Abstract
Muscle growth and regeneration are regulated through a series of spatiotemporally dependent signaling and transcriptional cascades. Although the transcriptional program controlling myogenesis has been extensively investigated, the full repertoire of transcriptional regulators involved in this process is far from defined. Various homeodomain transcription factors have been shown to play important roles in both muscle development and muscle satellite cell-dependent repair. Here, we show that the homeodomain factor Barx2 is a new marker for embryonic and adult myoblasts and is required for normal postnatal muscle growth and repair. Barx2 is coexpressed with Pax7, which is the canonical marker of satellite cells, and is upregulated in satellite cells after muscle injury. Mice lacking the Barx2 gene show reduced postnatal muscle growth, muscle atrophy, and defective muscle repair. Moreover, loss of Barx2 delays the expression of genes that control proliferation and differentiation in regenerating muscle. Consistent with the in vivo observations, satellite cell-derived myoblasts cultured from Barx2−/− mice show decreased proliferation and ability to differentiate relative to those from wild-type or Barx2+/− mice. Barx2−/− myoblasts show reduced expression of the differentiation-associated factor myogenin as well as cell adhesion and matrix molecules. Finally, we find that mice lacking both Barx2 and dystrophin gene expression have severe early onset myopathy. Together, these data indicate that Barx2 is an important regulator of muscle growth and repair that acts via the control of satellite cell proliferation and differentiation.
Keywords: Muscle stem cells, Adult stem cells, Homeobox genes, Muscular dystrophy, Skeletal muscle, Cell adhesion molecules, Tissue regeneration, Tissue-specific stem cells
Introduction
Adult mammalian muscle has the potential to regenerate by activation of undifferentiated myogenic precursor cells called satellite cells, which are normally quiescent and situated between the basal lamina and the myofiber plasmalemma [1–6]. On activation, satellite cells proliferate and divide asymmetrically producing one daughter cell that proceeds to myogenic differentiation and another that becomes quiescent and replenishes the satellite-cell pool [7, 8].
Satellite cells are estimated to account for approximately 30% of the nuclei in mouse limb muscle at birth; however, this pool diminishes as the cells are recruited for postnatal muscle growth [9]. In adults, substantial muscle growth ceases and the satellite cells, which account for less than 5% of the myonuclei, become quiescent until injury or exercise causes them to reenter the cell cycle [2]. Recent work shows that satellite cells are indispensable for adult skeletal muscle regeneration and cannot be compensated by other endogenous progenitor cells [10].
Several types of transcription factors control the specification, activation, proliferation, and differentiation of satellite cells, most prominently, members of the basic helix loop helix (bHLH) family and the paired and homeodomain families. Four bHLH myogenic regulatory factors (MRFs) act in a hierarchical fashion to orchestrate an embryonic muscle developmental program that involves myoblast determination, proliferation, migration, and differentiation [11–16]. The MRFs are also involved in various events downstream of satellite-cell activation during postnatal muscle growth and repair [17–19]. Homeobox transcription factors are key regulators of morphogenetic programs via activities such as the control of cell–cell and cell–substrate adhesion. Adhesion and signaling in turn influences cell activation, migration, and fusion [20]. Homeobox proteins play particularly important roles in embryonic and adult myogenesis. The best-characterized homeobox proteins in muscle development are the paired-homeodomain factors Pax3 and Pax7, which control the specification and migration of embryonic muscle precursors [21–23]. Pax7 is also expressed in all adult satellite cells, and Pax3 is present in a subset of muscle progenitor cells [24, 25]. Both Pax3 and Pax7 are involved in maintaining the myogenic identity of these cells and regulate entry into the myogenic program by activation of MyoD [25–27]. Pax7 is often considered the canonical marker for satellite cells and may promote satellite-cell survival and self-renewal [28, 29]. Other homeobox families involved in embryonic myogenesis include Pitx [30–32], Msx [33], Meox [34, 35], and Lbx [36, 37]. Some of these factors are also expressed in adult satellite cells where they may influence proliferation or differentiation [36, 38–40]. In general, however, the roles of non-paired homeodomain proteins in adult myogenesis are less well understood than in embryonic development.
In previous work, we showed that the antennapedia class Barx2 homeobox protein is a novel regulator of myogenesis. Barx2 interacts directly with MyoD and serum response factor (SRF) to regulate remodeling of the actin cytoskeleton and promote myoblast migration and differentiation in vitro [41, 42]. Barx2 also affects the morphological plasticity of nascent myofibres and controls expression of the cell-cycle factor cyclin-D1 [43]. Other known targets of Barx2 include genes involved in cell and substrate adhesion such as the neural cell adhesion molecule [44] and cadherins [45]. Moreover, the Barx2 gene promoter is activated by MyoD and myogenin suggesting that it may be subject to muscle-specific regulatory feedback [46].
Here, we report that Barx2 is coexpressed with Pax7 in muscle progenitor cells during embryonic and fetal development and in the satellite cells of postnatal and adult muscles. Mice lacking the Barx2 gene show reduced postnatal muscle growth, increased age-associated muscle atrophy, and impaired regeneration. Activation of genes involved in proliferation and differentiation are delayed after injury in Barx2−/− mice; this effect is recapitulated in cultured Barx2−/− myoblasts suggesting intrinsic impairment of myoblast function. Moreover, interbreeding of Barx2 null mice with dystrophic mdx mice leads to a striking exacerbation of the disease phenotype. Overall, our data indicate that Barx2 is a new marker of satellite cells and myoblasts and is an important regulator of muscle growth, maintenance, and regeneration.
Materials and Methods
Mice
Barx2 null mice were obtained from Dr. Geoff Rosenfeld, maintained by heterozygous crosses and genotyped according to [47]. C57BL/10ScSn-Dmdmdx/J (mdx) mice were obtained from Jackson Laboratories and interbred with Barx2 null mice. See supporting information Methods for more details. All animal studies were approved by The Scripps Research Institute and Flinders University animal welfare committees.
Histology and Myofibre Diameter Analysis
Various dissected muscles were fixed with 4% paraformaldehyde (PFA) in PBT (phosphate buffered saline supplemented with 0.05% Tween 20) and processed for paraffin embedding. Muscle sections (10 μm) were processed for hematoxylin and eosin (H&E), Masson’s Trichrome (to detect collagen deposition), and alizarin red (to detect calcification) staining at the Scripps Research Institute Histology core facility. To ensure that similar regions of muscle were compared between individuals, serial sections were cut from the midpoint of each fixed muscle specimen. Sections were examined using a Zeiss microscope. The maximal diameters of myofibers were measured in transverse sections from three different animals per genotype; approximately 400 myofibers were counted for each genotype. Myofiber diameters in micrometers were approximated using the scale bars on the micrographs.
Immunohistochemistry, Fluorescence Microscopy, and Image Analysis
Embryonic limbs and postnatal and adult muscles were fixed in 4% paraformaldehyde (PFA) washed in phosphate buffered saline (PBS) and frozen sections were prepared [48]. Sections were stained on slides with different combinations of primary antibodies: rabbit polyclonal anti-Barx2 (M-186, Santa Cruz Biotechnologies, Santa Cruz, CA, www.scbt.com), mouse monoclonal anti-skeletal myosin (Fast; clone MY-32, Sigma, St. Louis, http://www.sigmaaldrich.com), mouse monoclonal anti-MyoD, (clone MoAb5.8A, BD PharMingen, San Diego, CA, http://www.bdbiosciences.com/reagents/), mouse monoclonal anti-Myogenin (clone F5D BD Bioscience Pharmingen, San Diego, CA, http://www.bdbiosciences.com/reagents/), mouse monoclonal anti-Pax7 (R&D Systems, Minneapolis, MN, http://www.rndsystems.com/), goat polyclonal anti-Pax3 (Santa Cruz Biotechnologies, Santa Cruz, CA, www.scbt.com), rat anti-heparan sulfate (HS) proteoglycan monoclonal antibody (clone A7L6, Millipore, Billerica, MA, http://www.millipore.com/), mouse monoclonal anti-light meromyosin (clone MF20, Developmental Studies Hybridoma Bank, Iowa City, IA, http://dshb.biology.uiowa.edu/), and rabbit polyclonal Collagen I antibody (ab292) (Abcam). Fluorescent secondary antibodies for frozen sections were from Invitrogen (Molecular Probes, Grand Island, NY, http://www.invitrogen.com): Alexa Fluor-488 goat anti-mouse IgG1 (A21121), Alexa Fluor-633 goat anti-rat IgG (H+L) (A21094), Alexa Fluor-488 donkey anti-goat IgG (H+L) (A11055), Alexa Fluor-488 goat anti-mouse IgG (H+L) “highly cross-adsorbed” (A11029), Alexa Fluor-594 donkey anti-rabbit IgG (H+L) (A21207). For collagen staining on paraffin sections, we used horseradish peroxidise (HRP)-conjugated goat polyclonal secondary antibody to rabbit IgG (ab6721) (Abcam, Cambridge, UK, www.abcam.com), and the HRP reaction product was visualized using diaminobenzidine and counterstained with iron hematoxylin.
Single optical sections and Z-series were obtained using a Bio-Rad (Zeiss, Thornwood, NY, www.zeiss.com) Radiance 2100 Rainbow LSCM. The creation of three-dimensional images and animations using IMARIS software as well as calculation of colocalization are provided in the supporting information Methods [49].
Cardiotoxin Injection and Muscle Regeneration Analysis
Tibialis anterior (TA) muscles of anesthetized adult mice were injected with cardiotoxin or saline as described previously [50]. A detailed procedure is given in supporting information Methods. At various time points postinjection, muscles were harvested and either fixed with buffered 4% paraformaldehyde or processed in RNAlater (Ambion, Grand Island, NY, http://www.invitrogen.com) for preparation of RNA and gene expression analysis. Paraffin or frozen sections were prepared for H&E or antibody staining as described above. For gene expression analysis, n indicates the number of animals of each genotype, and the significance of the result was assessed using a Student’s t test.
