Abstract
Cupriavidus sp. are generally heavy metal tolerant bacteria with the ability to degrade a variety of aromatic hydrocarbon compounds, although the degradation pathways and substrate versatilities remain largely unknown. Here we studied the bacterium Cupriavidus gilardii strain CR3, which was isolated from a natural asphalt deposit, and which was shown to utilize naphthenic acids as a sole carbon source. Genome sequencing of C. gilardii CR3 was carried out to elucidate possible mechanisms for the naphthenic acid biodegradation. The genome of C. gilardii CR3 was composed of two circular chromosomes chr1 and chr2 of respectively 3,539,530 bp and 2,039,213 bp in size. The genome for strain CR3 encoded 4,502 putative protein-coding genes, 59 tRNA genes, and many other non-coding genes. Many genes were associated with xenobiotic biodegradation and metal resistance functions. Pathway prediction for degradation of cyclohexanecarboxylic acid, a representative naphthenic acid, suggested that naphthenic acid undergoes initial ring-cleavage, after which the ring fission products can be degraded via several plausible degradation pathways including a mechanism similar to that used for fatty acid oxidation. The final metabolic products of these pathways are unstable or volatile compounds that were not toxic to CR3. Strain CR3 was also shown to have tolerance to at least 10 heavy metals, which was mainly achieved by self-detoxification through ion efflux, metal-complexation and metal-reduction, and a powerful DNA self-repair mechanism. Our genomic analysis suggests that CR3 is well adapted to survive the harsh environment in natural asphalts containing naphthenic acids and high concentrations of heavy metals.
Introduction
Members of the genus Cupriavidus are Gram-negative β-proteobacteria that are commonly isolated from environmental and human clinical sources [1]. Cupriavidus has a complex taxonomic history, which was addressed by Vandamme and Coenye in 2004 [2]. The genus Cupriavidus currently is comprised of 11 species derived from diverse ecological niches [3]. These include several heavy metal tolerant bacteria such as C. metallidurans CH34 [4], C. necator N-1 [5], C. pinatubonensis JMP134 [6], and C. taiwanensis [7]. Several genetic determinants for heavy metal resistance have been identified in Cupriavidus [8–9] including the czc, pbr, and mer clsusters, and are present in several members of the genus [8–14]. The strain with the highest number of heavy metal resistance loci is CH34 tolerating up to milli-molar concentrations of over 20 different heavy metal ions [4, 9]. Various Cupriavidus sp. also have the ability to degrade a range of different aromatic hydrocarbons. Strain JMP134 is a well studied pollutant-degrading bacterium [6, 14], in which pathway predictions constructed for aromatic compounds are based in multiple peripheral ring cleavage pathways and associated central reactions [15]. Three other bacteria, C. eutropha H16, C. necator N-1 and C. taiwanensis are reported to effectively degrade bioplastics [5, 16–17]. Another representative of this taxon, C. metallidurans strain KUA-1, can degrade the standard reference naphthenic acid, cyclohexylacetic acid (CHAA) [18].
Naphthenic acids (NAs) are complex mixtures of hydrocarbons that are predominantly comprised of saturated aliphatic and alicyclic carboxylic acids [19]. As natural components of crude petroleum, NAs range from undetectable to up to 4% concentration by weight [20]. However, NAs are thought to be the most toxic components of both refinery effluent and oil sands-processing wastewaters, and pose significant environmental and human health risks [21–22]. Microbial biodegradation has been seen as a promising solution for treatment of NA mixtures [23], and a good biodegradation efficiency has been reported for trans-4-methyl-1-cyclohexane carboxylic acid (trans-4MCHCA) [24–25]. Prior studies have shown that diverse bacteria belonging to the genera Corynebactrium, Arthrobacter, Acinetobacter, Alcaligenes, Pseudomonas, and Bacillus [26–31] utilize different commercial NAs as sole sources of carbon and energy. In contrast, only a few strains of Cupriavidus that are capable of degrading NAs have been identified to date. To our knowledge, there is no pathway associated with degradation of this naphthenic acid in Cupriavidus. Although strain KUA-1 has been reported to degrade cyclohexylacetic acid [18], the degradation pathway and its substrate versatility remain largely unknown.
Here, we report the complete genome sequence and analysis of a novel C. gilardii strain CR3, which provides the foundation for elucidating the mechanisms for naphthenic acid biodegradation and heavy metal resistance. Trans-4MCHCA was chosen to elaborate the bacterial degradation pathways. Genomic analysis of C. gilardii CR3 indicated that this bacterium degraded the cyclohexanecarboxylic acid via a ring-cleavage pathway [23, 30]. The acyclic-open chain compound was then further oxidized by β-oxidation, which involves a fatty acid β-oxidation module. We propose a probable degradation pathway and corresponding versatility. Besides its ability to degrade naphthenic acid, metal resistance was also identified in CR3. This strain is tolerant to no less than ten heavy metals, which was mainly achieved by high-performance ion efflux, metal-complexation and metal-reduction, and a powerful self-repair mechanism.
Materials and Methods
Isolation of strain CR3
C. gilardii CR3 was isolated from the Rancho La Brea Tar Pits 91, Los Angeles, CA, USA. The geographical coordinates is Lat 34° 3' 46.1268'' N, Lon -118° 21' 21.1716'' W. Kim and Crowley completed a comprehensive survey of this same location earlier, which included a characterization of its chemical properties and microbial diversity [32]. Samples of asphalt-bearing soil were obtained by scraping away the upper surface of the asphalt with a sterile knife and were placed into sterile plastic tubes and transported to the laboratory where they were stored at 4°C until processing. For isolation of NA-biodegrading bacteria, 95 mL of sterilized Milli-Q water were added to 5 g of asphalt-bearing soil in a 250 mL Erlenmeyer flask and shaken for 24 h. The resulting cell suspension culture was spread onto MSM agar plates amended with trans-4-methyl-1-cyclohexane carboxylic acid (final concentrations of 150 mg L-1), and incubated for 5 days at 28°C. One strain, designated as strain CR3, was selected on the basis of its rapid growth on trans-4-methyl-1-cyclohexane carboxylic acid. Individual colonies were selected randomly and purified by single colony isolation after triple re-streaking on LB agar medium. The strain also grows on tryptic soy medium.
Isolation identification and extraction of genomic DNA
Identification of strain CR3 was performed based on morphological, physiological, and phylogenetic characteristics. The morphological and physiological characteristics of the strain were performed based on Bergey’s manual of systematic bacteriology (second edition, 2004). To determine the 16S rRNA gene sequences and to establish DNA libraries for genome sequencing, the genomic DNA of strain CR3 was extracted using a DNA extraction kit (TakaRa DNAiso Reagent code: D305A (TaKaRa Biotechnology Company Limited, Dalian, China)). To confirm the identities of the isolates, PCR amplification and sequencing of the 16S rRNA gene were performed. The 16S rRNA genes were PCR-amplified from the genomic DNA using the bacterial universal primer set 27f (5'-AGA GTT TGA TCC TGG CTC AG-3') and 1492r (5'-GGC TAC CTT GTT ACG ACT T-3'). The 16S rRNA gene sequence of strain CR3 was compared with those in EzTaxon-e database (http://www.ezbiocloud.net/eztaxon) [33] and GenBank database (http://www.ncbi.nlm.nih.gov/blast) using BLAST [34] to identify closely related bacteria.
Genome sequencing and genome assembly
In this study, CR3 cells were sequenced by Illumina HiSeq2000 (Illumina, San Francisco, USA) and Pacific Biosciences II (Pacific Biosciences, San Francisco, USA) sequencing technologies. Three DNA libraries for Illumina sequencing were constructed, including a 180 bp pair-end library, and 1–2 kb and 3–5 kb mate-pair libraries. We generated 50,065,980 pair-end reads (2×101 bp), and 128,546,539 mate-pair reads (2×101 bp). High quality of sequencing reads is a prerequisite for good genome assembly. To this end, we carried out a filtering process to filter low quality reads (reads with excess “N” and low quality score), duplication reads, and adaptor contamination reads. After filtering, three libraries yielded approximately 3126-fold coverage by high quality reads. High quality reads were assembled by SOAPdenovo v2.04 [35]. Inner gaps that emerged in the scaffolding were filled with GapCloser [36]. High quality HiSeq reads were assembled into 21 contigs and 4 scaffolds. We used Pacific Biosciences SMART analysis software 1.2 to generate long ‘filtered sub-reads’ from the instrument, which are high quality reads. Filtered sub-reads were generated through the following primary analysis: removal of adaptors, removal of low quality bases, and removal of short reads. Here, we call sub-reads as ‘reads’, and never refer to raw reads. After filtering, SMRT cell sequencing generated 52,939 long reads, and the sum of bases was 296,078,365 bases. Average read length was up to 5,592 bp. SMRT cells yielded approximately 44-fold coverage. All PacBio reads were assembled into 2 polished contigs using HGAP v3 [37]. One of them was able to be direct circularization. The other one contained a circularization gap. Circularization gap was filled with PBJelly [38]. The filled gap sequence errors were corrected using HiSeq assembly results. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession CP010516-CP010517.
