Abstract
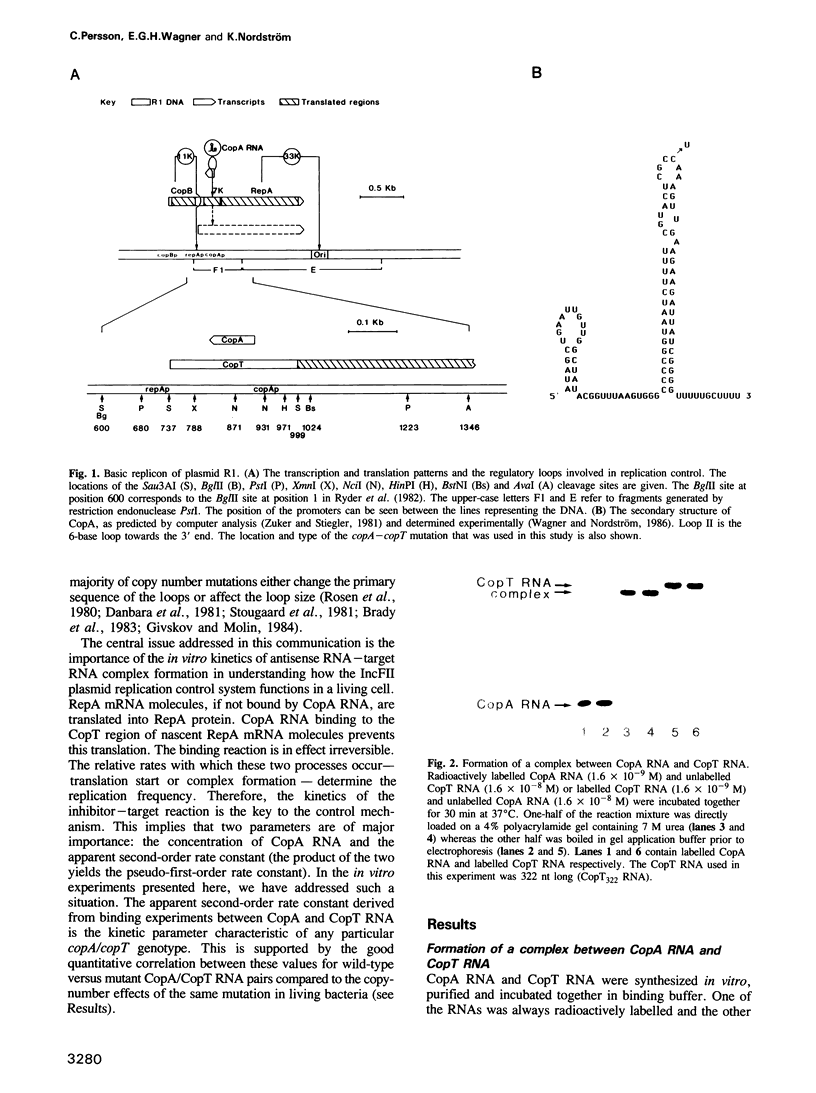
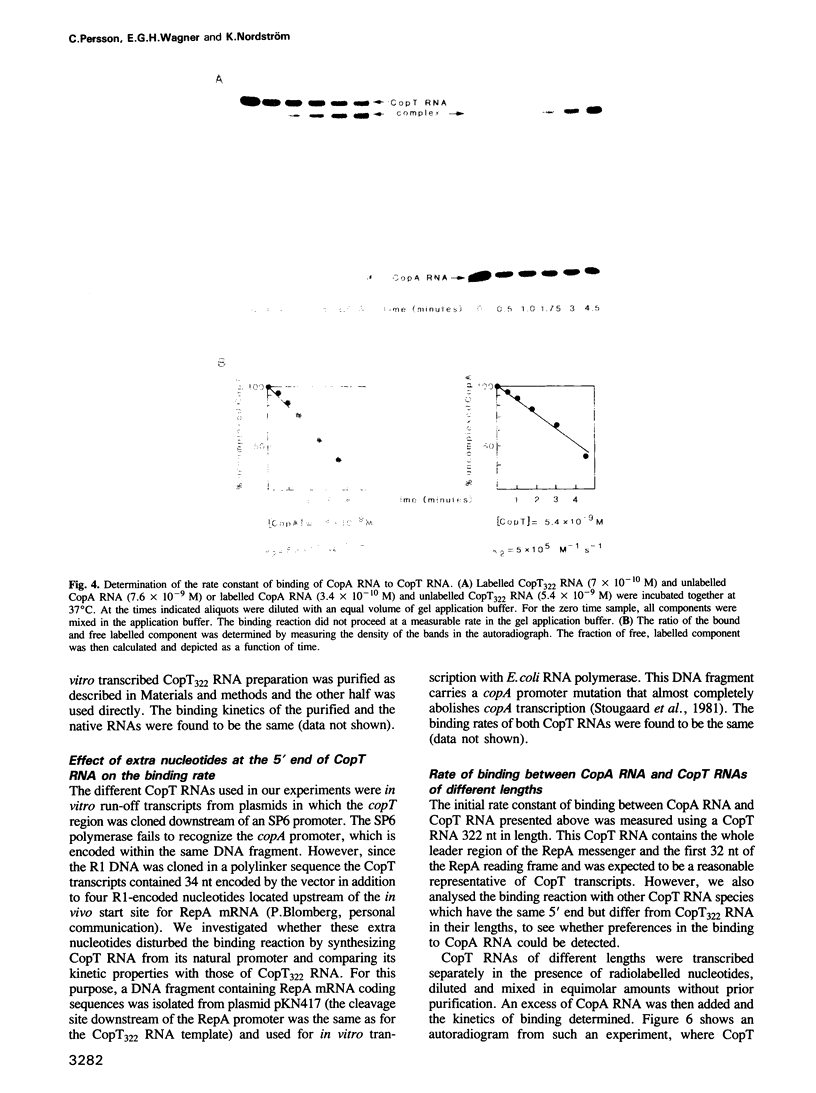
The frequency of replication of IncFII plasmids is regulated by the availability of a rate-limiting protein, RepA. This protein acts to promote initiation of replication and its synthesis is negatively controlled both at the transcriptional and translational level. The translational control is exerted by the binding of a small antisense RNA, CopA RNA, to its target, CopT, which is located in the leader region of the RepA mRNA. As a consequence, formation of RepA is inhibited. Here we demonstrate the binding of CopA RNA to CopT RNA in vitro; the rate constant of binding was determined to be approximately 1 X 10(6) M-1 s-1 at 37 degrees C. We have also shown that in vitro synthesized RepA mRNA molecules differing in length, but which contain the whole CopT region, are able to bind CopA RNA with similar rates. Analysis of the binding of CopA/CopT molecules derived from a copy-number mutant plasmid showed that the effect of the mutation on the rate of in vitro binding correlates well with its phenotypic effect in vivo, i.e. the binding rate constant is lowered in proportion to the increase in copy number. Likewise, the result of an in vitro incompatibility test is in agreement with in vivo data.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brady G., Frey J., Danbara H., Timmis K. N. Replication control mutations of plasmid R6-5 and their effects on interactions of the RNA-I control element with its target. J Bacteriol. 1983 Apr;154(1):429–436. doi: 10.1128/jb.154.1.429-436.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Bass B. L. Biological catalysis by RNA. Annu Rev Biochem. 1986;55:599–629. doi: 10.1146/annurev.bi.55.070186.003123. [DOI] [PubMed] [Google Scholar]
- Danbara H., Brady G., Timmis J. K., Timmis K. N. Regulation of DNA replication: "target" determinant of the replication control elements of plasmid R6-5 lies within a control element gene. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4699–4703. doi: 10.1073/pnas.78.8.4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey W. B. Transcript analysis of the plasmid R100 traJ and finP genes. Mol Gen Genet. 1987 Oct;209(3):533–544. doi: 10.1007/BF00331160. [DOI] [PubMed] [Google Scholar]
- Dong X. N., Womble D. D., Rownd R. H. Transcriptional pausing in a region important for plasmid NR1 replication control. J Bacteriol. 1987 Dec;169(12):5353–5363. doi: 10.1128/jb.169.12.5353-5363.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley T. P., Polisky B. Suppression of ColE1 RNA-RNA mismatch mutations in vivo by the ColE1 Rop protein. Plasmid. 1987 Jul;18(1):24–34. doi: 10.1016/0147-619x(87)90075-8. [DOI] [PubMed] [Google Scholar]