Preparation of Primary Myoblasts, Imaging, and Proliferation Analysis
Primary myoblast cultures were prepared using all muscles from the hind limbs of four to five pups (pooled and minced together) as described previously [51]. Barx2−/− and Barx2+/+ or Barx2+/− pups were age-matched (sibs or inbred cousins), and cultures were generated simultaneously using identical conditions. Cells were grown on plates or chamber slides coated with 50 μg/ml collagen type I and maintained in growth medium (1:1 Ham’s F10/Dulbecco’s modified Eagle’s medium [DMEM], supplemented with 20% fetal bovine serum (FBS) and 2.5 ng/ml of basic fibroblast growth factor (bFGF)). Differentiation medium was DMEM supplemented with 2% horse serum. Proliferation was assessed on cells grown on chamber slides using the Cell Proliferation Kit (Amersham, Scientifics) with antibodies to Bromodeoxyuridine (BrdU) followed by Alexa-conjugated secondary antibodies (Invitrogen) and imaging under a Leica MZ FLIII fluorescence microscope. At least five randomly selected fields were analyzed on each of several replicate slides to determine the average percentage of cells incorporating BrdU. Significance was assessed using the nonparametric Wilcoxon signed-rank test. Proliferation and morphology data are shown from one set of matched myoblast cultures (n indicates the total number of fields counted across several slides); however, similar results were seen in replicate myoblast isolates.
RNA and Reverse-Transcriptase Polymerase Chain Reaction (RT-PCR)
RNA was prepared from cell cultures and from muscle tissue using Trizol reagent (Gibco, Grand Island, NY, www.invitrogen.com). Cells were lysed directly in Trizol; muscle was ground with a pestle. RNA was DNAse-treated and reverse transcribed using random primers and MMluV reverse transcriptase (New England Biolabs, Ipswich, MA, www.neb.com). Quantitative RT-PCR reactions were performed on a Corbett Rotogene machine (Qiagen, Valencia, CA, www.qiagen.com) using GoTaq SYBR green reagents (Promega, Madison, WI, www.promega.com). Data were analyzed using the ΔΔCt method with comparison to a pool of housekeeping genes (18S ribosomal RNA, ribosomal protein S26, and glyceraldehyde-3-phosphate dehydrogenase [GAPDH]). Primer sequences are provided in the supporting information Methods.
Plate-Washing Assay
To assess the strength of cell adherence to a substrate, Barx2+/− and Barx2−/− primary myoblasts were cultured for 24 hours on 100-mm dishes that had been coated with 1 μg/ml fibronectin (Sigma, St. Louis, http://www.sigmaaldrich.com). Plates were then rinsed gently with PBS, covered with 7 ml of fresh PBS, and transferred to a slow-moving shaker for 15 min which allows weakly adherent cells to detach. The PBS was collected, centrifuged, and the detached cells were resuspended and counted. The experiment was repeated with three different myoblast isolates and all data were averaged. The significance of the results was assessed using the non-parametric paired test [52].
Results
Barx2 is Coexpressed with Pax7 in Embryonic and Adult Muscles
We previously showed that Barx2 is expressed in the ventral portion of embryonic limbs including putative early muscle masses [53]. To better understand the functions of Barx2 in muscle, we examined its expression pattern with respect to other muscle markers in embryonic, fetal, and adult mice.
Coimmunostaining of sections from E13.5 limbs with Barx2 and myosin heavy chain (MyHC) antibodies showed that Barx2 was expressed in a subset of nuclei within primary myofibers as well as in nuclei located between the fibers that are likely to represent undifferentiated myoblasts (Fig. 1A; Supporting Informtion Movie 1). Coimmunostaining with Barx2 and MyoD antibodies showed that these factors overlapped in many but not all nuclei (Fig. 1B, 1C). Pax7 is expressed in proliferating progenitors and differentiating myoblasts in embryonic and fetal muscle but is downregulated in myofibers [22, 54]. Coimmunostaining of fetal (E18) muscle with Barx2 and Pax7 antibodies revealed almost complete overlap in the expression of these two factors (Fig. 1D–1H), although Pax7-positive cells showed varying intensities of the Barx2 label (Fig. 1H, red arrowheads).
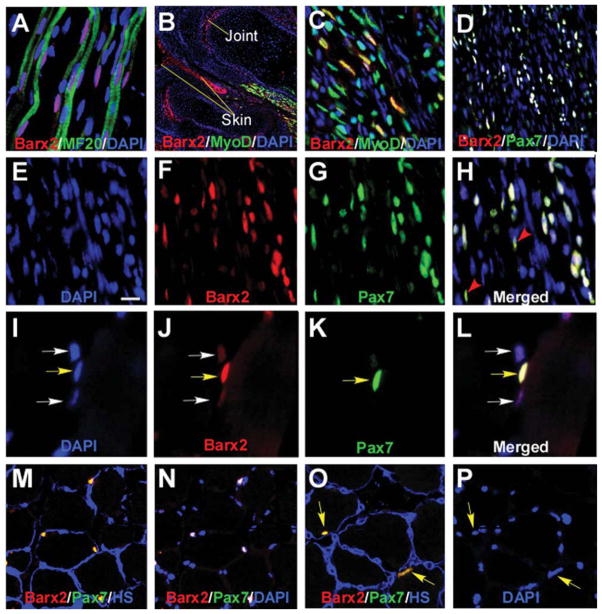
Figure 1.
Barx2 is expressed in embryonic and adult muscle. (A–C): E13.5 limb muscle. (A): Barx2 (red) is expressed in primary myofibers marked with the MF20 antibody (green) as well as between fibers. DAPI, blue. (B, C): Barx2 (red) and MyoD (green) coexpression is shown in a subset of muscle nuclei in the distal hind limb of E13.5 embryo. The image in (B) shows two adjacent digits, and Barx2 expression is also seen in skin and presumptive joint region (arrows). (C):Higher magnification of the muscle shown in (B). (D–H): E18 limb muscle; Barx2 expression overlaps extensively with Pax7 (Barx2, red; Pax7, green; DAPI, blue). Red arrowheads indicate Pax7-positive nuclei with low level of Barx2 expression. (I–P): Adult muscle; (I–L): Yellow arrows indicate nuclei in which Barx2 and Pax7 are coexpressed; White arrows indicate nuclei with weaker expression of Barx2 and no apparent expression of Pax7. (M–P): Barx2 is coexpressed with Pax7 in nuclei situated under the basal lamina. (M, O): Overlap of Barx2 (red) and Pax7 (green) staining is seen as yellow; the basal lamina is labeled with antibody to HS, blue. (N): The same section shown in (M) but labeled with Barx2 (red), Pax7 (green) and DAPI (blue), and without HS staining; nuclei that colocalize DAPI, Barx2, and Pax7 are white. (P): The same section shown in (O) but labeled with DAPI only (blue). Yellow arrows in (O) and (P) show nuclei expressing both Barx2 and Pax7 in (O). Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; HS, heparan sulfate.
In adult muscle, Pax7 is a marker for quiescent satellite cells as well as activated, proliferating satellite cells during muscle regeneration [22, 55, 56]. Coimmunostaining with Barx2 and Pax7 antibodies revealed that essentially all Pax7-positive nuclei also expressed Barx2 (an example of coexpression is shown in Fig. 1I–1L, yellow arrow). There was also expression of Barx2 in a population of apparently Pax7-negative cells that remain to be further characterized (Fig. 1I–1L, white arrows). To confirm the interpretation that the Barx2/Pax7-positive cells are satellite cells, we used an antibody to HS to mark the basal lamina. Essentially all Barx2/Pax7-positive cells were situated under the basal lamina (Fig. 1M–1P, yellow arrows). Thus the data indicate that Barx2 is a new marker for adult satellite cells.
Barx2 Knockout Mice Show Delayed Postnatal Muscle Growth and Adult Muscle Atrophy
To determine whether Barx2 is important for muscle development and growth, we examined the phenotype of embryonic and postnatal Barx2−/− mice. There were no obvious differences in body size, or muscle size, or morphology between wild-type, Barx2+/−, and Barx2−/− embryos at E12.5–18.0 (not shown) and no difference in body weight at birth (Fig. 2A). However, by postnatal day 4, Barx2−/− mice showed a 10% reduction in body weight relative to Barx2−/+ and Barx2+/+ littermates (Fig. 2A) indicating a growth delay. The delay became most pronounced at approximately 4 weeks of age when Barx2−/− mice were typically 20–25% smaller than their Barx2−/+ and wild-type littermates (Fig. 2A, 2B). The size difference was independent of sex. There was no overt sign of feeding difficulties in Barx2−/− mice relative to wild-type or heterozygous littermates and the stomachs of pups were full of milk. Older Barx2−/− mice (15–18 months) often showed musculoskeletal abnormalities such as spinal curvature, splayed stance, and “waddling” gait that were not apparent in wild-type or heterozygous mice at this age (Fig. 2E, supporting information Fig. 1 and supporting information Movies 2, 3).