Genome annotation and analysis
Open reading frames (ORFs) were predicted using GeneMarks [39] and Glimmer 3.0 [40]. The predicted ORFs were validated by BLAST [34], which was used to search for homologous genes. Protein domains were scanned by InterProScan [41]. Functional genes that were investigated as having possible roles in metabolic pathways were reconstructed by KAAS (a module of KEGG) [42]. Phylogenetic classification of proteins that are encoded in the CR3 genome were based on functions of clusters of orthologus groups (COG) [43]. Transfer RNA (tRNA) genes in the genomic sequence were detected by tRNAscan-SE [44]. Ribosomal RNAs were searched using RNAmmer [45].
Seven published genome sequences of Cupriavidus strains were collected (published before 2013) to analyze species phylogeny and to generate a synteny plot. These strains included C. pinatubonensis JMP134 [6], C. metallidurans strain CH34 [4], C. necator N-1 [5], C. taiwanensis [7], C. eutropha H16 [17], R. eutropha HPC(L) and C. basilensis OR16 [11]. We also gathered other public and completed 16S rRNA genes from NCBI to present a full taxonomic analysis. A rooted phylogenetic tree based on 16S rRNA gene sequence similarity of the genera Cupriavidus was performed using Mega 6.0 [46]. Cluster analysis was based on the neighbour-joining method with the closely related bacterium Ralstonia pickettii 12D [2] as the outgroup root. Synteny plots were analyzed by Mauve v2.3.1 [47]. Rooted phylogenetic tree based on gene gain and loss was performed by PGAP [48]. Phylogenetic trees were visualized by TreeView [49]. Ortholog gene clusters were identified by PGAP [48]. Core genes were identified as genes shared by all strains. Unique genes were identified as genes that were unique to one strain. Likewise, replicon chr1 core genes were identified as genes shared by all strains and limited in replicon chr1. Replicon chr1 unique genes were identified as genes that were unique to one strain and limited to replicon chr1. The same criteria were applied to replicon chr2 core genes and replicon chr2 unique genes. Functional distribution over chr1 and chr2 was reflected by the relative ratio of COG classes, which represented the normalized ratio of percentages of CDS numbers per replicon in a COG class [4]. Unique genes and genome genes in each GO category were counted. A statistical p value of unique genes enriched in each GO category was calculated by the chi-square test. GO functions having fewer than 3 genes were removed and GO functions in which the percent of unique genes was 1% or more less than the percent of the genome genes were also removed. The unique-gene percent was defined as the percent of unique genes in a GO function divided by the total of all unique genes. The genome gene percent was defined as the percent of genome genes in a GO function divided by the total of all genome genes. GO enrichment was considered significant when p values were <0.05. In the KEGG pathway enrichment analysis, pathways were removed if the gene count in CR3 was less than 5 or if the gene count discrepancy between CR3 and the maximum gene count of other 7 Cupriavidus strains was less than 5. Unique-gene pathway enrichment was selected as above by GO function enrichment analysis. COG enrichment analysis of unique genes was also performed as above by GO function enrichment analysis. Possible enzyme-catalyzed metabolic pathways for multi-step reactions involved in naphthenic acid degradation were predicted by PathPred [50], which was based on the local RDM pattern (KEGG atom type changes at the reaction center (R), the difference region (D), and the matched region (M) for each reactant pair.) match and the global chemical structure alignment. To analyze enzyme-encoding gene organization, we counted enzyme-encoding gene percent that genes located at gene clusters. Genes that genomic locations were adjacent formed a gene cluster.
Determination of minimum inhibitory concentration (MIC)
MIC values for nine heavy metals (Ag, Cd, Co, Cr, Cu, Hg, Ni, Pb and Zn) against C. gilardii CR3 were determined using broth cultures. The 1 M stock solutions of heavy metal salts were made from using analytical grade chemicals and were sterilized by filtration. MIC was firstly determined using a two-fold dilution method, which generated an appropriate MIC range. Afterwards serial continuous concentration dilution experiments were carried out to identify specific MIC values based on the pre-defined MIC range. The media used for MIC was Tris-buffered mineral salts medium containing 0.2% (m/m) sodium gluconate adopted by Mergeay et al. [51]. The optical density of cultures was measured at 600nm (OD600) using a UV-visible spectrometer (T6, Beijing Purkinje General Instrument Co. Ltd., Beijing, China). Pre-cultured C. gilardii CR3 (OD600 = 1) was inoculated to a tube with media, cultivated with shaking at 160rpm (30°C) for 12h. Heavy metal solutions (4% weight/volume) were added to the corresponding tubes, and the cells then further cultivated for 96h. Cultures containing media inoculated with C. gilardii CR3 but without heavy metals served as the controls. The OD600 of the cultures were measured from 0h to 96h at 12 h intervals.
Results and Discussion
Cupriavidus strains are well known for their ability to inhabit environments containing toxic heavy metals, especially those containing Cu2+, which lead to their genus name (i.e. “copper-loving”). Members of this genus also are able to degrade many different complex compounds [5–6, 15–18], such as polyhydroxybutyrate, which is degraded by strain H16 [17], chloroaromatics degraded by strain JMP134, mainly based on the β-ketoadipate pathway [15], and CHAA which can be degraded by strain KUA-1 [18]. However, the degradation pathways for these substances are unclear and their normal substrates are largely unknown. C. gilardii strain CR3, studied here, was isolated from a natural asphalt seep located at the Rancho La Brea Tar Pits (Los Angeles, CA), and was shown to use 4MCHCA and other similar molecules with branched alkyl groups (-(CH2)mCH3) as sole sources of carbon and energy for growth. This has never been shown for other strains of Cupriavidus. This establishes strain CR3 as a potential microbial agent to biodegrade either model NAs or commercially available NA compounds, which are among the most toxic components in both refinery effluent and oil sands process wastewaters, and also establishes this strain as a petroleum degrading bacterium with heavy metal resistance.
Isolation and description of Cupriavidus gilardii CR3
Four microorganisms capable of using trans-4MCHCA as a sole carbon and energy for growth were isolated from the Lar Brea Tar Pits (Los Aageles, CA). From these, we finally selected strain CR3 for determination of its naphthenic acid biodegradation pathway based on its high growth rate on this carboxylic acid. Cells of strain CR3 were observed to be aerobic, rod-shaped, and Gram-negative. The colonies produced by this bacterium (3–5 mm diameter) were smooth, circular convex, wet, and light yellowish in color for cells grown on LB agar medium for 2 days at 28°C. The strain was positive for catalase and oxidase.
The 16S rRNA gene sequence (1520 bp) of strain CR3 (NCBI GeneBank accession JX945578) was obtained. The BLAST search results based on EzTaxon and GenBank databases both indicated that strain CR3 belonged to Cupriavidus. The 16S rRNA gene sequence of strain CR3 was 100% identical to C. gilardii CIP 105966. Morphological, physiological and phylogenetic properties all suggested that strain CR3 was C. gilardii.
Genome structure and general features
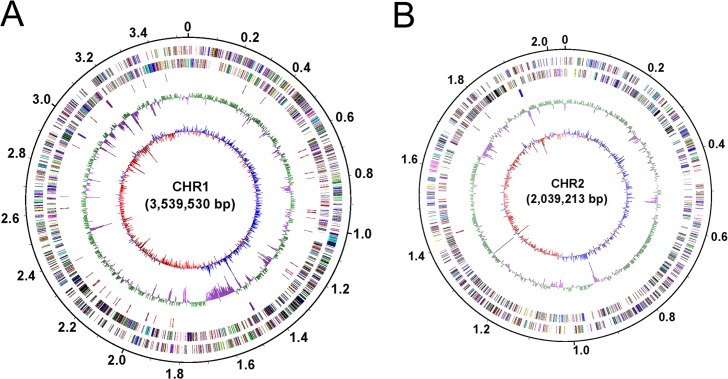
The complete genome of CR3 contained 2 circular chromosomes without any inner gaps. The size of chromosome 1 (chr1) was 3,539,530 bp and chromosome 2 (chr2) was 2,039,213 bp, and summed to 5,578,743 bp. The GC content of CR3 genome was 67.55%. A total of 4,502 putative coding sequences (CDSs) were validated by homology search. The complete genome contained 59 tRNA genes and 4 rrf-rrl-trnI-trnA-rrs operons (with rrf, rrl, and rrs coding for respective 5S, 23S, and 16S rRNA). There were 3,862 CDSs that were assigned to one or more function classes (Fig 1, Table 1). The GC content of two replicons was very similar (Fig 1). Approximately 86% of 4,501 CDSs could be assigned a putative function based on function annotation. The general features of two replicons were displayed in Table 1.
