Abstract
Motivation: The storage and transmission of high-throughput sequencing data consumes significant resources. As our capacity to produce such data continues to increase, this burden will only grow. One approach to reduce storage and transmission requirements is to compress this sequencing data.
Results: We present a novel technique to boost the compression of sequencing that is based on the concept of bucketing similar reads so that they appear nearby in the file. We demonstrate that, by adopting a data-dependent bucketing scheme and employing a number of encoding ideas, we can achieve substantially better compression ratios than existing de novo sequence compression tools, including other bucketing and reordering schemes. Our method, Mince, achieves up to a 45% reduction in file sizes (28% on average) compared with existing state-of-the-art de novo compression schemes.
Availability and implementation: Mince is written in C++11, is open source and has been made available under the GPLv3 license. It is available at http://www.cs.cmu.edu/∼ckingsf/software/mince.
Contact: carlk@cs.cmu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
The tremendous quantity of data generated by high-throughput sequencing experiments poses many challenges to data storage and transmission. The most common approach to reduce these space requirements is to use an ‘off-the-shelf’ compression program such as gzip (by Gailly and Adler, http://www.gnu.org/software/gzip/) or bzip2 (by Seward, http://www.bzip.org) to compress the raw read files. This approach can result in substantial savings during storage and transmission. These programs are general purpose, well-tested, and highly scalable. However, research over the past few years has demonstrated that approaches specifically tailored to compressing genomic data can achieve significantly better compression rates than general-purpose tools.
We introduce Mince, a compression method specifically designed for the compression of high-throughput sequencing reads, that achieves state-of-the-art compression ratios by encoding the read sequences in a manner that vastly increases the effectiveness of ‘off-the-shelf’ compressors. This approach, known as compression boosting, has been effectively applied in other contexts, and is responsible for the widely observed phenomenon that BAM files become smaller when alignments are ordered by genomic location. This places more similar alignments nearby in the file and results in more effective compression being possible. Mince is able to produce files that are 28% smaller than those of existing compression methods in a comparable amount of time.
Existing work on compressing sequencing reads falls into two main categories: reference-based and de novo compression. Reference-based methods most often, but not always (Bonfield and Mahoney, 2013; Kingsford and Patro, 2015; Rozov et al., 2014), attempt to compress aligned reads (e.g. BAM format files) rather than raw, unaligned sequences. They assume that the reference sequence used for alignment is available at both the sender and receiver. Most reference-based approaches attempt to take advantage of shared information between reads aligned to genomically close regions, and to represent the aligned reads via relatively small ‘edits’ with respect to the reference sequence (Bonfield and Mahoney, 2013; Campagne et al., 2013; Fritz et al., 2011; Kozanitis et al., 2011; Li et al., 2013; Popitsch and von Haeseler, 2013). These methods can, in general, be very effective at compressing alignments, but this does not necessarily imply effective compression of the original read sequences (Kingsford and Patro, 2015). Thus, if one is interested in the most efficient methods to compress the raw reads, reference-based methods can have drawbacks when compared with de novo approaches. They are generally slower, since they require that reads be mapped to a reference before being compressed. They assume that the sender and receiver have a copy of the reference (which, itself, would have to be transferred) and that the set of reads can be mapped with relatively high quality to this reference (such methods may perform poorly if there are many unmapped reads). Furthermore, since different types of analysis may require different types of alignments, recovering the original BAM file may not always be sufficient, in which case further processing, such as extracting the original sequences from the alignment file, may be required.
Conversely, de novo approaches compress the raw sequencing reads directly, and because they do not require aligning the reads to a reference, are often able to compress the reads much more quickly. De novo compression methods often work by trying to exploit redundancy within the set of reads themselves rather than between the reads and a particular reference (Adjeroh et al., 2002; Bhola et al., 2011; Bonfield and Mahoney, 2013; Brandon et al., 2009; Cox et al., 2012; Deorowicz and Grabowski, 2013; Hach et al., 2012; Jones et al., 2012; Tembe et al., 2010).
Although most approaches tend to fall into one or the other of these categories, some tools expose both reference-based and reference-free modes (Bonfield and Mahoney, 2013). Notably, Jones et al. (2012) introduced a novel approach for obtaining some of the benefits of reference-based compression, even when no reference is available, by constructing one ‘on-the-fly’.
Another similar area of research is the compression of collections of related genomes (Christley et al., 2009; Deorowicz and Grabowski, 2011; Pavlichin et al., 2013; Pinho et al., 2012; Rajarajeswari and Apparao, 2011; Wang and Zhang, 2011). These approaches are able to achieve a very high degree of compression, but generally rely on encoding a sparse and relatively small set of differences between otherwise identical sequences. Unfortunately, the reads of a sequencing experiment are much more numerous and diverse than a collection of related genomes, and hence, these methods do not apply to the compression of raw or aligned sequencing reads.
We focus on the problem of de novo compression of raw sequencing reads, since it is the most generally applicable. Mince was inspired by the approach of Hach et al. (2012) of compression ‘boosting’. Mince only compresses the actual sequences, because the compression of quality scores and other metadata can be delegated to other approaches (Cánovas et al., 2014; Ochoa et al., 2013; Yu et al., 2015) that are specifically designed for compressing those types of data.
