Abstract
Rare tautomeric and anionic nucleobases are believed to play fundamental biological roles but their prevalence and functional importance has remained elusive because they exist transiently, in low-abundance, and involve subtle movements of protons that are difficult to visualize. Using NMR relaxation dispersion, we show that wobble dG•dT and rG•rU mispairs in DNA and RNA duplexes exist in dynamic equilibrium with short-lived, low-populated Watson-Crick like mispairs that are stabilized by rare enolic or anionic bases. These mispairs can evade Watson-Crick fidelity checkpoints and form with probabilities (10−3-10−5) that strongly imply a universal role in replication and translation errors. Our results indicate that rare tautomeric and anionic bases are widespread in nucleic acids, expanding their structural and functional complexity beyond that attainable with canonical bases.
Nucleic acid bases exist predominantly in one neutral tautomeric form. This in turn gives rise to the strict Watson-Crick (WC) pairing rules (Fig. 1a) that govern how genetic information is replicated, transcribed, and translated. However, if bases adopt alternative energetically disfavored tautomeric or anionic forms (Fig. 1a), pairing rules can be violated and new functions can emerge. For example, although rarely observed, minor tautomeric and anionic bases can form WC-like dG•dT1-3, dA•dC4, and rG•rU5,6 mispairs that are believed to contribute to spontaneous mutations1,7-9 and translational errors10. Chemical modifications that stabilize or lock bases in their anionic or enol-like forms can be mutagenic11,12 or expand the decoding capacity of tRNAs13,14. In addition, anionic and tautomeric forms of the bases are believed to play crucial roles in nucleic acid catalysis15,16, RNA-ligand recognition17,18, and in the therapeutic mechanisms of nucleic acid base analogues19.
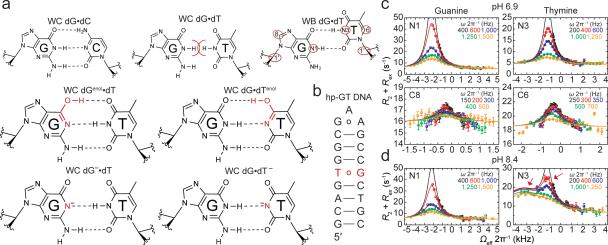
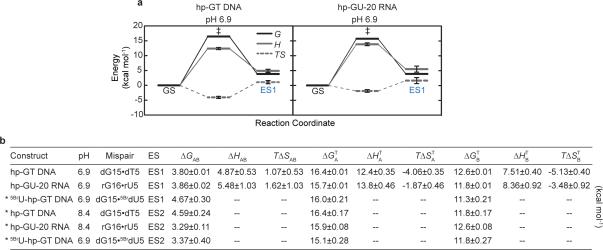
Figure 1. Chemical exchange in dG•dT mispairs.
a, WC dG•dC, sterically prohibited WC dG•dT, and WB dG•dT (R1ρ measured nuclei highlighted in red ovals). Below are four WC-like tautomeric and anionic (implied charge delocalization) bps. b, DNA duplex with a 13C/15N site-labeled dG•dT mispair. c, RD profiles for dG•dT (25 °C and pH 6.9) showing R2+Rex as a function of the spin lock offset (Ωeff 2π−1) and power (ωSL 2π−1, in insets) with global fits to dG-N1, dG-C8, dT-N3, and dT-C6. Error bars represent experimental uncertainty (one s.d., see Methods). d, RD profiles showing 3-state exchange (25 °C and pH 8.4) and global 3-state fit to dG-N1 and dT-N3.
Despite growing evidence that rare tautomeric and anionic bases play important roles in nucleic acids, their occurrence, stabilities, and biological significance has remained elusive. Characterizing rare tautomeric and anionic bases in polynucleotides is a longstanding problem because such energetically unfavorable species typically exist in low abundance, for short periods of time, and involve movements of protons that are difficult to visualize at the atomic level. NMR relaxation dispersion (RD) techniques20-22 are making it possible to characterize low-populated (populations of 0.1%-10%) transient (lifetimes of micro-to-milliseconds) states of nucleic acids23-25 that are often referred to as ‘excited states’ (ES). Here, we use NMR RD to characterize transient WC-like dG•dT and rG•rU mispairs in DNA and RNA that are stabilized by rare tautomeric and anionic bases and obtain evidence that they play universal roles in misincorporation during replication and translation.
Transient WC-like dG•dT tautomer mispair
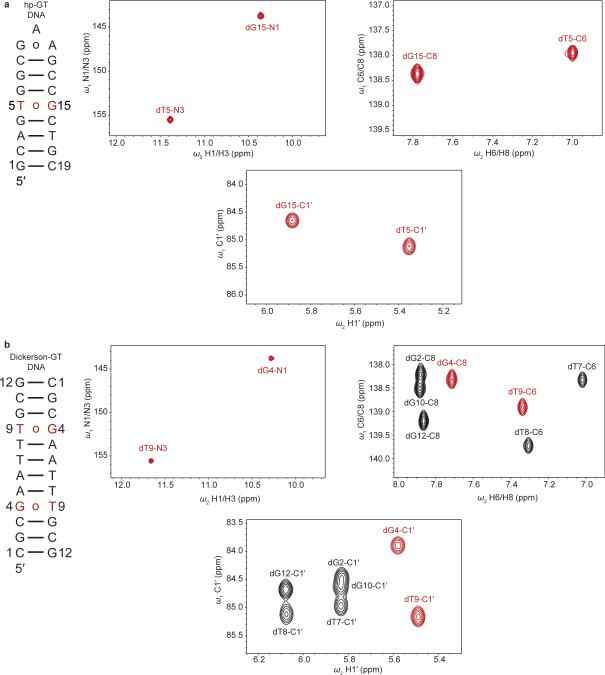
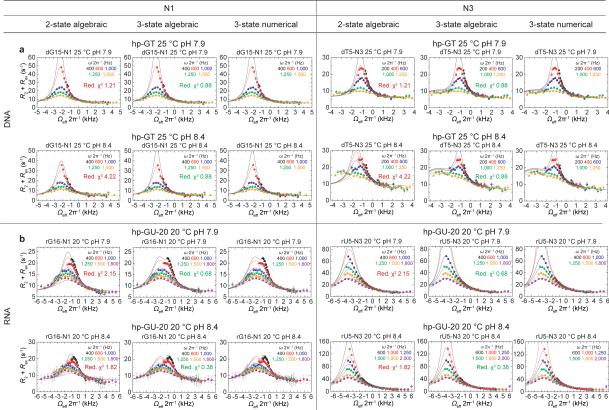
dG•dT mispairs generally adopt a distinct ‘wobble’ (WB) geometry (Fig. 1a) since a WC geometry results in a steric clash between imino protons (Fig. 1a). However, enol tautomers of dG or dT, or their anionic form, can alleviate this steric clash, and allow formation of WC-like dG•dT mispairs (Fig. 1a). Soon after the discovery of the DNA double helix, Watson and Crick hypothesized that such WC-like mispairs could provide a basis for spontaneous mutations7. We used NMR rotating frame spin relaxation (R1ρ) RD20,26,27 to examine whether wobble dG•dT mispairs can transiently morph into such WC-like dG•dT mispairs in canonical DNA duplexes. For these studies we used a hairpin DNA duplex (hp-GT DNA) containing a site-specifically 13C/15N-labeled dG•dT wobble mispair (Fig. 1b and Extended Data Fig. 1). Exchange between WB and WC dG•dT mispairs entails deprotonation of either dG-N1 or dT-N3 via tautomerization (neutral) or ionization (charged), both of which would induce large changes in N1/3 chemical shifts (CSs) and therefore give rise to significant 15N RD. In contrast, because a WB-to-WC transition preserves an anti base and C2′-endo sugar pucker, it is expected to induce smaller changes in the sugar (dG-C1′ and dT-C1′) and base (dG-C8 and dT-C6) carbon CSs and therefore induce more limited 13C RD. Indeed, we observed very significant 15N RD at base imino dG-N1 and to a lesser extent at dT-N3, much less significant 13C RD at base dG-C8 and dT-C6, and essentially no 13C RD at sugar dG-C1′ and dT-C1′ at pH 6.9 and 25 °C (Fig. 1c and Extended Data Fig. 2-3). This unique pattern of RD is consistent with exchange directed toward a transient WC-like mispair (Fig 1a). It is inconsistent with exchange directed toward other base pair (bp) geometries such as Hoogsteen24 or base opened states (Supplementary Discussion 1 and Extended Data Fig. 4). A second exchange process was apparent at pH 8.4 (Fig. 1d) and this will be discussed further below. Similar RD profiles were observed in a different DNA duplex (Extended Data Fig. 1-2), indicating that the observed dG•dT exchange occurs robustly in DNA duplexes.
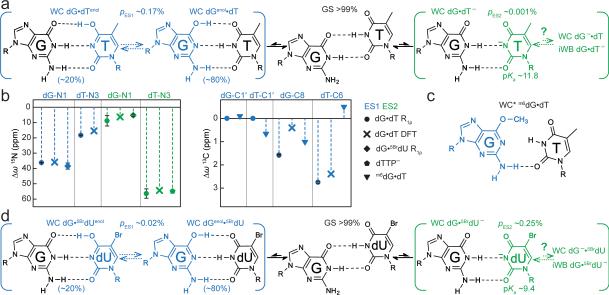
Figure 2. Characterizing WC-like transient states.
