Abstract
Recent studies have revealed the existence of numerous contact sites between the endoplasmic reticulum (ER) and endosomes in mammalian cells. Such contacts increase during endosome maturation and play key roles in cholesterol transfer, endosome positioning, receptor dephosphorylation, and endosome fission. At least 7 distinct contact sites between the ER and endosomes have been identified to date, which have diverse molecular compositions. Common to these contact sites is that they impose a close apposition between the ER and endosome membranes, which excludes membrane fusion while allowing the flow of molecular signals between the two membranes, in the form of enzymatic modifications, or ion, lipid, or protein transfer. Thus, ER–endosome contact sites ensure coordination of molecular activities between the two compartments while keeping their general compositions intact. Here, we review the molecular architectures and cellular functions of known ER–endosome contact sites and discuss their implications for human health.
Keywords: endoplasmic reticulum, endosome, membrane contact sites
Introduction
Endocytosis—uptake of material into the cell via inward budding of the plasma membrane—is crucial for membrane homeostasis, nutrient acquisition, and regulation of cell signalling (Conner & Schmid, 2003). Endocytosed material is initially found in early endosomes (EEs), and from these organelles, different cargoes are sorted to distinct destinations—back to the plasma membrane (PM) (recycling), into intraluminal vesicles (ILVs) for sorting to lysosomes, or to the biosynthetic pathway (Raiborg & Stenmark, 2009; Sorkin & von Zastrow, 2009). Early endosomes eventually mature into late endosomes (LEs), and when these fuse with lysosomes, most of their content is degraded by lysosomal hydrolases (Huotari & Helenius, 2011).
Several decades of work has revealed many of the components that regulate cargo sorting and endosome dynamics, including endosomal sorting complex required for transport (ESCRT) proteins that mediate cargo sorting into ILVs (Raiborg & Stenmark, 2009), Rab GTPases that regulate endosome fusion and motility (Stenmark, 2009), and motor proteins that power transport of endosomes along microtubules (Hirokawa & Noda, 2008). However, a new aspect of endosome biology has emerged recently, namely the occurrence of contact sites between endosomes and the endoplasmic reticulum (ER), the major membrane system of the biosynthetic pathway (Honscher & Ungermann, 2014; van der Kant & Neefjes, 2014) (Fig1). Such membrane contact sites, defined as sites of close (10–30 nm) apposition between two membranes, become increasingly abundant as endosomes mature, and it has been estimated that over 99% of all LEs in a cell may form contacts with the ER (Friedman et al, 2013). This begs the following questions: what are the molecular compositions of ER–endosome contact sites? And which are their functions? We will here highlight recent studies that shed light on these issues.
Figure 1.
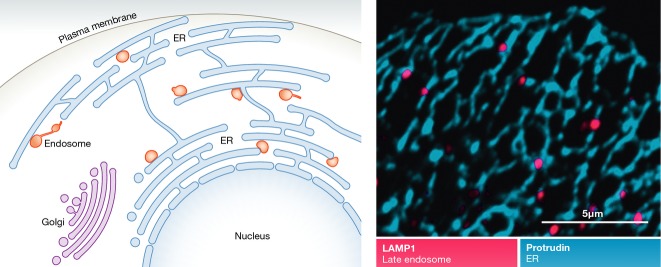
Endosomes make contact with the endoplasmic reticulum
(Left) Endosomes associate with tubular regions of the ER. (Right) Confocal micrograph showing how LEs (red) are juxtaposed to protrudin-positive ER (blue).
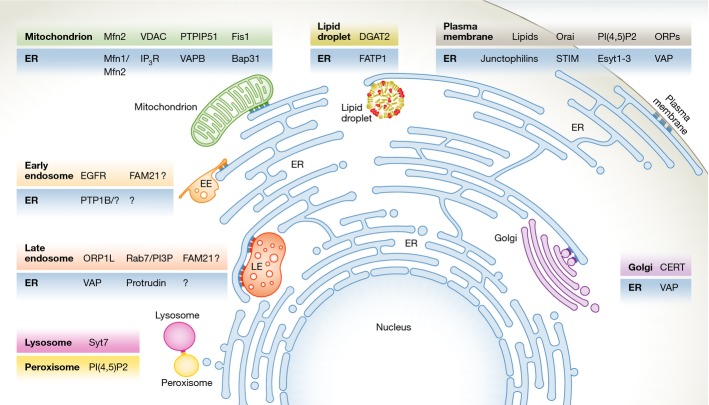
The ER makes contact sites with multiple membranes
As the most abundant membrane compartment of the cell, the ER is known to form contact sites with several other membranes, including mitochondria, the Golgi apparatus, the PM, and lipid droplets (LDs) (Fig2). For a detailed description of such sites, the reader is referred to other reviews (Elbaz & Schuldiner, 2011; Helle et al, 2013) as only a brief overview will be provided here.