At the core of Mince is the idea of bucketing, or grouping together, reads that share similar sub-sequences. After reads are assigned to buckets, they are reordered within each bucket to further expose similarities between nearby reads and deterministically transformed in a manner that explicitly removes a shared ‘core’ substring, which is the label of the bucket to which they have been assigned. The information encoding this reordered collection of reads is then written to a number of different output streams, each of which is compressed with a general-purpose compressor. Depending on the type of read library being compressed, we also take advantage of the ability to reverse complement reads to gain better compression.
In the presence of a reference, placing reads in the order in which they appear when sorted by their position in the reference reveals their overlapping and shared sequences. Without a reference, we cannot directly know the reference order. The bucketing strategy described here attempts to recover an ordering that works as well as a reference-based order without the advantage of being able to examine the reference.
We demonstrate that the bucketing scheme originally introduced by Hach et al. (2012), though very effective, can be substantially improved (on average > 15%) by grouping reads in a data-dependent manner and choosing a more effective encoding scheme. Choosing a better downstream compressor, lzip (by Diaz, http://www.nongnu.org/lzip/lzip.html) leads to a further reduction in size of 10%. Overall, Mince is able to obtain significantly better compression ratios than other de novo sequence compressors, yielding compressed sequences that are, on average, 28% smaller than those of SCALCE.
2 Algorithm
Mince’s general approach to compression is to group together similar sequences to make the underlying compression algorithm more effective. Thus, we want to re-order the set of reads so that those that share similar sub-sequences will appear close to each other in the file.
To achieve this goal Mince encodes a set of reads in a few phases, which are described later. The result of this processing is to place a transformed set of the original reads into a collection of buckets, each of which is designed to expose coherent sequence to the downstream compressor. The contents of these buckets, along with the information required to invert any transformations that have been applied, are written to a set of different output streams and compressed using a general-purpose compression tool.
Local bucketing. The first phase of Mince aggregates reads into buckets. A bucket b consists of a label and a collection of reads. The goal is to place within each bucket a collection of reads that are ‘similar’ to each other. In addition to being a difficult problem theoretically—bucketing is essentially clustering—the method we choose for bucket creation and assignment must be practically fast to handle the large number of reads we observe in most datasets. The approach we take to this problem is one of streaming bucket assignment based on a cost function that is designed to identify similar reads while simultaneously being quick to compute.
When a read, r is processed, we look through all k-mers (15-mers by default) in the read as well as its reverse complement rc(r) and check which k-mers, if any, correspond to the labels of existing buckets. Let buckets(r) be the set of existing buckets whose label matches some k-mer of r or rc(r). The set buckets(r) is a set of candidate buckets against which we will score the newly processed read r. We will then assign r to the bucket that satisfies
| (1) |
where denotes the set of all ℓ-mers in the read r and, by slight abuse of notation, we denote the set of all ℓ-mers in a bucket b by ℓ-mers ℓ-mers. By default . In Equation (1), we are measuring the score of read r with respect to each bucket b in which it can be placed. The score is simply the number of length- substrings (ℓ-mers) that are shared between the read r and the set of all reads currently in bucket b.
If is labeled by a k-mer of r, we place r in the bucket , while if is labeled by a k-mer of rc(r), we instead place rc(r) in the bucket . We also record whether r or its reverse complement is being assigned to a bucket (see later). If no k-mer in this read is the label for an existing bucket [], a new bucket is created. Initially, this new bucket will contain only this read, and its label will be the minimizer (Roberts et al., 2004) of this read—the lexicographically smallest k-mer among those in r and rc(r). We begin with no buckets.
Intuitively, we are trying to assign each read to the bucket whose contents share the most substrings with the read, with the hope that the compressor will be able to exploit the redundancy of these shared substrings. The set of ℓ-mers in a bucket acts as a lightweight model of the bucket contents and allows us to quickly estimate the relative benefit of each bucket assignment. Because we only consider placing r in the buckets labeled with some k-mer of r, we will only ever have to compute the score of assigning r to a relatively small number of buckets [never more than —the factor of 2 comes from considering both r and rc(r)]. This ensures that each read can be assigned to a bucket in time independent of the number of buckets or reads. When read r is assigned to a bucket, the set of ℓ-mers of this bucket is updated to include any new ℓ-mers present in r that did not previously exist in the bucket.
Reassigning singletons. The choice of the bucket into which a read r is placed is greedy, as the bucket contents themselves depend on the set of reads that have already been processed and the order in which they were observed. Thus, it is quite possible that, at the point r is observed, it will be bucketed using k-mer sr, but subsequently some other string contained within the read will actually correspond to a better bucket as determined by Equation (1). In the most extreme case, the bucket sr may contain only r. We call these reads that are alone in a bucket singleton reads and their buckets singleton buckets. These buckets are likely to be poorly compressed.
In an attempt to mitigate this effect, we perform a ‘rescue’ step after the initial bucketing in which we attempt to re-assign singleton reads to some other non-empty bucket. Specifically, let the set of singleton reads be denoted S. We attempt to re-bucket each singleton read in light of the buckets containing all other observed reads. We remove all the singleton buckets and then re-process the reads in S, assigning each to the remaining bucket that satisfies Equation (1). Because we now have a larger set of buckets than we did when initially processing the singleton reads, a number of these singletons can often be placed into non-empty buckets. This allows us to exploit shared sequence that occurs after the singleton read in the input file. If, during this rescue step, we are unable to place a read into any existing bucket, we place it into a special singleton bucket, which is labeled by the empty string. These remaining singletons will simply be 2-bit encoded and written at the beginning of the compressed file.
