Abstract
Motivation: We have created an R package named phylogeo that provides a set of geographic utilities for sequencing-based microbial ecology studies. Although the geographic location of samples is an important aspect of environmental microbiology, none of the major software packages used in processing microbiome data include utilities that allow users to map and explore the spatial dimension of their data. phylogeo solves this problem by providing a set of plotting and mapping functions that can be used to visualize the geographic distribution of samples, to look at the relatedness of microbiomes using ecological distance, and to map the geographic distribution of particular sequences. By extending the popular phyloseq package and using the same data structures and command formats, phylogeo allows users to easily map and explore the geographic dimensions of their data from the R programming language.
Availability and Implementation: phylogeo is documented and freely available http://zachcp.github.io/phylogeo
Contact: zcharlop@rockefeller.edu
1 Introduction
Deep sequencing of environmental metagenomes provides scientists with a way to assess the structure and function of microbial communities including the majority of microorganisms that cannot be cultured in the laboratory. In the course of a typical microbiome study, a number of datasets are generated that may include raw sequencing reads, tables of clustered reads, taxonomic tables, phylogenetic trees and sample collection information. This data can be organized and analyzed by a number of computational suites including QIIME (Caporaso et al., 2010), mothur (Schloss et al., 2009) and phyloseq (McMurdie and Holmes, 2013). While these are robust tools for processing data, none of them support mapping, a valuable tool for hypothesis generation that not only displays the physical location of samples but can also answer basic questions that have a spatial component (e.g. How are sample variable such as pH and carbon content distributed? Where are ecologically similar samples located? Are there sequences found in only one region?). To address this shortcoming we built phylogeo, an R package containing a set of functions for creating geography-centric plots of microbiome data. phylogeo was engineered as an extension of the phyloseq package, chosen for its simple design and its high-quality, programmable, ggplot-based (Wickham, 2009) figures. By adding mapping capabilities to a preexisting software package, phylogeo minimizes the effort needed to generate maps, and thereby facilitates the exploration of the geographic relationships in microbiome sequencing data.
2 Methods
phylogeo is written in R and extends the commonly used phyloseq package with only a single additional requirement: that the dataframe encoding sample information contain a latitude and longitude column. phylogeo’s plotting and mapping functions use a number of open source R packages for mapping, network and phylogenetic analyses. [maps (Richard et al, 2014), sp (Edzer and Pebesma, 2005), ggplot2 (Wickham, 2009), gridExtra (Auguie, 2012), igraph (Nepusz, 2006), ape (Strimmer, 2004)] By integrating many preexisting packages that process geographic and phylogenetic data, phylogeo facilates exploratory data analysis of microbiome data.
3 Biological applications
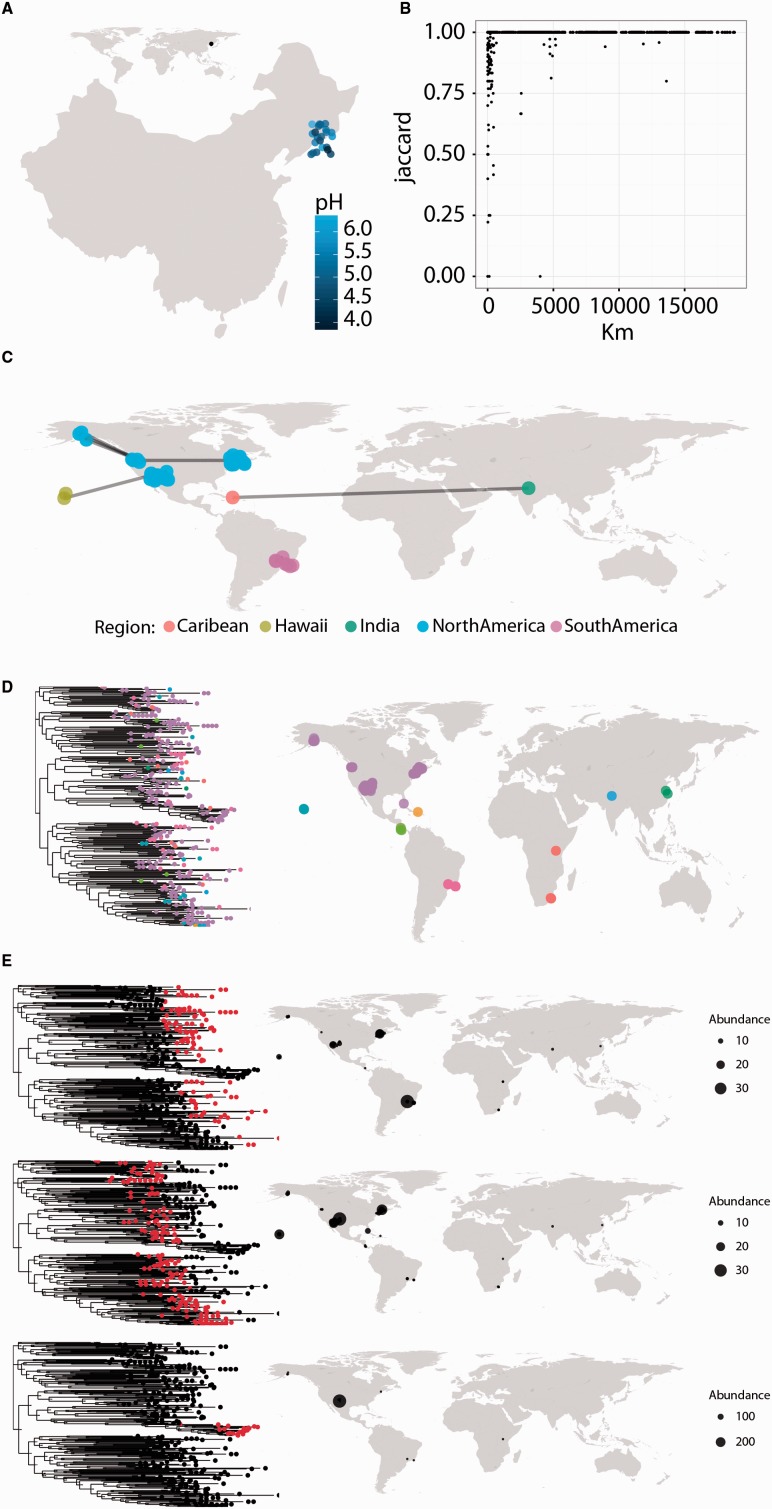
Microbial ecologists are interested in how microbial communities differ and what the functional significance and causes of those differences may be. Having a geographic perspective on the distribution of samples, their relationship with one another and the distribution of particular sequences can be an informative part of hypothesis generation. and phylogeo assists this process by providing a set of tools that can be used during the early stages of data analysis as well as to produce production-quality figures using the full customizability of ggplot. As illustrated in Figure 1, phylogeo’s functions allow users to map the intrinsic properties of samples (e.g. pH, nitrogen and carbon content) (Fig. 1A), to show how microbial populations vary from sample to sample (Fig. 1B,C), and to explore unusual distributions of particular metagenomic sequences (Fig. 1D, E). The most basic function of phylogeo, map_phyloseq, creates a customizable map of a metagenomic dataset that can zoom into a region of interest, offset crowded points, and use color and shape to highlight the data associated with each sample including a sample’s intrinsic properties and sequence-abundance (Fig. 1A). This functionality can be combined with the powerful subsetting abilities of phyloseq to map only relevant portions of the data (e.g. map only the distribution of reads belonging to Actinobacteria) by using phyloseq to subset the dataset prior to mapping it. map_phyloseq, also serves as the foundation for phylogeo’s other mapping functions which are tailored to look at the sample-sample relationships or to look at the geographic distribution of particular sequences.