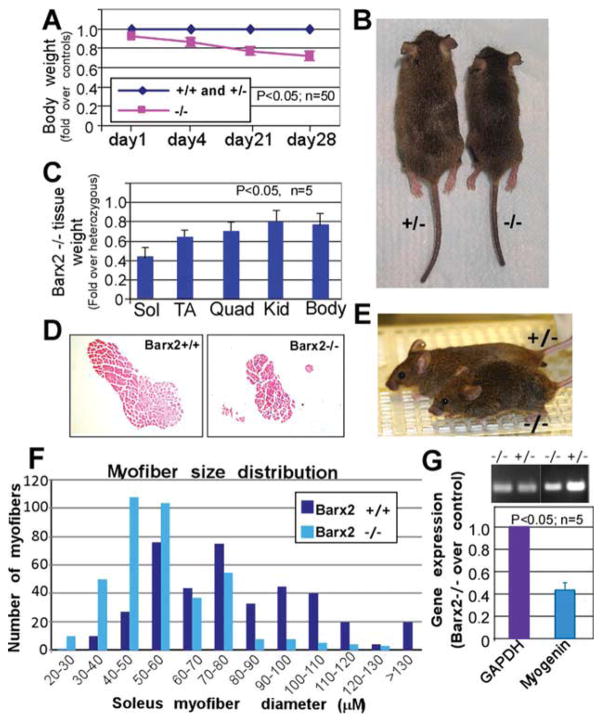
Figure 2.
Analysis of postnatal muscle growth in Barx2 mutant mice. (A): Between 1 and 28 days, Barx2−/− pups show significant reduction in growth as indicated by total body weight relative to Barx2+/+ and Barx2+/− littermates. The averaged weights of the wild-type and heterozygous mice were taken as the baseline. The statistical difference was observed at p < .05, n, number of mice per genotype. (B): Typical appearance of Barx2+/− and Barx2−/− mice at 28 days (wild-type and heterozygous mice are indistinguishable). (C): Sol, TA, and Quad muscles from four pairs of 4-week old Barx2−/− and Barx2+/− littermates were harvested and weighed (n, number of mice). (D): Transverse sections of Sol muscle from wild-type and Barx2−/− mice were stained with hematoxylin and eosin. (E): Example of a 16-month-old Barx2 null mouse displaying a hunched back and splayed stance relative to a heterozygous control mouse. (F): Histogram demonstrating the distribution of myofiber sizes in Barx2−/− and Barx2+/+ mice. Maximal myofiber diameters were measured in transverse sections of Sol muscle from three mice for each genotype; number of myofibers. n = 396 for Barx2+/+ and 392 for Barx2−/−. (G): Expression of the muscle differentiation marker myogenin is reduced in muscles from 4-week-old Barx2−/− mice relative to Barx2+/− littermates as indicated by semiquantitative RT-PCR (n, number of mice). GAPDH is used as a reference standard. Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; Kid, kidney; Quad, quadriceps; RT-PCR, reverse transcriptase polymerase chain reaction; Sol, Soleus; TA, tibialis anterior.
The masses of individual limb muscles were found to be reduced in adult Barx2−/− mice relative to Barx2+/− or wild-type mice. TA and quadriceps muscles from 4-week-old Barx2−/− mice weighed 20%–30% less than those of their Barx2+/− littermates (Fig. 2C), while the soleus muscle was reduced by almost 60% in Barx2−/− mice (Fig. 2C, 2D). The muscles were reduced in disproportion to overall body mass and the mass of other organs (Fig. 2C). Measurements of myofiber diameters in transverse sections of adult soleus muscles demonstrated a change in myofibre size distribution: Barx2−/− muscles contained fewer large fibers and a greater number of smaller myofibres relative to Barx2+/+ (Fig. 2F) or Barx2+/− (not shown) muscles. RT-PCR analysis revealed approximately 60% reduction in myogenin mRNA levels in adult Barx2−/− muscles relative to Barx2+/− muscles. (Fig. 2G).
Histological analysis of Barx2−/− and wild-type muscles at 6 and 12 months of age revealed striking differences in muscle appearance. TA, soleus, and diaphragm muscles from Barx2+/+ and Barx2-littermates were examined by H&E staining of paraffin-embedded serial sections. For each group, four to five mice were analyzed and representative images of 6-month-old mice are shown in Figure 3. Transverse sections of the Barx2−/− TA revealed narrower myofibers and greater variability in myofiber size and shape relative to the wild-type muscle (compare Fig. 3A, 3B and 3D, 3E). Masson’s trichrome staining for collagen expression revealed endomysial and perimysial collagen deposition suggesting fibrosis in Barx2−/− TA muscle (Fig. 3C, 3F).
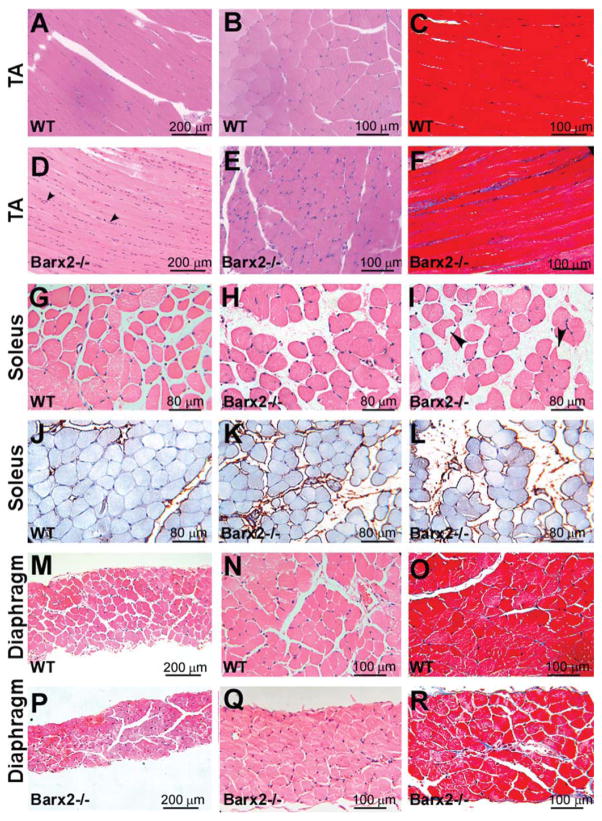
Figure 3.
Histological analysis of muscle in adult Barx2 mutant mice. (A–F): Comparison of TA muscle in 6-month-old WT (A–C) and Barx2−/− mice (D–F). Relative to wild-type muscle, Barx2−/− muscle shows narrower myofibers, an increased number of nuclei between myofibers, and increased fibrosis, as indicated by Masson’s trichrome staining (panels C and F). (G–L): Comparison of soleus muscle in 6-month-old wild-type (G) and Barx2−/− (H, I) mice. Relative to wild-type muscle, Barx2−/− muscle exhibits a larger distance between myofibers and increased collagen deposition between myofibers (as shown by collagen I immunostaining in J–L). Myofibers in Barx2−/− muscle were generally more rounded, although some showed an angulated morphology with cytoplasmic vacuoles (I, black arrowheads). (M–R): Comparison of diaphragm in 6-month-old wild-type (M–O) and Barx2−/− (P–R) mice. The diaphragm of Barx2−/− mice is much thinner than that of wild-type mice and moderately fibrotic as indicated by Masson’s trichrome staining (panels O and R). A, B, D, E, G, H, M, N, P, and Q, hematoxylin and eosin staining; (J–L), collagen I immunostaining (brown) with iron hematoxylin counterstain; C, F, O, and R, Masson’s trichrome staining. Abbreviations: TA, tibialis anterior; WT, wild type.
Barx2−/− soleus muscles exhibited narrower myofibers (Fig. 2F), more rounded myofibers, and an increased distance between myofibers compared to the soleus muscles of wild-type mice (Fig. 3G–3I). Variation in interstitial space may be due to changes in osmotic pressure during dissection, although sections of Barx2−/− soleus also showed increased staining with collagen I antibodies suggesting perimysial fibrosis (Fig. 3J–3L). We also found groups of angulated atrophic fibers (Fig. 3I, black arrowheads). Diaphragm muscle was also dramatically affected by loss of Barx2; diaphragms of 6-month-old Barx2−/− mice were substantially thinner than those of wild-type littermates (Fig. 3M–3R) and this was largely associated with thinner myofibers. There was only mild fibrosis of the diaphragm muscle at this age (Fig. 3O, 3R). Comparison of 6- and 12-month-old Barx2−/− muscle sections suggested that the muscle defects worsened as animals became older (not shown).
Barx2 Is an Important Regulator of Muscle Regeneration
To determine whether Barx2 expression is involved in regeneration after acute muscle injury, we performed intramuscular injection of cardiotoxin [50] in Barx2+/− mice and examined Barx2 expression by immunostaining. On day 4 after injection, the number of Barx2-expressing cells increased dramatically in the cardiotoxin-injected TA muscle but not in the control, saline-injected muscle (Fig. 4A, 4B). RT-PCR analysis of the injected muscles confirmed increased Barx2 mRNA expression 4 days after injury (Fig. 4C). Most cells that expressed Barx2 in regenerating muscle also expressed Pax7, suggesting that these are activated and proliferating satellite cells (Fig. 4D).
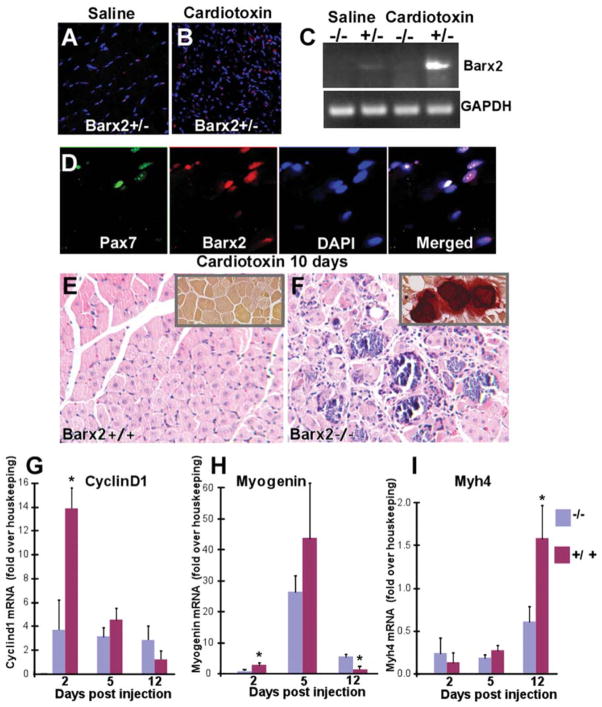
Figure 4.