- Gerhart E., Wagner H., Nordström K. Structural analysis of an RNA molecule involved in replication control of plasmid R1. Nucleic Acids Res. 1986 Mar 25;14(6):2523–2538. doi: 10.1093/nar/14.6.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givskov M., Molin S. Copy mutants of plasmid R1: effects of base pair substitutions in the copA gene on the replication control system. Mol Gen Genet. 1984;194(1-2):286–292. doi: 10.1007/BF00383529. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Söll D. G., Crothers D. M. Studies of the complex between transfer RNAs with complementary anticodons. I. Origins of enhanced affinity between complementary triplets. J Mol Biol. 1976 May 25;103(3):499–519. doi: 10.1016/0022-2836(76)90214-x. [DOI] [PubMed] [Google Scholar]
- Helmer-Citterich M., Anceschi M. M., Banner D. W., Cesareni G. Control of ColE1 replication: low affinity specific binding of Rop (Rom) to RNAI and RNAII. EMBO J. 1988 Feb;7(2):557–566. doi: 10.1002/j.1460-2075.1988.tb02845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Meyer R. J. Copy-number of broad host-range plasmid R1162 is regulated by a small RNA. Nucleic Acids Res. 1986 Oct 24;14(20):8027–8046. doi: 10.1093/nar/14.20.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. OOP RNA, produced from multicopy plasmids, inhibits lambda cII gene expression through an RNase III-dependent mechanism. Genes Dev. 1987 Nov;1(9):1005–1013. doi: 10.1101/gad.1.9.1005. [DOI] [PubMed] [Google Scholar]
- Kumar C. C., Novick R. P. Plasmid pT181 replication is regulated by two countertranscripts. Proc Natl Acad Sci U S A. 1985 Feb;82(3):638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacatena R. M., Cesareni G. Interaction between RNA1 and the primer precursor in the regulation of Co1E1 replication. J Mol Biol. 1983 Nov 5;170(3):635–650. doi: 10.1016/s0022-2836(83)80125-9. [DOI] [PubMed] [Google Scholar]
- Liao S. M., Wu T. H., Chiang C. H., Susskind M. M., McClure W. R. Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1987 Apr;1(2):197–203. doi: 10.1101/gad.1.2.197. [DOI] [PubMed] [Google Scholar]
- Light J., Molin S. Post-transcriptional control of expression of the repA gene of plasmid R1 mediated by a small RNA molecule. EMBO J. 1983;2(1):93–98. doi: 10.1002/j.1460-2075.1983.tb01387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light J., Molin S. Replication control functions of plasmid R1 act as inhibitors of expression of a gene required for replication. Mol Gen Genet. 1981;184(1):56–61. doi: 10.1007/BF00271195. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Reed R. The role of small nuclear ribonucleoprotein particles in pre-mRNA splicing. Nature. 1987 Feb 19;325(6106):673–678. doi: 10.1038/325673a0. [DOI] [PubMed] [Google Scholar]
- Masai H., Arai K. RepA and DnaA proteins are required for initiation of R1 plasmid replication in vitro and interact with the oriR sequence. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4781–4785. doi: 10.1073/pnas.84.14.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S., Nordström K. Control of plasmid R1 replication: functions involved in replication, copy number control, incompatibility, and switch-off of replication. J Bacteriol. 1980 Jan;141(1):111–120. doi: 10.1128/jb.141.1.111-120.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. F., Molin S. How the R1 replication control system responds to copy number deviations. Plasmid. 1984 May;11(3):264–267. doi: 10.1016/0147-619x(84)90034-9. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Projan S. J., Kumar C. C., Carleton S., Gruss A., Highlander S. K., Kornblum J. Replication control for pT181, an indirectly regulated plasmid. Basic Life Sci. 1985;30:299–320. doi: 10.1007/978-1-4613-2447-8_24. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Freundlich M. Mechanism for the autogenous control of the crp operon: transcriptional inhibition by a divergent RNA transcript. Proc Natl Acad Sci U S A. 1986 Jul;83(14):5000–5004. doi: 10.1073/pnas.83.14.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen P. B., Gerdes K., Molin S. Genetic analysis of the parB+ locus of plasmid R1. Mol Gen Genet. 1987 Aug;209(1):122–128. doi: 10.1007/BF00329846. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Ryder T. B., Davidson D. B., Rosen J. I., Ohtsubo E., Ohtsubo H. Analysis of plasmid genome evolution based on nucleotide-sequence comparison of two related plasmids of Escherichia coli. Gene. 1982 Mar;17(3):299–310. doi: 10.1016/0378-1119(82)90146-9. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Kleckner N. Translational control of IS10 transposition. Cell. 1983 Sep;34(2):683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- Stougaard P., Molin S., Nordström K. RNAs involved in copy-number control and incompatibility of plasmid R1. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6008–6012. doi: 10.1073/pnas.78.10.6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamm J., Polisky B. Characterization of the ColE1 primer-RNA1 complex: analysis of a domain of ColE1 RNA1 necessary for its interaction with primer RNA. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2257–2261. doi: 10.1073/pnas.82.8.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell. 1986 Oct 10;47(1):89–97. doi: 10.1016/0092-8674(86)90369-7. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T. Plasmid ColE1 incompatibility determined by interaction of RNA I with primer transcript. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6096–6100. doi: 10.1073/pnas.78.10.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Itoh T., Selzer G., Som T. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1421–1425. doi: 10.1073/pnas.78.3.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C., Borer P. N., Dengler B., Tinoco I., Jr Stability of RNA hairpin loops: A 6 -C m -U 6 . J Mol Biol. 1973 Feb 5;73(4):483–496. doi: 10.1016/0022-2836(73)90095-8. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., von Heijne J., Nordström K. Control of replication of plasmid R1: translation of the 7k reading frame in the RepA mRNA leader region counteracts the interaction between CopA RNA and CopT RNA. EMBO J. 1987 Feb;6(2):515–522. doi: 10.1002/j.1460-2075.1987.tb04783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Rownd R. H. Regulation of IncFII plasmid DNA replication. A quantitative model for control of plasmid NR1 replication in the bacterial cell division cycle. J Mol Biol. 1986 Dec 5;192(3):529–547. doi: 10.1016/0022-2836(86)90274-3. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]