Figure 2.
Organelles establish contact sites with a variety of cellular compartments
In mammalian cells, ER makes contact sites with endosomes, Golgi, mitochondria, lipid droplets, and the plasma membrane. Lysosomes and peroxisomes also establish contact sites. Examples of proteins known to mediate the contact sites in metazoans are depicted (Helle et al, 2013; Rowland et al, 2014; Chu et al, 2015; Raiborg et al, 2015).
The ER is the major Ca2+ storage compartment of the cell, with Ca2+ concentrations in the millimolar range, which is to be compared with the nanomolar Ca2+ concentrations in the cytosol (Helle et al, 2013). Because large-scale flux of Ca2+ to the cytosol may be toxic to cells, it is logical that Ca2+ export from the ER to other organelles occurs via membrane contact sites. The ER is also the major site of lipid biosynthesis in cells. Both cytosolic lipid transfer proteins and vesicular trafficking mediate interorganellar lipid transfer, and membrane contact sites have the advantage of providing high specificity and efficacy to such lipid transfer reactions. Indeed, many of the identified membrane contact sites between ER and other membranes are involved in either Ca2+ or lipid transfer (Helle et al, 2013).
Several different contact sites between ER and the outer membrane of mitochondria have been well characterized at the molecular level. ER–mitochondria contact sites that mediate Ca2+ transfer into mitochondria have been identified, and such influx is thought not only to ensure the functionality of Ca2+-containing mitochondrial proteins but also to play a role in the intrinsic (mitochondria-driven) apoptotic pathway (Helle et al, 2013). ER–mitochondria contact sites that mediate lipid transfer have been particularly well studied in yeast, but also mammalian cells harbour such sites. Certain lipid biosynthetic pathways require cooperations between enzyme complexes located in mitochondria and the ER, and contact site-mediated lipid transfer plays an important role in this context (Rowland & Voeltz, 2012). Other functions for ER–mitochondrial contact sites include control of mitochondrial biogenesis and fission, and biogenesis of autophagosome membranes (Friedman et al, 2011; Hamasaki et al, 2013).
Even though there is extensive vesicle trafficking between the ER and the Golgi complex, the two compartments are also connected by membrane contact sites. An important function of such sites is to coordinate sterol transfer from the ER with back-transfer of phosphatidylinositol 4-phosphate (PtdIns4P) from the Golgi, ensuring negative feedback control of sterol transfer (Mesmin et al, 2013). Other ER–Golgi contact sites mediate transfer of ceramide and glucosylceramide (Hanada et al, 2003; D’Angelo et al, 2007).
Contact sites between the ER (sarcoplasmic reticulum) and the PM are very abundant in muscle cells, in which they are involved in depolarization–contraction coupling. However, two other types of ER–PM contact sites are found in a variety of cell types, one that promotes Ca2+ influx into the ER after Ca2+ depletion, and one that may be involved in lipid transfer (Giordano et al, 2013; Helle et al, 2013).
LDs serve as major compartments for lipid storage in a variety of cell types. Unlike other organelles, these compartments have a micellar structure with a core consisting of neutral lipid esters and a surface consisting of a phospholipid monolayer. LDs are formed from ER membranes, and contact sites between ER and LDs, which can be observed in both yeast and mammalian cells, are thought to control LD size. The composition of ER–LD contact sites has been characterized in yeast, and conservation of key components in mammalian cells suggests a similar architecture in such cells (Wang et al, 2014).