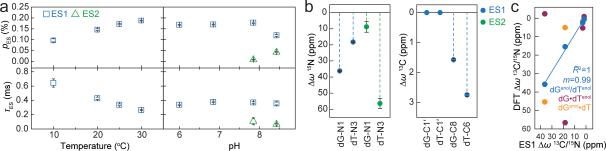
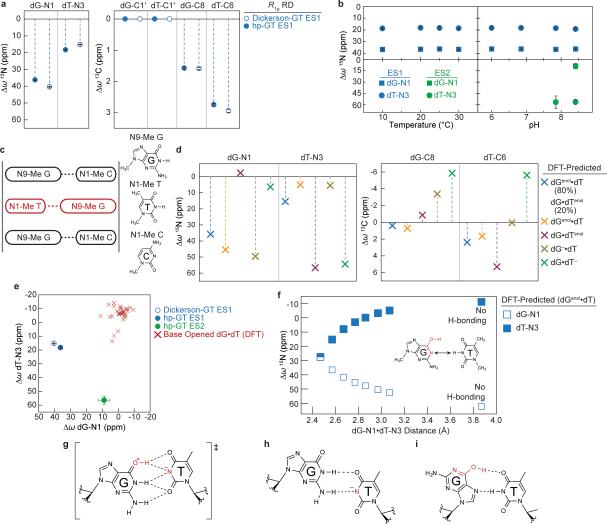
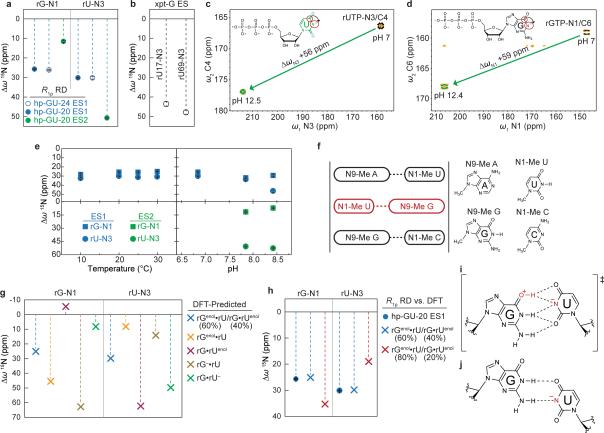
a, Population and lifetime of dG•dT ES1/ES2 measured in hp-GT DNA as a function of temperature (at pH 6.9) and pH (at 25 °C). Errors in fitted parameters denote s.e. from the weighted global fit. b, Differences between the GS (referenced to 0 ppm) and ES CSs (Δω=ωES-ωGS) for hp-GT DNA. c, Measured CSs for ES1 are plotted against DFT-predicted values.
The RD data measured at dG-N1, dT-N3, dG-C8 and dT-C6 could be globally fitted (Supplementary Table 1 and Extended Data Fig. 2-3) to a single exchange process directed toward an excited state (ES1) that has a population (pES1) of ~0.17% and a lifetime (tES1) of ~0.38 ms (Fig. 2a). ES1 is characterized by 15N CSs that are significantly downfield shifted for dG-N1 (ΔωN1 +36 ppm) and to a lesser extent dT-N3 (ΔωN3 +18 ppm)(Fig. 2b, Extended Data Fig. 4). The downfield shifted imino nitrogen CSs are unprecedented for nucleic acids, and are directed toward the CSs of bases (dG and dT) that have been deprotonated due to ionization or modifications that lock an enol-like form (Δω ~50-60 ppm)(Extended Data Fig. 5)28-30. On the other hand, ES1 features much smaller changes in carbon CSs (Fig. 2b), consistent with a WB-to-WC transition.
Figure 3. Mutate-and-CS fingerprinting ES1 and ES2.
a, Multi-state equilibrium between WB and WC-like dG•dT mispairs. ES1/ES2 populations and weights are shown (25 °C and pH 6.9). pES2 estimated based on the observed apparent pKa. b, CS fingerprinting dG•dT ES1/ES2 using chemical modifications and structure-based DFT predictions of CSs. ES1 DFT CSs are given for 80:20 dGenol•dT:dG•dTenol weighting. c, m6dG•dT structure12,30. d, dG•5BrdU– ES2 stabilized relative to dG•dT– ES2. Populations and weights (10 °C and pH 6.9) are shown.
It would be highly energetically disfavored to simultaneously deprotonate dG-N1 and dT-N3 when forming ES1. Moreover, though the magnitude of the ES1 N1/N3 downfield CSs strongly suggests deprotonation, it is not as far downfield shifted as expected based on deprotonation of nucleotides in free solution (Extended Data Fig. 5). Therefore, the strongly but incompletely downfield shifted dG-N1 and dT-N3 CSs suggest that ES1 consists of at least two WC-like species in rapid exchange on the NMR timescale in which either dG-N1 (dGenol•dT or dG–•dT) or dT-N3 (dG•dTenol or dG•dT–) is deprotonated (Fig. 1a). The ES1 population and CSs are largely independent of pH within the pH range of 6.0-7.9 (Fig. 2a and Supplementary Discussion 2). This is inconsistent with exchange directed toward ionic dG–•dT and dG•dT– (Extended Data Fig. 5). Rather, the population of ES1 increases with temperature (Fig. 2a) as expected for tautomeric species dGenol•dT and dG•dTenol.
Based on the measured pES1 and tES1, the free energy difference (ΔG) between GS and ES1 is ~3.8 kcal/mol and the forward free energy barrier (ΔG‡) is ~16.4 kcal/mol (Extended Data Fig. 6). These values are in good agreement with computationally predicted parameters (2.8-5.6 kcal/mol31 and ~17-21 kcal/mol31,32, respectively) for dG•dT WB-to-WC tautomer transitions.
These computational studies also predict that dGenol•dT likely exists in fast exchange on the NMR timescale (free energy barrier ~5-6 kcal/mol31) with a minor dG•dTenol (20%) species (Fig. 1a and Supplementary Discussion 3-4). Under these conditions, the measured ES1 CSs would represent a population-weighted average of the two tautomeric states (Methods). We find that all ES1 CSs (dG-N1, dT-N3, dG-C8 and dT-C6) are in quantitative agreement with values predicted by density functional theory (DFT)33 calculations for a weighted dGenol•dT(80%)⇌dG•dTenol(20%) equilibrium (Fig. 2c, 3a).
Transient WC-like dG•dT– anionic mispair
Interestingly, upon increasing the pH to 8.4, we observed evidence for a second excited state (ES2), which is seen as a second peak in the off-resonance RD profile of dT-N3 (Fig. 1d). Global fitting of this RD data (Extended Data Fig. 2-3) revealed two excited states (ES1 and ES2) that are most likely arranged in a linear topology (ES1⇌dG•dT⇌ES2).
Compared to ES1, ES2 (ΔG‡ 16.4 and ΔG 4.59 kcal/mol) has a considerably lower population (pES2 ~0.04%) and lifetime (tES2 ~70 μs) at pH 8.4 (Fig. 2a). The dG-N1 and dT-N3 ES2 CSs are not only ‘swapped’ relative to ES1 such that dT-N3 experiences the larger downfield shift (ΔωN3 +56 ppm) while dG-N1 experiences a smaller downfield shift (ΔωN1 +9 ppm)(Fig. 2b); they are also more asymmetric in favor of a deprotonated dT species. In addition, unlike pES1, pES2 increases significantly with pH, consistent with ionization and the formation of dG•dT– (Fig. 3a). The ES2 CSs are in excellent agreement with values computed using DFT assuming a WC-like dG•dT– (100%) species (Fig. 3a-b). However, we cannot rule out that dG•dT– is in rapid equilibrium with a WC-like dG–•dT or an inverted wobble (iWB) geometry3,34,35 (Extended Data Fig. 4 and Supplementary Discussion 1) that falls outside detection limits (Fig. 3a).
Fingerprinting the dG•dT excited states
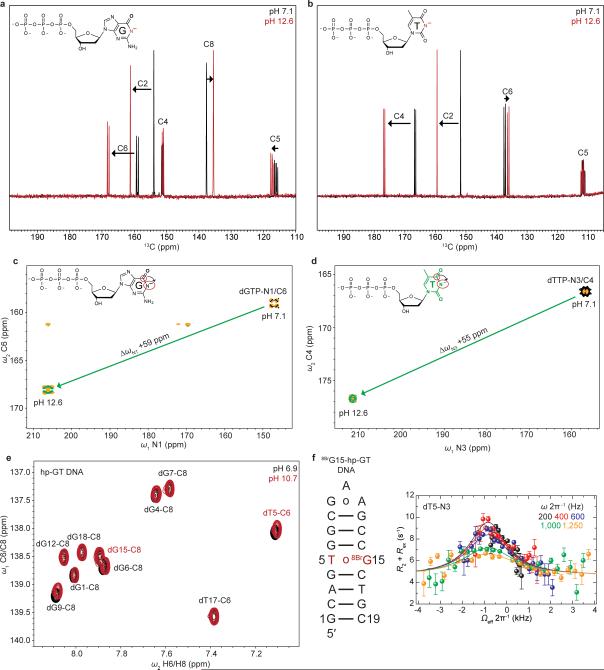
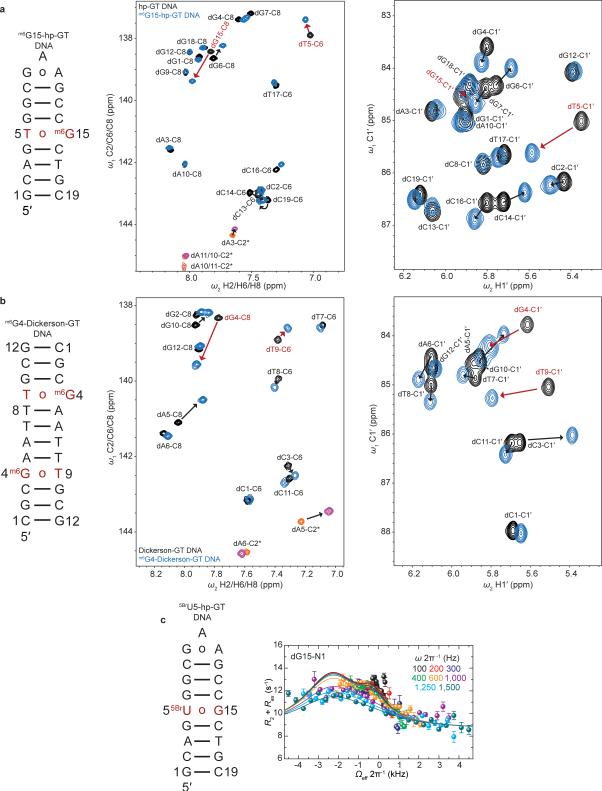
We adapted a mutate-and-CS fingerprint strategy24,25 to test the proposed ES1 and ES2 (Fig. 3a). Here, chemical modifications are used to trap an ES, or induce specific perturbations to the GS⇌ES equilibrium. We trapped ES1 (dGenol•dT) using the mutagenic base O6-methyl-2′-deoxyguanosine (m6dG)(Fig. 3c and Extended Data Fig. 7) which is known to adopt a distorted WC dGenol•dT-like mispair12,30. Relative to the WB, this modification resulted in negligible changes in dG-C1′ (ΔωC1′ −0.1 ppm) and dT-C1′ (ΔωC1′ +0.7 ppm) CSs, and a small downfield shift in dG-C8 (ΔωC8 +1 ppm), consistent with the RD-derived ES1 CSs (Fig. 3b and Extended Data Fig. 7). The modification induced a small upfield shift in dT-C6 (ΔωC6 −0.5 ppm) that is inconsistent with the downfield dT-C6 CS (Fig. 3b) observed by RD. However, such a deviation is expected based on DFT calculations (ΔωC6 −2.3 ppm for dT-C6 in the m6dG•T pair)(Supplementary Discussion 5) and can be attributed to minor deviations from an ideal dGenol•dT WC-like mispair geometry (Fig. 3c)12,30. Severe line broadening did not permit measurement of the dT-N3 CSs in these non-isotopically enriched samples.