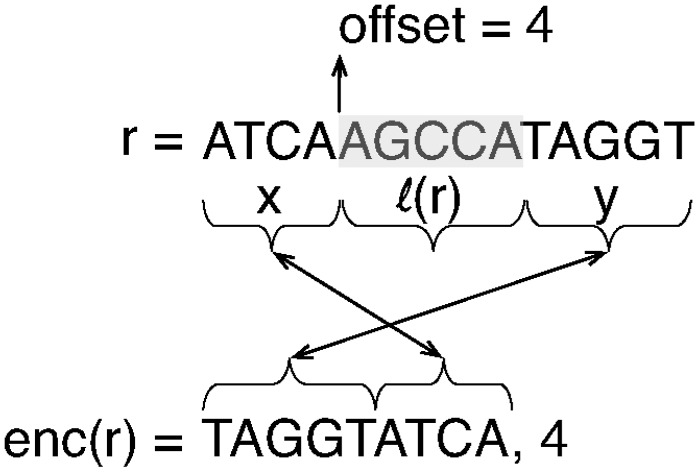
Read transformation. When a read r is placed in a bucket, it is encoded using a transformation , illustrated in Figure 1 that was initially described in Hach (2013). This transformation partitions the read r, conceptually, into three regions such that , where · represents string concatenation and is the label of the bucket into which this read is being placed. Given this partition, where o is the offset into the original read where the first occurrence of appears. We call this the split-swap read transformation, since it splits the read at a particular offset and swaps the second and first substrings produced by this split. Given and , it is possible to reconstruct r. The purpose of this transformation is twofold. First, it removes explicit redundancy [i.e. ] that exists among the reads that have been bucketed together. Second, it moves to the front of each read in the bucket the region of the read that directly follows the shared substring. Prefixes of these regions are more likely to share similar sequence, and placing them at the front of every read may improve the ability of the downstream compressor to discover and exploit these shared substrings. Within each bucket, the reads are sorted by the offset of the first occurrence of the label string within the read, with ties being broken lexicographically.
Fig. 1.
When a read r is placed into a bucket, it is encoded by splitting it at the first occurrence of the bucket label in r, removing this substring and placing the proceeding substring at the front of the encoded read
Sub-bucketing and bucket ordering. Once each read has been assigned to a final bucket, the buckets are encoded and written to file. Because most buckets are small, we avoid using a relatively large 4 or 8 byte integer to record the size of each bucket. Rather, large buckets are broken up into sub-buckets, each with a maximum size of 256 reads. The sub-buckets belonging to a single bucket are written to file sequentially, and the order of the reads in the concatenation of these sub-buckets is the same as the order of the reads within the original bucket.
As it leads to improved compression, we choose to record the transformed read sequences, concatenated together, using 2-bit encoding. The raw bitstream contains sequences of bits encoding the read sequences, separated by segments of control information encoding the label string for a bucket, its length and the number of elements in the sub-bucket to follow.
Because the reads in each bucket are sorted according to the offset of the bucket label, these offsets will then be a non-decreasing list of positive integers. This allows us to encode them using delta encoding, which we find leads to improved compression. The lexicographic tie-breaking is performed with respect to the reads after they have been encoded, as described earlier.
2.1 Mince file format
The bucket information output by Mince consists of two required files and two optional files. The required files are fseq and foffset. fseq consists of the read sequence and ‘control’ information. This stream begins by recording the necessary meta-data about the entire set of reads such as the read length and the read library type, which signifies which types of read transformations may be performed lossily (Section 2.2). This information is then followed by the count of singletons (encoded as a 32-bit unsigned integer) and a bitstream containing all singleton reads, sorted reverse lexicographically and 2-bit encoded. The singleton reads are then followed by a collection of sub-buckets that constitute the remainder of the file.
Each sub-bucket contains the following control information: the size of the bucket label, the sequence of the bucket label (2-bit encoded), the number of elements, m, in the sub-bucket minus one (this is encoded with an 8-bit unsigned integer and thus has a maximum value of 255), and a sequence of m encoded reads. Each read is written as the 2-bit encoding of split-swap(r,o). If a sub-bucket has the same core string as the preceding sub-bucket, we record a length of 0 for the bucket label, and we do not re-record this string for the current sub-bucket.
The file foffset simply consists of a list of positions of the bucket labels within each read, where the read order is the same as in fseq. These offsets are delta-encoded within each sub-bucket. Because the reads within each sub-bucket are sorted by the bucket label offset, this often exposes long runs of 0s in the offset stream that are encoded particularly well.
The two optional files are the reverse complement file frc and the file fN containing the location of Ns in the original reads. frc consists of a simple binary stream of 0s and 1s that encode, for each read in fseq whether we encoded the original read (in which case we record a 0) or the reverse-complement of the read (in which case we record a 1). Because it may not be necessary to recover the original strand of the raw reads, which is often arbitrary, this stream can sometimes be discarded, though it is often of a negligible size. For example, when dealing with single-end reads that do not originate from a known strand of DNA or RNA, a given read r and its reverse complement are often equivalent for the purpose of most analyses—the same is also true of non-strand-specific paired-end reads, though the relative orientation of these reads should be preserved (Section 2.2).