Fig 1. Circular representation of the two replicons of C. gilardii CR3.
(A). Chromosome 1; (B). Chromosome 2. Circles display from the inside outwards, (1) circle 1: GC-skew (G-C/G+C ratio) using a 3 kb window; (2) circle 2: GC-content using a 3 kb window; (3) circle 3: RNA genes; (4) circle 4: COG assignments for predicted CDSs on the minus strand; (5) circle 5: COG assignments for predicted CDSs on the plus strand; (6) circle 6: scale in Mb.
Table 1. General features of the C. gilardii CR3 genome.
| Chromosome 1 | Chromosome 2 | Genome | |
|---|---|---|---|
| Size (bp) | 3,539,530 | 2,039,213 | 5,578,743 |
| GC content | 67.40% | 67.81% | 67.55% |
| tRNA | 52 | 7 | 59 |
| rRNA operons | 3 | 1 | 4 |
| Total number of CDSs | 2,874 | 1,627 | 4,501 |
| Overlapping CDSs | 545 | 342 | 887 |
| CDSs with assigned function | 2,504 | 1,358 | 3,862 |
| Hypothetical proteins | 370 | 269 | 639 |
| CDSs assigned to COGs | 2,130 | 1,104 | 3,234 |
| CDSs assigned to KEGG Ontology | 1,687 | 698 | 2,385 |
| CDSs assigned to GO function | 1,740 | 923 | 2,663 |
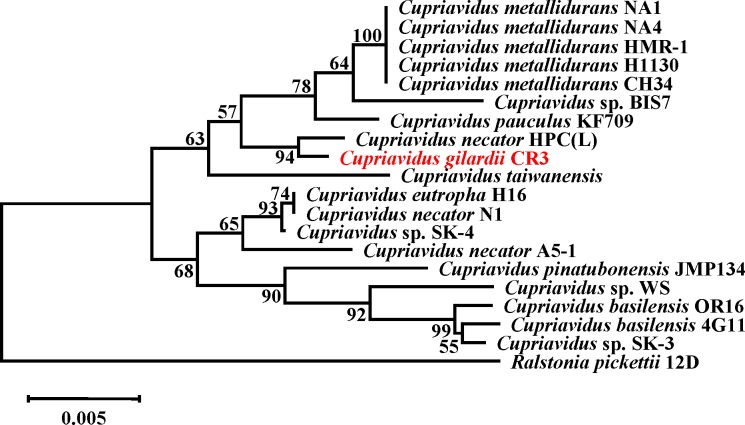
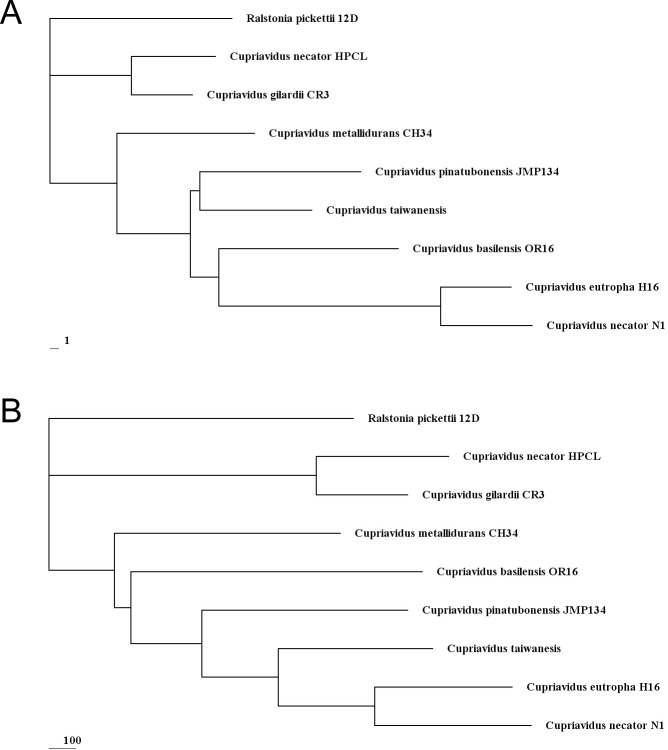
Phylogenetic analyses based on 16S rRNA gene sequences suggested C. gilardii CR3 fit firmly into the genus Cupriavidus strain. The result indicated that its closest relative was C. necator HPC (L) (Fig 2). The relationship between the CR3 and HPC (L) strains was also shown in their genome composition analysis. As shown in Table 2, other Cupriavidus genome sizes [4–7, 11, 17] range from 5.49 Mb to 8.55 Mb, and the smallest genome is HPC (L); CR3 comes second. Whereas, GC content of all these other members of Cupriavidus varies from 63.53% to 67.54%, and the highest GC content genome is HPC (L). And strain CR3 has the GC content at 67.55%. Gene number in other genomes ranges from 4,990 to 7,915, and the fewest gene number genomes is HPC (L). And strain CR3 contained only 4,572 genes. Both results of GC content and gene number indicate the close relationship between CR3 and HPC (L). Moreover, the CR3 genome was composed of only two chromosomes, while other finished genomes are composed of two chromosomes and at least one plasmid.
Fig 2. Rooted phylogenetic tree based on 16S rRNA gene sequence similarity of the genus Cupriavidus.
Cluster analysis was based upon the neighbour-joining method with Ralstonia pickettii 12D as the outlier. The scale bar represents 5‰ sequence divergence. Numbers at branch-points are percentages of 1000 bootstrap resamplings that support the tree topology.
Table 2. Genome structure comparison between CR3 and seven other published Cupriavidus bacteria.
| Bacterium | Status | Genome Size (M) | GC content (%) | Chr | Plasmid | Protein | rRNA | tRNA | Total | Reference or download source |
|---|---|---|---|---|---|---|---|---|---|---|
| Cupriavidus eutropha H16 | Finished | 7.42 | 66.34 | 2 | 1 | 6,626 | 15 | 58 | 6,699 | [17] |
| Cupriavidus taiwanensis | Finished | 6.48 | 66.99 | 2 | 1 | 5,896 | 15 | 63 | 5,974 | [7] |
| Cupriavidus necator N-1 | Finished | 8.48 | 65.47 | 2 | 2 | 7,832 | 15 | 68 | 7,915 | [5] |
| Cupriavidus metallidurans CH34 | Finished | 6.91 | 63.53 | 2 | 2 | 6,477 | 12 | 62 | 6,551 | [4] |
| Cupriavidus pinatubonensis JMP134 | Finished | 7.26 | 64.43 | 2 | 2 | 6,446 | 18 | 66 | 6,530 | [6] |
| Cupriavidus basilensis OR16 | Draft | 8.55 | 65.39 | - | - | 7,534 | 5 | 61 | 7,600 | [11] |
| Cupriavidus necator HPC(L) | Draft | 5.49 | 67.54 | - | - | 4,931 | 6 | 53 | 4,990 | ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria_DRAFT/Cupriavidus_necator_HPC_L__uid180939/ |
| Cupriavidus gilardii CR3 | Finished | 5.58 | 67.55 | 2 | 0 | 4,501 | 12 | 59 | 4,572 | This study |
To explore the genome structure of strain CR3, we carried out synteny analysis on CR3 and all other reported Cupriavidus genomes. CR3 genome sequences clearly showed synteny in chr1 with other finished strains (Fig 3A) but lack of synteny in chr2 (S1 Fig). The synteny phenomenon is similar to JMP134 [6]. The sequence in the region chr1:1,518,182–1,620,123 of CR3 showed some successive synteny with a plasmid of CH34 (pMOL30) (Fig 3B), which has not been previously reported in other Cupriavidus strains. The synteny analysis reveals conservation of chr1 sequence composition since it's an ancestral replicon [4, 6]. Plasmid fragment recombination into chr1 was generally incompact, whereas the synteny region between CR3 chr1 and CH34 plasmid pMOL30 tended to be relatively concentrated. Moreover, CH34 was the closest relative to CR3 among the five finished Cupriavidus strains to which it was compared. The lack of synteny in the smaller replicon chr2 probably originated from ancestral plasmids during the evolution of Cupriavidus and appeared to have evolved with frequent recombination, gene duplication and large numbers of transfered genes [4, 6]. The chr2 replicon displayed conserved repA-parA-parB (CR3_2936, 2938, 2939) gene structure, implying that it may have derived from plasmid-like partitioning [4–7, 17]. This indicates that the plasmid fragment recombination mechanism in replicon chr1 and replicon chr2 is quite different, not only in recombination frequency, but also in the source of plasmid fragments.
Fig 3. Synteny plot analysis of Cupriavidus replicons.
(A) Synteny plot analysis of Chr1 from C. galardii CR3 and from 5 other finished Cupriavidus strains; (B) Synteny plot analysis of Chr 1 from C. galardii CR3 and pMOL30 from C. metallidurans CH34.