To test the proposed dG•dT– ES2, we measured the difference in dT-N3 CS between neutral and anionic dTTP (ΔωN3 +55 ppm) and found them to be in excellent agreement with the dT-N3 CS differences measured by RD (ΔωN3 +56 ppm)(Fig. 3b and Extended Data Fig. 5). In addition, we used the mutagenic thymidine-analogue, 5-bromo-2′-deoxyuridine (5BrdU) to push the equilibrium toward dG•5BrdU– (Fig. 3d). This modification lowers the pKa of 5BrdU-N3 (~8.6) and favors a WC-like dG•5BrdU– geometry at high pH11. This modification increased the population of ES2 (ΔG‡ 15.1 and ΔG 3.37 kcal/mol) by over two orders of magnitude at the expense of ES1 (ΔG‡ 16.0 and ΔG 4.67 kcal/mol) while minimally affecting the ES1 and ES2 CSs (Fig. 3b). The consistencies in ES2 CSs between dG•dT– and dG•5BrdU– mispairs further support a dominant WC-like ES2, rather than an iWB bp, in naked DNA. The unaffected ES1 CSs indicate that 5BrdU does not significantly impact the dGenol•5BrdU⇌dG•5BrdUenol equilibrium (Fig. 3d) relative to dG•dT (Fig. 3a), consistent with prior computational studies31,36.
Transient WC-like rG•rU mispairs in RNA
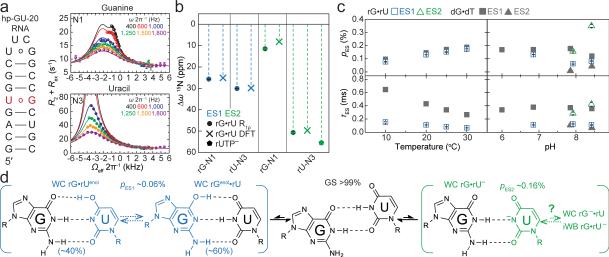
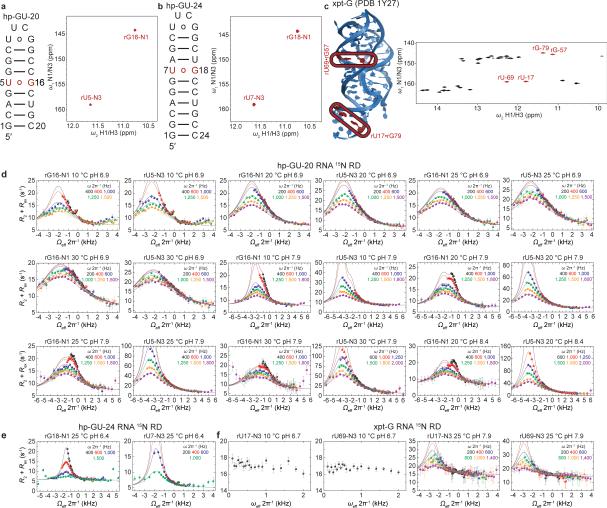
If the observed ESs correspond to WC-like dG•dT mispairs, one would predict that similar ESs should arise in rG•rU mispairs in RNA where WC bps are also readily accommodated within the A-form helix. To test this hypothesis, we carried out analogous pH- and temperature-dependent RD measurements on two RNA duplexes. RD profiles measured for rG•rU in A-form RNA (Fig. 4a and Extended Data Fig. 8) are very similar to those measured for dG•dT in B-form DNA (Fig. 1c-d). Global analyses of the RD data (Supplementary Table 1) revealed an apparent 3-state exchange process at pH ≥7.9 (Extended Data Fig. 3). The RD-derived CSs (Fig. 4b and Extended Data Fig. 9), together with the pH and temperature dependence of the populations and lifetimes (Fig. 4c), are consistent with rG•rUenol⇌rGenol•rU as ES1 and rG•rU– as ES2 (Fig. 4d) as observed in DNA. The rG•rU ES1 forward free energy barrier (ΔG‡ 15.7 and ΔG 3.86 kcal/mol) is consistent with barriers measured for dG•dT ES1 (16.4 kcal/mol) and computationally predicted barriers for G•U tautomerization (17.1 kcal/mol)32.
Figure 4. Transient tautomeric and anionic WC-like mispairs in A-form RNA.
a, RNA duplex and R1ρ RD profiles with 3-state global fits to rG-N1 and rU-N3 (20 °C and pH 7.9). b, CSs for rG•rU ES1/ES2 compared to structure-based DFT predictions and rUTP ionization. c, Population and lifetime of rG•rU ES1/ES2 measured as a function of temperature (at pH 6.9) and pH (at 20 °C). dG•dT ES1 and ES2 shown (in grey) for comparison. d, Multi-state equilibrium between WB and WC-like rG•rU mispairs (20 °C and pH 7.9).
Computational studies36 show that dG•dUenol is stabilized relative to dG•dTenol. This is predicted to tilt the rapid rGenol•rU⇌rG•rUenol equilibrium in favor of rG•rUenol (40%) in RNA as compared to dG•dTenol (20%) in DNA (Methods). We find that in RNA, the ES1 rU-N3 CSs are slightly more downfield shifted (ΔωN3 +30 ppm) than rG-N1 (ΔωN1 +26 ppm)(Fig. 4b). Reweighting the DFT-predicted CSs assuming 60:40 ratios of rGenol•rU:rG•rUenol gives an excellent fit to RD-derived values (Fig. 4b), and are in better agreement than 80:20 rGenol•rU:rG•rUenol (Extended Data Fig. 9). We note that we observe changes in the ES1 CSs at higher pH that suggest a potentially more complex exchange process (Supplementary Discussion 6). As with dG•dT–, the WC-like rG•rU– may exist in equilibrium with both a WC-like rG–•rU and/or an iWB rG•rU– (Fig. 4d).
rG•rU wobbles are widespread in RNA where they play important structural and functional roles37. We therefore examined whether the ESs observed here would occur in more complex RNA structural contexts. Indeed, the rU-N3 15N RD profiles measured for two wobble rG•rU mispairs in a 69-nt Bacillus subtilis guanine riboswitch (Extended Data Fig. 8) provide initial evidence (ΔωN3 +44-47 ppm and pES ~0.04% at pH 7.9) for WC-like rG•rU– mispairs in more complex RNA structures (Extended Data Fig. 9 and Supplementary Discussion 7). Therefore, we can expect that transient WC-like rG•rU mispairs exist robustly across the RNA transcriptome.
Biological implications
Initial selection of NTPs during replication, and tRNAs during translation, strongly relies on WC stereochemical geometry as a means of discriminating against mispairs38,39. The low error rate (10−3-10−6) during initial selection accounts for most of the overall fidelity of replication (~10−6-10−10)40-42 and translation (~10−3-10−5)43-45. By stereochemically mimicking the WC geometry, the ES WC-like dG•dT and rG•rU mispairs observed here can provide a mechanism for evading WC fidelity checks during initial substrate selection1,8,10. The intrinsic probabilities with which WC-like mispairs form have long been suspected as important determinants of substitution mutation probability8,10. By carrying out the first measurements of the intrinsic probabilities with which WC-like dG•dT and rG•rU mispairs form in native DNA/RNA systems, we are able to obtain unique insights into the mechanisms of misincorporation and the potential roles of ES1/ES2.
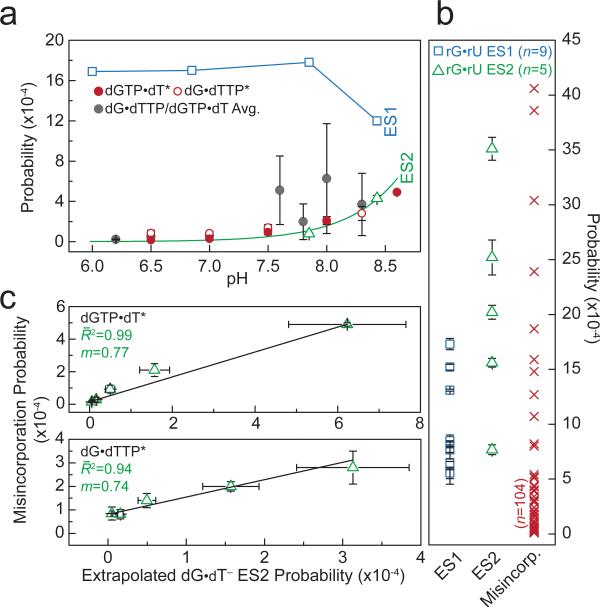
We find that the probabilities with which WC-like dG•dT ES1 and ES2 form in duplex DNA (10−3-10−5) span the dGTP•dT/dG•dTTP misincorporation and base substitution probabilities seen during replication using high-fidelity polymerases with little/no proof-reading capabilities (Fig. 5a, Extended Data Fig. 10 and Supplementary Discussion 8)46-48. Similarly, the WC-like rG•rU ES1 and ES2 probabilities (10−3-10−4) span the majority of amino acid misincorporation probabilities arising specifically due to rG•rU pairing at any codon position (10−3-10−5)43,44 (Fig. 5b and Supplementary Discussion 8). Note that some of the amino acid misincorporation probabilities (10−5) are lower than the measured rG•rU ES2 probability, which could be due to translational proofreading45 and/or lower pH conditions that destabilize ES2. These results, together with previous structural studies showing that WB and WC-like mispairs can exist within polymerase1-3 and ribosome5,6,13 active sites, strongly suggest that energetic competition between WB and WC-like mispairs is robust and is a key determinant of misincorporation probability during replication and translation (Supplementary Discussion 9). However, additional studies are needed to determine the probabilities with which WC-like mispairs form within the unique environment of polymerases and ribosomes. A recent MD study found that rGenol•rU tautomers can be stabilized in a ribosome context, but challenges their involvement in decoding errors based on predicted tRNA binding energies49.