Finally, fN consists of the positions of all of the nucleotides that were recorded as N in the original sequencing reads. This file is necessary because we rely, in fseq, on a 2-bit encoding of the sequences. To allow this, we transform all Ns into a 2-bit representable character (we chose As) when encoding the reads. fN is written in a binary format where each entry consists of the index of the next read containing encoded Ns, the number of Ns in this read and then the positions, within this read, where the Ns occur. The fN file is often optional because, if we are encoding a FASTQ file and maintain the quality values, they can be used to recover the positions in each read where Ns have been called (Hach et al., 2012).
All of these output files—fseq, foffset and optionally frc and fN —are subsequently compressed independently using the lzip compressor as part of the Mince program.
2.2 Handling paired-end reads
There is significant diversity in the type of information that may be represented by a set of read sequences. For example, reads can be paired-end or single-ended; they can have a prescribed strandedness, or originate from either strand. Paired-end read libraries, additionally, are prepared in a way that results in the mate pairs having a prescribed relative orientation—that is, they may face in the same direction, away from each other or toward each other.
Mince handles paired-end reads by first concatenating the left and right ends of the pair together in accordance with a user-provided library type. The library type specifies the relative orientation of the two reads as well as whether or not one of the reads is prescribed to originate from a particular strand (e.g. as in a stranded library preparation protocol). Specifically, Mince reverse complements one of the ends of the paired-end read, if necessary, to ensure that both pairs are oriented with respect to the same strand. For example, if the reads are sequenced according to the standard paired-end Illumina protocol, they will face toward each other and come from complementary strands of the molecule being sequenced. In this case, reverse complementing the second read of the pair will reverse its orientation and strand to be consistent with that of the first read in the pair. The library type is encoded as a 1-byte number and placed at the beginning of the fseq file. This allows the relative orientation of paired-end reads to be properly recovered during decoding. After this transformation, the resulting sequences are then encoded simply as if they were single-end reads. By encoding the lengths of the left and right mates, the reads can then be separated into two streams during or after decoding to recover the original mated reads. Because Mince, like SCALCE (Hach et al., 2012), re-orders the reads, if the mates were encoded separately, it would have to either re-order one end according to the order induced by the other, or record, explicitly, the permutation between the two encoded files. The first of these strategies usually results in a larger encoded size, while the latter limits the ability to perform streaming decompression, since the positions of the mates of a pair may be arbitrarily different in their respective encoded files. These considerations led us to choose the strategy of concatenating the mate pairs to handle paired-end reads.
3 Results
3.1 Mince produces smaller files than other de novo compressors
We compared Mince against fastqz (in reference-free mode) and SCALCE, both of which were among the top de novo compression tools in a recent survey (Bonfield and Mahoney, 2013). We also experimented with the fqzcomp (Bonfield and Mahoney, 2013) program, but it performed worse than fastqz in all of our tests, and so the results are not reported here. We used SCALCE version 2.7, and encoded the read sets with the default options. Paired-end reads were encoded by SCALCE using the -r option. We used fastqz version 1.5, and compressed reads using the c command. Fastqz does not handle paired-end reads in a special way, so we provided fastqz with a single file of the concatenated paired reads, prepared as described in Section 2.2. Finally, we used Mince version 0.6 and encoded reads with the default options (except for the ‘no rc’ sizes which were generated using the -n flag) and the default k-mer size of k = 15.
For a fair comparison, we extracted the sizes of the various encodings that represent the sequence only, ignoring sections of encoded files corresponding to quality values and names. This is straightforward since the top de novo compressors against which we compared write different parts of the encoded data (i.e. sequences, qualities and names) to different files, because data of a similar type tend to share more patterns and be more easily compressed than more heterogeneous data. For SCALCE, we report the size of the.scalcer file for single-end reads or the sum of the two appropriate.scalcer files for paired-end reads. For fastqz, we report the size of the.fxb.zpaq files and for Mince we report the sum of the.seqs, .offs and .nlocs files, which correspond to fseq, foffset and fN.
We measured compression performance on the diverse array of sequence files listed in Table 1. This collection of data represents sequences from a mix of different organisms and types of experiments. Further, there is substantial technical diversity among this set of files; the sequences vary significantly in read length and paired-endedness. We selected this set of data to explore the relative performance of these different de novo compression techniques on data of varying type, quality and redundancy.
Table 1.
Different read sets used in the experiments. ‘PE’ indicates paired-end reads while ‘SE’ indicates single-end reads
| Dataset | Read length (bp) | Read type | Description | No. reads |
|---|---|---|---|---|
| SRR034940 | 100 × 2 | PE | Whole genome (H. sapiens) | 18 037 535 |
| ERR233214 | 92 × 2 | PE | Whole genome P. falciparum | 7 578 837 |
| SRR037452 | 35 | SE | RNA-seq H. sapiens brain tissue | 11 712 885 |
| SRR445718 | 100 | SE | RNA-seq H. sapiens oocyte | 32 943 665 |
| SRR490961 | 100 | SE | RNA-seq H. sapiens ES cell | 49 127 668 |
| SRR635193 | 108 | PE | RNA-seq H. sapiens pooled placental amnion | 27 265 881 |
| SRR1294122 | 101 | SE | RNA-seq H. sapiens ES cell line UCLA6 | 39 666 314 |
| SRR689233 | 90 × 2 | PE | RNA-seq M. muculus | 16 407 945 |
| SRR519063 | 51 × 2 | PE | RNA-seq P. aeruginosa | 26 905 342 |
The resulting compressed file sizes are recorded in Table 2. Over the nine different test files, Mince always produces the smallest encoding. This result holds regardless of the read-length, single/paired-endedness of the file or the organism from which the reads were sequenced. In most cases, the Mince-encoded files are substantially smaller than those produced by competing methods, in some cases achieving up to a 66% reduction in file size of fastqz and a 45% reduction in file size over SCALCE, which is generally the next best method.
