Abstract
The human cerebral cortex controls complex cognitive behaviors. During mammalian evolution, the number of neurons increased in many lineages, requiring a larger cortical surface area to fit into a skull that did not scale proportionally. This space problem was solved by cortical folding, resulting in gyrencephalic (folded) brains. While several hypotheses have been proposed to explain cortical gyrification, we lack mechanistic insights to understand the process itself and in particular its underlying genomic changes, that lead to the appearance of cortical folds. In this issue of The EMBO Journal, de Juan Romero et al (2015) tackle the question using a transcriptomics approach to identify gene expression changes in the developing ferret brain prior to the onset of gyrification.
See also: C de Juan Romero et al (July 2015)
Brain size changes remarkably across mammalian species, and this is mostly due to a disproportionate increase in size of the cerebral cortex. While the thickness of the cerebral cortex varies little (by less than one order of magnitude) during evolution, the cortical surface increased dramatically (> 1,000-fold) between certain species, requiring effective packaging into limited skull volume. Folding solves this problem, creating convolutions of the cortical surface called gyri and fissures called sulci. Although folded brains may erroneously be assumed to be a trait distinctive of primates, gyrencephaly is actually present in essentially all mammalian orders. In fact, it is thought that gyrencephaly already existed in the ancestor of all mammals and that the smooth (lissencephalic) cortex seen in some mammalian species evolved secondarily (Kelava et al, 2013; Lewitus et al, 2014).
Even though the folds and fissures are easy to observe, their appearance is not so easy to explain. The pattern of the major gyri and sulci is largely conserved among members of the same order of mammals, suggesting a genetic component in their specification (Zilles et al, 2013). Several theories of gyrification have been proposed (discussed in Kelava et al, 2013). The neural tension hypothesis proposes that axons of the underlying white matter may influence gyrification by pulling together strongly interconnected regions of the cortex. Kriegstein et al (2006) suggest that differential growth rates of upper versus lower neuronal layers result in cortical folding. Moreover, the orientation of neurons, their arborization, and incoming fibers might contribute to gyrification. Novel hypotheses have been inspired recently by progress in the characterization of cortical progenitors and the appreciation of their diversity.
The generation of cortical neurons during development is the result of proliferative and differentiative divisions of neural stem and progenitor cells that form two germinal layers: the ventricular zone (VZ) and the subventricular zone (SVZ) (Borrell & Götz, 2014; Florio & Huttner, 2014; Taverna et al, 2014). Apical radial glia (aRG) are progenitors, the nuclei of which reside in the VZ and that have the ability to self-renew and to generate neurons and basal progenitors, the latter of which migrate to, and form, the SVZ. In the mouse, basal intermediate progenitors (bIPs) usually divide symmetrically to produce two postmitotic neurons, whereas in primates, bIPs also frequently undergo symmetric proliferative divisions, an essential mechanistic determinant of neocortical expansion (Lewitus et al, 2014). Recently, a new progenitor type has been identified that is particularly abundant in gyrencephalic mammals with an expanded SVZ that is subdivided into an inner (iSVZ) and outer (oSVZ) SVZ, the basal radial glia (bRG) (Borrell & Götz, 2014; Florio & Huttner, 2014; Taverna et al, 2014). In addition to their ability to directly generate neurons, bRG exhibit extensive self-renewal and proliferative capacities (Betizeau et al, 2013), thus providing a basis for cortical expansion. The evolutionary expansion of the neocortex is thought to be based on differences in the abundance of these neural progenitor cell types and in their lineages (Lewitus et al, 2014). Moreover, it is reasonable to assume that the balance of proliferation versus differentiation of progenitor types, which is a determinant of germinal layer thickness, contributes to cortical folding. Indeed, in the fetal monkey cortex, the SVZ is relatively large in areas where gyri subsequently develop and relatively thin in areas of prospective sulci (Kriegstein et al, 2006).