Fig. 1.
phylogeo facilitates the geographic exploration of microbiome sequencing datasets. phylogeo functions can display (A) a zoomed-in map that displays sample properties (pH) with map_phyloseq; (B) pairwise geographic and ecological distances of all samples using plot_distance; (C) ecological relatedness of samples with map_network; (D) plotting sequences location with map_tree; and (E) the location of sequence subgroups as identified by k-means clustering with map_clusters. Code to recreate these figures is part of the web-based documentation on the phylogeo homepage
Sample similarity, an important component of microbiome studies, is calculated using an ecological distance metric such as the Jaccard, Bray-Curtis and UniFrac distances (Lozupone and Knight, 2005; Oksanen, 2013). phylogeo provides two tools for exploring how intersample comparisons correlate with geographic distance. The plot_distance function calculates the ecological and geographic distance between every set of samples and creates a scatter plot that provides a global overview of the relationship between geographic proximity and ecological similarity within a dataset (Fig. 1B). The second function, map_network, produces a map in which sample sites are connected by lines if they are more ecologically similar than a threshold value (Fig. 1C). As in phyloseq’s network-based utilities, the user can specify the distance metric and cutoff values, allowing a user to quickly assess ecological similarity across samples.
Finally, if a phylogenetic tree of sequences is available, phylogeo provides two functions that allow a user to look at the distribution of these sequences in space. This sort of analysis can be of particular use to microbiologists studying enzymes where different subclades may have unique activities, allowing them to visualize where these subsets are located. map_tree plots a phylogenetic tree of sequences along with a map of sample locations, allowing the user to easily locate sequences on the map (Fig. 1D) while map_clusters uses k-means clustering to divide the phylogenetic tree into k similarity groups, and individually map those groups to show the location and abundance of the sequences. (Fig. 1E)
4 Conclusion
The geographic component of environmental microbiomes has been underexplored in microbial ecology studies due, in part, to the difficulty of combining microbiome data with geographic plotting tools. phylogeo makes it possible for any user of R to easily and reproducibly generate maps showing the geographic patterns in their microbiome data.
Funding
This work was supported by National Institutes of Health grant number GM077516 (S.F.B.), and AI110029 (Z.C.P.). S.F.B. is a Howard Hughes Medical Institute Early Career Scientist.
Conflict of Interest: none declared.
References
- Auguie B. (2012) gridExtra: functions in Grid graphics. R package version 0.9.1 http://CRAN.R-project.org/package=gridExtra. [Google Scholar]
- Caporaso J.G., et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edzer J., Pebesma R.S.B. (2005) Classes and methods for spatial data in R. R News, 5, 9–13. [Google Scholar]
- Jari,O. et al. (2015) vegan: Community Ecology Package. R package version 2.2-1. http://CRAN.R-project.org/package=vegan.
- Lozupone C., Knight R. (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol, 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One, 8, e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepusz G.C.A.T. (2006) The igraph software package for complex network research, InterJournal, Complex Systems, 1695. [Google Scholar]
- Richard A., et al. (2014) maps: Draw Geographical Maps. R package version 2.3-9 http://CRAN.R-project.org/package=maps. [Google Scholar]
- Schloss P.D., et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol., 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimmer E.P.A.J.C.A.K. (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: elegant graphics for data analysis. Springer, New York. [Google Scholar]