Table 2.
Sizes (in bytes) of the compressed sequences from a number of different sequencing experiments, using both lzip and gzip compression as the downstream compressor
| Using gzip |
Using lzip |
||||||
|---|---|---|---|---|---|---|---|
| Read set | fastqz | SCALCE | Mince | frc | Mince no RC | Mince | frc |
| SRR034940 | 761 004 012 | 773 713 270 | 742 714 887 | 2 206 070 | 763 594 066 | 2 224 625 | |
| ERR233214 | 110 774 782 | 108 400 240 | 96 358 342 | 934 495 | 114 197 621 | 946 677 | |
| SRR037452 | 85 510 908 | 66 629 150 | 58 819 463 | 1 323 208 | 62 823 740 | 1 304 612 | |
| SRR445718 | 325 231 326 | 252 989 630 | 191 556 289 | 3 665 690 | 213 776 989 | 3 659 251 | |
| SRR490961 | 444 636 843 | 300 176 804 | 211 414 052 | 5 536 382 | 241 571 742 | 5 531 911 | |
| SRR635193 | 355 334 940 | 294 524 184 | 261 228 305 | 3 137 672 | 297 856 270 | 3 162 639 | |
| SRR1294122 | 441 798 609 | 299 329 596 | 230 388 405 | 4 284 749 | 260 421 919 | 4 208 551 | |
| SRR689233 | 247 811 387 | 233 812 318 | 199 160 825 | 1 945 419 | 225 423 423 | 1 944 118 | |
| SRR519063 | 162 308 902 | 100 399 410 | 66 749 829 | 3 347 952 | 78 356 214 | 3 386 879 | |
The numbers that appear in the frc columns are the sizes of the file that encodes which reads were reverse-complemented during encoding, which is required if the original strand of the read needs to be preserved
3.2 Exploiting reverse complementation leads to improved compression
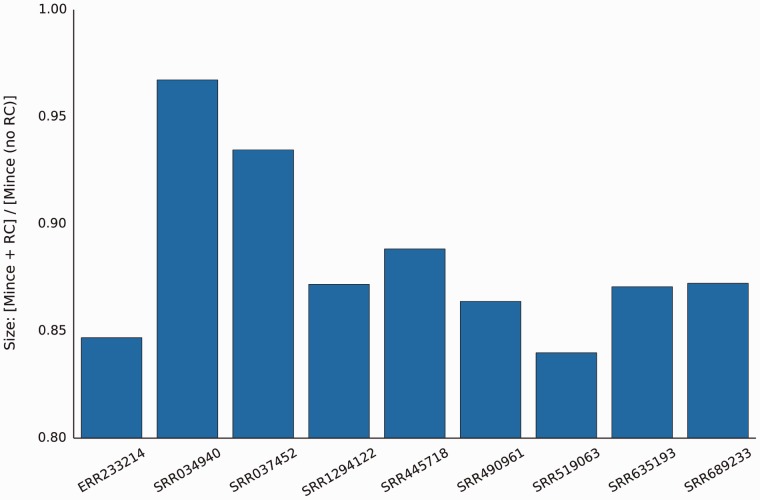
If the reverse complement of reads is not considered during bucketing, Mince produces larger files (Fig. 2) than when reverse complement sequences are considered. In fact, for each read set we use here, the sum of the sizes of the Mince encoded file and frc, the file which encodes whether or not each read was reverse complemented, is smaller—usually by a substantial amount—than the encoding size that we would be able to achieve if we did not allow reverse complementing of the reads in the first place. This is due to the fact that the frc file is very small—typically only a few megabytes (Table 2, frc columns). Further, if we do not need to recover the original orientation of the reads, the frc file can be discarded completely. These results suggest that, even if a transformation cannot be performed lossily, such as reordering the reads, it may still prove beneficial to perform the lossy transformation and additionally encode the sideband information necessary to recover the original data completely.
Fig. 2.
Even when the record of which reads were reverse-complemented needs to be maintained, Mince produces smaller files when it is allowed to consider both a read and its reverse complement
3.3 Mince is better able to exploit k-mer redundancy
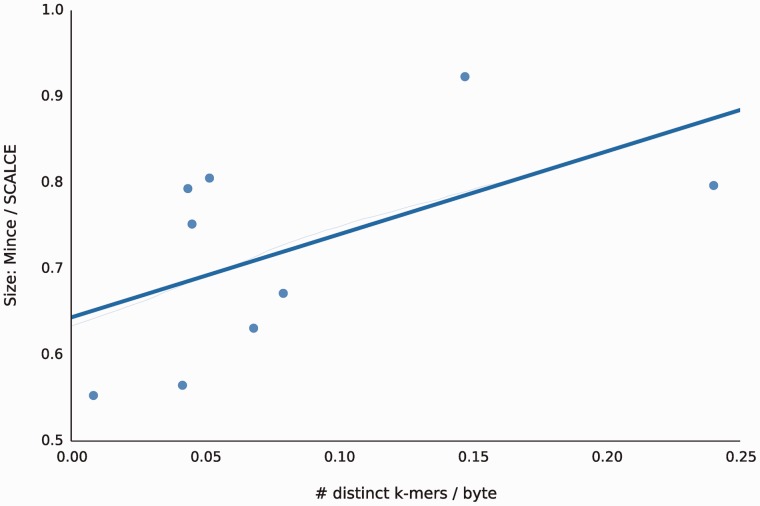
The number of duplicated k-mers (identical k-mers that appear multiple times) in a file is a strong indicator of the benefit of Mince over other methods. This indicates that Mince is better able to identify and exploit sequence similarity between the reads than other approaches. The lower the number of distinct k-mers per byte, the greater was Mince’s compression ratio relative to SCALCE (Fig. 3). Files with highly diverse sequences contain little redundancy and are thus more difficult to compress in a de novo setting (although Mince is still able to compress them more effectively than other methods).
