Abstract
Summary: A key to understanding RNA function is to uncover its complex 3D structure. Experimental methods used for determining RNA 3D structures are technologically challenging and laborious, which makes the development of computational prediction methods of substantial interest. Previously, we developed the iFoldRNA server that allows accurate prediction of short (<50 nt) tertiary RNA structures starting from primary sequences. Here, we present a new version of the iFoldRNA server that permits the prediction of tertiary structure of RNAs as long as a few hundred nucleotides. This substantial increase in the server capacity is achieved by utilization of experimental information such as base-pairing and hydroxyl-radical probing. We demonstrate a significant benefit provided by integration of experimental data and computational methods.
Availability and implementation: http://ifoldrna.dokhlab.org
Contact: dokh@unc.eu
1 Introduction
The iFoldRNA is a physical simulation-based automated RNA structure prediction webserver designed to predict 3D RNA structure based on available sequence data. The previous version of iFoldRNA allowed prediction of the structure of short RNA molecules (<50 nt) within 4 Å Root-Mean-Square deviation (RMSD) from corresponding experimental structures (Sharma et al., 2008). The limitation on RNA size is due to inaccuracies in the force-field and insufficient sampling. Increasing sequence size quickly becomes computationally prohibitive, since the RNA conformational space grows exponentially. We have previously shown that sampling conformational space can be drastically reduced by use of experimental constraints allowing us to correctly predict the tertiary structure of RNA up to a few hundred nucleotides long (Ding et al., 2012; Gherghe et al., 2009; Lavender et al., 2010). Here, we present a second version of iFoldRNA, which allows automated inclusion of two categories of constraints: base-pairing and nucleotide solvent accessibility. The source of base-pairing information can be generated either by sequence covariation analysis (Gutell et al., 1992) or by using a number of chemical probing techniques. The SHAPE technique, in particular, has proven to be a quick and effective method for determining RNA secondary structure (Low et al., 2010). To infer solvent accessibility iFoldRNA v2 uses data from hydroxyl radical probing (HRP) experiments. In HRP, RNA in solution is treated with reagents that generate hydroxyl radicals, which cleave RNA strands. By identifying cleavage frequency for specific nucleotide bonds, solvent exposure can be determined. Incorporating these data as a burial force in simulations imposes long-range constraints on folded structures (Ding et al., 2012).
2 Methods
The prediction of RNA structure in iFoldRNA v2 is accomplished using a coarse-grained 3-bead RNA model (Ding et al., 2008). Each bead in this model represents a phosphate, sugar or nucleobase. Simulations are performed using the Discrete Molecular Dynamics (DMD) simulation engine (Dokholyan et al., 1998). Base-pairing information is implemented as an additional potential promoting tertiary contacts between corresponding nucleotides. An ensemble of RNA molecules at different temperatures that undergo replica exchange is used to enhance conformation sampling (Sugita and Okamoto, 1999). Following DMD simulation, one hundred of the lowest energy structures are selected and clustered according to RMSD between pairs of selected structures. The centroids of the resulting clusters are retained for all-atom reconstruction.
2.1 HRP constraints
If HRP data are available, an additional force field, effectively biasing RNA towards the correct structure, is applied. The structures selected for clustering are also required to have high structure-reactivity correlations (Ding et al., 2012).
2.2 All-atom reconstruction
Conversion to an all-atom representation is performed by replacing each 3-bead nucleotide by randomly selected rotamers of corresponding nucleotides in an all-atom representation. The initial all-atom structure is run through a short DMD simulation to connect bonds and remove clashes. This simulation is performed with a high heat exchange coefficient, which allows for rapid dissipation of excess heat generated. A user is allowed to change the values of temperatures used to run replica exchange simulations and the number of DMD time steps.
Simulations are performed on the Kure computational cluster at the University of North Carolina at Chapel Hill, which consist of 122 blade servers, each with 8-cores 2.80 GHz processors running RHEL 5.6 operating system. The users are able to check the status of submissions through the server interface and are notified by email of job completion.
