in PDGFB results in leaky BBB due to markedly increased transcytosis14,15,16. However, these pericyte deficient models do not have microcephaly. In addition, Mfsd2a was reported to be reduced by ~ 40 % in brain capillaries of a pericyte deficient model as determined by immunofluorescence microscopy, and suggested to be a cause for leaky BBB in pericyte deficiency5. However, Mfsd2a is reduced in Mfsd2a heterozygous mice by ~ 50 %, without exhibiting microcephaly5,7. Together these findings suggest that disruption of BBB through increased transcytosis is unlikely to cause microcephaly in Mfsd2a KO mice. Nevertheless, leaky BBB in Mfsd2a KO mice could be a result of defective transport of LPCs into the BBB endothelium, and raises the possibility that LPCs play a role in membrane integrity and function at the BBB. It will be interesting to discern if patients with MFSD2A mutations display telltale evidence of BBB breakdown, such as small petechial microlesions23, but brain samples were not available from deceased patients. Our study establishes a mechanistic association between LPC transport by MFSD2A and human brain growth and function.
ONLINE METHODS
Patient Ascertainment
Patients were enrolled and sampled according to standard practice in approved human subject protocols at the University of California. Patients were recruited from developmental child neurology clinics throughout the Middle East, North Africa and Central Asia presenting with features of neurodevelopmental delay or regression, intellectual disability, autism, epilepsy or structural brain malformations between 2004 and 2012. Clinical records, radiographs, and past history were reviewed and patients were examined by one or more of the authors. Blood and/or saliva were collected on all potentially informative family members, upon informed consent approval and consistent with IRB guidelines. DNA was extracted and subject to quality control measures, and subsequent genetic investigation.
Animals
Mfsd2a-knockout mice were generated as described previously7. Experimental protocols were approved by the SingHealth Institutional Animal Care and Use Committee. Zebrafish experiments were performed in compliance with IACUC at University of California San Diego.
Exome sequencing was performed on both parents and affected member from each family. Genomic DNA was subject to Agilent Human All Exon 50 Mb kit library preparation, then paired-end sequencing (2 × 150 bp) on the Illumina HiSeq 2,000 instrument. For each patient sample, > 90 % of the exome was covered at > 30x. GATK17 was used for variant identification. We tested for segregating rare structural variants using XHMM18. We then filtered for homozygous variants using custom Python scripts (available upon request), to remove alleles with > 0.1 % frequency in the population19, not occurring in homozygous intervals, or without high scores for likely damage to protein function. Novel mutations were identified in the MFSD2A gene in families 1422 and 1825, where no other members of the cohort displayed putative deleterious variants.
Sanger sequencing
Primers were designed using the Primer3 program and tested for specificity using NIH BLAST software. PCR products were treated with Exonuclease I (Fermentas) and Shrimp Alkaline Phosphatase (USB Corporation) and sequenced using Big Dye terminator cycle sequencing Kit v.3.1 on an ABI 3,100 DNA analyzer (Applied Biosystems). Sequence data was analyzed by Sequencher 4.9 (Gene Codes).
Mutagenesis of MFSD2A
The human MFSD2A from Sport6 (OpenBiosystems) was amplified using primers hMfsd2aBamHI and hMfsd2aXbaI (Supplementary Table 6) and cloned into pcDNA3.1 into BamHI and XbaI sites. For mutagenesis of MFSD2A p.Thr159Met and p.Ser166Leu by PCR, specific primers were used (Supplementary Table 6). The mutated PCR products of p.Thr159Met and p.Ser166Leu were subsequently cloned into pcDNA3.1 and sequence verified.
PNGase F treatment
WT MFSD2A, p.Thr159Met and p.Ser166Leu mutated MFSD2A expressed in HEK293 cells was performed as previous described2 with the exception that incubation time was 3 h. Western blot was performed with previously described MFSD2A antibody3,7.
Modeling of MFSD2A
The 3D structure of MFSD2A was modeled using i-Tasser program. The best fit model for MFSD2A is the bacterial melibiose permease (MelB) for which atomic structure was recently solved4. The transmembrane domains and residues of modeled MFSD2A were subsequently viewed using PyMol.