Fig. 3.
The more redundant the k-mer (15-mer) content of the file (as measured in distinct k-mers -per-byte along the x-axis), the better able Mince is to exploit this redundancy and produce smaller files. Even in read sets with high k-mer diversity, Mince produces smaller files than SCALCE. However, as the level of redundancy increases, the data-driven bucketing scheme employed by Mince is better able to take advantage of sequence similar reads, resulting in better overall compression ratios and a larger marginal gain over the simpler bucketing approach of SCALCE
3.4 Effect of bucket label size
The single user-tunable parameter to Mince is the size of the k-mer used to label the buckets into which reads are placed. Reads are only considered for placement in a bucket if they contain the k-mer that labels that bucket. Thus, using shorter bucket labels will, in general, increase the number of buckets we examine when trying to assign a read, potentially leading to a better match between the read and bucket. Conversely, because all of the reads belonging to a bucket have the bucket label explicitly removed when they are encoded and written to file, using longer labels may result in better compression because more redundancy is explicitly removed from the reads.
We use a default bucket label size of 15 in Mince and find that it works well over a wide range of different files. We investigated the effect of this parameter to ensure that the results we observe are robust to its setting. We encoded the same set of files from Table 2 using a smaller bucket label size of 12. This size was chosen to match the length of the ‘core’ strings used to label buckets in SCALCE. We find that, though length 15 bucket labels generally result in better compression than length 12 labels, the difference is fairly minor. In fact, files encoded using length 12 bucket labels are, on average, only 1.5% larger than those encoded using length 15 bucket labels. The single largest relative difference occurred with the read set SRR490961, where the Mince encoding using length 15 bucket labels was 5.8% smaller than the encoding using length 12 bucket labels. We also observed that the file SRR037452 actually compressed better using length 12 bucket labels and was 2.6% smaller than its counterpart encoded with the default bucket label size. This indicates Mince’s advantage over SCALCE is not due to simply choosing larger ‘core’ strings. It is likely that small changes in the size of the label string will have only a small effect on the set of reads which appear close together in the final ordering, and despite, the fact that a small change in the label length will change the number of buckets, we expect that it will have a substantially smaller change on the distance in the final order among sets of similar reads.
3.5 Effect of read order on compression size
Because the bucketing schemes used by Mince and SCALCE are both heuristic in nature, they are, in theory, affected by the order in which the reads are observed. We expect that in most files, the order of the reads will be random. However, a particularly beneficial or adversarial read order might result in significantly different compression ratios.
To explore the effect of read order on the ability of Mince and SCALCE to compress reads, we performed two different tests. First, we tested the ability of Mince and SCALCE to compress a given file (SRR1294122) under 10 random permutations of the read order within the file. We find that, across 10 trials, neither Mince nor SCALCE appears sensitive to the order of reads in the file. For Mince, the maximum difference in the compressed file size between the largest and smallest files over the 10 trials was 43, 410 bytes or 0.02% of the average compressed file size. For SCALCE, the maximum difference was 30 379 bytes, or 0.01% of the average compressed file size.
To demonstrate that Mince and SCALCE both have relatively effective bucketing heuristics that result in compression rates which are robust to the order in which reads are observed in the input, we attempted to create a particularly beneficial read order. Using the same file, SRR1294122, we aligned the reads against the Ensembl human transcriptome (Flicek et al., 2013) using the STAR aligner (Dobin et al., 2013). The resulting BAM file was then sorted by alignment location and converted back into a FASTQ file, which was then encoded with Mince, SCALCE and lzip.
Similar to the randomization tests described earlier, presenting the reads to Mince and SCALCE in this favorable order has little effect on the size of the resulting compressed files. Specifically, compared with the size of the file compressed in the given order (Table 2), the size of the compressed file produced by Mince when the reads were given in alignment-sorted order was only 0.2% smaller, while the SCALCE file was only 3.2% smaller.
However, if we simply extract the sequences from the original FASTQ and compress them using lzip, the size difference between the random and alignment-ordered files is very large. Specifically, the size of the randomly ordered sequences when compressed by lzip is 826 708 745 bytes while the size of the alignment-ordered raw sequences when compressed by lzip is only 309 463 739. Thus, reordering the reads before compressing them with lzip reduced the size of the file by 63% percent. In this case, the re-ordered reads, simply compressed with lzip approaches the size of the original file as compressed with SCALCE; it is only ∼9.6 Mb or 3.4% larger. However, this file is still ∼106.3 Mb or 56.3% larger than the Mince compressed file. This indicates that Mince is better able to recover an ordering as good as the ‘reference-based’ ordering.