3 Results
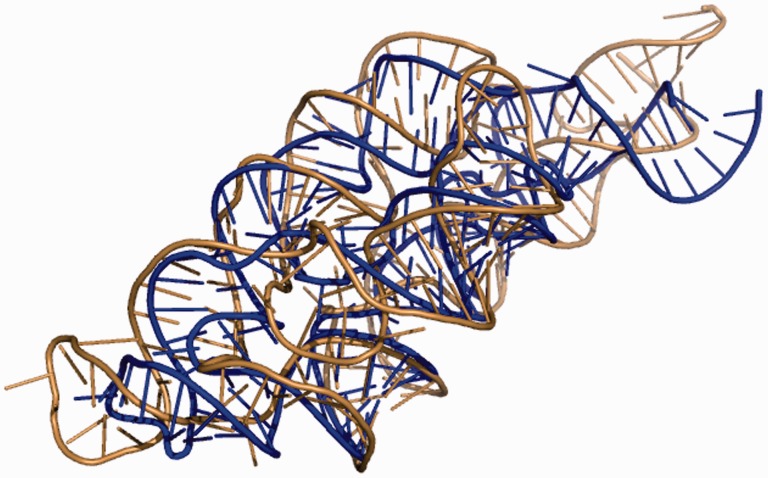
The iFoldRNA v2 webserver generates accurate predictions of RNA tertiary structure using DMD. Predicted structures are typically within 10–20 Å RMSD from crystal structures for 200 nt RNA. The average processing time for an RNA of this size is ∼1 day. We demonstrate how the inclusion of constraints increases the performance of the server on the example of M-box riboswitch (161 nt). The structure of RNA of this size is challenging to predict de novo. Without constraints the quality of the predicted structure is poor. RMSD between the predicted and the crystal structures calculated through phosphate atoms is 32 Å. The interaction network fidelity (INF) is 0.363 (Parisien et al., 2009). The inclusion of the base-pairing constraints improves the prediction (RMSD = 25 Å, INF = 0.692). The resulting structure has correct base pairing while most of the higher order tertiary contacts are missing. The addition of HRP data significantly increases the quality of the prediction (RMSD = 7.7 Å, INF = 0.725) (Fig. 1). We observe formation of the correct secondary and tertiary structures. Closer examination of the predicted structure using MC Annotate (Gendron et al., 2001) reveals that it has 39 out of 46 canonical base pairs (AU or CG; A-adenine, U - uracil, C - cytosine, G-guanine) and 5 out of 6 non-canonical UG base pairs as compared with the crystal structure. As a benchmark we used two other servers RNAComposer (Popenda et al., 2012) and 3dRNA (Zhao et al., 2012). The structure predicted by RNAComposer (RMSD = 25.8 Å, INF = 0.792) has correct base pairing; however, most of the higher order tertiary contacts are missing. The structure predicted by 3dRNA (RMSD = 22.4 Å, INF = 0.621) only partially reproduces correct base pairing and does not recapitulate most of the higher order tertiary contacts.
Fig. 1.
The structure of M-box riboswitch predicted by iFoldRNA v2 (sand color) is superimposed on the top of the crystal structure (PDB ID: 3pdr) (blue). RMSD between the predicted and the crystal structures is 7.7 Å. P-value, showing statistical significance of the prediction (Hajdin et al., 2010), is less than 10−6. RMSD was calculated using phosphate atoms only. INF = 0.725 (Parisien et al., 2009). Experimental HRP data and base-pairing information were used (Ding et al., 2012) (Color version of this figure is available at Bioinformatics online.)
4 Conclusions
The iFoldRNA v2 webserver offers a platform that combines experimental data and molecular dynamics simulations to predict tertiary structures of RNA as long as a few hundred nucleotides with atomic level detail. The comparison with other servers demonstrates the superior performance of iFoldRNA v2 for predicting the structure of long RNA molecules, provided HRP data is available. Currently, the server operates on two types of experimental constraints that include base-pairing information and reactivities derived from HRP experiments. However, there is growing number of approaches used to translate data from chemical probing experiments to constraints on RNA structure. For example, the recently proposed RING-MaP technique (Homan et al., 2014) uses chemical probing, massive parallel sequencing and correlation analysis to find nucleotides located close in space. To keep pace with experimental innovations we plan to introduce a new interface that allows users to set up constraints between any two nucleotides of their choice. The iFoldRNA v2 webserver is freely accessible at http://iFoldRNA.dokhlab.org for academic and non-profit users.
Acknowledgements
We thank the University of North Carolina IT Services for providing hardware support.
Funding
This work was supported by National Institutes of Health Grant GM064803 (PI: Kevin M. Weeks).
Conflict of Interest: none declared.
References
- Ding F., et al. (2008) Ab initio RNA folding by discrete molecular dynamics: from structure prediction to folding mechanisms. RNA, 14, 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding F., et al. (2012) Three-dimensional RNA structure refinement by hydroxyl radical probing. Nat. Methods, 9, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokholyan N.V., et al. (1998) Discrete molecular dynamics studies of the folding of a protein-like model. Fold. Des., 3, 577–587. [DOI] [PubMed] [Google Scholar]
- Hajdin C.E., et al. (2010) On the significance of an RNA tertiary structure prediction. RNA, 16, 1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homan P.J., et al. (2014) Single-molecule correlated chemical probing of RNA. Proc. Natl Acad. Sci. U.S.A., 111, 13858–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron P., et al. (2001) Quantitative analysis of nucleic acid three-dimensional structures. J. Mol. Biol., 308, 919–936. [DOI] [PubMed] [Google Scholar]
- Gherghe C.M., et al. (2009) Native-like RNA tertiary structures using a sequence-encoded cleavage agent and refinement by discrete molecular dynamics. J. Am. Chem. Soc., 131, 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R.R., et al. (1992) Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res., 20, 5785–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender C.A., et al. (2010) Robust and generic RNA modeling using inferred constraints: a structure for the hepatitis C virus IRES pseudoknot domain. Biochemistry, 49, 4931–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.T., et al. (2010) SHAPE-directed RNA secondary structure prediction. Methods, 52, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisien M., et al. (2009) New metrics for comparing and assessing discrepancies between RNA 3D structures and models. RNA, 15, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M., et al. (2012) Automated 3D structure composition for large RNAs. Nucleic Acids Res., 40, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., et al. (2008) iFoldRNA: three-dimensional RNA structure prediction and folding. Bioinformatics, 22, 1951–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y., Okamoto Y. (1999) Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett., 314, 141–151. [Google Scholar]
- Zhao Y., et al. (2012) Automated and fast building of three-dimensional RNA structures. Sci. Rep., 2. [DOI] [PMC free article] [PubMed] [Google Scholar]