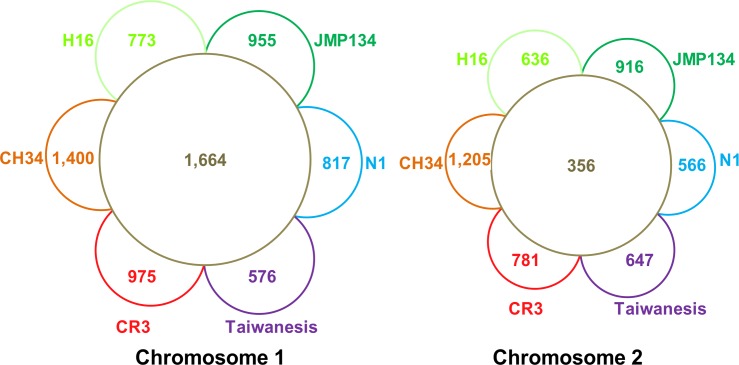
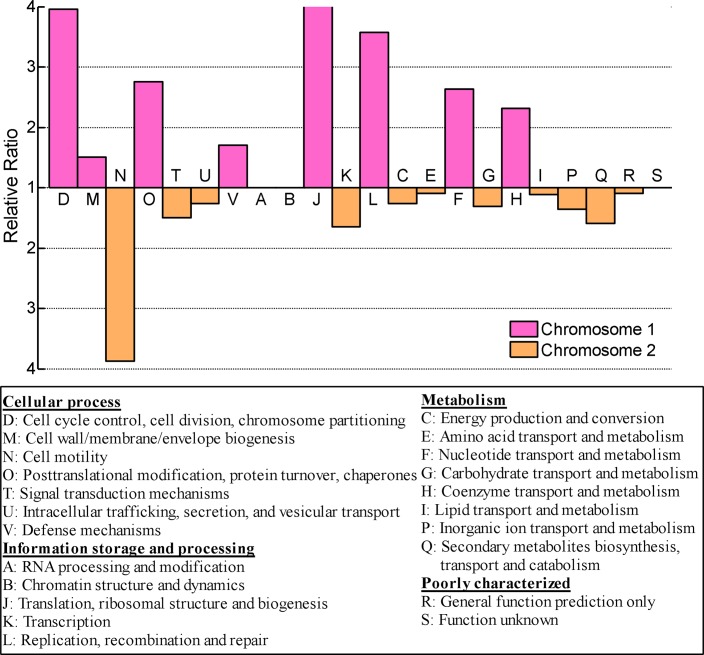
Gene distribution analyses also indicated that CR3 chr1 constitutes the ancestral replicon. Compared with five other finished Cupriavidus strains, the core gene number in chr1, 1,672, was almost 5X larger than chr2, 356 (Fig 4). For each replicon in CR3, replicon core genes accounted for 58.91% chr1 genes and 22.28% chr2 genes respectively. Replicon unique genes accounted for 32.66% chr1 genes and 46.81% chr2 genes. In addition, chr2 replicon contained several genes paralogous to chr1 genes, such as membrane protein ompC (CR3_4033), cobalt-zinc-cadmium resistance protein czcB (CR3_3629), ABC transporter tauB (CR3_3580), aromatic amino acid aminotransferase tyrB (CR3_3642), and others. The core genes were generally associated with housekeeping gene functions, while the unique genes were associated with special features. The diversity in core gene number and unique gene number distribution in the two replicons also reflected the diversity of gene function distribution. In strain CR3, the main replicon chr1 carried most of the essential housekeeping gene functions, including those for translation, ribosome production, DNA replication, DNA repair, protein processing, cell cycle, cell division and cell component etc. (Fig 5). In contrast, the smaller replicon chr2 carried functions mainly related to adaptation and survival, including signal transduction, energy metabolism and ion transduction. The bias of replicon function distribution in CR3 is more obvious than in CH34. CR3 chr2 consists of more genes relevant to energy metabolism than chr1 (classes C, E, G), while CH34 does not [4].
Fig 4. Comparative analysis of gene distribution in two chromosomes.
Inner ring shows the number of ortholog gene clusters that are shared by all species based on PGAP result. Outer semi-ring shows the number of ortholog gene clusters that are unique to one strain based on PGAP result. Abbreviation: CR3, C. gilardii CR3; CH34, C. metallidurans CH34; H16, C. eutropha H16; JMP134, C. pinatubonensis JMP134; N1, C. necator N1; Taiwanesis, C. taiwanesis.
Fig 5. Functional distribution over two replicons based on COG classification.
The scale represents the relative ratio of CDS numbers per gene source (i.e. the ratio of percentages for each class per replicon).
Compared with seven other Cupriavidus strains, 897 genes were unique in CR3. To explore the specific character of CR3, we carried out GO function enrichment analysis (S2 Fig), KEGG pathway analysis (S3 Fig), and COG function classification enrichment analysis (S4 Fig) of whole genome genes and unique genes in CR3. Function annotation analysis revealed many of these unique genes in CR3 were enriched in genes relevant to biodegradation of complex compounds, signal interaction with its surroundings, and environmental adaption, which were speculated to be associated with its NAs biodegradation potential and heavy metal resistance.
Naphthenic acid degradation
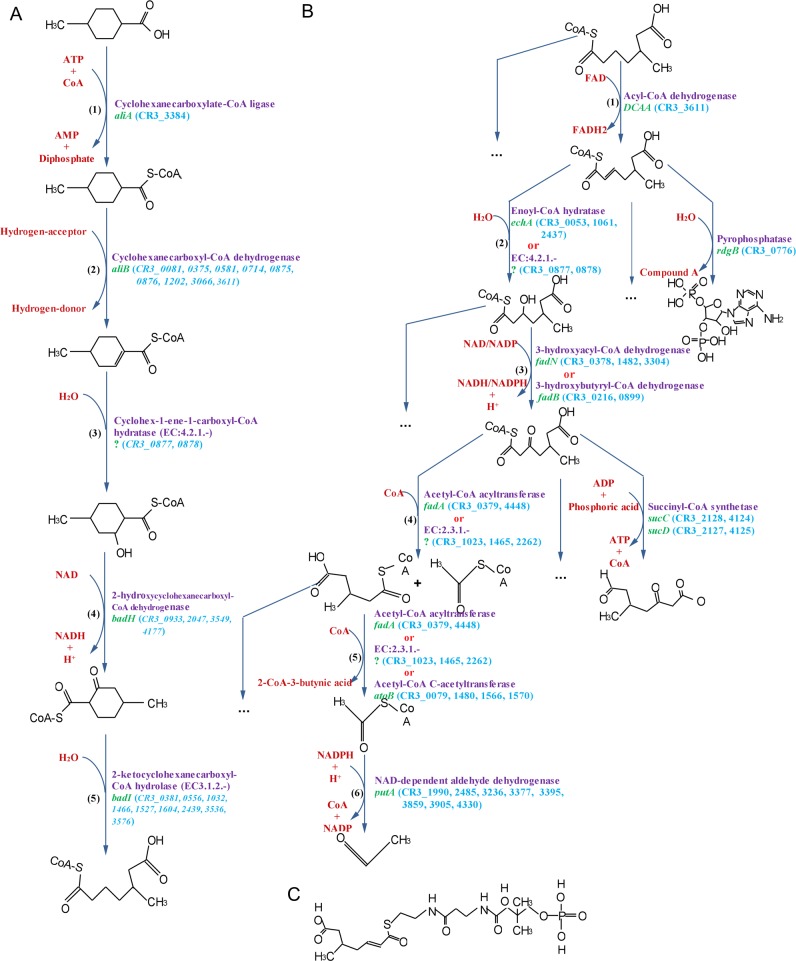
Strain CR3 could use 4MCHCA and other compounds having different numbers of branched alkyl groups (-(CH2)mCH3) as sole carbon and energy sources for growth. 4MCHCA is a monocyclic saturated carboxylic acid and a general representative of nonaromatic naphthenic acids (NAs). NAs biodegradation by CR3 was observed to be an aerobic process. The metabolic pathways for 4MCHCA degradation in strain CR3 were predicted by PathPred [50], which uses a reactant pair library to obtain plausible pathways of multi-step reactions. There were 17 matched aerobic degradation pathways, which contained two main processes: the peripheral ring-cleavage step and central oxidative degradation. The peripheral ring-cleavage process in CR3 was similar to the proposed process for aerobic oxidation of cyclohexane carboxylic acid by aerobic oxidation pathways in Pseudominas putida [30] (Fig 6A). The central pathways functioned for transformation of acyclic-open chain compounds into a few volatile or unstable compounds, yielding products such as acetaldehyde and adenosine compounds, which were subsequently oxidized via core metabolic pathways. The degradation mechanism was consistent for NAs with different branched alkyl groups in CR3.