Transport assay
Transport assay using HEK293 cells was performed as previously described4. Briefly, WT MFSD2A, p.Thr159Met (p.T159M) and p.Ser166Leu (p.S166L) mutated MFSD2A, zebrafish mfsd2aa and mfsd2ab plasmids were transfected into HEK293 cells. Uptake assay was performed after 24 h of transfection with a range of LPC [14C]-DHA, LPC [14C]-oleate, LPC [3H]-palmitate concentrations. Experiments were repeated twice with triplicates in 12 well plates. Uptake activity was expressed as DPM/well. Radiolabeled LPC [14C]-DHA, LPC [14C]-oleate, LPC [3H]-palmitate was purchased from ARC. Non-radiolabeled LPC-DHA was synthesized as described previously3. LPC-oleate and LPC-palmitate were obtained from Avanti Polar Lipids, Inc.
TLC analysis of phospholipids
HEK293 cells overexpressing pcDNA3.1hMfsd2a (WT), pcDNA3.1Mfsd2aT159M (p.T159M), pcDNA3.1Mfsd2aS166L (p.S166L), zebrafish mfsd2aa, mfsd2ab, or pcDNA3.1 (mock) plasmids were washed once with serum-free DMEM medium before incubation with 100 μM radiolabeled LPC [14C]oleate and incubated for 30 min. Wells were washed three times with DMEM containing 0.5% BSA. Lipids were extracted twice with HIP (Hexanes/Isopropanol, ratio 3:2) buffer for 30 min, dried with nitrogen stream, reconstituted in chloroform and spotted on TLC plates (Millipore). Solvent for phospholipid separation was chloroform/methanol/ammonia solution (25%) (50:25:6, per volume). TLC plates of radiolabeled phospholipids were dried for 30 min and exposed overnight to Phosphor screens and scanned with Typhoon FLA 9,000 scanner (Agilent). Phospholipid bands were quantified using ImageQuant software and expressed as fold change to mock.
Lipidomic analysis of plasma samples
For human plasma samples, single plasma sample of the father, mother and affected (1.5 years old) from family 1825, and duplicated plasma samples from the father, mother and affected (4.5 years old) from family 1422 were used for LPC analysis. Age matched human plasma samples from 15 healthy children (male and female) between 0.1 and 18 years old (mean 4.3 years old; s.d. ±4.4 years) were used as controls. Lysophospholipids were extracted using a methanol-based protocol described previously5,20. Briefly, plasma samples (2 μL) were re-suspended in 200μL of methanol containing 100 pmol/mL of LPC 20:0 as an internal standard (Avanti Polar Lipids, USA), followed by 30s vortex and 30 min sonication on ice. Samples were centrifuged at 14,000rpm for 10 min at 4°C to remove debris. The supernatants were diluted 5x with methanol (total volume 25 μL) prior to injection in LC-MS/MS. For mouse plasma samples, lysophospholipids were extracted using activated charcoal. Briefly, plasma (150 μL) from 5 WT and 5 KO littermates aged 3.5 months, both male and female, were first diluted with 650 μL PBS and then with 800 μL activated charcoal solution (1g/50ml PBS). No randomization was performed. Samples were rotated for 1h at 25°C followed by a centrifugation for 5 min at 10,000rpm to collect the charcoal pellets. The pellets were washed three times with PBS and then resuspended in 500 μL PBS. An equal amount of chloroform/methanol (2:1) was added to the samples and rigorously vortexed for 30 min at 25°C. Organic phase was separated by centrifugation, and lipid extraction was performed twice with choloform/methanol (2:1) and dried with N2 gas. Prior to lipidomic analysis, dried lipid extracts were re-suspended in 150μL of chloroform/methanol 1/1, and further diluted with 200μL of methanol containing 0.91nmol/mL of LPC 20:0 as an internal standard. These solutions were used for injection into LC/MSMS.
Mass spectrometry analysis
Samples were randomized for injection into LC/MSMS. Each sample was analyzed in technical triplicates. Each sample analysis was followed by a blank injection to avoid carry-over. Stability of signal throughout the analysis was monitored by regular injection of a QC sample. Chromatographic analysis was undertaken on a 1290 Liquid Chromatography System (Agilent Technologies, USA) using a Kinetex HILIC stationary phase (150×2.1mm, 2.6μm, 100Å, Phenomenex, USA). Gradient elutions were performed with solvents were A: 95% Acetonitrile/5% 10mM ammonium formate/0.1% formic acid, and B: 50% Acetonitrile/50% 10mM ammonium formate/0.1% formic acid, with gradient range from 0.1% to 75% B in 6 min, to 90% B in 1 min, to 0.1%B in 0.1 min, kept at 0.1% B for 3 min (total runtime 10.1 min). Under these conditions, LPC species elute ~4.9 min with flow rate 0.5mL/min. LPC species quantified using Multiple Reaction Monitoring (MRM) on a 6460 triple quadruple mass spectrometer (Agilent Technologies, USA) with gas temperature 300°C, gas flow 5L/min, sheath gas flow 11L/min, and capillary voltage 3500V. MRM transitions were from precursor ions to the choline head fragment (m/z 184) with collision energy of 29V. 36 transitions were monitored simultaneously with a dwell time of 20ms. Quantification data were extracted using MassHunter Quantitative Analysis (QQQ) software, data manually curated to ensure correct peak integration. Areas under curve (AUC) of the extracted ion chromatogram peaks for each MRM transition and lipid species were normalized to internal standard. Total and individual LPC species from human and mouse samples were expressed as mM.