Barx2 is important for muscle regeneration. (A, B): Regenerating cardiotoxin (CTX)-injected muscle (B) shows a dramatic increase in Barx2-expressing nuclei (red) when compared to saline-injected muscle (A). (C): RT-PCR shows that Barx2 expression is upregulated in regenerating CTX-injected muscle relative to saline-injected muscle. A representative result is shown; similar upregulation was seen in replicate experiments. (D): Barx2-expressing cells also express Pax7 (green). (E): Hematoxylin-eosin-stained paraffin sections of Barx2+/− TA muscles harvested 10 days after CTX injection show efficient muscle regeneration indicated by centrally located nuclei in almost all myotubes. (F): In contrast, sections of Barx2−/− muscle harvested 10 days after injection show disorganized morphology with necrotic fibers, myotubes of different sizes, and undifferentiated myoblasts. Alizarin red staining also reveals calcium deposits in Barx2−/− muscle (insets). (G, H, I): Quantitative RT-PCR analysis of cyclin-D1 (G), myogenin (H), and myosin heavy chain (I) expression at 2, 5, and 12 days after CTX injection in the TA muscle. Gene expression data from the cardiotoxin-treated limb was normalized to that from the saline (vehicle)-treated contralateral limb and to a pool of housekeeping genes. Three mice of each genotype were used per time point. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; RT-PCR, reverse transcriptase polymerase chain reaction; TA, tibialis anterior.
Barx2+/+, Barx2+/−, and Barx2−/− TA muscles were examined histologically 10 days after cardiotoxin injury. Barx2+/+ (Fig. 4E) and Barx2+/− (not shown) muscle showed efficient regeneration as indicated by many new myofibers with centrally located nuclei. In contrast, in Barx2−/− muscle, necrosis was still evident; there were more mononucleate cells, and regenerating myofibers were more irregular in shape and size than in Barx2+/− muscle (Fig. 4F). Alizarin red staining, which identifies fibers with high intracellular calcium in dystrophic muscles [57], revealed frequent calcium deposits in the regenerating muscles of Barx2−/− but not Barx2+/− mice (Fig. 4E, 4F insets).
We next examined whether loss of Barx2 altered the normal temporal pattern of gene expression during muscle repair. The TA muscles of Barx2−/− or control (Barx2+/+ and Barx2+/−) mice were injected with cardiotoxin or saline (in the contralateral limb); muscles were harvested at 2, 5, and 12 days after injury for isolation of RNA. At day 2 after injection, the expression of cyclin-D1 was induced nearly 14-fold in cardiotoxin-treated muscle relative to saline-treated muscle in Barx2+/+ and Barx2+/− mice (controls). In contrast, Barx2−/− mice showed less than fourfold induction of cyclin-D1 at this time. The difference in cyclin-D1 expression between genotypes was no longer significant at day 5 (Fig. 4G). Myogenin showed nearly threefold induction by day 2 after cardiotoxin treatment in control mice suggesting that differentiation begins while myoblast proliferation is ongoing. This is in agreement with previous studies [58]. In contrast, there was no induction of myogenin at day 2 in Barx2−/− mice (Fig. 4H). Myogenin expression was induced nearly 40-fold at day 5 in control mice, and while apparently lower in Barx2−/− mice this was not statistically significant (Fig. 4H). By day 12 after treatment, myogenin expression was reduced to baseline in controls suggesting that the differentiation phase is complete. However, myogenin expression remained significantly elevated in Barx2−/− mice at this time. This pattern suggests a temporal shift in myogenin expression after injury and is similar to that reported previously for mice lacking the Mnf1 forkhead factor [58]. Finally, expression of MyHC 4 (Myh4), which is a marker for myofibers, is lower in Barx2−/− mice than control mice at day 12 after injury (Fig. 4I). This is consistent with the appearance of fewer regenerated myofibers in Barx2−/− muscle sections (Fig. 4F).
Barx2−/− Myoblasts Show Altered Morphology and Reduced Proliferation in Culture
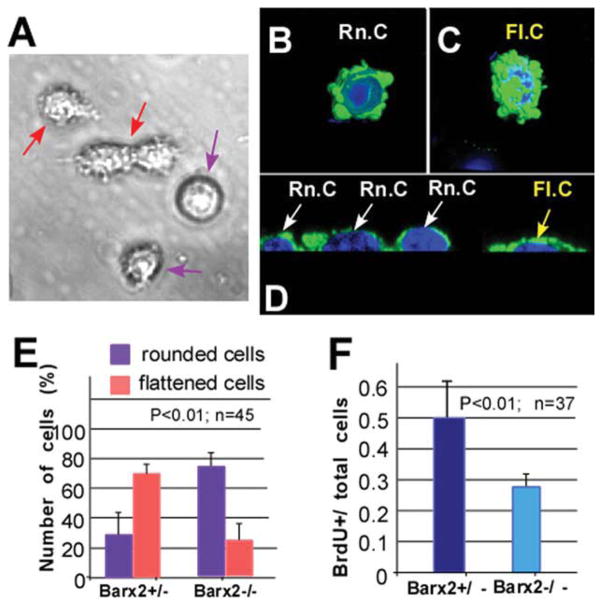
We prepared cultures of satellite-cell derived myoblasts from postnatal day 4 (P4) muscles [51]; these cultures contain primarily proliferating myoblasts expressing Pax7 and MyoD. In line with previous report [59], we found that cultured myoblasts displayed two different morphologies: rounded cells with smooth membranes (Fig. 5A, black arrows) and more elongated and flattened cells with processes (Fig. 5A, red arrows). The “rounded” and “flattened” myoblasts were examined in more detail by confocal imaging after staining with antibodies to smooth muscle actin (SMA), which is expressed transiently in undifferentiated myoblasts [41, 60]. The flattened morphology was associated with cell spreading, filamentous actin redistribution, and increased numbers of cell surface protrusions (Fig. 5B–5D). Both rounded and flattened cells expressed Pax7 (not shown), consistent with undifferentiated myoblasts.
Figure 5.
Primary myoblast cultures from Barx2−/− muscle show altered morphology and reduced proliferation. (A): Myoblast cultures maintained in growth media contain a mixture of two types of cells: relatively rounded cells with smooth membranes (violet arrows) and slightly elongated cells with more processes (red arrows). (B–D): Confocal microscopy and serial 3D reconstruction performed using IMARIS software show that these two populations correspond to rounded (Rn.c; white arrows) (B, D) or elongated/flattened cells (Fl.c; yellow arrows) as is most apparent in transverse optical sections (C, D). Green, smooth muscle actin antibody staining; blue, DAPI. (E): Cells displaying rounded or flattened morphologies were counted in confocal microscopy images, revealing that cells with the flattened morphology predominate in Barx2+/− cultures, whereas cells with rounded morphology predominate in Barx2−/− cultures. (F): Reduced proliferation in Barx2−/− cultures relative to Barx2+/− cultures as measured by BrdU incorporation. In panels (E) and (F), n indicates number of fields per genotype used for quantification. Replicate experiments with independent myoblast isolates gave similar results. Abbreviation: BrdU, Bromodeoxyuridine.
When we compared cultures derived from Barx2−/− muscle with cultures derived from Barx2+/− (Fig. 5E) or wild-type (not shown) muscle, we found that Barx2−/− cells were more likely to be rounded. Specifically, only 20% of cells showed the flattened morphology in Barx2−/− cultures whereas greater than 60% of cells had this morphology in Barx2+/− cultures (Fig. 5E). This suggests that absence of Barx2 results in decreased spreading and thus a reduced area of cell/substrate interaction. This result is in agreement with our previous findings that Barx2 regulates SMA expression, cell adhesion, and cell migration [41, 53, 61].
It has been previously reported that rounded myoblasts proliferate more slowly than flattened myoblasts in culture [59]. Consistent with this, analysis of BrdU incorporation showed that Barx2−/− myoblast cultures had decreased proliferation relative to Barx2+/− cultures (Fig. 5F).
Barx2−/− Myoblasts Show Delayed Differentiation and Barx2−/− Myotubes Have Poor Substrate Adhesion
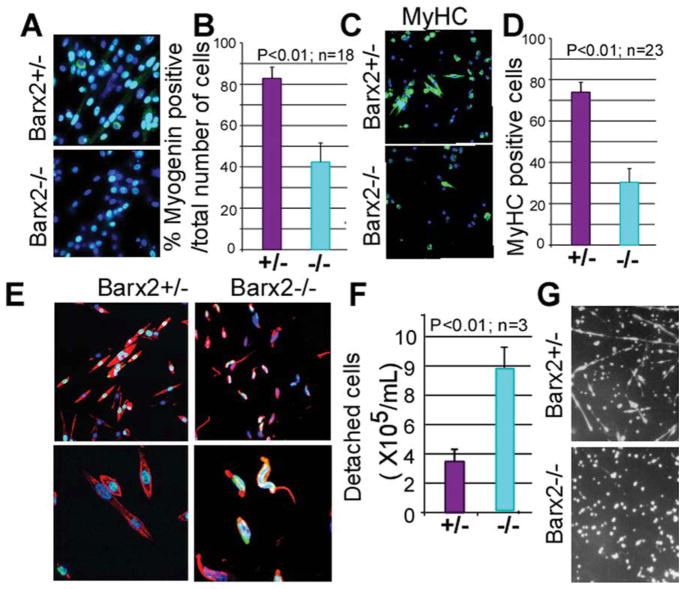
On serum withdrawal, myoblasts in culture cease to divide and begin to express differentiation markers [41, 62]. It has been reported that rounded myoblasts are less differentiation-competent than flattened myoblasts [59]. We examined the expression of the differentiation markers myogenin and heavy chain myosin in Barx2−/− and Barx2+/− myoblast cultures after serum withdrawal. After 6 hours of serum withdrawal, Barx2−/− cultures contained fewer myogenin-expressing cells than Barx2+/− cultures (Fig. 6A, 6B) and at 24 hours Barx2−/− cultures contained fewer fast MyHC-expressing myotubes (Fig. 6C, 6D). Thus loss of Barx2 expression impairs the response of myoblasts to terminal differentiation signals and delays morphological differentiation.