Fig 6. Predicted naphthenic acid degradation pathway in CR3.
(A) Peripheral ring-cleavage pathway; (B) Central oxidative degradation pathways; (C) The structure of compound A. Compounds in red are substrates. Enzyme genes are marked on the right of arrows, including gene description (purple color), gene symbol (green color) and gene identifications (light blue color). If CR3 genome only encoded isoenzyme gene, gene identifications were marked in italic. Isoenzyme gene were defined as a gene assigned to a same enzyme class with the same EC NO. And enzyme genes with unknown gene symbol were marked as ‘?’.
The peripheral ring-cleavage pathway was proposed to include five main reactions (Fig 6A, Table 3) as follows: 1) Acylation: acyl-CoA connected to 4MCHCA, and substituted carboxylic acid in the catalysis of cyclohexanecarboxylate-CoA ligase aliA (CR3_3384). 2) Vinylation: the saturated bond at the α position can be destroyed under the action of the cyclohexanecarboxyl-CoA dehydrogenase aliB (isoenzyme genes CR3_0081, 0375, 0581, 0714, 0875, 0876, 1202, 3066, 3611), forming an unsaturated ethylenic bond with double bonds. 3) Alcoholization: 4-methyl-cyclohex-1-ene-carboxyl-CoA can be transformed to 4-methyl-2-hydroxycyclohexane-carboxyl-CoA by the enzyme cyclohex-1-ene-carboxyl-CoA hydratase (EC: 4.2.1.-, isoenzyme genes CR3_0877, 0878), which destroys the ethylenic bond and attaches a hydroxyl group to the molecule. The production of a hydroxylated intermediate is a general metabolic process that is commonly involved in aerobic degradation of NAs. 4) Aldehyde: 4-methyl-2-hydroxycyclohexane-carboxyl-CoA can be transformed to yield 4-methyl-2-ketocyclohexane-carboxyl-CoA under the action of the 2-hydroxycyclohexane-carboxyl-CoA dehydrogenase badH (isoenzyme genes CR3_0933, 2047, 3549, 4177), which oxidized the hydroxyl group into an aldehyde. 5) Ortho-ring cleavage: 4-methyl-2-ketocyclohexane-carboxyl-CoA can be transformed to the compound 5-methyl-6-carboxy-hexanoyl-CoA under the action of the enzyme 2-ketocyclohexanecarboxyl-CoA hydrolase badI (isoenzyme genes CR3_0381, 0556, 1032, 1466, 1527, 1604, 2439, 3536, 3576), then the saturated ring can be converted into an acyclic-open chain. The participation of a variety of enzymes through a series of reactions would cleave the saturated NA ring to an acyclic-open chain. Subsequently, the acyclic-open chain can be further oxidized into a few volatile and unstable compounds.
Table 3. NAs degradation genes.
| Process | Step | Gene symbol | EC No. | Definition | Gene(s) identification NO. | Isoenzyme gene(s) identification NO. 1 |
|---|---|---|---|---|---|---|
| Ring-cleavage | Acylation | aliA | EC:6.2.1.- | Cyclohexanecarboxylate-CoA ligase | CR3_3384 | CR3_0434, 0592, 1107, 1488, 2610, 3988 |
| Vinylation | aliB | EC:1.3.99.- | Cyclohexanecarboxyl-CoA dehydrogenase | / | CR3_0081, 0375, 0581, 0714, 0875, 0876, 1202, 3066, 3611 | |
| Alcoholization | - 2 | EC:4.2.1.- | Cyclohex-1-ene-1-carboxyl-CoA hydratase | / | CR3_0877, 0878 | |
| Aldehyde | badH | EC:1.1.1.- | 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase | / | CR3_0933, 2047, 3549, 4177 | |
| Ortho-ring cleavage | badI | EC:3.1.2.- | 2-ketocyclohexanecarboxyl-CoA hydrolase | / | CR3_0381, 0556, 1032, 1466, 1527, 1604, 2439, 3536, 3576 | |
| Acyclic-open chain degradation | Vinylation | DCAA | EC:1.3.99.- | Acyl-CoA dehydrogenase | CR3_3611 | CR3_0081, 0375, 0581, 0714, 0875, 0876, 1202, 3066 |
| Alcoholization | echA | EC:4.2.1.17 | Enoyl-CoA hydratase | CR3_0053, 1061, 2437 | / | |
| - 2 | EC:4.2.1.- | - | CR3_0877 CR3_0878 | / | ||
| Aldehyde | fadN 3 | EC:1.1.1.35 | 3-hydroxyacyl-CoA dehydrogenase | CR3_0378, 1482, 3304 | / | |
| fadB | EC:1.1.1.157 | 3-hydroxybutyryl-CoA dehydrogenase | CR3_0216, 0899 | / | ||
| Deacetylation | fadA | EC:2.3.1.16 | Acetyl-CoA acyltransferase | CR3_0379, 4448 | / | |
| - 2 | EC:2.3.1.- | - | CR3_1023, 1465, 2262 | CR3_0137, 0138, 0139, 1571, 2147, 2925, 2964, 4321 | ||
| Further deacetylation | fadA | EC:2.3.1.16 | Acetyl-CoA acyltransferase | CR3_0379, 4448 | / | |
| - 2 | EC:2.3.1.- | - | CR3_1023, 1465, 2262 | CR3_0137, 0138, 0139, 1571, 2147, 2925, 2964, 4321 | ||
| atoB | EC:2.3.1.9 | Acetyl-CoA C-acetyltransferase | CR3_0079, 1480, 1566, 1570 | / | ||
| Deacylation | putA 4 | EC:1.2.1.3 3 | NAD-dependent aldehyde dehydrogenase 4 | CR3_1990, 2485, 3236, 3377, 3395, 3859, 3905, 43304 | / |
1. Isoenzyme genes were defined as a gene assigned to a same enzyme class with the same EC NO.
2. Gene symbol was unknown.
3. The fadN gene was assigned to a same enzyme class and definition as HADH gene, except KEGG ko orthology.
4. The CR3 genome did not encode an acetaldehyde dehydrogenase (EC1.2.1.10), but encoded week specific enzyme of aldehyde dehydrogenase (EC1.2.1.3).
We identified several possible central pathways to transform 5-methyl-6-carboxy-hexanoyl-CoA into a few volatile and unstable compounds. For example, the end-product of acetaldehyde metabolism in CR3 was predicted to pass through following reactions (Fig 6B and 6C, Table 3): 1) Vinylation: 2-position saturated bond of 5-methyl-6-carboxy-hexanoyl-CoA can be destroyed under the action of the acyl-CoA dehydrogenase DCAA (CR3_3611), forming an unsaturated ethylenic bond with double bonds. 2) Alcoholization: involving either enoyl-CoA hydratase echA (CR3_0053, 1061, 2437) or a type of hydro-lyases enzyme (EC:4.2.1.-, CR3_0877 CR3_0878), the double ethylenic bond in 2-ene-5-methyl-6-carboxy-hexanoyl-CoA can be destroyed, while an hydroxyl was connected. 3) Aldehyde: 2-position hydroxyl can be oxidized into aldehyde, which may be mediated by 3-hydroxyacyl-CoA dehydrogenase fadN (isoenzyme gene of HADH gene, CR3_0378, 1482, 3304) or 3-hydroxybutyryl-CoA dehydrogenase fadB (CR3_0216, 0899). 3-hydroxyl-5-methyl-6-carboxy-hexanoyl-CoA can be oxidized into 3-aldehyde-5-methyl-6-carboxy-hexanoyl-CoA. 4) Deacetylation: acetyl-CoA can be removed from the long chain compound 3-aldehyde-5-methyl-6-carboxy-hexanoyl-CoA under a catalytic reaction mediated by acetyl-CoA acyltransferase fadA (CR3_0379, 4448) or a type of acyltransferases enzyme (EC:2.3.1.-, CR3_1023, 1465, 2262) to generate 3-methyl-4-carboxy-hexanoyl-CoA and acetyl-CoA. Acetyl-CoA could then be degraded in step 6). 5) Further deacetylation: acetyl-CoA can be further broken from 3-methyl-4-carboxy-hexanoyl-CoA via the enzyme acetyl-CoA acyltransferase fadA (CR3_0379, 4448), or a type of acyltransferases enzyme (EC:2.3.1.-, CR3_1023, 1465, 2262), or acetyl-CoA C-acetyltransferase atoB (CR3_0079, 1480, 1566, 1570). 6) Deacylation: acetyl-CoA could be degraded into acetaldehyde by the enzyme NAD-dependent aldehyde dehydrogenase putA (CR3_1990, 2485, 3236, 3377, 3395, 3859, 3905, 4330), which is a highly volatile compound. The end product was not toxic to CR3. The acyclic-open chain degradation pathway is similar to β-oxidation module in fatty acid metabolism. Multiple plausible pathways and multiple genes encoding this suite of enzymes permit CR3 to degrade NAs for use as a carbon and energy source for growth.