Blood LPC [14C]-oleate Analysis
Mfsd2a KO and WT mice were injected intravenously with 100mM radioactive labeled LPC [14C]-oleate. 10ml of blood samples were collected after 2 min (initial dose) and after 2 h from tail vein, and radioactivity was quantified by scintillation counting. The amount of plasma LPC [14C]-oleate in the KO mice was expressed as ratio to the WT at each time point.
In situ hybridization
Whole-mount in situ hybridization was performed on 24, 48 and 96hr post fertilization (hpf) zebrafish embryos as described previously6, using mfsd2aa, mfsd2ab and huc (neuronal marker) RNA probes as well as mfsd2aa sense (negative control) RNA probe. The RNA probes were generated by PCR, using specific primers (Supplementary Table 6).
Morpholino knockdown and mfsd2a rescue studies
Adult male and female zebrafish (<18 months old) from wild-type (AB Tübingen) and transgenic strains were maintained under standard laboratory conditions. At least three adult pairs were used to generate embryos at 0–4 dpf for each experiment. No randomization was performed. Translational blocking antisense morpholino oligonucleotides (MO) for mfsd2aa and mfsd2ab and control (scramble) MO were injected into one-cell stage embryos. Lethality (count of immobile embryos) and microcephaly (head size measured as distance between the eyes) phenotypes were assessed. For mRNA rescue experiments, full-length zebrafish wild-type mfsd2aa mRNA (50 ng; n=118), human WT MFSD2A mRNA (50 ng; n=135), as well as mutated p.Thr159Met and p.Ser166Leu human MFSD2A mRNA (50 ng; n=120 and 107 respectively) were co-injected with the MO (n=592) as described previously6. Treated embryos that survived were counted and scored as rescued. MO binding site in human and zebrafish mRNA can be found in Supplementary Fig. 14.
Blood Brain Barrier integrity assay
For zebrafish vascular phenotypes, Tg(kdrl:mcherry-ras)s896 embryos were used to visualize cranial vessels7. One-cell stage embryos were injected with 1 ng of mfsd2aa MO (n=467) as described6. After 3 days post fertilization (dpf), 10 kD and 2, 000kD 488-Alexa dextran was injected into the heart, and fluorescence was visualized 10 min and 40 min after injection. Bright field and confocal fluorescence microscopy were used to determine whether the experimental embryos exhibited vascular dextran extravasation.
Statistical analysis
All in vitro experiments were performed in quadruplicate. Data are expressed as means with standard errors. We used the Student t-test to perform between group comparisons (two-tailed) and Tukey’s test in conjunction with analysis of variance for multiple comparisons. Kaplan–Meier curves were calculated for survival and survival curves were compared using Log-rank (Mantel-Cox) test. All p-values were tested as two-sided and p-value <0.05 was considered indicative of statistical significance after appropriate correction for multiple comparisons. No statistical methods were used to predetermine sample sizes. Experimental sample size was determined empirically from previous experimental experience with similar assays and/or from sizes generally employed in the field. Observers were not blinded to conditions.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health P01HD070494 (to J.G.G. and N.C.C.), R01NS048453 (to J.G.G.) and K99NS089943 (to A.G-G.), Broad Institute grant U54HG003067, the Yale Center for Mendelian Disorders U54HG006504 (to M.G.), Singapore Ministry of Health’s National Medical Research Council CBRG/025/2014 (to D.L.S.), the Singapore National Research Foundation Competitive Research Program grants 2007-04 (to M.R.W.), National University of Singapore’s Life Sciences Institute (to M.R.W.). Sequencing provided in part by a gift from BGI to Rady Children’s Hospital for undiagnosed patients. Human brain samples provided by Subhojit Roy.