Figure 6.
Primary myoblasts from Barx2−/− muscle have poor adhesion and show delayed differentiation. (A–D): Differentiation was induced in myoblast cultures by serum withdrawal, and gene expression was examined after 6 (A, B) and 24 (C, D) hours. At 6 hours after serum withdrawal, Barx2−/− cultures show almost twofold reduction in the number of myogenin-expressing cells relative to Barx2+/− cultures suggesting a delay in the onset of differentiation (A, B); green, myogenin; blue, DAPI. At 24 hours after serum withdrawal, Barx2−/− cultures show fewer myotubes and reduced expression of MyHC relative to Barx2+/− cultures (C, D); green, MyHC; blue, DAPI. In panels (B) and (D), n indicates number of fields per genotype used for quantification. Replicate experiments with independent myoblast isolates gave similar results. (E): At 72 hours after serum withdrawal, myotubes in Barx2−/− cultures begin to detach from the plate. Red, F-actin, blue, DAPI. (F): Barx2−/− cells plated on fibronectin detach more readily than Barx2+/− cells in a plate-washing assay (n, number of independent experiments). (G): Barx2+/− myoblasts plated on fibronectin form myotubes in approximately 72 hours whereas Barx2−/− myoblasts do not. Abbreviations: DAPI, 4′,6-diamidino-2-phenylindole; MyHC, myosin heavy chain.
Although Barx2−/− myoblasts were delayed in differentiation over the first 24 hours after serum withdrawal, by 48 hours most of Barx2−/− myoblasts had formed short myotubes. However, between 48 and 72 hours many nascent Barx2−/− myotubes assumed an irregular curve shape in contrast to the uniformly straight elongated Barx2+/− myotubes (Fig. 6E). The irregular shape appeared to be due to partial detachment of the myotubes from the collagen substrate.
To determine whether Barx2−/− and Barx2+/− myoblasts have reduced adhesion properties, we seeded Barx2+/− and Barx2−/− myoblasts onto dishes coated with fibronectin instead of collagen. Our experience with wild-type myoblasts indicated that fibronectin mediates only weak attachment of myoblasts and delays myotube formation by several days relative to collagen. It has also been recently reported by others that myoblasts plated on fibronectin adhere weakly [63]. Cells were grown for 48 hours and then transferred to differentiation medium for 7–8 days. Barx2+/− cells formed myotubes after 1 week whereas Barx2−/− myoblasts failed to form myotubes and remained rounded and poorly attached (Fig. 6G). We also performed a plate-washing assay to determine how readily myoblasts detached from the fibronectin coated plate. Plates were rinsed five times with PBS, the rinsates were combined, and the numbers of cells that they contained were counted. Nearly three times more Barx2−/− cells were detached from the plates than Barx2+/− cells (Fig. 6F), indicating that their adhesion to a fibronectin substrate is substantially reduced.
To better understand the mechanism of reduced substrate adhesion in Barx2−/− myoblast cultures, we examined gene expression profiles of cultured wild-type and Barx2−/− myoblasts using a PCR array focused on extracellular matrix and adhesion molecule genes (SABiosciences). A total of 15 genes involved in regulation of cell adhesion and cell matrix remodeling showed greater than twofold decrease in expression in Barx2−/− myoblasts relative to wild-type myoblasts (supporting information Table 1). This result correlates with our previous findings that Barx2 is involved in regulation of cell adhesion in mesenchymal progenitor cells [44, 46, 53] and that ectopic Barx2 expression could induce matrix metalloprotease (MMP) expression in MCF7 cells [61]. Downregulation of cell adhesion and matrix remodeling molecules in Barx2−/− myoblasts could contribute to impaired substrate attachment, migration, and fusion [41], thus delaying differentiation.
Loss of Barx2 Exacerbates the Dystrophic Phenotype in mdx Mice
To better understand the role of Barx2 in muscle repair, we examined the effects of Barx2 deletion in the mdx mouse model of human Duchenne muscular dystrophy (DMD) and Becker muscular dystrophy [64]. Mdx mice carry a loss-of-function point mutation in the X-linked dystrophin gene. Although mdx mice display extensive necrosis of muscle fibers at 2 weeks of age, they maintain muscle integrity due to a high regenerative capacity, which leads to hypertrophy [65–67]. Except in the diaphragm, adult mdx mice do not display the muscle fiber loss and extensive interstitial fibrosis observed in human DMD patients. Moreover, mdx mice display normal external appearance and only moderately reduced life spans [68].
We crossed Barx2 mutant and mdx mice and inbred the “Barx2:mdx” double mutants for up to nine generations to reduce genetic variability. By six generations, crosses between mice that were both heterozygous for Barx2 and null for dystrophin (i.e., Barx2+/−:mdx) produced litters in which Barx2−/−:mdx mice were significantly under-represented by weaning age (supporting information Table 2). This was in contrast to Barx2−/− mice on the C57Bl/6 background that retained normal mendelian ratios. It is notable that MyoD−/−:mdx double mutant mice become nonviable around the ninth generation [69]. Our subsequent analyses were performed on eighth to tenth generation mice.
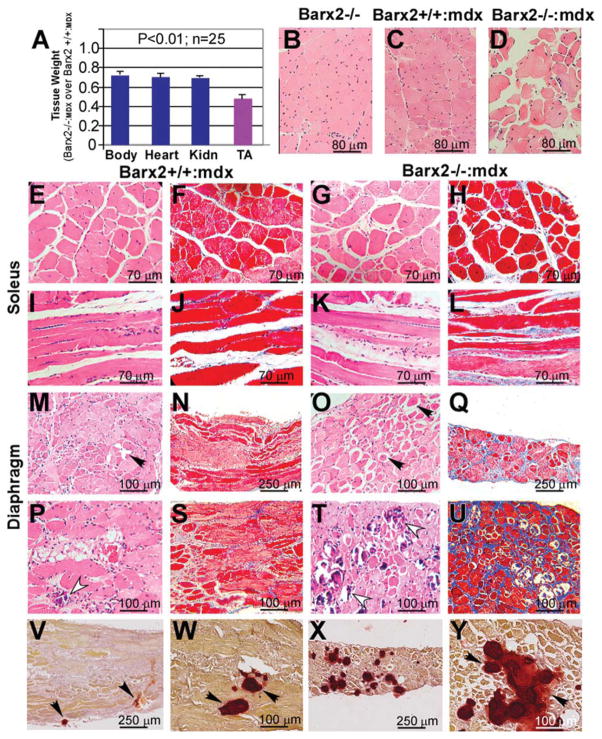
Barx2+/+:mdx and Barx2−/−:mdx sibling pairs (all of which were null for dystrophin) were collected and body, muscle, and organ weights were measured. Barx2−/−:mdx were at approximately 30% smaller than Barx2+/+:mdx mice at 4 weeks of age and organs such as kidney were reduced proportionately (~ 30%), However, the TA muscle in Barx2−/−:mdx mice weighed on average 50% less than that in Barx2+/+:mdx mice (Fig. 7A). suggesting that muscle is specifically wasted in Barx2−/−:mdx mice (Fig. 7A). This reduction in TA weight is also more dramatic than that observed in Barx2−/− mice that have a functional dystrophin gene (Fig. 2).
Figure 7.
Phenotype of Barx2/mdx double null mice. (A): Comparison of total body, heart, Kidn, and TA weights in Barx2−/−:mdx (shown as a ratio of Barx2−/−:mdx over Barx2+/+:mdx weights). TA muscle shows a dramatic reduction in weight relative to other organs. Tissues were harvested and weighed from 4-week-old Barx2+/+:mdx and Barx2−/−:mdx sibling pairs (n, number of pairs studied). (B–D): Hematoxylin-eosin (H&E)-stained paraffin sections of TA muscles obtained from 6-month-old Barx2−/−, Barx2+/+:mdx, and Barx2−/−:mdx mice. Double knockout TA muscle shows variability of fiber size suggesting presence of atrophy and hypertrophy of myofibers (D). (E–L): Soleus muscle of Barx2−/−:mdx mice (G, K, H&E staining; H, L, trichrome staining) shows greater variability in myofiber size and shape and marked endomysial and perimysial fibrosis relative to soleus muscle of Barx2+/+:mdx mice (E, I, H&E staining; F, J, trichrome staining). (M–U): Diaphragm muscles of 6-month-old (M–Q) and 12-month-old (P–U) Barx2+/+:mdx (M, P, H&E staining; N, S, trichrome staining) and Barx2−/−:mdx (O, T, H&E staining; Q, U, trichrome staining) mice. Diaphragms of 6-month-old Barx−/−:mdx mice appear much thinner than diaphragms of Barx2+/+:mdx mice (compare N and Q). In addition, diaphragms of 6-month-old Barx2−/−:mdx mice show frequent rounded atrophic fibers (black arrows). Atrophic fibers are dark red and glassy (they represent hypercontracted fibers). Trichrome staining shows more extensive fibrosis in six Barx2−/−:mdx diaphragms relative to Barx2+/+:mdx diaphragms (compare N and Q). The diaphragms of 12-month-old Barx2−/−:mdx mice show extensive fibrosis and greater instance of focal myofiber fibrosis (compare S and U), and necrotic myofibers undergoing myophagocytosis (T, white arrowheads) relative to Barx2+/+:mdx mice (P, white arrowhead). (V–Y): Alizarin red staining reveals increased calcium deposits in the Barx2−/−:mdx diaphragm muscle (X, Y) relative to Barx2+/+:mdx muscle (V, W). Abbreviations: Kidn, kidney; TA, tibialis anterior.