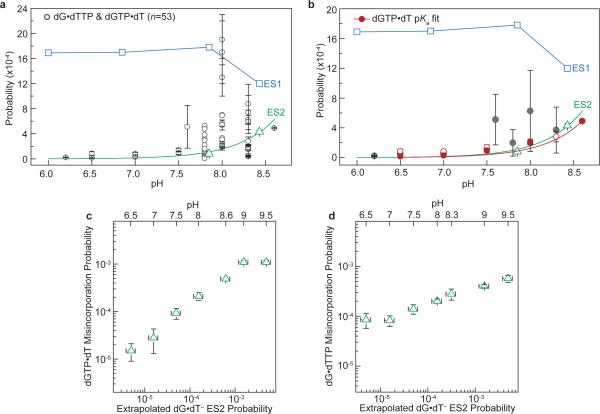
Figure 5. Correlation between WC-like bps and misincorporation probabilities.
a, dG•dT ES1 (blue square) and ES2 (green triangle) probabilities. pH-dependent dGTP•dT*/dG•dTTP* misincorporation probabilities (errors as published)47. Averaged dGTP•dT/dG•dTTP misincorporation and base substitution probabilities; error bars reflect the s.d. (Supplementary Discussion 8). ES2 fit to Henderson-Hasselbalch equation shown as green line (see Methods). A blue line connects ES1 points for visualization purposes. b, rG•rU ES1 (blue square) and ES2 (green triangle) probabilities measured at varied conditions (Supplementary Table 1), and amino acid misincorporation probabilities (red ‘X’) due to rG•rU mispairs (Zhang et al.44). c, The probabilities of dGTP•dT/dG•dTTP misincorporation (error as given) by AMV RT (Yu et al.47) versus the pKa-predicted probability (s.e.) of forming a WC-like dG•dT– mispair as a function of pH.
The pH-dependent dG•dT misincorporation probability47,48,50 points to the involvement of an anionic species in misincorporation1. Our results strongly suggest that this species is most likely ES2 dG•dT– and not the energetically disfavored dG–•dT. We observe excellent agreement between the pKa-predicted probability of forming ES2 dG•dT– and pH-dependent dGTP•dT/dG•dTTP misincorporation probabilities measured for a reverse transcriptase47 which lacks any proofreading ability (Fig. 5c and Extended Data Fig. 10). We note that the correlation is reduced at more extreme pH, near the pKas of other ionizable groups in proteins and DNAs (Extended Data Fig. 10 and Supplementary Discussion 10). We also find that dG•5BrdU– enhances the probability of forming a WC-like ES2 (Fig. 3d) and correspondingly results in an ~8-fold increase dG•5BrdU misincorporation47. These data suggest that for this polymerase, misincorporation proceeds predominantly via a WC-like dG•dT–. It is very likely that parameters such as polymerase types, DNA sequence, and the chemical environment can affect the relative stabilities and lifetimes of the anionic, tautomeric, and WB mispairs3. Therefore we can expect this to affect the flux through distinct misincorporation pathways involving different WC and WB conformations, which may help to explain the broad range of misincorporation probabilities.
Our findings suggest that unconventional enol tautomeric and anionic bases exist robustly in genomes. We anticipate these rare tautomeric and anionic bases play unique roles in DNA damage induction and repair, nucleic acid recognition, chemical modifications of nucleic acids, and catalysis. The NMR methods outlined here can immediately be applied to characterize tautomeric and anionic species, which we believe will not be restricted to dG•dT and rG•rU mispairs, but rather will be widespread across diverse nucleic acid motifs.
METHODS
Sample Preparation
NMR buffer
All duplex DNA and RNA samples were buffer exchanged using a centrifugal concentrator (EMD Millipore) into a solution containing 25 mM sodium chloride (100 mM sodium chloride for Dickerson-GT DNA), 15 mM sodium phosphate, 0.1 mM EDTA, and 10% D2O with variable pH (6.0, 6.4, 6.8, 6.9, 7.9). pH values of 8.4 were obtained for hp-GT DNA and hp-GU-20 RNA samples by direct titration of pH 7.9 samples with filtered 0.5 M NaOH solution. Monovalent ion concentration subsequently increased by a small amount proportional to the NaOH titrated in but did not affect DNA/RNA conformation as confirmed by NMR. Natural isotopic abundance oligonucleotide sample concentrations ranged from 2-3.5 mM. 13C/15N-labeled oligonucleotide sample concentrations ranged from 0.7-3.5 mM. xpt-G riboswitch sample was diluted to ~30 μM in a solution containing saturated guanine, denatured, and annealed on ice. Sample was buffer exchanged against either potassium acetate (pH 6.7) or sodium phosphate (pH 7.9) buffer and concentrated to 0.7-1.7 mM. Mg2+ was titrated in until total concentration was ~5 mM.
Site-specifically 13C/15N-labeled DNA samples
Selectively 13C/15N-labeled DNA samples (hp-GT DNA, 5BrU5-hp-GT and 8BrG15-hp-GT DNA) were purchased from the Yale Keck Oligonucleotide Synthesis Facility and were synthesized using commercially available 2′-deoxyguanosine DMT-phosphoramidite (98% 13C10, 98% 15N5) and 2′-deoxythymidine phosphoramidite (98% 13C10, 98% 15N2) purchased from Cambridge Isotope Labs. hp-GT DNA was selectively 13C/15N-labeled at dT5 and dG15, 5BrU5-hp-GT DNA was 13C/15N-labeled at dG15, and 8BrG15-hp-GT was 13C/15N-labeled at dT5. Samples were purified using RP-HPLC prior to buffer exchange. NMR experiments were used to confirm native folding of hp-GT, 5BrU5-hp-GT, and 8BrG15-hp-GT DNA constructs.
Enzymatic synthesis of 13C/15N-labeled DNA samples
The Dickerson-GT DNA sample was synthesized in vitro using uniformly enriched 13C/15N dGTP and dTTP (Silantes GmbH) as previously described51. Reaction mixture was centrifuged and filtered to remove excess pyrophosphate and concentrated down to 1 mL in a 3 kDa molecular weight cut-off centrifugal concentrator (EMD Millipore). Sample was mixed with 1 mL of a formamide-based denaturing loading dye, denatured at 95 °C for 5 min, and loaded onto a 33×102 cm sequencing gel (20% polyacrylamide/8M urea) and run for 12 hours to resolve target oligonucleotide from template and other nucleic acid species. Target band was shadowed briefly using a UV hand-lamp and excised prior to gel electroelution (Whatman, GE Healthcare), followed by ethanol precipitation. Sample purity was confirmed using gel electrophoresis (20% polyacrylamide/8M urea) stained with SybrGOLD prior to buffer exchange.
Enzymatic synthesis of 13C/15N-labeled RNA samples
The hp-GU-20, hp-GU-24 and xpt-G riboswitch RNA samples were prepared using in vitro transcription as previously described25 using uniformly enriched 13C/15N ribonucleotide triphosphates (hp-GU-20 RNA: rGTP & rUTP only, hp-GU-24: All and xpt-G: All). Purification was carried out as described above for 13C/15N-labeled DNA construct.
Unlabeled and unlabeled-modified DNA samples
hp-GT and Dickerson-GT DNA constructs at natural isotopic abundance were purchased from Integrated DNA Technologies. The O6-Methyl-2′-deoxyguanosine mismatch constructs (m6G15-hp-GT and m6G4-Dickerson-GT DNA) were purchased from the Yale Keck Oligonucleotide Synthesis Facility. hp-GT and Dickerson-GT DNA constructs were desalted prior to buffer exchange. Unlabeled-modified samples were purified using RP-HPLC prior to buffer exchange.
Isotopically enriched dNTP and rNTP samples
Uniformly 13C/15N enriched dGTP, dTTP, rGTP and rUTP samples were purchased (Silantes GmbH) and added to an NMR buffer (25 mM sodium chloride, 15 mM sodium phosphate, 0.1 mM EDTA and 10% D2O at pH 6.9). Sample was adjusted to pH ~12.5 directly using 5 M NaOH.
NMR experiments
Resonance assignment
The CS assignment for DNA and RNA constructs were obtained using aromatic [13C, 1H], aliphatic [13C, 1H], imino [15N, 1H] heteronuclear and [1H, 1H] NOESY homonuclear correlation experiments. The data for all DNA constructs were acquired on an 18.8T Agilent spectrometer equipped with a triple resonance HCN cryogenic probe, for the uniformly 13C/15N-labeled hp-GU-24 RNA construct on a 14.1T Bruker Avance spectrometer equipped with a triple-resonance HCN cryogenic probe, and for the xpt-G RNA riboswitch on a 14.1T Agilent spectrometer equipped with a Bruker HCPN cryogenic probe. All data were processed and analyzed using the software NMRpipe52 and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco), respectively. Resonance assignment for exchangeable and non-exchangeable 1H was performed using the 2D [1H, 1H] WATERGATE NOESY experiment53 (mixing time 250 ms) as described previously27,54, and their covalently bonded 13C/15N nuclei were assigned using heteronuclear single/multiple quantum coherence correlation experiments (HSQC or HMQC). For the labeled RNA constructs conventional HSQC experiments53 were acquired for all spins, while for unlabeled DNA constructs conventional HSQC was used for the aliphatic C1′ spins and SOFAST-HMQC55,56 were employed for the imino and aromatic spins.
15N R1ρ relaxation dispersion
1D 15N R1ρ RD experiments27,57 targeting imino nitrogen resonances of interest were carried out at 14.1T (hp-GT, 5BrU5-hp-GT, Dickerson-GTDNA and hp-GU-24 RNA) and 16.4T (hp-GU-20 RNA) as previously described57. Raw data were processed using NMRpipe52 to generate a series of peak intensities. On- and off-resonance R1ρ RD profiles were recorded using spinlock powers (ωSL 2π−1) ranging from 100-2000 Hz, with the absolute offset frequencies (Ω 2π−1 Hz) ranging from 0-3.5× the applied spinlock power (Supplementary Table 1). Offset frequencies greater than 3.5× the given spinlock power were not used owing to significant R1 relaxation contributions27. Magnetization of the spins of interest was allowed to relax under an applied spinlock for the following durations: [0 – 120 ms] for N1/N3 of hp-GT, 5BrU5-hp-GT, Dickerson-GT DNA and hp-GU-24 RNA and [0 – 100 ms] for N1/N3 of hp-GU-20 RNA, and [0 – 80 ms], [0 – 74 ms], [0 – 68 ms] for N3 of the xpt-G riboswitch.
13C R1ρ relaxation dispersion
13C R1ρ RD experiments targeting carbon resonances of interest were carried out at 14.1T as previously described24,27. On- and off-resonance R1ρ RD profiles were measured using spinlock powers (ωSL) ranging from 150-3500 Hz, with the absolute offset frequencies (Ω 2π−1, Hz) ranging from 0-3.5× the applied spinlock power (Supplementary Table 1). Magnetization of the spins of interest were allowed to relax under an applied spinlock for the following durations: [0 – 60 ms] for C1′/C6/C8 of hp-GT DNA and C1′ of Dickerson-GT, [0 – 50 ms] for C6/C8 Dickerson-GT DNA.
13C/15N CSs of ionized dNTPs and rNTPs
Carbon and nitrogen CSs of neutral and deprotonated dNTPs and rNTPs were collected at 25 °C and pH ~6.9 and ~12.5 using a 2D [15N, 13C] HMQC experiment on a 14.1T Agilent spectrometer equipped with a Bruker HNCP cryoprobe.