Likewise, there were 18 plausible reaction pathways to degrade 4-pentyl-cyclohexane carboxylic acid. The distinction between the two broad groups of metabolic pathways mainly involved different enzymes that are used to catalyze branched alkyl groups levels for different NAs. The central pathway degraded nonaromatic NAs consisted of two processes: ring cleavage and further oxidation to volatile compounds. These two processes both included four similar steps, which were dehydrogenation to form an ethylenic bond, a hydratase to produce alcohol, oxidation of the alcohol to an aldehyde, and finally a bond break to release the aldehyde. The ring-cleavage pathways identified here were consistent with the previously proposed cyclohexanecarboxylic acid degradation pathway [23, 30], in which multiple genes encoded the same reaction enzymes or isoenzymes. However, the enzyme specificity might be not particularly specific. In summary, multiple pathways and a variety of optional enzymes for degradation of NAs by strain CR3 guarantee its metabolic capacities in environments where a variety of hydrocarbons may serve as primary substrates for growth.
Cupriavidus species are able to degrade many different complex compounds [5–6, 15–18]. However, the degradation pathways have been unclear and the substrate is largely unknown. C. gilardii strain CR3 was able to use 4MCHCA and other similar molecules with branched alkyl groups (-(CH2)mCH3) as sole sources of carbon and energy for growth. This has never been found in any other strains of Cupriavidus. In an evolutionary tree based on gene gain and-loss, the branch length reflects the occurrence frequency of gene gain and-loss. The difference of occurrence frequency (gain and-loss) of genes related to NAs biodegradation (Fig 7A) was far more significant than that of genome-wide genes (Fig 7B). We suggest that NAs biodegradation capabilities in Cupriavidus have changed dramatically during evolution. The occurrence of gene gain and-loss in CR3 related to genes for enzymes participating in NAs biodegradation were less than in other Cupriavidus (Fig 7A). Moreover, enzyme-encoding genes in CR3 frequently occurred in gene clusters (two or more genes are adjacent) and involved 25.00% of these genes, while in other Cupriavidus strains, only 20.41% ~ 26.15% of enzyme encoding genes were clustered. Conserved enzyme genes in CR3 might increase the NAs degradation rate through a more compact gene organization to achieve genome structure optimization. Altogether strain CR3 is a good candidate for study of degradation pathways for model NAs or commercially available NA compounds.
Fig 7. Rooted phylogenetic tree based on gene gain and loss.
Cluster analysis was based upon the neighbour-joining method with Ralstonia pickettii 12D as the outlier. (A) Rooted phylogenetic tree based on NAs associated enzyme genes gain and loss; (B) Rooted phylogenetic tree based on genome genes gain and loss.
Metal resistance
Cupriavidus and other closely related bacteria possess a common characteristic of metal resistance [4–5, 8–9, 12–14]. Many genetic determinants support that CR3 strain would have potential resistance to heavy metals, including Cd2+, Zn2+, Co2+, Cu2+, Cu+, Hg2+, Pb2+, Ni2+, CrO4 2-, Ag+, and possibly other heavy metals (Table 4). Some metal cations (e.g. Zn2+, Cu2+) are essential to living cells and function in enzyme-catalyzed reactions [52–53]. In contrast, other heavy metal cations as Ag+, Hg2+ are non-essential and considered toxic. Comparing to the model metal resistant strain CH34, strain CR3 carried many typical metal resistance loci such as czcABC, copCBA etc [4]. However, metal resistance mechanisms among diverse bacteria are slightly different for different metal tolerant strains. For instance, pbrD gene contained in CH34 [13] is absent in the CR3 pbr operon, while the other pbrTRAB/C genes are all present in CR3. PbrD gene is a Pb(II) binding protein involved in Pb(II). The absence of this gene would inhibit free export of Pb2+ and disable cyclical Pb2+ import and export, which would reduce accumulation of Pb2+ in CR3 cells [8]. In the case of resistance to mercury, the Hg2+ resistance system in CR3 consisted of the merR gene that regulates the operon, and the merT and merP genes that function to transport Hg2+ into the cell [8, 54]. The Ag+ resistance system was comprised of the central four sil genes, including silA, silB, silR, silS, while silCEP genes are absent. Here, strain CR3 presented two possible Ag+ resistance mechanisms: one involving an ATPase efflux pump, the other employing a chemiosmotic efflux pump [52, 55–56]. CR3 contained some classical ABC three-component resistance systems, most of them involving ATPase efflux pump systems, such as czcABC [10], cusCBA [57]. Other types of ion efflux systems were also identified in CR3 that export CrO4 2-, Ni2+ etc. [4, 58–60]. Details on the major constitutive genes with putative roles in metal resistance are shown in Table 4. Based on current reports, merP, pbrT, and chrF genes are not required [61–63], but the merT gene is important in metal resistance [61, 64]. Comparing to other Cupriavidus strains, we find that czc, pbr, chr, cus genetic resistance determinants are common in all Cupriavidus bacteria. Mer is only present in CH34, OR16 and CR3, while the required merT gene was absent in the other five Cupriavidus bacteria. ABC.PE is present in CH34, H16, HPC (L), JMP134, N1 and CR3, while the required ABC.PE.A gene was absent in the taiwanensis and OR16 strains. Cop functions in CH34 and CR3, but it is not clear that whether the absent genes in the cop genetic determinant are essential. Likewise, mercury resistance is functional in the strains with no silC gene [65], which it is absent in C. necator HPC (L) strain, C. pinatubonensis JMP134 strain, and C. taiwanensis strain.
Table 4. Metal resistance gene operons in C. gilardii CR3.
| Operon | Gene cluster | Gene identification number | Metal(s) |
|---|---|---|---|
| czc | czcMNCBADRSE 1 ; czcCBA 2 ; czcI tolC czcBA 2 ; czcABC 2 | CR3_1349,1350,1352–1358; CR3_3402–3404; CR3_3627–3630; CR3_4522–4524 | Cd2+, Zn2+, Co2+ |
| cop | copSRABCD 1 ; copQLFGJIDCBARS ompC copKBA 1 ; copKHFIDCBARS 1 | CR3_0172–0177; CR3_1309,1312–1315,1317–1323,1325–1328; CR3_1370–1372, 1374–1380 | Cu2+, Cu+ |
| mer | merRTP 1 | CR3_2714–2716 | Hg2+ |
| pbr | pbrAB/CRT 1 | CR3_1340–1342,1345 | Pb2+ |
| sil | silABRS 1 | CR3_1305,1306,1322,1323 | Ag+ |
| chr | chrFAB 1 | CR3_1391–1393 | CrO4 2- |
| cus | cusFAB 1 | CR3_1304–1306 | Cu+, Ag+ |
| ABC.PE | ABC.PE.A1AP1S 1 ; ABC.PE. A1AP1S 1 ; ABC.PE.SPP1A 2 ; ABC.PE.A1ASP1P 2 ; ABC.PE.AA1P1PS 2 | CR3_0010–0013; CR3_2425–2428; CR3_3225–3228; CR3_3295–3299; CR3_3708–3712 | Ni2+ |
1. Gene operons were located at chr1 replicon.
2. Gene operons were located at chr2 replicon.
To validate metal resistance capacity in CR3, MIC tests of the predicted metals (Ag+, Cd2+, Co2+, Cr6+, Cu2+, Hg2+, Ni2+, Pb2+ and Zn2+) were performed. For comparison, the published MICs of the model metal resistant strain C. metallidurans CH34 [65] are also summarized at Table 5. C. gilardii CR3 showed resistance to all nine of the tested heavy metals. CR3 exhibited very high tolerance to Ag+, Cr6+, Hg2+, Pb2+ and Zn2+ compared with the model metal resistant strain C. metallidurans CH34. It is interesting to note that there is only one Mer gene cluster in CR3, while 4 Mer gene clusters in CH34. CR3 had the same MIC value for Cd2+ and Cu2+ as C. metallidurans CH34. Although there were 4 czc operons in CR3, the MIC values of Co2+ in CR3 were lower than those in CH34. It can be concluded that heavy metal resistance is a salient feature in CR3 and gene number is not proportional to the capacity of metal resistance.
Table 5. The minimum inhibitory concentration (MIC) of nine heavy metals in C.gilardii CR3 and C.metallidurans CH34.
| MIC(mM) a | |||
|---|---|---|---|
| Metal Ionic form | Formulation Compound | C. gilardii CR3 | C. metallidurans CH34 |
| Ag + | AgNO3 | 0.06 | 0.0005 |
| Cd 2+ | CdCl25H2O | 4 | 4 |
| Co 2+ | CoCl26H2O | 2 | 25 |
| Cr 6+ | K2CrO4 | 2 | 0.4 |
| Cu 2+ | CuSO45H2O | 3 | 3 |
| Hg 2+ | HgCl2 | >0.04 | 0.0027 |
| Ni 2+ | NiCl26H2O | 3 | 13 |
| Pb 2+ | Pb(NO3)2 | 4 | 1 |
| Zn 2+ | ZnSO47H2O | >24 | 12 |
a MIC value of C.gilardii CR3 was from this study, MIC value of C. metallidurans CH34 was from reference [51].