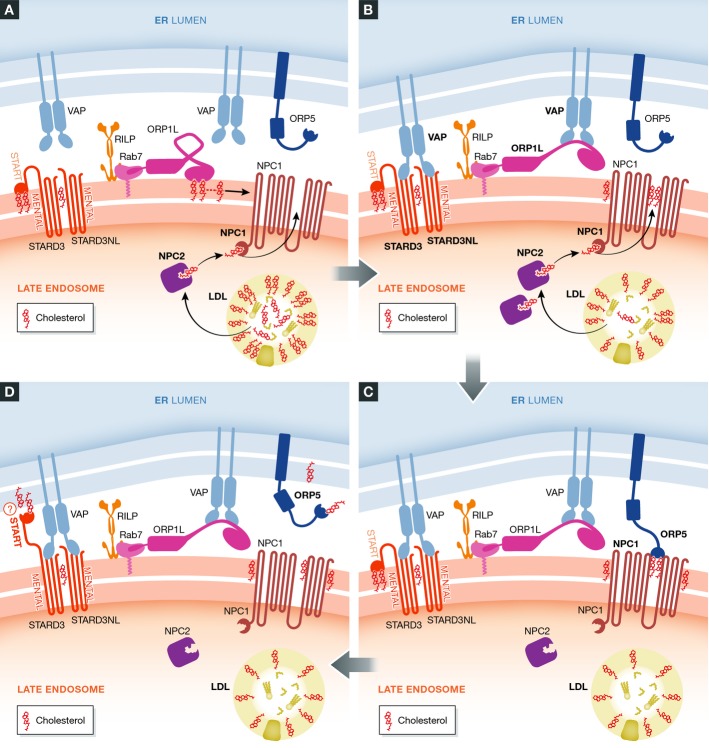
Contact sites mediate cholesterol transfer from LEs to ER
The first ER–endosome contact site to be characterized in mammalian cells was the one formed when the integral ER protein VAP-A interacts with the peripheral late-endosomal cholesterol-binding protein ORP1L (Rocha et al, 2009) (Fig3). This occurs when there is little cholesterol in the endosome membrane, which leaves the cholesterol-binding domain of ORP1L unoccupied and favours a conformation that allows interaction between the FFAT (diphenylalanine in an acidic tract) motif of ORP1L and VAP-A. Two other late-endosomal proteins, STARD3/MLN64 and STARD3NL/MENTHO, also contain FFAT motifs that enable them to interact with VAP-A to form ER–endosome contact sites (Alpy et al, 2013) (Fig3). STARD3 and STARD3NL can dimerize and are attached to endosomes by their cholesterol-binding transmembrane MENTAL domain (Alpy et al, 2005). STARD3 additionally contains a steroidogenic acute regulatory-related lipid transfer (START) domain that is able to bind cholesterol and can transfer cholesterol between membranes (Kallen et al, 1998).
Figure 3.
Cholesterol transfer in ER–endosome contact sites
(A) Prior to contact formation, cholesterol-binding protein complexes define cholesterol-rich patches on the endosome membrane. Cholesterol, internalized into endosomes via LDL particles, is transferred to the cholesterol transporter NPC1 via the carrier NPC2. (B) Cholesterol accumulates in NPC1. Upon reduction in free cholesterol in the endosome membrane, ORP1L undergoes a conformational change which initiates binding to the ER protein VAP. The MENTAL domain of STARD3/NL can also bind to VAP in the ER. (C) The ER–endosome contact initiated by ORP1L-VAP-A might facilitate the interaction of ORP5 with NPC1. (D) When contact is established, cholesterol is transferred from the endosome to the ER via ORP5 and possibly also via the START domain of STARD3.