These experiments suggest that the order in which reads are observed by Mince and SCALCE has little effect on their ability to successfully compress a file. Overall, the difference in resulting file sizes when the underlying read order is permuted is very small.
3.6 The choice of downstream compressor can have a significant effect
Mince uses plzip, a parallel implementation of lzip, as its downstream compressor. This is different from SCALCE, which by default boosts gzip compression. Our choice was motivated by the fact that lzip generally produces smaller files than either gzip or bzip2. Though lzip tends to be somewhat slower than gzip in terms of compression speed, it is still reasonably fast and has comparable speed during decompression, which is the more important factor in our case, as reads will generally only be compressed once, but may be decompressed many times. The choice of lzip as the downstream compressor leads to an improvement in the compression ratio of Mince over what might be obtained if we relied on the same downstream compressor, gzip, as SCALCE.
To test the overall effect of boosting lzip rather than gzip compression, we ran Mince on all of the files from Table 2, but compressed the resulting files with gzip instead. This resulted in the file sizes reported in Table 2 in the middle columns. On average, the lzip-compressed files are 13% smaller than their gzip-compressed counterparts. We note that the gzip-compressed Mince files are still much smaller than their SCALCE counterparts, providing evidence that Mince is generally a more effective compression booster regardless of downstream compressor.
In addition, Mince can be used to boost the compression of other read compression techniques. Supplementary Table S1 shows the improved compression achieved by fastqz when it is provided read files that have first been reordered using Mince. Although Mince combined with lzip provides the best compression, Mince combined with fastqz improves the fastqz compression by a significant amount. This further indicates the usefulness of the Mince reordering strategy and is evidence that the Mince reordering may be useful to boost the compression of other tools.
3.7 Computational resources required
Supplementary Tables S2 and S3 provide detailed timing and memory usage for both SCALCE and Mince for the compression and decompression phases. When encoding, Mince is slower than SCALCE when using four threads. It also uses more memory than SCALCE, although its memory usage is still within practical limits (3–16 Gb). This is the tradeoff needed to achieve the significantly better compression of Mince. (When run with 20 threads, Mince runs on the order of a few minutes (3–15 min/file), making multi-core compression significantly more practical; see Supplementary Table S4). When decompressing, however, Mince is often faster and uses less memory than SCALCE. This is a reasonable tradeoff (slower, better compression but faster decompression) since decompression is the more common task.
4 Discussion
We introduced Mince, a de novo approach to sequence read compression that outperforms existing de novo compression techniques and works by boosting the already impressive lzip general purpose compressor. Rather than rely on a set of pre-specified ‘core substrings’ like SCALCE (Hach et al., 2012), Mince takes a data-driven approach, by considering all k-mers of a read before deciding the bucket into which it should be placed. Further, Mince improves on the ‘heaviest bucket’ heuristic used by SCALCE, and instead defines a more representative model for the marginal benefit of particular bucket assignment. This model takes into account the ℓ-mer composition of the read and how similar it is to the set of ℓ-mers of reads that have already been placed in this bucket. Early on in the processing of a file, when little information exists about the relative abundance of different k-mers, ties between buckets are broken consistently by preferring to bucket a read based on its minimizer.
This approach allows the selection of core substrings that are among the most frequent k-mers in the provided set of reads, and the improved model for bucket assignment leads to more coherent buckets and better downstream compression. In the rare situations where a specific order is required for the reads, Mince is not the most appropriate compression approach. Further, in addition to re-ordering, Mince exploits other transformations of a read, such as reverse complementing, that may or may not be performed in a lossy fashion. Regardless of whether or not these transformations need to be reversed during decoding, they lead to improvements in compression that overcome the cost of storing the ‘sideband’ information necessary to reverse them.
Mince only compresses the sequence portion of FASTQ files, requires all reads to have the same length, and ignores the quality values. The challenges associated with compression of quality values are quite different than those associated with compressing sequence data, and other approaches (e.g. Yu et al., 2015) have been developed that can be used to compress quality values well. Furthermore, quality values are often not needed for many analyses—tools such as BWA (Li, 2013), STAR (Dobin et al., 2013) and Sailfish (Patro et al., 2014) routinely ignore them during all or some phases of their operation—while sequence data are, of course, the primary and central reason for the FASTQ file to exist to begin with. For these reasons, we have focused on developing better methods for sequence compression, leaving the problem of improving quality value compression to future work.
Additional future work includes speeding up the compression and decompression approaches presented here. Although the current speed of Mince (particularly when decoding) is practical for many applications, it is always desirable to minimize the time spent on data manipulation tasks such as compression and decompression. This is the reason, for example, that we have used a near-greedy heuristic for selecting buckets and assigning reads to them—this is fast and produces reasonable results (differing by over various random read orderings). However, interesting directions for future work include both speeding up the implementation of this near-greedy approach and designing faster, equally performant approaches for read bucketing. This will be especially important for larger files, such as produced by high-coverage whole-genome human sequencing, where the 2x–11x difference in Mince and SCALCE runtimes will become more significant.