Analysis of R1ρ data
Fitting of 13C and 15N R1ρ data
R1ρ values were calculated by fitting the decay of peak intensity versus relaxation delay to a monoexponential58. Errors in R1ρ were estimated using spectral noise and duplicate R1ρ data points using a suite of Mathematica notebooks58. Measured on- and off-resonance R1ρ data were globally fit to algebraic equations describing N-site chemical exchange using a Levenberg-Marquardt method, weighted to the experimental error in the R1ρ data. For 2-state exchange, data was fit to the Laguerre equation (1)59 under the valid assumption that the populations are highly asymmetric, such that pGS >> pES where pES < 0.3. For the 3-state chemical exchange model where kBC = kCB = 0, data was fit to both the 3-state Laguerre equation (2) as well as the general 3-state equation (3)59, where pGS >> pES and pES < 0.1. Fitted parameters derived from Eqn. 2 and Eqn. 3 are in excellent agreement with one another (Supplementary Table 1). Statistical tests, F-Test and Akaike information criterion (AIC), were used to select the best-fit N-state exchange model24 (Extended Data Fig. 3). 13C and 15N RD data from the dG•dT or rG•rU mispair resonances for each construct, at each temperature and pH condition were fitted globally (where kex and pB are the shared-parameters) when possible. In the cases where 15N RD data was available but 13C RD data showed no chemical exchange, did not exhibit 3-state exchange, or was not collected, the 15N N1/N3 RD data were globally fitted as described above. Bloch-McConnell60 (B-M) numerical simulations were used to validate the algebraic approximations’ for 2- and 3-state exchange scenarios.
- (1)
-
(2)3-state Laguerre equation with no minor exchange25:
- (3)
in which R1 and R2 are the intrinsic longitudinal and transverse relaxation rates (s−1). The exchange rates are defined as kexi = kGS→ESi + kESi→GS, where kGS→ESi = pESikexi and kESi→GS = pGSkexi and where i = 1 or 2. The CS difference between the GS and ESs is given by ΔωESi = ΩESi - ΩGS, where Ω = Ωobs - ωrf defines the resonance offset from the carrier frequency (ωrf), Ωobs = (ΩGSpGS + ΩES1pES1) or Ωobs = (ΩGSpGS + ΩES1pES1 + ΩES2pES2), and where Ωobs ≈ ΩGS when pGS >> pESi, as is the case in the ESs measured. The average effective spinlock field in the rotating frame is given by ω2eff = Ω2 + ω2SL and ω2GS = (ΩGS – ωrf)2 + ω2SL, ω2ESi = (ΩESi – ωrf)2 + ω2SL and ωSL is the spinlock power. The tilt angle in the rotating frame is given by θ = arctan(ωSL/Ω).
Analysis of the 3-state exchange model
We repeated measurements of RD profiles for hp-GT dG-N1 and dT-N3 at pH 8.4 and 25 °C on a different spectrometer and also obtained data that is consistent with 3-state exchange (Extended Data Fig. 2 and Supplementary Table 1). In addition, we collected one additional spinlock power (800 Hz) for both resonances, and find that the inclusion of this extra spinlock power has negligible effect on the fitted exchange parameters indicating that they are robustly determined by the measured data. We note that differences in the RD profiles and fitted parameters between spectrometers are largely within error, with minor differences likely arising due to small differences in temperature and/or spinlock calibrations.
The dT-N3 resonance of hp-GT DNA at 25 °C and pH 7.9 was also individually refit assuming both 2-state and 3-state exchange. The individual dT-N3 3-state fit gave very similar fitted parameters as the dT-N3 in the 3-state global fit with dG-N1. The F-test (at 0.05 significance level) favored the 3-state individual fit model over the 2-state individual fit model. AIC also favored the 3-state model, estimating the 3-state model to be 3.1×1015 times more likely to be the correct model than the 2-state model. The individual 2-state and 3-state fits to dG-N1 RD profiles give similar agreement (reduced χ2 ~0.83) when the ES2 CS is fixed based on the globally fitted value; however, statistical tests slightly favor the simpler model (AIC favors 2-state by approximately two-fold).
The 3-state model is supported by statistical tests (F-test and AIC favor the 3-state over 2-state exchange model) and also by B-M simulations (data not shown) for the hp-GT dG-N1 and dT-N3 data at pH 7.9. Here, R1ρ values were simulated, noise corrupted, and evaluated for the ability to report on the 3-state exchange following the procedure reported in Bothe et al.63.
Bloch-McConnell 3-state numerical simulations
Parameters derived from the algebraic 3-state fits, along with the ωrf and ωSL, were used to simulate numerical solutions to the 3-state B-M equations60 (Extended Data Fig. 3). The B-M simulations were carried out using a procedure similar to that described recently for 2-state exchange63. Simulations were carried out assuming a 0.25 second relaxation delay under the applied ωSL.
Thermodynamic analysis of R1ρ RD-derived parameters
Temperature-dependent analysis of forward and reverse exchange rates in the site-labeled (dG15•dT5 13C/15N) hp-GT DNA and G/U labeled hp-GU-20 RNA samples were carried out as previously described24. This analysis employed the ES populations and exchange rates obtained based on 2-state global fitting of dG15-N1 and dT5-N3 RD data measured in hp-GT DNA at 10, 20, 25 and 30 °C. For hp-GU-20 RNA, the analysis employed populations and exchange rates obtained based on 2-state and 3-state global fitting of rG16-N1 and rU5-N3 RD data measured at 10, 20, 25 and 30 °C at pH 6.9. Errors in the fitted thermodynamic parameters are given by weighted fits of the modified van't Hoff equation24 to the RD-derived forward and reverse exchange rates and their errors. The NMR sample temperature was calibrated using 99.8% methanol-d4 (Cambridge Isotope Laboratories) using the equation T = −16.7467(Δδ)2 – 52.5130(Δδ) + 419.1381, where Δδ is the difference in CS (ppm) between the hydroxyl and methyl proton64.
Density functional theory geometry optimizations and CS calculations
All DFT calculations33 were performed using Gaussian 09c (Gaussian, Inc.)65 and carried out on the University of Michigan's Advanced Research Computing HPC cluster, Flux, as previously described24 with minor modifications to DFT method and basis set used. Geometry optimizations were carried out using the empirical exchange-correlation functional, M06-2X66, with the 6-31+G(d,p) basis-set. The 13C and 15N isotropic magnetic shielding (σ13C and σ15N) were calculated using the GIAO method with M06-2X/6-31+G(d,p). CSs of the nucleobases (δ13C and δ15N) were calculated by δ13C/15N = σ13C/15N – σTMS/NH3, where σTMS and σNH3 are the isotropic magnetic shieldings calculated for the reference compounds trimethylsilane (13C) and NH3 (15N), respectively.
Input structures for DFT calculations
We generated idealized B-/A-form helices corresponding to our sequence contexts (hp-GT DNA and hp-GU-24 RNA) using make-NA (J. Stroud, make-NA, http://structure.usc.edu/make-na/server.html 2011). The duplexes were truncated to the trinucleotide step centered on the target mispair (GTG/CGC for hp-GT DNA and GUA/UGC for hp-GU-24 RNA). The sugar and phosphate moieties were removed and replaced with a methyl for i (dG•dT or rG•rU mispair), i+1, and i−1 base pairs to save on computational time. Although the structures lack a 2′-deoxyribose or ribose sugar moieties, they will be denoted as dG•dT or rG•rU to avoid confusion. All heavy atoms were frozen for the i+1 and i−1 base pairs while geometry optimizations were carried out for protons and heavy atoms of the central dG•dT or rG•rU base pair as well as the protons of the i+1/i−1 pairs. We performed full geometry optimizations on: dG•dT and rG•rU GS WB pairs, dGenol•dT, dG•dTenol, rGenol•rU, rG•rUenol, and dG–•dT and rG–•rU ES WC-like pairs. All converged to the expected WB or WC-like geometries. In the instance where the geometry optimizations of dG•dT– and rG•rU– starting states failed to converge to a stable WC-like dG•dT–/rG•rU– geometry (and instead converged to an iWB geometry, in vacuum), the WC-like dG•dTenol/rG•rUenol states were converted to dG•dT–/rG•rU– and geometry optimizations were carried out on the protons only. CSs for each state were calculated and later used in calculating population weighted CSs assuming different populations of these mispair species. In addition, while the sugar moieties were truncated to methyl groups to save on computation time, it should be noted that previous DFT studies of nucleotides have shown that tautomerization, primarily of pyrimidines, can have an affect on the sugar conformation67. However, we can rule out large changes in sugar pucker arising in ES1 based on the negligibly small chemical exchange contributions to both dG-C1′ and dT-C1′ (see Extended Data Fig. 2) and only very small changes in dG-C1′ and dT-C1′ CSs upon locking the enol-like form with m6dG•dT (|ΔωC1′| ≤ 0.7 ppm, see Fig. 3b and Extended Data Fig. 7). Finally, while the CSs of the anionic pairs are predicted assuming planar pair geometry, prior computational studies of G•T– and G–•T nucleobase pairs in isolation have shown that they can favor non-planar and non-WC geometry35.
dGenol•dT distance dependent DFT calculations
We carried out distance dependent DFT calculations68 on a pair of WC-like dGenol•dT N1/N9-methyl nucleobases in vacuum using the M06-2X method and 6-31+G(d,p) basis-set, as described above. The geometry of a dG•dT wobble pair and dGenol•dT WC-like pair was optimized with no constraints prior to CS calculations. The ideal N1-N3 distance of the dGenol•dT WC-like mispair was then manually varied from 2.44Å to 3.8Å in increments of 0.1Å from 2.44-3.04Å and then to 3.8Å (Extended Data Fig. 4). At every increment the proton positions alone were optimized and CSs were calculated relative to an optimized dG•dT wobble base pair.
Population-weighted average DFT-predicted CS calculations
Based on the computationally predicted energetic differences between interconverting dGenol•dT and dG•dTenol base mispairs in water (ΔG 0.7-0.8 kcal/mol)31 and in a weakly polar medium (ΔG 0.99 kcal/mol)69, we can predict that the dG•dT ES1 CSs represent a population weighted average between interconverting dGenol•dT(~80%)⇌dG•dTenol(~20%) states. Thus, the DFT-predicted CSs for dGenol•dT and dG•dTenol were summed in a population-weighted manner. It is noted that the computationally predicted energetic stabilities of the tautomeric states differ when calculated in water versus vacuum, or a weakly polar medium, with the values predicted in water giving the greatest agreement with our experimental results.