In addition to de novo cholesterol synthesis, cells acquire cholesterol through receptor-mediated endocytosis of cholesterol-containing particles, mostly in the form of low density lipoprotein (LDL) particles produced by the liver (Ikonen, 2008). Endocytosis of LDL is followed by dissociation from its receptor and hydrolysis into unesterified cholesterol by lipases in the lumen of LEs. Here, a soluble cholesterol carrier, Niemann–Pick disease type C2 (NPC2), delivers cholesterol to a membrane-associated cholesterol transporter, NPC1. As much as 30% of LE-associated cholesterol is transferred to the ER (Neufeld et al, 1996), and ER–LE contact sites are thought to play a major role in such transfer (van der Kant & Neefjes, 2014) (Fig3). An integral ER membrane protein, a member of the oxysterol-binding protein-related protein (ORP) family, ORP5 (Olkkonen & Li, 2013), which contains a cytosolic cholesterol-binding domain, forms contact sites with LEs through interaction with NPC1 (Du et al, 2011). This is accompanied by cholesterol transfer from the LE membrane to the ER membrane, presumably mediated by shuttling of the cholesterol-binding domain of ORP5 between the two membranes (Du et al, 2011). Despite their cholesterol-sensing ability, a direct role of ORP1L or STARD3/STARD3NL in cholesterol transport has not been demonstrated. It has been speculated that these proteins can define cholesterol-containing patches on endosomes and initiate ER–endosome contact prior to cholesterol transfer mediated by other proteins like NPC1–ORP5 (Alpy & Tomasetto, 2006; van der Kant & Neefjes, 2014).
Even though the exact functional mechanisms of the NPC1–ORP5 contact sites in cholesterol transfer still need to be characterized, a possible parallel exists in transfer of cholesterol from ER to the Golgi mediated by the VAP-A-binding Golgi ORP family member OSBP. Like ORP5, OSBP contains a sterol-binding domain, and this domain mediates transfer of cholesterol from the ER to the trans-Golgi membrane once a contact site has been formed through the interaction of the FFAT motif of OSBP with VAP-A. The binding of OSBP to the Golgi membrane is mediated by coincident detection of the small GTPase Arf1 and PtdIns4P via a PH domain (Levine & Munro, 2002). The fact that ORP5 and ORP1L, like OSBP, contain a PH domain, raises the possibility that these proteins could also recognize a phosphoinositide in the LE membrane (Du & Yang, 2013). Recently, a subfamily of ORP proteins, including ORP5, was shown to play a role in non vesicular transfer of phosphatidylserine from the ER to the plasma membrane, indicating that ORP proteins have additional functions other than cholesterol transfer (Maeda et al, 2013).
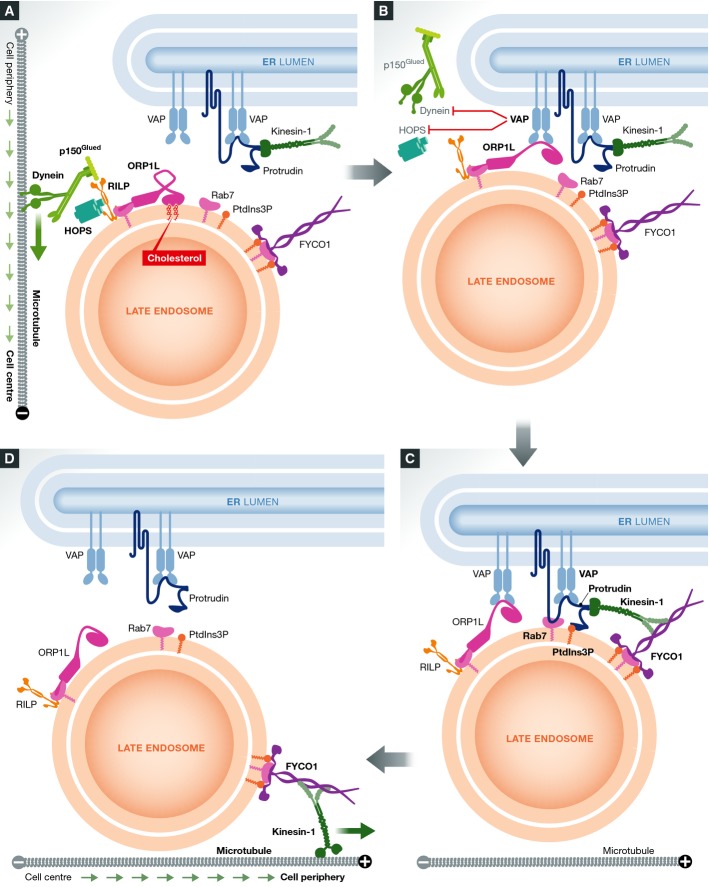
The ER controls association of endosomes with the cytoskeleton
While it still remains to be established whether ORP1L plays any role in cholesterol transfer, its functions in endosome positioning and fusion are well characterized. The p150Glued subunit of the minus-end-directed microtubule motor dynein/dynactin complex interacts with the late-endosomal Rab7–RILP complex as does the endosomal tethering and fusion complex HOPS. This mediates transport of LEs to the cell centre, where they can fuse in a HOPS-dependent manner. The establishment of ER–LE contact by ORP1L and VAP under low cholesterol conditions favours a secondary interaction between VAP and the RILP–HOPS–p150Glued complex, which leads to the dissociation of the dynein motor and the HOPS complex (Rocha et al, 2009; van der Kant et al, 2013a). Thus, the net effect of forming ORP1L-containing ER–endosome contact sites is that only LEs devoid of dynein and HOPS remain in contact with the ER (Fig4).