By capitalizing on an efficient, novel encoding of reads that leads to improved compression boosting, Mince is able to compress sets of read sequences more effectively than exiting de novo approaches. The compressed read sequences can be decompressed efficiently and in a streaming fashion. As the size and number of datasets that we analyze continues to grow, Mince will prove an effective tool for mitigating the ever-increasing cost of storage and transmission. Mince is written in C++11, it is open source and has been made available under the GPLv3 license at http://www.cs.cmu.edu/∼ckingsf/software/mince.
Supplementary Material
Acknowledgements
We would like to thank Geet Duggal, Darya Filippova, Emre Sefer, Brad Solomon and Hao Wang for useful discussions relating to this work and for comments on the initial manuscript. We would also like to thank the anonymous reviewers for their helpful feedback on the manuscript and testing of the software.
Funding
This work has been partially funded by the US National Science Foundation (CCF-1256087, CCF-1319998) and US National Institutes of Health (R21HG006913 and R01HG007104). C.K. received support as an Alfred P. Sloan Research Fellow. This work is also funded in part by the Gordon and Betty Moore Foundation’s Data-Driven Discovery Initiative through Grant GBMF4554 to Carl Kingsford.
Conflict of Interest: none declared.
References
- Adjeroh D., et al. (2002) DNA sequence compression using the Burrows-Wheeler Transform. In: Proceedings of the IEEE Computer Society on Bioinformatics Conference, 2002. IEEE Computer Society, Washington, DC, USA, pp. 303–313. [PubMed] [Google Scholar]
- Bhola V., et al. (2011) No-reference compression of genomic data stored in fastq format. In: Bioinformatics and Biomedicine (BIBM), 2011. IEEE, pp. 147–150. [Google Scholar]
- Bonfield J.K., Mahoney M.V. (2013) Compression of FASTQ and SAM format sequencing data. PLoS One, 8, e59190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M.C., et al. (2009) Data structures and compression algorithms for genomic sequence data. Bioinformatics, 25, 1731–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campagne F., et al. (2013) Compression of structured high-throughput sequencing data. PLoS One, 8, e79871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas R., et al. (2014) Lossy compression of quality scores in genomic data. Bioinformatics, 30, 2130–2136. [DOI] [PubMed] [Google Scholar]
- Christley S., et al. (2009) Human genomes as email attachments. Bioinformatics, 25, 274–275. [DOI] [PubMed] [Google Scholar]
- Cox A.J., et al. (2012) Large-scale compression of genomic sequence databases with the Burrows-Wheeler transform. Bioinformatics, 28, 1415–1419. [DOI] [PubMed] [Google Scholar]
- Deorowicz S., Grabowski S. (2011) Robust relative compression of genomes with random access. Bioinformatics, 27, 2979–2986. [DOI] [PubMed] [Google Scholar]
- Deorowicz S., Grabowski S. (2013) Data compression for sequencing data. Algorithms Mol. Biol., 8, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., et al. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics, 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., et al. (2013) Ensembl 2014. Nucleic Acids Res., 42(Database issue), D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz M.H.-Y., et al. (2011) Efficient storage of high throughput DNA sequencing data using reference-based compression. Genome Res., 21, 734–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hach F. (2013) Scalable mapping and compression of high throughput genome sequencing data. Ph.D. Thesis, Simon Fraser University. [Google Scholar]
- Hach F., et al. (2012) SCALCE: boosting sequence compression algorithms using locally consistent encoding. Bioinformatics, 28, 3051–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.C., et al. (2012) Compression of next-generation sequencing reads aided by highly efficient de novo assembly. Nucleic Acids Res., 40, e171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford C., Patro R. (2015) Reference-based compression of short-read sequences using path encoding. Bioinformatics, 31, 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozanitis C., et al. (2011) Compressing genomic sequence fragments using SlimGene. J. Comput. Biol., 18, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v1 [q-bio.GN]. [Google Scholar]
- Li P., et al. (2013) HUGO: Hierarchical mUlti-reference Genome cOmpression for aligned reads. J. Am. Med. Inform. Assoc., 21, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa I., et al. (2013) Qualcomp: a new lossy compressor for quality scores based on rate distortion theory. BMC Bioinformatics, 14, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R., et al. (2014) Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol., 32, 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlichin D.S., et al. (2013) The human genome contracts again. Bioinformatics, 29, 2199–2202. [DOI] [PubMed] [Google Scholar]
- Pinho A.J., et al. (2012) Green: a tool for efficient compression of genome resequencing data. Nucleic Acids Res., 40, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popitsch N., von Haeseler A. (2013) NGC: lossless and lossy compression of aligned high-throughput sequencing data. Nucleic Acids Res., 41, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarajeswari P., Apparao A. (2011) DNABIT compress–genome compression algorithm. Bioinformation, 5, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M., et al. (2004) Reducing storage requirements for biological sequence comparison. Bioinformatics, 20, 3363–3369. [DOI] [PubMed] [Google Scholar]
- Rozov R., et al. (2014) Fast lossless compression via cascading bloom filters. BMC Bioinformatics, 15 (Suppl. 9), S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tembe W., et al. (2010) G-SQZ: compact encoding of genomic sequence and quality data. Bioinformatics, 26, 2192–2194. [DOI] [PubMed] [Google Scholar]
- Wang C., Zhang D. (2011) A novel compression tool for efficient storage of genome resequencing data. Nucleic Acids Res., 39, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.W., et al. (2015) Quality score compression improves genotyping accuracy. Nat. Biotechnol., 33, 240–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.