Barx2−/−:mdx mice also developed spine deformation resembling kyphosis by 6 months of age and became progressively weak and less ambulatory with a waddling gait. Although similar characteristics were observed in some older Barx2−/− mice (Fig. 2), this pathology appeared earlier and with greater penetrance in Barx2−/−:mdx mice.
Sections of TA muscles from 6-month-old Barx2−/−, Barx2+/+:mdx and Barx2−/−:mdx mice were examined histologically and compared to those from Barx2−/− mice (Fig. 7B–7D). Barx2+/+:mdx muscle showed variability in myofiber size as well as many central nuclei indicating ongoing repair of muscle damage as would be expected in mdx muscle at this age (Fig. 7C). In contrast, the organization of Barx2−/−:mdx TA muscle appeared extremely aberrant with greater variability in myofiber size and fewer central nuclei relative to muscle from Barx2+/+:mdx mice suggesting less repair (Fig. 7D). Barx2−/−:mdx soleus muscles also showed greater variability in myofiber size and shape as well as more endomysial and perimysial fibrosis relative to the soleus muscles of Barx2+/+:mdx mice (Fig. 7F–7L).
The diaphragm muscles (Fig. 7Q) of 6-month-old Barx−/−:mdx mice appeared much thinner than the diaphragms of Barx2+/+:mdx mice (Fig. 7N). In addition, diaphragms of Barx2−/−:mdx mice showed more frequent rounded opaque fibers (dark red and glassy, black arrows) often with clear vacuoles (compare Fig. 7M and 7O). These are hypercontracted atrophic fibers. At 12 months, diaphragms of Barx2−/−:mdx mice show a greater instance of focal myofiber necrosis and necrotic myofibers undergoing myophagocytosis (compare Fig. 7P and 7T) as well as much more fibrosis (compare Fig. 7S and 7U), relative to Barx2+/+: mdx mice. Alizarin red staining revealed greatly increased calcium deposits in Barx2−/−:mdx diaphragm muscles relative to Barx2+/+:mdx mice (compare Fig. 7V, 7W and 7X, 7Y). While some of the histological features of Barx2−/−:mdx muscles were also observed in Barx2−/− muscles (Fig. 3), the latter do not show the degenerative character of the mdx model, and fibrosis and calcium deposition is less pronounced.
Discussion
Our studies identify Barx2 as an important new regulator of myogenesis that is expressed in embryonic myoblasts as well as adult satellite cells and is required for normal muscle growth, maintenance, and regeneration.
Overlapping Functions for Barx2 and Pax7 in Muscle Progenitor Cells
Barx2 is expressed in muscle progenitor cells at embryonic, juvenile, and adult stages in a pattern that overlaps extensively with Pax7 expression, suggesting that Barx2 is a new marker for embryonic and fetal myoblasts and adult satellite cells. We also observed a population of Barx2-positive/Pax7-negative mononucleate cells in adult muscle suggesting that in addition to satellite cells, Barx2 is expressed in some interstitial cells or another muscle progenitor type [70–73]. Studies of Barx2-expressing cell populations in muscle are ongoing.
The Barx2 null mouse shows several parallels with the Pax7 germline null model. Embryonic muscle development in Pax7 germline null mice is grossly normal [74]; however, postnatal muscle growth and regeneration in these mice is severely impaired [28, 74, 75]. Similarly, Barx2 null mice show apparently normal embryonic development but moderately impaired postnatal muscle growth and maintenance, and severely impaired regeneration. We observed no alteration of the Pax7 expression pattern in Barx2 null mice by immunostaining (not shown), although Pax7 expression was slightly reduced in Barx2 null muscle by RT-PCR and in cultured Barx2 null myoblasts (supporting information Fig. 2). Currently, it is unclear whether this has any role in the phenotype of Barx2 null mice; Barx2 and Pax7 may function in parallel pathways. It was recently shown that conditional Pax7 null mice in which Pax7 is deleted after 4 weeks of age have no defects in muscle maintenance or regeneration after acute injury [76]. This suggests compensation by other factors potentially working in parallel with Pax7 in adult muscle; Barx2 might be one such factor.
Barx2 Regulates the Proliferation and Differentiation of Satellite Cells in Culture
A previous study described populations of satellite cell-derived myoblasts with “round” and “thick” morphologies that grow clonally on isolated myofibers [59]. The round cells were shown to be directly descended from satellite cells and were described as having “stem cell-like characteristics” being slow-dividing and self-renewing. The thick cells were the rapidly dividing, differentiation competent progeny of the round cells [59]. Our satellite cell-derived myoblast cultures (from mice of all genotypes) contained “rounded and flattened cells” that appeared to correspond to the “round and thick” morphologies described by Hashimoto et al. [59]. However, Barx2−/− myoblasts showed a predominantly rounded morphology in culture and consistent with this, they were less proliferative and markedly delayed in differentiation when compared to wild-type or heterozygous cells. These data indicate that myoblasts lacking Barx2 tend to retain a primitive state marked by a reduced capacity for proliferation and differentiation.
The presence of fewer flattened cells in Barx2−/− cultures also suggests defects in cell spreading and is consistent with our previous observations that Barx2−/− primary myoblasts show delayed cytoskeletal remodeling and SMA upregulation after serum withdrawal [41]. This SMA-associated remodeling is an important early step in myoblast differentiation that facilitates the migration and alignment of myoblasts for fusion. Overall, the delay in fusion of Barx2−/− myoblasts in this study agrees with our previous data showing that Barx2 cooperates with MRFs and that its overexpression accelerates differentiation of C2C12 myoblasts [41, 46].
Nascent Barx2−/− myofibers showed reduced substrate adhesion leading to a curved shape and Barx2−/− myoblasts had reduced expression of cell adhesion and matrix molecules, including integrins and cell adhesion molecules (CAMs) that are known to be important for cell spreading and differentiation in culture [77–79]. Moreover, reduced expression of particular CAMs could compromise crosstalk between the satellite cell and the myofiber in vivo [80, 81]. Barx2 expression is downregulated in mature myofibers [43]. Presumably, therefore, the effect of Barx2 gene deletion would be more acute in nascent than in mature myofibers. Consistent with this, we recently found that forcing a high level of Barx2 expression in young but not mature myofibers affects their morphological stability [43]. Overall, our data suggest that Barx2 facilitates the emergence of fast dividing, differentiation-competent myoblasts and promotes differentiation by altering the expression of both regulatory and mechanochemical factors.
The Role of Barx2 in Muscle Growth, Maintenance, and Regeneration
Satellite cells are required for postnatal muscle growth, are activated when the muscle is overloaded [82], and mediate repair of muscle injury [83]. Moreover, diseases involving accelerated muscle atrophy such as congenital myotonic dystrophy are suggested to involve impaired satellite cell function [84]. Based on our observations that Barx2 is predominantly expressed in satellite cells and their myoblast progeny and that lack of Barx2 delays myoblast proliferation and differentiation in vitro, it is most likely that delayed muscle growth in Barx2−/− pups, the atrophic phenotype of adult Barx2−/− muscle, and lack of repair after injury are due to satellite cell/myoblast dysfunction.
Regarding the precise nature of this dysfunction, our muscle injury studies show misregulation of cell cycle and myogenic genes in regenerating Barx2 null muscle. Lower cyclin-D1 expression in Barx2−/− mice at day 2 after cardiotoxin injury is consistent with delay in satellite cell activation and/or reduced proliferation of their progeny myoblasts. Delayed upregulation of myogenin at day 2 and sustained expression of this gene at day 12, together with reduced expression of the myofibre marker Myh4 at day 12 in Barx2−/− mice, are consistent with delayed onset and progression of differentiation. While it remains to be determined whether these gene expression changes are a cause or consequence of impaired proliferation and differentiation, the observation that Barx2 overexpression [43] or loss of expression (this study) in myoblast culture also alters expression of these genes suggests that Barx2 may be a direct effector of their regulation.
Impaired satellite cell and myoblast function in the absence of Barx2 is also likely to underlie the phenotype of the Barx2/mdx mouse model. The muscle of homozygous mdx mice is characterized by ongoing degeneration and regeneration of myofibers leading eventually to accumulation of fibrous and fatty infiltrates [85]. However, muscle function is maintained in young mdx mice, likely because repair is able to keep pace with injury. In contrast, double mutant Barx2−/−:mdx mice display increased penetration of the disease with phenotypic similarity to human DMD. Even young Barx2−/−:mdx mice showed extremely aberrant repair and extensive fibrous infiltration and calcification, which are consistent with impaired satellite cell/myoblast function. The mice also display gross musculoskeletal abnormalities including changes in gait, the latter resembling the waddling gait of boys with DMD due to contracture of the Achilles tendons [86]. The rapid and severe decline in muscle integrity in Barx2−/−:mdx mice suggests that they may be useful model for studies of DMD.