The heavy metal tolerance of CR3 strain was mainly achieved by efficient ion efflux, metal-complexation and metal-reduction. The robust characteristics of this strain apparently result not only from its high-performance transport and detoxification systems for metal ions, but also from effective self repair mechanisms. The CR3 genome encoded direct repair protein genes, and genes that play roles in base-excision repair, mismatch repair (lack of mutH gene), nucleotide excision repair (a complete pathway). It also carried the complete RecFOR pathway to repair double strand DNA breaks by homologous recombination. The CR3 genome does not encode strand discrimination factor mutH gene as in the CH34 genome [4], nor a known DNA adenine methylase and DNA patch repair protein. The supplementary genes of other mismatch excision repair factors, ssb (CR3_0318) and uvrD (CR3_0231, CR3_1976), might complement mismatch repair function in CR3. Both direct resistance mechanisms and self repair mechanisms safeguard the ability of strain CR3 to tolerate the harsh environment presented by life in natural asphalt deposits.
Supporting Information
(TIF)
(A). GO function distribution of CR3 genes based on level 1 classification; (B). Significantly enriched GO function of CR3 unique genes based on level 3 classification. The significance is characterized by p_value, based on chi-square test.
(TIF)
(A). KEGG pathways that CR3 genes mainly participate in; (B). KEGG pathway enrichment analysis of CR3 unique genes. The significance is characterized by p value, based on chi-square test.
(TIF)
(A). Primary COG classification; (B). Significant secondary COG features. The significance is characterized by p_value, based on chi-square test.
(TIF)
Acknowledgments
We are thankful to the anonymous reviewers for their helpful comments on the manuscript. We also thank colleagues (Core Genomic Facility, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing,China) for excellent construction of sequencing libraries.
Data Availability
The 16S rRNA gene sequence of strain CR3 is available from the GenBank database (accession number JX945578). The genome sequence of Cupriavidus gilardii CR3 is available from the GenBank database (accession numberCP010516-CP010517).
Funding Statement
XW received support from National Natural Science Foundation of China (No.31170476), http://www.nsfc.gov.cn/. XW received support from Jilin Provincial Science and Technology Department Project (No. 20110745 and No. 20150414046GH), http://kjt.jl.gov.cn/kjt/4/tindex.shtml. MH received support from National Natural Science Foundation of China, No.51238001.
References
- 1. Cuadrado V, Gomila M, Merini L, Giulietti AM, Moore ER. Cupriavidus pampae sp. nov., a novel herbicide-degrading bacterium isolated from agricultural soil. Int J Syst Evol Microbiol. 2010;60(Pt 11):2606–12. Epub 2009/12/22. 10.1099/ijs.0.018341-0 . [DOI] [PubMed] [Google Scholar]
- 2. Vandamme P, Coenye T. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol. 2004;54(Pt 6):2285–9. Epub 2004/11/17. 10.1099/ijs.0.63247-0 . [DOI] [PubMed] [Google Scholar]
- 3. Coenye T, Goris J, De Vos P, Vandamme P, LiPuma JJ. Classification of Ralstonia pickettii-like isolates from the environment and clinical samples as Ralstonia insidiosa sp. nov. Int J Syst Evol Microbiol. 2003;53(Pt 4):1075–80. Epub 2003/08/02. . [DOI] [PubMed] [Google Scholar]
- 4. Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One. 2010;5(5):e10433 Epub 2010/05/14. 10.1371/journal.pone.0010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Poehlein A, Kusian B, Friedrich B, Daniel R, Bowien B. Complete genome sequence of the type strain Cupriavidus necator N-1. J Bacteriol. 2011;193(18):5017 Epub 2011/07/12. 10.1128/JB.05660-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lykidis A, Perez-Pantoja D, Ledger T, Mavromatis K, Anderson IJ, Ivanova NN, et al. The complete multipartite genome sequence of Cupriavidus necator JMP134, a versatile pollutant degrader. PLoS One. 2010;5(3):e9729 Epub 2010/03/27. 10.1371/journal.pone.0009729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amadou C, Pascal G, Mangenot S, Glew M, Bontemps C, Capela D, et al. Genome sequence of the beta-rhizobium Cupriavidus taiwanensis and comparative genomics of rhizobia. Genome Res. 2008;18(9):1472–83. Epub 2008/05/21. 10.1101/gr.076448.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mergeay M, Monchy S, Vallaeys T, Auquier V, Benotmane A, Bertin P, et al. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol Rev. 2003;27(2–3):385–410. Epub 2003/06/28. 10.1016/S0168-6445(03)00045-7 . [DOI] [PubMed] [Google Scholar]
- 9. Monchy S, Benotmane MA, Janssen P, Vallaeys T, Taghavi S, van der Lelie D, et al. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J Bacteriol. 2007;189(20):7417–25. Epub 2007/08/07. 10.1128/JB.00375-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol. 1995;14(2):142–53. Epub 1995/02/01. . [DOI] [PubMed] [Google Scholar]
- 11. Cserhati M, Kriszt B, Szoboszlay S, Toth A, Szabo I, Tancsics A, et al. De novo genome project of Cupriavidus basilensis OR16. J Bacteriol. 2012;194(8):2109–10. Epub 2012/03/31. 10.1128/JB.06752-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hong KW, Thinagaran DA, Gan HM, Yin WF, Chan KG. Whole-Genome Sequence of Cupriavidus sp. Strain BIS7, a Heavy-Metal-Resistant Bacterium. J Bacteriol. 2012;194(22):6324 Epub 2012/11/02. 10.1128/JB.01608-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borremans B, Hobman JL, Provoost A, Brown NL, van Der Lelie D. Cloning and functional analysis of the pbr lead resistance determinant of Ralstonia metallidurans CH34. J Bacteriol. 2001;183(19):5651–8. Epub 2001/09/07. 10.1128/JB.183.19.5651-5658.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trefault N, De la Iglesia R, Molina AM, Manzano M, Ledger T, Perez-Pantoja D, et al. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ Microbiol. 2004;6(7):655–68. Epub 2004/06/10. 10.1111/j.1462-2920.2004.00596.x . [DOI] [PubMed] [Google Scholar]
- 15. Perez-Pantoja D, De la Iglesia R, Pieper DH, Gonzalez B. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiol Rev. 2008;32(5):736–94. Epub 2008/08/12. 10.1111/j.1574-6976.2008.00122.x . [DOI] [PubMed] [Google Scholar]
- 16. Wei YH, Chen WC, Huang CK, Wu HS, Sun YM, Lo CW, et al. Screening and Evaluation of Polyhydroxybutyrate-Producing Strains from Indigenous Isolate Cupriavidus taiwanensis Strains. Int J Mol Sci. 2011;12(1):252–65. Epub 2011/02/23. 10.3390/ijms12010252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, et al. Genome sequence of the bioplastic-producing "Knallgas" bacterium Ralstonia eutropha H16. Nat Biotechnol. 2006;24(10):1257–62. Epub 2006/09/12. 10.1038/nbt1244 . [DOI] [PubMed] [Google Scholar]
- 18. Iwaki H, Nakai E, Nakamura S, Hasegawa Y. Isolation and characterization of new cyclohexylacetic acid-degrading bacteria. Curr Microbiol. 2008;57(2):107–10. Epub 2008/04/09. 10.1007/s00284-008-9158-0 . [DOI] [PubMed] [Google Scholar]
- 19.Brient JA, Wessner PJ, Doyle MN. Naphthenic acids. Kirk-Othmer Encyclopedia of Chemical Technology. 1995:1017–29.