Figure 4.
ER–endosome contacts regulate microtubule-dependent endosome transport
(A) Prior to contact formation, under cholesterol-rich conditions, ORP1L-RILP is bound to the minus-end-directed microtubule motor dynein and the HOPS complex. This facilitates transport of endosomes to the cell centre where they can fuse in a HOPS-dependent manner. (B) When cholesterol levels are reduced, ORP1L undergoes a conformational change that initiates binding to the ER protein VAP-A. VAP-A dissociates dynein and HOPS from RILP. The ER protein protrudin is also localized to VAP-A sites of the ER membrane, where it concentrates the plus-end-directed microtubule motor kinesin-1. (C) Protrudin initiates contact with endosomes by binding to Rab7-GTP and PtdIns3P. The endosomal motor adaptor FYCO1 receives kinesin-1 from protrudin. (D) Endosomes are transported to the cell periphery by kinesin-1 coupled to FYCO1.
Recently, a new VAP-A-binding protein, protrudin, was found to form ER–LE contact sites (Raiborg et al, 2015). Like ORP1L, STARD3 and STARD3NL, protrudin contains a FFAT motif that binds VAP-A, but in contrast to these proteins, protrudin is an integral ER membrane protein (Shirane & Nakayama, 2006). Association of protrudin with the ER membrane is mediated by two juxtaposed membrane helices and a membrane-inserted hairpin loop, so VAP-A is not strictly needed for the ER association of this protein (Chang et al, 2013). Protrudin forms contact sites with LEs through coincident detection of Rab7-GTP and phosphatidylinositol 3-phosphate (PtdIns3P) on the endosome membrane (Raiborg et al, 2015) (Fig4). At least one of the functions of such contact sites is to charge LEs with the plus-end-directed microtubule motor kinesin-1. Protrudin binds to the heavy chain of this motor and hands it over to the late-endosomal protein FYCO1, which binds kinesin-1 light chain. Like protrudin, FYCO1 binds to Rab7-GTP and PtdIns3P on the endosome membrane (Pankiv et al, 2010), and this probably ensures that FYCO1 is present in the same endosomal microdomains as the protrudin-containing contact sites. Loading of FYCO1 with kinesin-1 promotes motility of FYCO1-containing LEs to the cell periphery where they can undergo synaptotagmin VII-dependent fusion with the plasma membrane. This in turn promotes formation of cellular protrusions or neurites (Raiborg et al, 2015).
The exact functions of the STARD3 and STARD3NL contact sites remain to be identified, but it is interesting that overexpression of STARD3 induces perinuclear clustering followed by increased formation of Arp2/3-positive actin patches on LEs, whereas its depletion causes their peripheral dispersion and loss of actin association (Holtta-Vuori et al, 2005). Thus, STARD3 is likely to modulate late endosome positioning through regulating their association with the actin cytoskeleton. Further, overexpression of STARD3NL, which increases the number of STARD3NL-VAP contact sites, prevents tubulation of LEs (Alpy et al, 2013), an activity that might be associated with microtubule-based motors or actin dynamics (Derivery et al, 2009; Skjeldal et al, 2012). It is interesting to note that the function of both ORP1L and STARD3 in endosome positioning is dependent on their cholesterol-binding ability, arguing for a regulatory role of cholesterol in the association of endosomes with cytoskeletal elements (Holtta-Vuori et al, 2005; Rocha et al, 2009).