Conclusion
Based on our current studies and previous work [41, 46], we conclude that Barx2 is a key regulator of proliferation and differentiation competence in satellite cell-derived myoblasts; in the absence of the vital activities of Barx2, satellite cell-mediated muscle maintenance and regeneration is grossly impaired.
Supplementary Material
Acknowledgments
We thank Kathryn Crossin and Bruce Cunningham for critical reading of the manuscript. This work was supported by National Institutes of Health Grant 5R01AR053163 (to H.P.M. and R.M.) and by a grant from the Association Francaise contre les Myopathies (to H.P.M.). This study was also supported by the Neurosciences Support Foundation (to H.P.M).
Footnotes
Disclosure of Potential Conflicts of Interests
The authors indicate no potential conflicts of interests.
Author contributions: R.M.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript; K.G., M.B.,A.G.,J.Z., and J.-A.H.: collection and/or assembly of data; H.M.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of manuscript.
References
- 1.Mauro A. Satellite cell of skeletal muscle fibers. J Cell Biol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schultz E, Gibson MC, Champion T. Satellite cells are mitotically quiescent in mature mouse muscle: An EM and radioautographic study. J Exp Zool. 1978;206:451–456. doi: 10.1002/jez.1402060314. [DOI] [PubMed] [Google Scholar]
- 3.Asakura A, Rudnicki MA, Komaki M. Muscle satellite cells are multi-potential stem cells that exhibit myogenic, osteogenic, and adipogenic differentiation. Differentiation. 2001;68:245–253. doi: 10.1046/j.1432-0436.2001.680412.x. [DOI] [PubMed] [Google Scholar]
- 4.Gargioli C, Slack JMW. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- 5.Montarras D, Morgan J, Collins C, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 6.Meeson AP, Hawke TJ, Graham S, et al. Cellular and molecular regulation of skeletal muscle side population cells. Stem Cells. 2004;22:1305–1320. doi: 10.1634/stemcells.2004-0077. [DOI] [PubMed] [Google Scholar]
- 7.Shinin V, Gayraud-Morel B, Gomès D, et al. Asymmetric division and cosegregation of template DNA strands in adult muscle satellite cells. Nat Cell Biol. 2006;8:677–682. doi: 10.1038/ncb1425. [DOI] [PubMed] [Google Scholar]
- 8.Kuang S, Kuroda K, Le Grand F, et al. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007;129:999–1010. doi: 10.1016/j.cell.2007.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardasis CA, Cooper GW. An analysis of nuclear numbers in individual muscle fibers during differentiation and growth: A satellite cell-muscle fiber growth unit. J Exp Zool. 1975;191:347–358. doi: 10.1002/jez.1401910305. [DOI] [PubMed] [Google Scholar]
- 10.Sambasivan R, Yao R, Kissenpfennig A, et al. Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development. 2011;138:3647–3656. doi: 10.1242/dev.067587. [DOI] [PubMed] [Google Scholar]
- 11.Rudnicki MA, Schnegelsberg PN, Stead RH, et al. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 12.Tajbakhsh S, Bober E, Babinet C, et al. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev Dyn. 1996;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Kassar-Duchossoy L, Gayraud-Morel B, Gomès D, et al. Mrf4 determines skeletal muscle identity in Myf5:Myod double-mutant mice. Nature. 2004;431:466–471. doi: 10.1038/nature02876. [DOI] [PubMed] [Google Scholar]
- 14.Hinterberger TJ, Sassoon DA, Rhodes SJ, et al. Expression of the muscle regulatory factor MRF4 during somite and skeletal myofiber development. Dev Biol. 1991;147:144–156. doi: 10.1016/s0012-1606(05)80014-4. [DOI] [PubMed] [Google Scholar]
- 15.Brennan TJ, Edmondson DG, Olson EN. Aberrant regulation of MyoD1 contributes to the partially defective myogenic phenotype of BC3H1 cells. J Cell Biol. 1990;110:929–937. doi: 10.1083/jcb.110.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pownall ME, Gustafsson MK, Emerson CP. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu Rev Cell Dev Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 17.Fuchtbauer EM, Westphal H. MyoD and myogenin are coexpressed in regenerating skeletal muscle of the mouse. Dev Dyn. 1992;193:34–39. doi: 10.1002/aja.1001930106. [DOI] [PubMed] [Google Scholar]
- 18.Cooper RN, Tajbakhsh S, Mouly V, et al. In vivo satellite cell activation via Myf5 and MyoD in regenerating mouse skeletal muscle. J Cell Sci. 1999;112:2895–2901. doi: 10.1242/jcs.112.17.2895. [DOI] [PubMed] [Google Scholar]
- 19.Charge SBP, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 20.Morgan BA, Tabin C. Hox genes and growth: Early and late roles in limb bud morphogenesis. Dev Suppl. 1994:181–186. [PubMed] [Google Scholar]
- 21.Bober E, Franz T, Arnold HH, et al. Pax-3 is required for the development of limb muscles: A possible role for the migration of dermomyotomal muscle progenitor cells. Development. 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 22.Relaix F, Rocancourt D, Mansouri A, et al. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature. 2005;435:948–953. doi: 10.1038/nature03594. [DOI] [PubMed] [Google Scholar]
- 23.Kassar-Duchossoy L, Giacone E, Gayraud-Morel B, et al. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005;19:1426–1431. doi: 10.1101/gad.345505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkpatrick LJ, Yablonka-Reuveni Z, Rosser BW. Retention of Pax3 expression in satellite cells of muscle spindles. J Histochem Cytochem. 2010;58:317–327. doi: 10.1369/jhc.2009.954792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckingham M. Skeletal muscle progenitor cells and the role of Pax genes. C R Biol. 2007;330:530–533. doi: 10.1016/j.crvi.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 26.Relaix F, Montarras D, Zaffran S, et al. Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J Cell Biol. 2006;172:91–102. doi: 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zammit PS, Relaix F, Nagata Y, et al. Pax7 and myogenic progression in skeletal muscle satellite cells. J Cell Sci. 2006;119:1824–1832. doi: 10.1242/jcs.02908. [DOI] [PubMed] [Google Scholar]
- 28.Oustanina S, Hause G, Braun T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004;23:3430–3439. doi: 10.1038/sj.emboj.7600346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFarlane C, Hennebry A, Thomas M, et al. Myostatin signals through Pax7 to regulate satellite cell self-renewal. Exp Cell Res. 2008;314:317–329. doi: 10.1016/j.yexcr.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 30.Dong F, Sun X, Liu W, et al. Pitx2 promotes development of splanchnic mesoderm-derived branchiomeric muscle. Development. 2006;133:4891–4849. doi: 10.1242/dev.02693. [DOI] [PubMed] [Google Scholar]
- 31.Shih HP, Gross MK, Kioussi C. Muscle development: Forming the head and trunk muscles. Acta Histochem. 2008;110:97–108. doi: 10.1016/j.acthis.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shih HP, Gross MK, Kioussi C. Expression pattern of the homeodomain transcription factor Pitx2 during muscle development. Gene Expr Patterns. 2007;7:441–451. doi: 10.1016/j.modgep.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Bendall AJ, Ding J, Hu G, et al. Msx1 antagonizes the myogenic activity of Pax3 in migrating limb muscle precursors. Development. 1999;126:4965–4976. doi: 10.1242/dev.126.22.4965. [DOI] [PubMed] [Google Scholar]
- 34.Mankoo BS, Skuntz S, Harrigan I, et al. The concerted action of Meox homeobox genes is required upstream of genetic pathways essential for the formation, patterning and differentiation of somites. Development. 2003;130:4655–4664. doi: 10.1242/dev.00687. [DOI] [PubMed] [Google Scholar]
- 35.Fukada S, Uezumi A, Ikemoto M, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe S, Kondo S, Hayasaka M, et al. Functional analysis of homeodomain-containing transcription factor Lbx1 in satellite cells of mouse skeletal muscle. J Cell Sci. 2007;120:4178–4187. doi: 10.1242/jcs.011668. [DOI] [PubMed] [Google Scholar]
- 37.Gross MK, Moran-Rivard L, Velasquez T, et al. Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development. 2000;127:413–424. doi: 10.1242/dev.127.2.413. [DOI] [PubMed] [Google Scholar]
- 38.Ono Y, Boldrin L, Knopp P, et al. Muscle satellite cells are a functionally heterogeneous population in both somite-derived and branchiomeric muscles. Dev Biol. 2010;337:29–41. doi: 10.1016/j.ydbio.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brohmann H, Jagla K, Birchmeier C. The role of Lbx1 in migration of muscle precursor cells. Development. 2000;127:437–45. doi: 10.1242/dev.127.2.437. [DOI] [PubMed] [Google Scholar]
- 40.Ochi H, Westerfield M. Lbx2 regulates formation of myofibrils. BMC Dev Biol. 2009;9:13. doi: 10.1186/1471-213X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarenkova HP, Gonzalez KN, Kiosses WB, et al. Barx2 controls myoblast fusion and promotes MyoD-mediated activation of the smooth muscle alpha actin gene. J Biol Chem. 2009;284:14866–14874. doi: 10.1074/jbc.M807208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herring BP, Kriegel AM, Hoggatt AM. Identification of Barx2b, a serum response factor-associated homeodomain protein. J Biol Chem. 2001;276:14482–14489. doi: 10.1074/jbc.M011585200. [DOI] [PubMed] [Google Scholar]