In the case of rG•rU ES1, computational studies have shown that dG•dUenol is ~1 kcal/mol more stable than dG•dTenol in a DNA fragment36, suggesting that an rGenol•rU⇌rG•rUenol equilibrium should be titled slightly more towards rG•rUenol than dG•dTenol in DNA. We can qualitatively estimate the relative stability between rGenol•rU and rG•rUenol to be 60:40 based on a best fit to the RD-derived CSs.
pKa fitting and probability estimation
The apparent pKas for hp-GT ES2, 5BrU-hp-GT ES2, and dGTP•dT misincorporation (pH 6.5-8.6) were fit to the Henderson-Hasselbalch equation using a Monte-Carlo (MC) approach. Here, 106 pBs at pH 7.9 and/or 8.4 were selected from a Gaussian distribution with mean pB value and standard deviation representing the uncertainty in pB based on fitting of the RD data. 106 fits to Eqn. 4 were then carried out assuming these pB values to generate 106 pKas.
| (4) |
Where pB is the probability of forming ES2 or dGTP•dT misincorporation probability at a given pH. The resulting fitted pKa values were fitted to a Gaussian distribution. The mean value of the Gaussian distribution is the reported pKa value and the standard deviation is assumed to be the error. An analogous approach was used to back-calculate predicted pBs at a given pH using the pKa derived by the above method.
Extended Data
Extended Data Figure 1. NMR spectra of site- and selectively-labeled dG•dT mispair DNA constructs.
a-b Shown are the a, hp-GT DNA and b, Dickerson-GT constructs with 13C/15N labeled dG•dT mispairs highlighted in red along with 2D imino [15N, 1H] HSQC, 2D aromatic [13C, 1H] HSQC and 2D C1′ [13C, 1H] HSQC spectra (pH 6.9, 25 °C).
Extended Data Figure 2. Rotating frame relaxation dispersion profiles of dG•dT mispairs in hp-GT and Dickerson-GT DNA constructs.
RD profiles showing chemical exchange (R2+Rex) in the dG•dT mispair as a function of the spin lock offset (Ωeff 2π−1) and spin lock power (ωSL 2π−1, color coded in insets). Shown are a, 15N and b, 13C RD profiles in hp-GT DNA in hp-GT. On-resonance profiles showing solid and dashed black lines indicate fits assuming no chemical exchange (solid) and simplified 2-state exchange process (dash). The hp-GT dG15-N1 and dT5-N3 in brackets denote duplicate profiles (with an additional 800 Hz spinlock power for each) collected at pH 8.4 and 25 °C collected on a different spectrometer from the preceding profiles. c, 15N and 13C RD profiles for Dickerson-GT. Sample conditions are indicated on each profile. Error bars represent experimental uncertainty (one s.d., see Methods).
Extended Data Figure 3. Multiple site exchange comparison and numerical solutions.
Global fitting of a, hp-GT DNA and b, hp-GU-20 RNA N1 and N3 RD profiles to 2-state algebraic equation (Eqn. 1, fit reduced χ2 shown in inset) and 3-state algebraic equation (Eqn. 2, fit reduced χ2 shown in inset). Numerical solutions to the Bloch-McConnell 3-state equations assuming no minor exchange and input exchange parameters obtained based on the 3-state algebraic fit are also shown to establish the validity of the 3-state expression under these exchange scenarios (Eqn. 2, see Methods). Sample conditions are indicated on each profile. Error bars represent experimental uncertainty (one s.d., see Methods).
Extended Data Figure 4. Chemical shift fingerprinting dG•dT excited states.
a, RD-derived dG15-N1 and dT5-N3 CSs (CSs) (referenced to GS WB) for ES1 (25 °C and pH 6.9) and ES2 (25 °C and pH 8.4) of hp-GT and ES1 of Dickerson-GT (25 °C and pH 6.9) are shown. Errors in all RD-derived fitted parameters (eg. Δω) denote s.e. from the weighted global fit (see Methods). b, RD-derived hp-GT dG•dT ES1 (blue) and ES2 (green) 15N CSs are shown as a function of temperature and pH for both dG15-N1 (square) and dT5-N3 (circle). c, Scheme used to calculate CSs using DFT (see Methods). Shown is a schematic representation of scenario used to for calculating CSs using DFT. Idealized B-form DNA helix is generated to give a central dG•dT mispair (red) that is flanked by canonical dG•dC pairs, analogous to the hp-GT construct. Residues are trimmed to 1-/9-methyl bases and i+1/i−1 pairs are frozen in place for subsequent geometry optimizations and NMR CS calculations. d, DFT-calculated CSs (referenced to an energy optimized WB geometry) are shown for various tautomeric and anionic configurations, where dGenol•dT/dG•dTenol represents population weighted average over dGenol•dT (80%) and dG•dTenol (20%). e, RD-derived ES1 and ES2 CSs are plotted against DFT-calculated CSs of base opened dG•dT mispairs, taken from X-ray structures and pruned to 1-/9-methyl bases. f, DFT-calculated CSs (referenced to an energy optimized WB geometry) are plotted as a function of dG-N1- -dT-N3 inter-atomic distance for a WC-like dGenol•dT tautomeric pair. g, Computational studies31,32,70 predict that the tautomeric pathway proceeds via a planar dG+•dT− ion pair (charge delocalization is implied) that is highlighted by a network of five H-bonds. h, Predicted pair geometry of an anionic dG•dT– inverted wobble. Deprotonated dT-N3 is highlighted in red (charge delocalization is implied). i, Predicted pair geometry of a dGenol•dT Hoogsteen mispair.
Extended Data Figure 5. Attempts to trap anionic dG.
a, 1D 13C spectra (without 13C-13C homonuclear decoupling) of the aromatic carbon region of protonated dGTP (black) and anionic dGTP (red) showing CS perturbations induced upon deprotonation of dGTP-N1. b, 13C spectra (without 13C-13C homonuclear decoupling) of the aromatic carbon region of protonated dTTP (black) and anionic dTTP (red) showing CS perturbations induced upon deprotonation of dTTP-N3. c, 2D [15N, 13C] HMQC spectra of dGTP showing CS of dGTP-N1 induced upon deprotonation. The spectra is rotated by 90°, to depict 15N CS along x-axis for visualization purposes. Red circles on inset structure highlight measured resonances (C6 and N1). d, 2D [15N, 13C] HMQC spectra of dTTP showing CS perturbation of dTTP-N3 induced upon deprotonation. The spectra is rotated by 90°, to depict 15N CS along x-axis for visualization purposes. Red circles on inset structure highlight measured resonances (C4 and N3). e, hp-GT DNA spectra of the dG/dT aromatic carbons upon increase in pH from 6.9 (black) to 10.7 (red). Minor upfield CSs are observed for dT5-C6 and dG9-C8, but not dG15-C8, indicating the dT5 in the dG•dT mispair is likely undergoing deprotonation and not the paired dG15. f, 8BrG15-hp-GT DNA construct bearing a 13C/15N site-labeled dT5 paired with a 8-bromo-2′-deoxyguanosine is shown (left) along with the 15N RD profile for the paired dT5-N3. Error bars represent experimental uncertainty (one s.d., see Methods).
Extended Data Figure 6. Kinetic-thermodynamic plots and parameters.
a, Kinetic-thermodynamic diagram for exchange between GS and ES1 via a transition state for hp-GT DNA ES1 (left) and hp-GU-20 RNA ES1 (right), showing activation (G‡) and net free energy (G), enthalpy (H), and entropy (TS) changes (referenced to 0). b, Kinetic-thermodynamic parameters derived from RD data. Asterisk denotes parameters calculated using only a single temperature (see Methods), wherein enthalpic and entropic parameters cannot be derived. Here, dG15•dT5 ES2 values were calculated at 25.05 °C, rG16•rU5 ES2 values were calculated at 20.05 °C, and dG15•5BrdU5 ES1 and ES2 values were calculated at 10.05 °C. Error is given by the s.e. of the weighted global fits of the corresponding RD profiles. Error is propagated using the respective uncertainties in kex and pES.
Extended Data Figure 7. Trapping or stabilizing dG•dT ES1 and ES2.
a, m6G15-hp-GT DNA construct is shown (left) where dG15 is methylated at the O6 position to trap a near-WC “dGenol•dT”-like geometry (Fig. 3c). CS perturbations induced in the aromatic (center) and sugar (right) resonances upon O6-methylation (blue) with the hp-GT DNA spectra (black) with the resonances for the dG•dT mispair from hp-GT DNA in red. m6dG•dT mispair and CSs are highlighted in red. b, Similarly, m6G4-Dickerson-GT DNA construct is shown (left) where dG4 is O6-methylated to trap a WC-like state, with similar color scheme as a. c, 5BrU5-hp-GT DNA construct bearing a 13C/15N site-labeled dG15 paired with a 5-bromo-2′-deoxythymidine is shown (left) along with the 15N RD profile for the paired dG15-N1. Error bars represent experimental uncertainty (one s.d., see Methods).
Extended Data Figure 8. Rotating frame relaxation dispersion profiles for rG•rU mispairs in hp-GU-20, hp-GU-24 and xpt-G RNA constructs.
a-b RNA constructs and the imino [15N, 1H] HSQC zoomed into the rG•rU wobble region of the spectra for hp-GU-20 and hp-GU-24. rG•rU mispair resonances are shown in red. c, The Bacillus subtilis guanine binding riboswitch (xpt-G RNA)71 construct and full imino [15N, 1H] HSQC of folded and guanine ligand-bound riboswitch. rG•rU mispair resonances are shown in red. d-f, 15N RD profiles for d, hp-GU-20 e, hp-GU-24 and f, xpt-G riboswitch RNA. Error bars represent experimental uncertainty (one s.d., see Methods).