- 20. Headley JV, McMartin DW. A review of the occurrence and fate of naphthenic acids in aquatic environments. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2004;39(8):1989–2010. Epub 2004/08/31. . [DOI] [PubMed] [Google Scholar]
- 21. Dokholyan B, Magomedov A. Effect of sodium naphthenate on survival and some physiological-biochemical parameters of some fishes. Journal of Ichthyology. 1983;23(6):125–32. [Google Scholar]
- 22. Scarlett AG, West CE, Jones D, Galloway TS, Rowland SJ. Predicted toxicity of naphthenic acids present in oil sands process-affected waters to a range of environmental and human endpoints. Sci Total Environ. 2012;425:119–27. Epub 2012/04/03. 10.1016/j.scitotenv.2012.02.064 . [DOI] [PubMed] [Google Scholar]
- 23. Whitby C. Microbial naphthenic Acid degradation. Adv Appl Microbiol. 2010;70:93–125. Epub 2010/04/03. 10.1016/S0065-2164(10)70003-4 . [DOI] [PubMed] [Google Scholar]
- 24. Paslawski JC, Headley JV, Hill GA, Nemati M. Biodegradation kinetics of trans-4-methyl-1-cyclohexane carboxylic acid. Biodegradation. 2009;20(1):125–33. Epub 2008/07/18. 10.1007/s10532-008-9206-2 . [DOI] [PubMed] [Google Scholar]
- 25. Huang J, Nemati M, Hill G, Headley J. Batch and continuous biodegradation of three model naphthenic acids in a circulating packed-bed bioreactor. J Hazard Mater. 2012;201–202:132–40. Epub 2011/12/14. 10.1016/j.jhazmat.2011.11.052 . [DOI] [PubMed] [Google Scholar]
- 26. Tokuyama T, Kaneda T. Corynebacterium cyclohexanicum n. sp.: a cyclohexanecarboxylic acid utilizing bacterium. Can J Microbiol. 1973;19(8):937–42. Epub 1973/08/01. . [DOI] [PubMed] [Google Scholar]
- 27. Blakley E. The microbial degradation of cyclohexanecarboxylic acid: a pathway involving aromatization to form p-hydroxybenzoic acid. Canadian Journal of Microbiology. 1974;20(10):1297–306. [Google Scholar]
- 28. Rho EM, Evans WC. The aerobic metabolism of cyclohexanecarboxylic acid by Acinetobacter anitratum. Biochem J. 1975;148(1):11–5. Epub 1975/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blakley ER. The microbial degradation of cyclohexanecarboxylic acid by a beta-oxidation pathway with simultaneous induction to the utilization of benzoate. Can J Microbiol. 1978;24(7):847–55. Epub 1978/07/01. . [DOI] [PubMed] [Google Scholar]
- 30. Blakley ER, Papish B. The metabolism of cyclohexanecarboxylic acid and 3-cyclohexenecarboxylic acid by Pseudomonas putida. Can J Microbiol. 1982;28(12):1324–9. Epub 1982/12/01. . [DOI] [PubMed] [Google Scholar]
- 31. Johnson RJ, Smith BE, Sutton PA, McGenity TJ, Rowland SJ, Whitby C. Microbial biodegradation of aromatic alkanoic naphthenic acids is affected by the degree of alkyl side chain branching. The ISME journal. 2011;5(3):486–96. 10.1038/ismej.2010.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim JS, Crowley DE. Microbial diversity in natural asphalts of the Rancho La Brea Tar Pits. Appl Environ Microbiol. 2007;73(14):4579–91. Epub 2007/04/10. 10.1128/AEM.01372-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716–21. Epub 2011/12/06. 10.1099/ijs.0.038075-0 . [DOI] [PubMed] [Google Scholar]
- 34. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. Epub 1997/09/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1(1):18 Epub 2012/01/01. 10.1186/2047-217X-1-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–72. Epub 2009/12/19. 10.1101/gr.097261.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods. 2013;10(6):563–9. Epub 2013/05/07. 10.1038/nmeth.2474 . [DOI] [PubMed] [Google Scholar]
- 38. English AC, Richards S, Han Y, Wang M, Vee V, Qu J, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7(11):e47768 Epub 2012/11/28. 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001;29(12):2607–18. Epub 2001/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23(6):673–9. Epub 2007/01/24. 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–8. Epub 2001/10/09. . [DOI] [PubMed] [Google Scholar]
- 42. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–5. Epub 2007/05/29. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278(5338):631–7. Epub 1997/10/24. . [DOI] [PubMed] [Google Scholar]
- 44. Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. Epub 1997/03/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–8. Epub 2007/04/25. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. Epub 2011/05/07. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–403. Epub 2004/07/03. 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. PGAP: pan-genomes analysis pipeline. Bioinformatics. 2012;28(3):416–8. Epub 2011/12/02. 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Page RDM. Visualizing Phylogenetic Trees Using TreeView Current Protocols in Bioinformatics: John Wiley & Sons, Inc.; 2002. [DOI] [PubMed] [Google Scholar]
- 50. Moriya Y, Shigemizu D, Hattori M, Tokimatsu T, Kotera M, Goto S, et al. PathPred: an enzyme-catalyzed metabolic pathway prediction server. Nucleic Acids Res. 2010;38(Web Server issue):W138–43. Epub 2010/05/04. 10.1093/nar/gkq318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Monsieurs P, Moors H, Van Houdt R, Janssen PJ, Janssen A, Coninx I, et al. Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals. 2011;24(6):1133–51. Epub 2011/06/28. 10.1007/s10534-011-9473-y . [DOI] [PubMed] [Google Scholar]
- 52. Gupta A, Phung LT, Taylor DE, Silver S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology. 2001;147(Pt 12):3393–402. Epub 2001/12/12. . [DOI] [PubMed] [Google Scholar]
- 53. Ngendahimana VM. Identification of a Putative P-type ATPase Involved in Zinc and Cadmium Resistance in Enterobacter Sp. YSU: Youngstown State University; 2012. [Google Scholar]
- 54.Rebello RCDL, Gomes KM, Duarte RS, Rachid CTCDC, Rosado AS, Regua-Mangia AH. 2013. Diversity of mercury resistant Escherichia coli strains isolated from aquatic systems in Rio de Janeiro, Brazil. International Journal of Biodiversity.
- 55. Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5(2):183–8. Epub 1999/02/04. 10.1038/5545 . [DOI] [PubMed] [Google Scholar]
- 56. Silver S. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev. 2003;27(2–3):341–53. Epub 2003/06/28. 10.1016/S0168-6445(03)00047-0 . [DOI] [PubMed] [Google Scholar]
- 57. Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185(13):3804–12. Epub 2003/06/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Juhnke S, Peitzsch N, Hubener N, Grosse C, Nies DH. New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch Microbiol. 2002;179(1):15–25. Epub 2002/12/10. 10.1007/s00203-002-0492-5 . [DOI] [PubMed] [Google Scholar]
- 59. Nies A, Nies DH, Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265(10):5648–53. Epub 1990/04/05. . [PubMed] [Google Scholar]
- 60. Grass G, Fan B, Rosen BP, Lemke K, Schlegel HG, Rensing C. NreB from Achromobacter xylosoxidans 31A Is a nickel-induced transporter conferring nickel resistance. J Bacteriol. 2001;183(9):2803–7. Epub 2001/04/09. 10.1128/JB.183.9.2803-2807.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sone Y, Nakamura R, Pan-Hou H, Itoh T, Kiyono M. Role of MerC, MerE, MerF, MerT, and/or MerP in resistance to mercurials and the transport of mercurials in Escherichia coli. Biol Pharm Bull. 2013;36(11):1835–41. Epub 2013/08/30. 10.1248/bpb.b13-00554 . [DOI] [PubMed] [Google Scholar]
- 62. Hynninen A, Touze T, Pitkanen L, Mengin-Lecreulx D, Virta M. An efflux transporter PbrA and a phosphatase PbrB cooperate in a lead-resistance mechanism in bacteria. Mol Microbiol. 2009;74(2):384–94. Epub 2009/09/10. 10.1111/j.1365-2958.2009.06868.x . [DOI] [PubMed] [Google Scholar]
- 63. Branco R, Chung AP, Johnston T, Gurel V, Morais P, Zhitkovich A. The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium(VI) and superoxide. J Bacteriol. 2008;190(21):6996–7003. Epub 2008/09/09. 10.1128/JB.00289-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bersch B, Derfoufi KM, De Angelis F, Auquier V, Ekende EN, Mergeay M, et al. Structural and metal binding characterization of the C-terminal metallochaperone domain of membrane fusion protein SilB from Cupriavidus metallidurans CH34. Biochemistry. 2011;50(12):2194–204. Epub 2011/02/09. 10.1021/bi200005k . [DOI] [PubMed] [Google Scholar]
- 65. Mergeay M, Nies D, Schlegel HG, Gerits J, Charles P, Van Gijsegem F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J Bacteriol. 1985;162(1):328–34. Epub 1985/04/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(A). GO function distribution of CR3 genes based on level 1 classification; (B). Significantly enriched GO function of CR3 unique genes based on level 3 classification. The significance is characterized by p_value, based on chi-square test.
(TIF)
(A). KEGG pathways that CR3 genes mainly participate in; (B). KEGG pathway enrichment analysis of CR3 unique genes. The significance is characterized by p value, based on chi-square test.
(TIF)
(A). Primary COG classification; (B). Significant secondary COG features. The significance is characterized by p_value, based on chi-square test.
(TIF)
Data Availability Statement
The 16S rRNA gene sequence of strain CR3 is available from the GenBank database (accession number JX945578). The genome sequence of Cupriavidus gilardii CR3 is available from the GenBank database (accession numberCP010516-CP010517).