Footnotes
Accession numbers. The exome sequencing data from individuals in this study have been deposited to dbGAP (phs000288.v1.p1).
Supplementary Information is linked to the manuscript.
AUTHOR CONTRIBUTIONS
M.S.Z., M.K., T.B.-O., K.K.V. and O.R.O recruited subjects and analyze the clinical data; E.S., J.L.S. and B.C. interpreted exome results. J.G.G. and D.L.S. conceived and designed the project. A.G.-G. performed genetic analysis to identify MFSD2A mutations; L.N.N. performed lipid transport studies, lipidomic, TLC, and confocal microscopy; H.Y. and N.C.C performed zebrafish morpholino studies and A.G.-G. analyze and interpret the data; B.R., D.Q.Q, B.H.W, and B.C.T assisted with cloning, western blots, and imaging; A.C.-G. and M.R.W provided expertise in mass spectroscopy; D.L.S, L.N.N., A.G.-G., and J.G.G. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests
References
- 1.Svennerholm L. Distribution and fatty acid composition of phosphoglycerides in normal human brain. J Lipid Res. 1968;9:570–579. [PubMed] [Google Scholar]
- 2.Soderberg M, Edlund C, Kristensson K, Dallner G. Fatty acid composition of brain phospholipids in aging and in Alzheimer’s disease. Lipids. 1991;26:421–425. doi: 10.1007/BF02536067. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen LN, et al. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509:503–506. doi: 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- 4.Engle PL, et al. Strategies to avoid the loss of developmental potential in more than 200 million children in the developing world. Lancet. 2007;369:229–242. doi: 10.1016/S0140-6736(07)60112-3. [DOI] [PubMed] [Google Scholar]
- 5.Ben-Zvi A, et al. Mfsd2a is critical for the formation and function of the blood-brain barrier. Nature. 2014;509:507–511. doi: 10.1038/nature13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daneman R, et al. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PloS one. 2010;5:e13741. doi: 10.1371/journal.pone.0013741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger JH, Charron MJ, Silver DL. Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PloS one. 2012;7:e50629. doi: 10.1371/journal.pone.0050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakanishi H, et al. Cloning and characterization of mouse lung-type acyl-CoA:lysophosphatidylcholine acyltransferase 1 (LPCAT1). Expression in alveolar type II cells and possible involvement in surfactant production. J Biol Chem. 2006;281:20140–20147. doi: 10.1074/jbc.M600225200. [DOI] [PubMed] [Google Scholar]
- 9.Ethayathulla AS, et al. Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordat E, Leblanc G, Mus-Veteau I. Evidence for a role of helix IV in connecting cation- and sugar-binding sites of Escherichia coli melibiose permease. Biochemistry. 2000;39:4493–4499. doi: 10.1021/bi991852i. [DOI] [PubMed] [Google Scholar]
- 11.Chi NC, et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 2008;22:734–739. doi: 10.1101/gad.1629408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He C, Qu X, Cui L, Wang J, Kang JX. Improved spatial learning performance of fat-1 mice is associated with enhanced neurogenesis and neuritogenesis by docosahexaenoic acid. Proc Natl Acad Sci U S A. 2009;106:11370–11375. doi: 10.1073/pnas.0904835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawakita E, Hashimoto M, Shido O. Docosahexaenoic acid promotes neurogenesis in vitro and in vivo. Neuroscience. 2006;139:991–997. doi: 10.1016/j.neuroscience.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 14.Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood-brain barrier integrity during embryogenesis. Nature. 2010;468:562–566. doi: 10.1038/nature09513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell RD, et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron. 2010;68:409–427. doi: 10.1016/j.neuron.2010.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Armulik A, et al. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- 17.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromer M, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Gen. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shui G, et al. Comparative plasma lipidome between human and cynomolgus monkey: are plasma polar lipids good biomarkers for diabetic monkeys? PloS one. 2011;6:e19731. doi: 10.1371/journal.pone.0019731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleming A, Diekmann H, Goldsmith P. Functional characterization of the maturation of the blood-brain barrier in larval zebrafish. PloS one. 2013;8:e77548. doi: 10.1371/journal.pone.0077548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolk FO, et al. Reverse Genetic Screening Reveals Poor Correlation between Morpholino-Induced and Mutant Phenotypes in Zebrafish. Dev Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harata N, Iwasaki Y. The blood-brain barrier and selective vulnerability in experimental thiamine-deficiency encephalopathy in the mouse. Metab Brain Dis. 1996;11:55–69. doi: 10.1007/BF02080931. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.