Extended Data Figure 9. CS fingerprinting rG•rU excited states.
a, RD-derived rG16-N1 and rU5-N3 CSs (referenced to GS WB) are shown for ES1 (20 °C and pH 6.9) and ES2 of hp-GU-20 (20 °C and pH 7.9) and ES1 rG18-N1 and rU7-N3 CSs of hp-GU-24 (25 °C and pH 6.9). Errors in all RD-derived fitted parameters (eg. Δω) denote s.e. from the weighted global fit (see Methods). b, RD-derived CSs (referenced to GS WB) are shown for the ES of xpt-G riboswitch (rU17-N3 and rU69-N3) at 25 °C and pH 7.9. c, 2D [15N, 13C] HMQC spectra of rUTP showing CS of rUTP-N3 induced upon deprotonation. The spectra is rotated by 90°, to depict 15N CS along x-axis for visualization purposes. Red circles on inset structure highlight measured resonances (C4 and N3). d, 2D [15N, 13C] HMQC spectra of rGTP showing CS of rGTP-N1 induced upon deprotonation. The spectra is rotated by 90°, to depict 15N CS along x-axis for visualization purposes. Red circles on inset structure highlight measured resonances (C6 and N1). e, RD-derived hp-GU-20 rG•rU ES1 (blue) and ES2 (green) CSs are shown as a function of temperature and pH for both rG16-N1 (square) and rU5-N3 (circle). f, Scheme used to calculate CSs using DFT. Idealized A-form RNA helix is generated to give a central rG•rU mispair (red) that is flanked by canonical rG•rC and rA•rU pairs, analogous to the hp-GU-24 construct. Residues are trimmed to 1-/9-methyl bases and i+1/i−1 pairs are frozen in place for subsequent geometry optimizations and CS calculations (see Methods). g, DFT-predicted CSs (referenced to an energy optimized WB geometry) are shown for various tautomeric and anionic configurations, where rGenol•rU/rG•rUenol represents population weighted average CSs of rGenol•rU (60%) and rG•rUenol (40%). h, 15N rG-N1 and rU-N3 CS comparison between RD-derived ES1 CSs and population weighted DFT-predicted CSs (60:40 vs. 80:20). i, Computational studies32 predict that the tautomeric pathway for a rG•rU pair can proceed via a planar rG+•rU− ion pair (charge delocalization is implied) that is highlighted by a network of five H-bonds. j, Pair geometry of an anionic rG•rU– inverted wobble. Deprotonated rU-N3 is highlighted in red (charge delocalization is implied).
Extended Data Figure 10. dG•dT Misincorporation probabilities and correlation to WC-like excited states.
a, Explicit dGTP•dT and dG•dTTP kinetic misincorporation and base substitution probabilities (n=53) and associated errors46-48,72-77 (see Supplementary Discussion 8) are plotted against hp-GT dG•dT ES1 (blue squares) and ES2 (green triangles). The pKa fit of ES2 probabilities to the Henderson-Hasselbalch equation (Eqn. 4, see Methods) is shown as the green trend line. b, Red trend line shows the pKa fit to dGTP•dT misincorporation probabilities47 from pH 6.5-8.6 to the Henderson-Hasselbalch equation. The fit was weighted using reported experimental errors and gave a reduced χ2 of 3.56. c-d, Extrapolated dG•dT– ES2 probability (s.e. from the weighted global fit) is plotted against dGTP•dT (left) and dG•dTTP (right) misincorporation probabilities (errors as given)47 from pH 6.5–9.5.
Supplementary Material
Acknowledgements
We thank S. Horowitz, H. Zhou, J. Lee, A.M. Mustoe, and E.N. Nikolova for assistance and critical comments. We are grateful for technical support and resources from the Duke Magnetic Resonance Spectroscopy Center and University of Michigan Flux HPC Cluster. This work was supported by an NIH grant (R01GM089846) and an Agilent Thought Leader Award given to H.M.A.
Footnotes
Author Contributions I.J.K and H.M.A. conceived the project and experimental design. I.J.K prepared NMR samples as well as performed and analyzed all NMR RD experiments. I.J.K assigned resonances in all nucleic acid constructs with assistance from B.S. K.P. prepared the hp-GU-24 sample and carried out additional NMR RD experiments. I.J.K. performed all DFT calculations. Z.W.S. assisted I.J.K. with numerical Bloch-McConnell simulations. I.J.K. and H.M.A. wrote the manuscript with critical input from B.S. and K.P.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
Supplementary Information is available online at www.nature.com/nature.
REFERENCES
- 1.Bebenek K, Pedersen LC, Kunkel TA. Replication infidelity via a mismatch with Watson–Crick geometry. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1862–1867. doi: 10.1073/pnas.1012825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koag M-C, Nam K, Lee S. The spontaneous replication error and the mismatch discrimination mechanisms of human DNA polymerase β. Nucleic Acids Res. 2014;42:11233–11245. doi: 10.1093/nar/gku789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia S, Konigsberg WH. Mispairs with Watson-Crick base-pair geometry observed in ternary complexes of an RB69 DNA polymerase variant. Protein Sci. 2014;23:508–513. doi: 10.1002/pro.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Hellinga HW, Beese LS. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:17644–17648. doi: 10.1073/pnas.1114496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogle JM, Murphy Iv FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the Ribosome Requires a Transition from an Open to a Closed Form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- 6.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 7.Watson JD, Crick FHC. The Structure of DNA. Cold Spring Harbor Symp. Quant. Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 8.Topal MD, Fresco JR. Complementary base pairing and the origin of substitution mutations. Nature. 1976;263:285–289. doi: 10.1038/263285a0. [DOI] [PubMed] [Google Scholar]
- 9.Harris VH, et al. The effect of tautomeric constant on the specificity of nucleotide incorporation during DNA replication: support for the rare tautomer hypothesis of substitution mutagenesis. J. Mol. Biol. 2003;326:1389–1401. doi: 10.1016/s0022-2836(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 10.Topal MD, Fresco JR. Base pairing and fidelity in codon-anticodon interaction. Nature. 1976;263:289–293. doi: 10.1038/263289a0. [DOI] [PubMed] [Google Scholar]
- 11.Sowers L, Goodman M, Eritja R, Kaplan B, Fazakerley G. Ionized and wobble base-pairing for bromouracil-guanine in equilibrium under physiological conditions: A nuclear magnetic resonance study on an oligonucleotide containing a bromouracil-guanine base-pair as a function of pH. J. Mol. Biol. 1989;205:437–447. doi: 10.1016/0022-2836(89)90353-7. [DOI] [PubMed] [Google Scholar]
- 12.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weixlbaumer A, et al. Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat. Struct. Mol. Biol. 2007;14:498–502. doi: 10.1038/nsmb1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantara WA, Murphy FV, Demirci H, Agris PF. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. U. S. A. 2013;110:10964–10969. doi: 10.1073/pnas.1222641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bevilacqua PC, Yajima R. Nucleobase catalysis in ribozyme mechanism. Curr. Opin. Chem. Biol. 2006;10:455–464. doi: 10.1016/j.cbpa.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Cochrane JC, Strobel SA. Catalytic Strategies of Self-Cleaving Ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive Ligand Binding by the Purine Riboswitch in the Recognition of Guanine and Adenine Analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh V, et al. Direct Observation of Multiple Tautomers of Oxythiamine and their Recognition by the Thiamine Pyrophosphate Riboswitch. ACS Chem. Biol. 2013;9:227–236. doi: 10.1021/cb400581f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D, et al. Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2′-deoxycytidine. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E3252–E3259. doi: 10.1073/pnas.1405635111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korzhnev DM, Orekhov VY, Kay LE. Off-Resonance R1ρ NMR Studies of Exchange Dynamics in Proteins with Low Spin-Lock Fields: An Application to a Fyn SH3 Domain. J. Am. Chem. Soc. 2004;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 21.Palmer AG., III Chemical exchange in biomacromolecules: Past, present, and future. J. Magn. Reson. 2014;241:3–17. doi: 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen AL, Kay LE. Measurement of histidine pKa values and tautomer populations in invisible protein states. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E1705–E1712. doi: 10.1073/pnas.1400577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoogstraten CG, Wank JR, Pardi A. Active Site Dynamics in the Lead-Dependent Ribozyme. Biochemistry. 2000;39:9951–9958. doi: 10.1021/bi0007627. [DOI] [PubMed] [Google Scholar]
- 24.Nikolova EN, et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Visualizing transient low-populated structures of RNA. Nature. 2012;491:724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massi F, Johnson E, Wang C, Rance M, Palmer AG., III NMR R1ρ Rotating-Frame Relaxation with Weak Radio Frequency Fields. J. Am. Chem. Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 27.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the Range of Microsecond-to-Millisecond Chemical Exchange Detected in Labeled and Unlabeled Nucleic Acids by Selective Carbon R1ρ NMR Spectroscopy. J. Am. Chem. Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 28.Duthaler RO, Roberts JD. Effects of solvent, protonation, and N-alkylation on the nitrogen-15 chemical shifts of pyridine and related compounds. J. Am. Chem. Soc. 1978;100:4969–4973. [Google Scholar]
- 29.Cho BP, Kadlubar FF, Culp SJ, Evans FE. Nitrogen-15 nuclear magnetic resonance studies on the tautomerism of 8-hydroxy-2′-deoxyguanosine, 8-hydroxyguanosine, and other C8-substituted guanine nucleosides. Chem. Res. Toxicol. 1990;3:445–452. doi: 10.1021/tx00017a010. [DOI] [PubMed] [Google Scholar]
- 30.Goswami B, Gaffney BL, Jones RA. Nitrogen-15-labeled oligodeoxynucleotides. 5. Use of 15N NMR to probe H-bonding in an O6MeG•T base pair. J. Am. Chem. Soc. 1993;115:3832–3833. [Google Scholar]
- 31.Nomura K, et al. DFT Calculations on the Effect of Solvation on the Tautomeric Reactions for Wobble Gua-Thy and Canonical Gua-Cyt Base-Pairs. J. Mod. Phys. 2013;4:422–431. [Google Scholar]
- 32.Brovarets OO. Effect of a modification of uracil on the tautomerization barrier of the wobble Gua•5XUra base pair into the Gua*•5XUra base pair with the Watson-Crick geometry: quantum-chemical study. Rep. Natl. Acad. Sci. Ukraine. 2013;4:154–158. [Google Scholar]
- 33.Xu X-P, Au-Yeung SCF. Investigation of Chemical Shift and Structure Relationships in Nucleic Acids Using NMR and Density Functional Theory Methods. J. Phys. Chem. B. 2000;104:5641–5650. [Google Scholar]