- 43.Meech R, Gomez M, Woolley C, et al. The homeobox transcription factor Barx2 regulates plasticity of young primary myofibers. PLoS ONE. 2010;5:e11612. doi: 10.1371/journal.pone.0011612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edelman DB, Meech R, Jones FS. The homeodomain protein Barx2 contains activator and repressor domains and interacts with members of the CREB family. J Biol Chem. 2000;275:21737–21745. doi: 10.1074/jbc.M909998199. [DOI] [PubMed] [Google Scholar]
- 45.Sellar GC, Li L, Watt KP, et al. BARX2 induces cadherin 6 expression and is a functional suppressor of ovarian cancer progression. Cancer Res. 2001;61:6977–6981. [PubMed] [Google Scholar]
- 46.Meech R, Makarenkova H, Edelman DB, et al. The homeodomain protein Barx2 promotes myogenic differentiation and is regulated by myogenic regulatory factors. J Biol Chem. 2003;278:8269–8278. doi: 10.1074/jbc.M207617200. [DOI] [PubMed] [Google Scholar]
- 47.Olson LE, Zhang J, Taylor H, et al. Barx2 functions through distinct corepressor classes to regulate hair follicle remodeling. Proc Natl Acad Sci USA. 2005;102:3708–3713. doi: 10.1073/pnas.0500519102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Makarenkova H, Becker DL, Tickle C, et al. Fibroblast growth factor 4 directs gap junction expression in the mesenchyme of the vertebrate limb bud. J Cell Biol. 1997;138:1125–1137. doi: 10.1083/jcb.138.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Costes SV, Daelemans D, Cho EH, et al. Automatic and quantitative measurement of protein–protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couteaux R, Mira J-C, d’Albis A. Regeneration of muscles after cardiotoxin injury. I. Cytological aspects. Biol Cell. 1988;62:171–182. [PubMed] [Google Scholar]
- 51.Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol. 1994;125:1275–1287. doi: 10.1083/jcb.125.6.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ostle B, Malone LC, editors. Statistics in Research: Basic Concepts and Techniques for Research Workers: 7. Inferences concerning two populations. 4. Iowa State University Press; Iowa 50010: 1988. pp. 145–175. [Google Scholar]
- 53.Meech R, Edelman DB, Jones FS, et al. The homeobox transcription factor Barx2 regulates chondrogenesis during limb development. Development. 2005;132:2135–2146. doi: 10.1242/dev.01811. [DOI] [PubMed] [Google Scholar]
- 54.Zammit PS, Golding JP, Nagata Y, et al. Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J Cell Biol. 2004;166:347–357. doi: 10.1083/jcb.200312007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halevy O, Piestun Y, Allouh MZ, et al. Pattern of Pax7 expression during myogenesis in the posthatch chicken establishes a model for satellite cell differentiation and renewal. Dev Dyn. 2004;231:489–502. doi: 10.1002/dvdy.20151. [DOI] [PubMed] [Google Scholar]
- 56.Buckingham M, Bajard L, Chang T, et al. The formation of skeletal muscle: From somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bodensteiner JB, Engel AG. Intracellular calcium accumulation in Duchenne dystrophy and other myopathies: A study of 567,000 muscle fibers in 114 biopsies. Neurology. 1978;28:439–46. doi: 10.1212/wnl.28.5.439. [DOI] [PubMed] [Google Scholar]
- 58.Garry DJ, Meeson A, Elterman J, et al. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc Natl Acad Sci. 2000;97:5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hashimoto N, Murase T, Kondo S, et al. Muscle reconstitution by muscle satellite cell descendants with stem cell-like properties. Development. 2004;131:5481–5490. doi: 10.1242/dev.01395. [DOI] [PubMed] [Google Scholar]
- 60.Lancioni H, Lucentini L, Palomba A, et al. Muscle actin isoforms are differentially expressed in human satellite cells isolated from donors of different ages. Cell Biol Int. 2007;31:180–185. doi: 10.1016/j.cellbi.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 61.Stevens TA, Meech R. BARX2 and estrogen receptor-α (ESR1) coordinately regulate the production of alternatively spliced ESR1 isoforms and control breast cancer cell growth and invasion. Oncogene. 2006;25:5426–5435. doi: 10.1038/sj.onc.1209529. [DOI] [PubMed] [Google Scholar]
- 62.Stern-Straeter J, Bran G, Riedel F, et al. Characterization of human myoblast cultures for tissue engineering. Int J Mol Med. 2008;21:49–56. [PubMed] [Google Scholar]
- 63.Siegel AL, Atchison K, Fisher KE, et al. 3D timelapse analysis of muscle satellite cell motility. Stem Cells. 2009;27:2527–2538. doi: 10.1002/stem.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sicinski P, Geng Y, Ryder-Cook AS, et al. The molecular basis of muscular dystrophy in the mdx mouse: A point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 65.Anderson JE, Ovalle WK, Bressler BH. Electron microscopic and autoradiographic characterization of hindlimb muscle regeneration in the mdx mouse. Anat Rec. 1987;219:243–257. doi: 10.1002/ar.1092190305. [DOI] [PubMed] [Google Scholar]
- 66.Carnwath JW, Shotton DM. Muscular dystrophy in the mdx mouse: histopathology of the soleus and extensor digitorum longus muscles. J Neurol Sci. 1987;80:39–54. doi: 10.1016/0022-510x(87)90219-x. [DOI] [PubMed] [Google Scholar]
- 67.Coulton GR, Morgan JE, Partridge TA, et al. The mdx mouse skeletal muscle myopathy. I. A histological, morphometric and biochemical investigation. Neuropathol Appl Neurobiol. 1988;14:53–70. doi: 10.1111/j.1365-2990.1988.tb00866.x. [DOI] [PubMed] [Google Scholar]
- 68.Chamberlain JS, Metzger J, Reyes M, et al. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 69.Inanlou MR, Dhillon GS, Belliveau AC, et al. A significant reduction of the diaphragm in mdx:MyoD−/− (9th) embryos suggests a role for MyoD in the diaphragm development. Dev Biol. 2003;261:324–336. doi: 10.1016/s0012-1606(03)00319-1. [DOI] [PubMed] [Google Scholar]
- 70.Ferrari G, Cusella-De Angelis G, Coletta M, et al. Muscle Regeneration by bone marrow-derived myogenic progenitors. Science. 1998;279:1528–1530. doi: 10.1126/science.279.5356.1528. [DOI] [PubMed] [Google Scholar]
- 71.Gussoni E, Soneoka Y, Strickland CD, et al. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 72.Polesskaya A, Seale P, Rudnicki MA. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell. 2003;113:841–852. doi: 10.1016/s0092-8674(03)00437-9. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell KJ, Pannérec A, Cadot B, et al. Identification and characterization of a non-satellite cell muscle resident progenitor during postnatal development. Nat Cell Biol. 2010;12:257–266. doi: 10.1038/ncb2025. [DOI] [PubMed] [Google Scholar]
- 74.Mansouri A, Stoykava A, Torres M, et al. Dysgenesis of cephalic neural crest derivatives in Pax7−/− mutant mice. Development. 1996;122:831–838. doi: 10.1242/dev.122.3.831. [DOI] [PubMed] [Google Scholar]
- 75.Olguin HC, Olwin BB. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: A potential mechanism for self-renewal. Dev Biol. 2004;275:375–88. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lepper C, Conway SJ, Fan C-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460:627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mege RM, Goudou D, Diaz C, et al. N-cadherin and N-CAM in myoblast fusion: Compared localisation and effect of blockade by peptides and antibodies. J Cell Sci. 1992;103:897–906. doi: 10.1242/jcs.103.4.897. [DOI] [PubMed] [Google Scholar]
- 78.Peck D, Walsh FS. Differential effects of over-expressed neural cell adhesion molecule isoforms on myoblast fusion. J Cell Biol. 1993;123:1587–1595. doi: 10.1083/jcb.123.6.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeschnigk M, Kozian D, Kuch C, et al. Involvement of M-cadherin in terminal differentiation of skeletal muscle cells. J Cell Sci. 1995;108:2973–2981. doi: 10.1242/jcs.108.9.2973. [DOI] [PubMed] [Google Scholar]
- 80.Irintchev A, Zeschnigk M, Starzinski-Powitz A, et al. Expression pattern of M-cadherin in normal, denervated, and regenerating mouse muscles. Dev Dyn. 1994;199:326–337. doi: 10.1002/aja.1001990407. [DOI] [PubMed] [Google Scholar]
- 81.Wróbel E, Brzóska E, Moraczewski J. M-cadherin and β-catenin participate in differentiation of rat satellite cells. Eur J Cell Biol. 2007;86:99–109. doi: 10.1016/j.ejcb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 82.Dangott B, Schultz E, Mozdziak PE. Dietary creatine monohydrate supplementation increases satellite cell mitotic activity during compensatory hypertrophy. Int J Sports Med. 2000;21:13–16. doi: 10.1055/s-2000-8848. [DOI] [PubMed] [Google Scholar]
- 83.Schultz E, Jaryszak DL, Valliere CR. Response of satellite cells to focal skeletal muscle injury. Muscle Nerve. 1985;8:217–222. doi: 10.1002/mus.880080307. [DOI] [PubMed] [Google Scholar]
- 84.Furling D, et al. Defective satellite cells in congenital myotonic dystrophy. Human Mol Genet. 2001;10:2079–2087. doi: 10.1093/hmg/10.19.2079. [DOI] [PubMed] [Google Scholar]
- 85.Lefaucheur JP, Pastoret C, Sebille A. Phenotype of dystrophinopathy in old mdx mice. Anat Rec. 1995;242:70–76. doi: 10.1002/ar.1092420109. [DOI] [PubMed] [Google Scholar]
- 86.Parker AE, Robb SA, Chambers J, et al. Analysis of an adult Duchenne muscular dystrophy population. QJM. 2005;98:729–736. doi: 10.1093/qjmed/hci113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.