In this issue of The EMBO Journal, Victor Borrell and colleagues (de Juan Romero et al, 2015) present an elegant approach to understand the mechanism underlying the folding process. They used the ferret, a gyrencephalic carnivore, to analyze transcriptional differences within a given germinal zone, comparing regions prospective of the splenial gyrus and the lateral sulcus. Specifically, they microdissected the VZ, iSVZ, and oSVZ at postnatal day 2 (P2), that is, one week prior to the first morphological distinction of these folds, and analyzed the transcriptomes using a ferret-specific microarray. Remarkably, although the two regions are nearly adjacent and have a similar cytoarchitecture and cellular composition, de Juan Romero et al (2015) identified 2,218 differentially expressed genes (DEGs) of which the majority was differentially expressed in only one of the germinal zones. The oSVZ contained the largest number of DEGs, which is interesting in light of the notion that bRG located in this germinal layer play central roles in the expansion of the cerebral cortex (Borrell & Götz, 2014; Florio & Huttner, 2014; Taverna et al, 2014).
These findings prompted the idea that in the developing ferret cerebral cortex, gene expression in the germinal layers may occur in modular patterns corresponding to gyri and sulci. The authors set out to test this hypothesis using in situ hybridization of DEGs with known roles in progenitor proliferation/differentiation or cortical patterning. As predicted, changes in gene expression levels often occurred quite abruptly, rather than in smooth gradients, delineating distinct, gyral or sulcal, gene expression domains. Some genes, including Fgfr2, Lhx2, Eomes, and Cdk6, showed obvious domains within the oSVZ, while others, like Cdh8, were specific for the VZ. In contrast, the same genes were expressed homogeneously or in long-range shallow gradients in the mouse. Interestingly, one of the genes that was expressed in specific domains in ferret was Trnp1, which has previously been shown to exhibit regional differences in human fetal cerebral cortex (Stahl et al, 2013). Moreover, knockdown of Trnp1 in the mouse induced greater proliferation of basal progenitors, leading to radial growth and subsequent folding of the cortex, whereas forced overexpression of Trnp1 had the opposite effect, inducing selective aRG self-amplification and decreased generation of basal progenitors (Stahl et al, 2013). It will be interesting to address in the future whether some of the DEGs identified by de Juan Romero et al (2015) have similar instructive roles.
The initial signals that induce folding, and how the differential expression patterns are generated, remain unclear. To address the age at which modular expression can first be observed, de Juan Romero et al (2015) analyzed expression of Eomes, Fgfr2, Fgfr3, and Cdk6 at three different time points spanning the neurogenic period (E30, E34, and P2). All of the selected genes showed homogeneous expression at E34, and only later during early postnatal development did expression domains become distinguishable. Eomes displayed the greatest contrast between modules at P6, the age of onset of gyrus formation. Moreover, Eomes expression levels matched precisely to several of the emerging folds and fissures, suggesting that this could be an important gene in patterning of the cortical folds. In this regard, it is interesting to note that mutations of EOMES in humans cause microcephaly with polymicrogyria (Baala et al, 2007).
Examination of public databases revealed that 81% of genes mutated in human cortical malformation syndromes (see Sun & Hevner, 2014) are among the ferret DEGs. The authors therefore tested whether such genes might also be expressed in modular patterns in the human developing cortex. Indeed, they found variations in expression levels in brain sections from human embryos at 16 and 21 gestational weeks (the latter stage being immediately prior to the formation of the first folds and fissures) for all genes tested, including EOMES, TRNP1, and GPR56. In this context, it is worth mentioning that our laboratory has recently used transcriptomics to identify genes that are preferentially expressed in human apical and basal progenitors. Among these was the human-specific gene ARHGAP11B, which when overexpressed in mouse promotes basal progenitor generation and self-renewal and can induce gyrification (Florio et al, 2015). In light of the observation by de Juan Romero et al (2015), it would be very interesting to examine whether ARHGAP11B is expressed in modular patterns during human cortical development.
The data reported by de Juan Romero et al (2015) (Fig 1) are consistent with the cortical protomap concept proposed by Rakic (1988) to pattern the cerebral cortex primordium into prospective anatomical and functional regions. For cortical folding, modular patterns of gene expression, with combinations of genes possibly different depending on the specific gyrus or sulcus concerned, may impose differential growth, eventually leading to the evagination of cortical tissue and formation of folds. Specific enhancer elements may serve as readout of combinatorial transcription factor expression, especially since enhancers have recently been shown to drive reporter gene expression in discrete regions, or protomaps, in the cerebral cortex (Visel et al, 2013). The transcriptome dataset generated by Borrell and colleagues provides a rich resource and opens novel avenues for future investigations into the genetic regulation of cortical folding. It will remain a challenge to explain the regulation of stereotyped formation of folds and to identify the initial signals upstream of the transcriptional differences described here, so more exciting research lies ahead.
