Abstract
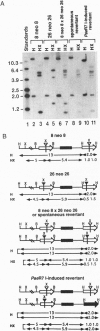
We have derived Chinese hamster ovary (CHO) cell hybrids containing herpes simplex virus thymidine kinase (tk) heteroalleles for the study of spontaneous and restriction enzyme-induced interchromosomal recombination. These lines allowed us to make a direct comparison between spontaneous intrachromosomal and interchromosomal recombination using the same tk heteroalleles at the same genomic insertion site. We find that the frequency of interchromosomal recombination is less by a factor of at least 5000 than that of intrachromosomal recombination. Our results with mammalian cells differ markedly from results with Saccharomyces cerevisiae, with which similar studies typically give only a 10-to 30-fold difference. Next, to inquire into the fate of double-strand breaks at either of the two different Xho I linker insertion mutations, we electroporated PaeR7I enzyme, an isoschizomer of Xho I, into these hybrids. A priori, these breaks can be repaired either by recombination from the homology or by end-joining. Despite a predicted bias against recovering end-joining products in our system, all cells characterized by enzyme-induced resistance to hypoxanthine/aminopterin/thymidine were, in fact, due to nonhomologous recombination or end-joining. These results are in agreement with other studies that used extrachromosomal sequences to examine the relative efficiencies of end-joining and homologous recombination in mammalian cells, but are in sharp contrast to results of analogous studies in S. cerevisiae, wherein only products of homologous events are detected.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ager D. D., Phillips J. W., Columna E. A., Winegar R. A., Morgan W. F. Analysis of restriction enzyme-induced DNA double-strand breaks in Chinese hamster ovary cells by pulsed-field gel electrophoresis: implications for chromosome damage. Radiat Res. 1991 Nov;128(2):150–156. [PubMed] [Google Scholar]
- Benjamin M. B., Little J. B. X rays induce interallelic homologous recombination at the human thymidine kinase gene. Mol Cell Biol. 1992 Jun;12(6):2730–2738. doi: 10.1128/mcb.12.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin M. B., Potter H., Yandell D. W., Little J. B. A system for assaying homologous recombination at the endogenous human thymidine kinase gene. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6652–6656. doi: 10.1073/pnas.88.15.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Liskay R. M. Direct-repeat analysis of chromatid interactions during intrachromosomal recombination in mouse cells. Mol Cell Biol. 1991 Oct;11(10):4839–4845. doi: 10.1128/mcb.11.10.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag R. J., Waldman A. S., Liskay R. M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- Chang X. B., Wilson J. H. Modification of DNA ends can decrease end joining relative to homologous recombination in mammalian cells. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4959–4963. doi: 10.1073/pnas.84.14.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson R. L., O'Malley K. A., Wheeler T. B. Polyethylene glycol-induced mammalian cell hybridization: effect of polyethylene glycol molecular weight and concentration. Somatic Cell Genet. 1976 May;2(3):271–280. doi: 10.1007/BF01538965. [DOI] [PubMed] [Google Scholar]
- Engels W. R., Johnson-Schlitz D. M., Eggleston W. B., Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990 Aug 10;62(3):515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin A. R., Liskay R. M. The effects of insertions on mammalian intrachromosomal recombination. Genetics. 1994 Feb;136(2):607–617. doi: 10.1093/genetics/136.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J. A., Fink G. R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981 Jul 23;292(5821):306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- Johnson-Schlitz D. M., Engels W. R. P-element-induced interallelic gene conversion of insertions and deletions in Drosophila melanogaster. Mol Cell Biol. 1993 Nov;13(11):7006–7018. doi: 10.1128/mcb.13.11.7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H. Replication-dependent sister chromatid recombination in rad1 mutants of Saccharomyces cerevisiae. Genetics. 1993 Mar;133(3):469–487. doi: 10.1093/genetics/133.3.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk L. C., Hartwell L. H. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992 Oct;132(2):387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman C. W., Trautman J. K., Carroll D. Illegitimate recombination in Xenopus: characterization of end-joined junctions. Nucleic Acids Res. 1994 Feb 11;22(3):434–442. doi: 10.1093/nar/22.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Haber J. E. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989 Oct;123(2):261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liskay R. M., Stachelek J. L., Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harb Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- Mulligan R. C., Berg P. Selection for animal cells that express the Escherichia coli gene coding for xanthine-guanine phosphoribosyltransferase. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2072–2076. doi: 10.1073/pnas.78.4.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairn R. S., Adair G. M., Humphrey R. M. DNA-mediated gene transfer in Chinese hamster ovary cells: clonal variation in transfer efficiency. Mol Gen Genet. 1982;187(3):384–390. doi: 10.1007/BF00332616. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988 Feb 11;16(3):907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Chang X. B., Wilson J. H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989 Jul;9(7):3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas K. R., Folger K. R., Capecchi M. R. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986 Feb 14;44(3):419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- Winegar R. A., Lutze L. H., Rufer J. T., Morgan W. F. Spectrum of mutations produced by specific types of restriction enzyme-induced double-strand breaks. Mutagenesis. 1992 Nov;7(6):439–445. doi: 10.1093/mutage/7.6.439. [DOI] [PubMed] [Google Scholar]
- Zheng H., Chang X. B., Wilson J. H. Primary cells and established cell lines join DNA ends with the same efficiency relative to homologous recombination. Plasmid. 1989 Sep;22(2):99–105. doi: 10.1016/0147-619x(89)90019-x. [DOI] [PubMed] [Google Scholar]