- 34.Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- 35.Brovarets OO, Zhurakivsky RO, Hovorun DM. Is there adequate ionization mechanism of the spontaneous transitions? Quantum-chemical investigation. Biopolym. Cell. 2010;26:398–405. [Google Scholar]
- 36.Orozco M, Hernández B, Luque FJ. Tautomerism of 1-Methyl Derivatives of Uracil, Thymine, and 5-Bromouracil. Is Tautomerism the Basis for the Mutagenicity of 5-Bromouridine? J. Phys. Chem. B. 1998;102:5228–5233. [Google Scholar]
- 37.Varani G, McClain WH. The G•U wobble base pair. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunkel TA, Bebenek K. DNA Replication Fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 39.Voorhees RM, Ramakrishnan V. Structural Basis of the Translational Elongation Cycle*. Annu. Rev. Biochem. 2013;82:203–236. doi: 10.1146/annurev-biochem-113009-092313. [DOI] [PubMed] [Google Scholar]
- 40.Kunkel TA. DNA Replication Fidelity. J. Biol. Chem. 2004;279:16895–16898. doi: 10.1074/jbc.R400006200. [DOI] [PubMed] [Google Scholar]
- 41.Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- 42.Xia S, Konigsberg WH. RB69 DNA Polymerase Structure, Kinetics, and Fidelity. Biochemistry. 2014;53:2752–2767. doi: 10.1021/bi4014215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogle JM, Ramakrishnan V. Structural Insights Into Translational Fidelity. Annu. Rev. Biochem. 2005;74:129–177. doi: 10.1146/annurev.biochem.74.061903.155440. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z, Shah B, Bondarenko PV. G/U and Certain Wobble Position Mismatches as Possible Main Causes of Amino Acid Misincorporations. Biochemistry. 2013;52:8165–8176. doi: 10.1021/bi401002c. [DOI] [PubMed] [Google Scholar]
- 45.Gromadski KB, Rodnina MV. Kinetic Determinants of High-Fidelity tRNA Discrimination on the Ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- 46.Kunkel TA, Alexander PS. The base substitution fidelity of eucaryotic DNA polymerases. Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J. Biol. Chem. 1986;261:160–166. [PubMed] [Google Scholar]
- 47.Yu H, Eritja R, Bloom LB, Goodman MF. Ionization of bromouracil and fluorouracil stimulates base mispairing frequencies with guanine. J. Biol. Chem. 1993;268:15935–15943. [PubMed] [Google Scholar]
- 48.Eckert KA, Kunkel TA. Effect of reaction pH on the fidelity and processivity of exonuclease-deficient Klenow polymerase. J. Biol. Chem. 1993;268:13462–13471. [PubMed] [Google Scholar]
- 49.Satpati P, Åqvist J. Why base tautomerization does not cause errors in mRNA decoding on the ribosome. Nucleic Acids Res. 2014;42:12876–12884. doi: 10.1093/nar/gku1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Driggers PH, Beattie KL. Effect of pH on the base-mispairing properties of 5-bromouracil during DNA synthesis. Biochemistry. 1988;27:1729–1735. doi: 10.1021/bi00405a052. [DOI] [PubMed] [Google Scholar]
- 51.Zimmer DP, Crothers DM. NMR of Enzymatically Synthesized Uniformly 13C15N-Labeled DNA Oligonucleotides. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3091–3095. doi: 10.1073/pnas.92.8.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 53.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR spectroscopy: principles and practice. Academic Press; 1995. [Google Scholar]
- 54.Wijmenga SS, van Buuren BNM. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 55.Farjon J, et al. Longitudinal-Relaxation-Enhanced NMR Experiments for the Study of Nucleic Acids in Solution. J. Am. Chem. Soc. 2009;131:8571–8577. doi: 10.1021/ja901633y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sathyamoorthy B, Lee J, Kimsey I, Ganser L, Al-Hashimi H. Development and application of aromatic [13C, 1H] SOFAST-HMQC NMR experiment for nucleic acids. J. Biomol. NMR. 2014:1–7. doi: 10.1007/s10858-014-9856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nikolova EN, Gottardo FL, Al-Hashimi HM. Probing Transient Hoogsteen Hydrogen Bonds in Canonical Duplex DNA Using NMR Relaxation Dispersion and Single-Atom Substitution. J. Am. Chem. Soc. 2012;134:3667–3670. doi: 10.1021/ja2117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Spyracopoulos L. A suite of Mathematica notebooks for the analysis of protein main chain 15N NMR relaxation data. J. Biomol. NMR. 2006;36:215–224. doi: 10.1007/s10858-006-9083-0. [DOI] [PubMed] [Google Scholar]
- 59.Palmer AG, III, Massi F. Characterization of the Dynamics of Biomacromolecules Using Rotating-Frame Spin Relaxation NMR Spectroscopy. Chem. Rev. 2006;106:1700–1719. doi: 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- 60.McConnell HM. Reaction Rates by Nuclear Magnetic Resonance. The Journal of Chemical Physics. 1958;28:430–431. [Google Scholar]
- 61.Miloushev VZ, Palmer AG., III R1ρ relaxation for two-site chemical exchange: General approximations and some exact solutions. J. Magn. Reson. 2005;177:221–227. doi: 10.1016/j.jmr.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 62.Trott O, Palmer AG., III Theoretical study of R1ρ rotating-frame and R2 free-precession relaxation in the presence of n-site chemical exchange. J. Magn. Reson. 2004;170:104–112. doi: 10.1016/j.jmr.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Bothe JR, Stein ZW, Al-Hashimi HM. Evaluating the uncertainty in exchange parameters determined from off-resonance R1ρ relaxation dispersion for systems in fast exchange. J. Magn. Reson. 2014;244:18–29. doi: 10.1016/j.jmr.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Findeisen M, Brand T, Berger S. A 1H-NMR thermometer suitable for cryoprobes. Magn. Reson. Chem. 2007;45:175–178. doi: 10.1002/mrc.1941. [DOI] [PubMed] [Google Scholar]
- 65.Gaussian 09. Gaussian, Inc.; Wallingford, CT, USA: 2009. [Google Scholar]
- 66.Zhao Y, Truhlar D. The M06 suite of density functionals for main group thermochemistry, thermochemical kinetics, noncovalent interactions, excited states, and transition elements: two new functionals and systematic testing of four M06-class functionals and 12 other functionals. Theor. Chem. Acc. 2008;120:215–241. [Google Scholar]
- 67.Kochina OS, Zhurakivsky RO, Hovorun DM. Effect of the nucleotide bases tautomerization on the conformational properties of the nucleosides: quantum-mechanical investigation by the DFT method. Rep. Natl. Acad. Sci. Ukraine. 2008;1:181–186. [Google Scholar]
- 68.Czernek J, Fiala R, Sklenář V. r. Hydrogen Bonding Effects on the 15N and 1H Shielding Tensors in Nucleic Acid Base Pairs. J. Magn. Reson. 2000;145:142–146. doi: 10.1006/jmre.2000.2091. [DOI] [PubMed] [Google Scholar]
- 69.Brovarets OO, Hovorun DM. The nature of the transition mismatches with Watson–Crick architecture: the G*•T or G•T* DNA base mispair or both? A QM/QTAIM perspective for the biological problem. J. Biomol. Struct. Dyn. 2014:1–21. doi: 10.1080/07391102.2014.924879. [DOI] [PubMed] [Google Scholar]
- 70.Brovarets OO, Hovorun DM. Physicochemical mechanism of the wobble DNA base pairs Gua•Thy and Ade•Cyt transition into the mismatched base pairs Gua*•Thy and Ade•Cyt* formed by the mutagenic tautomers. Ukr. Bioorg. Acta. 2009;8:12–18. [Google Scholar]
- 71.Serganov A, et al. Structural Basis for Discriminative Regulation of Gene Expression by Adenine- and Guanine-Sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 73.Mendelman LV, Boosalis MS, Petruska J, Goodman MF. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 74.Singer B, Chavez F, Goodman MF, Essigmann JM, Dosanjh MK. Effect of 3′ flanking neighbors on kinetics of pairing of dCTP or dTTP opposite O6-methylguanine in a defined primed oligonucleotide when Escherichia coli DNA polymerase I is used. Proc. Natl. Acad. Sci. U. S. A. 1989;86:8271–8274. doi: 10.1073/pnas.86.21.8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dosanjh MK, Essigmann JM, Goodman MF, Singer B. Comparative efficiency of forming m4T•G versus m4T•A base pairs at a unique site by use of Escherichia coli DNA polymerase I (Klenow fragment) and Drosophila melanogaster polymerase α-primase complex. Biochemistry. 1990;29:4698–4703. doi: 10.1021/bi00471a026. [DOI] [PubMed] [Google Scholar]
- 76.Singer B, Dosanjh MK. Site-directed mutagenesis for quantitation of base-base interactions at defined sites. Mutat. Res. Fund. Mol. Mech. Mut. 1990;233:45–51. doi: 10.1016/0027-5107(90)90150-3. [DOI] [PubMed] [Google Scholar]
- 77.Dosanjh MK, Galeros G, Goodman MF, Singer B. Kinetics of extension of O6-methylguanine paired with cytosine or thymine in defined oligonucleotide sequences. Biochemistry. 1991;30:11595–11599. doi: 10.1021/bi00113a015. [DOI] [PubMed] [Google Scholar]
- 78.Nikolova EN, Goh GB, Brooks CL, III, Al-Hashimi HM. Characterizing the Protonation State of Cytosine in Transient G•C Hoogsteen Base Pairs in Duplex DNA. J. Am. Chem. Soc. 2013;135:6766–6769. doi: 10.1021/ja400994e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Várnai P, Canalia M, Leroy J-L. Opening Mechanism of G•T/U Pairs in DNA and RNA Duplexes: A Combined Study of Imino Proton Exchange and Molecular Dynamics Simulation. J. Am. Chem. Soc. 2004;126:14659–14667. doi: 10.1021/ja0470721. [DOI] [PubMed] [Google Scholar]
- 80.Yin Y, et al. Dynamics of spontaneous flipping of a mismatched base in DNA duplex. Proc. Natl. Acad. Sci. U. S. A. 2014;111:8043–8048. doi: 10.1073/pnas.1400667111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giudice E, Várnai P, Lavery R. Base pair opening within B - DNA: free energy pathways for GC and AT pairs from umbrella sampling simulations. Nucleic Acids Res. 2003;31:1434–1443. doi: 10.1093/nar/gkg239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Padermshoke A, Katsumoto Y, Masaki R, Aida M. Thermally induced double proton transfer in GG and wobble GT base pairs: A possible origin of the mutagenic guanine. Chem. Phys. Lett. 2008;457:232–236. [Google Scholar]
- 83.Koag M-C, Kou Y, Ouzon-Shubeita H, Lee S. Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 2014;42:8755–8766. doi: 10.1093/nar/gku554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Patel DJ, Shapiro L, Kozlowski SA, Gaffney BL, Jones RA. Structural studies of the O6meG•T interaction in the d(C-G-T-G-A-A-T-T-C-O6meG-C-G) duplex. Biochemistry. 1986;25:1036–1042. doi: 10.1021/bi00353a013. [DOI] [PubMed] [Google Scholar]
- 85.Zaher HS, Green R. Quality control by the ribosome following peptide bond formation. Nature. 2009;457:161–166. doi: 10.1038/nature07582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Parker J. Errors and alternatives in reading the universal genetic code. Microbiological Reviews. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jang YH, et al. pKa Values of Guanine in Water: Density Functional Theory Calculations Combined with Poisson-Boltzmann Continuum-Solvation Model. J. Phys. Chem. B. 2002;107:344–357. [Google Scholar]
- 88.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G•C+ Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.