Figure 1.
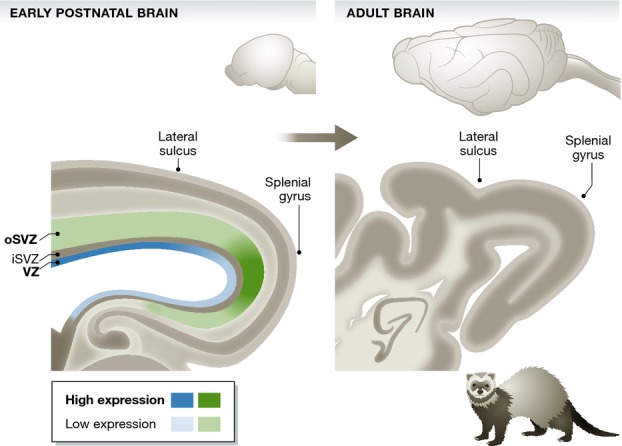
Gene expression occurs in modular patterns in areas prospective of folds and fissures
Abrupt changes in gene expression levels within germinal layers delineate areas prospective of the lateral sulcus and the splenial gyrus in the early postnatal ferret brain prior to the onset of gyrification. Model based on de Juan Romero et al (2015).
References
- Baala L, Briault S, Etchevers HC, Laumonnier F, Natiq A, Amiel J, Boddaert N, Picard C, Sbiti A, Asermouh A, Attie-Bitach T, Encha-Razavi F, Munnich A, Sefiani A, Lyonnet S. Homozygous silencing of T-box transcription factor EOMES leads to microcephaly with polymicrogyria and corpus callosum agenesis. Nat Genet. 2007;39:454–456. doi: 10.1038/ng1993. [DOI] [PubMed] [Google Scholar]
- Betizeau M, Cortay V, Patti D, Pfister S, Gautier E, Bellemin-Ménard A, Afanassieff M, Huissoud C, Douglas RJ, Kennedy H, Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- Borrell V, Götz M. Role of radial glial cells in cerebral cortex folding. Curr Opin Neurobiol. 2014;27:39–46. doi: 10.1016/j.conb.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, Guhr E, Klemroth S, Prufer K, Kelso J, Naumann R, Nusslein I, Dahl A, Lachmann R, Paabo S, Huttner WB. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- de Juan Romero C, Bruder C, Tomasello U, Sanz-Anquela JM, Borrell V. Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J. 2015;34:1859–1874. doi: 10.15252/embj.201591176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelava I, Lewitus E, Huttner WB. The secondary loss of gyrencephaly as an example of evolutionary phenotypical reversal. Front Neuroanat. 2013;7:16. doi: 10.3389/fnana.2013.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martinez-Cerdeno V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lewitus E, Kelava I, Kalinka AT, Tomancak P, Huttner WB. An adaptive threshold in mammalian neocortical evolution. PLoS Biol. 2014;12:e1002000. doi: 10.1371/journal.pbio.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Stahl R, Walcher T, de Juan Romero C, Pilz GA, Cappello S, Irmler M, Sanz-Aquela JM, Beckers J, Blum R, Borrell V, Götz M. Trnp1 regulates expansion and folding of the Mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153:535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Sun T, Hevner RF. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci. 2014;15:217–232. doi: 10.1038/nrn3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna E, Götz M, Huttner WB. The cell biology of neurogenesis: toward an understanding of the development and evolution of the neocortex. Annu Rev Cell Dev Biol. 2014;30:465–502. doi: 10.1146/annurev-cellbio-101011-155801. [DOI] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, Hansen DV, Nord AS, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, Kaplan T, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilles K, Palomero-Gallagher N, Amunts K. Development of cortical folding during evolution and ontogeny. Trends Neurosci. 2013;36:275–284. doi: 10.1016/j.tins.2013.01.006. [DOI] [PubMed] [Google Scholar]