The involvement of VAP-A in several different ER–endosome contact sites raises the question whether these sites co-localize. ORP1L- and STARD3-containing contact sites occupy distinct domains of the ER membrane (van der Kant et al, 2013b). It will be interesting to explore whether protrudin- and ORP1L-containing contact sites are formed in the same regions. It is tempting to speculate that formation of ORP1L-containing contact sites, which is associated with dynein dissociation from LEs, might be coordinated with formation of protrudin-containing contact sites, which is associated with kinesin-1 recruitment (Fig4). This might allow a tight spatiotemporal control of motor dissociation and loading in directional endosome motility and might explain why LEs need to recruit kinesin-1 from ER–endosome contact sites rather than from cytosol.
Dephosphorylation of endosomal receptors by an ER-associated phosphatase
Ligand-dependent dimerization and autophosphorylation of receptor tyrosine kinases is a widespread signalling mechanism in higher eukaryotes (Schlessinger, 2000). Such signalling is known to be attenuated by protein tyrosine phosphatases that dephosphorylate activated receptors (Ostman & Bohmer, 2001). While some of these phosphatases are cytosolic, PTP1B, which dephosphorylates several receptors including epidermal growth factor receptors (EGFRs) and granulocyte colony-stimulating factor receptors (G-CSFRs), is a peripheral membrane protein of the ER (Feldhammer et al, 2013) (Fig5). The seemingly paradoxical localization of PTP1B with respect to activated receptors was solved by the finding that a “substrate-trap” mutant of ER-bound PTP1B is present in contact sites with EGFR-containing multivesicular endosomes (MVEs) that are positive for the early-endosomal GTPase Rab5 (Eden et al, 2010). Contacts formed between wild-type PTP1B and activated EGFR are likely to be much more short-lived, and it is plausible that they may be stabilized by additional factors. In this respect, it is interesting to note that contacts between MVEs and the ER can be observed even when EGFRs are not ligand activated (and therefore not internalized into endosomes), indicating that additional molecules mediate ER–MVE contact sites.
Figure 5.
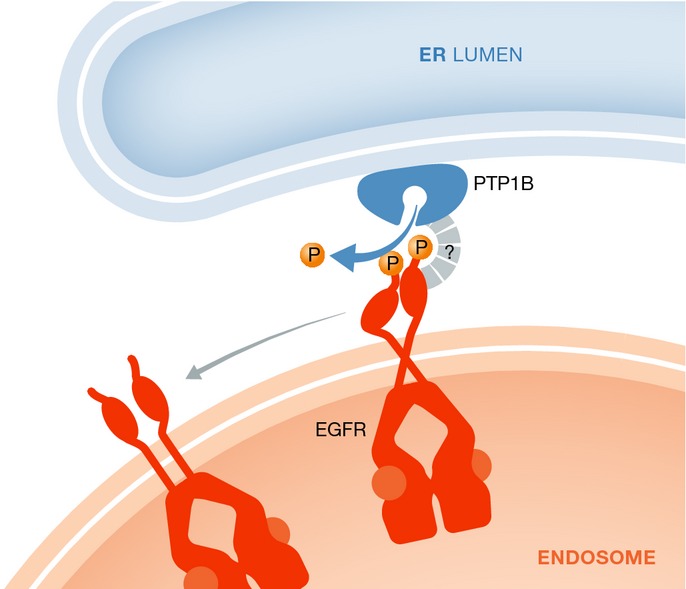
Dephosphorylation of endosomal receptors by an ER-associated phosphatase
The ER-localized phosphatase PTP1B dephosphorylates activated EGFRs in the endosomal membrane. The ER–endosome association is likely to be stabilized by additional factors.
In a similar manner, G-CSFRs localized to early endosomes following G-CSF stimulation of cells have been found to interact with PTP1B and also with the ER-associated peroxiredoxin PRDX4 (Palande et al, 2011). The exact function of PRDX4 in regulation of PTP1B and G-CSFR remains unresolved, but one possibility is that its antioxidant activity could serve to keep PTP1B active as it is known that PTP1B is inhibited by reactive oxygen species (ROS). Another possibility is that PRDX4 may function as a tether that stabilizes the interaction between G-CSFR and PTP1B (Palande et al, 2011).
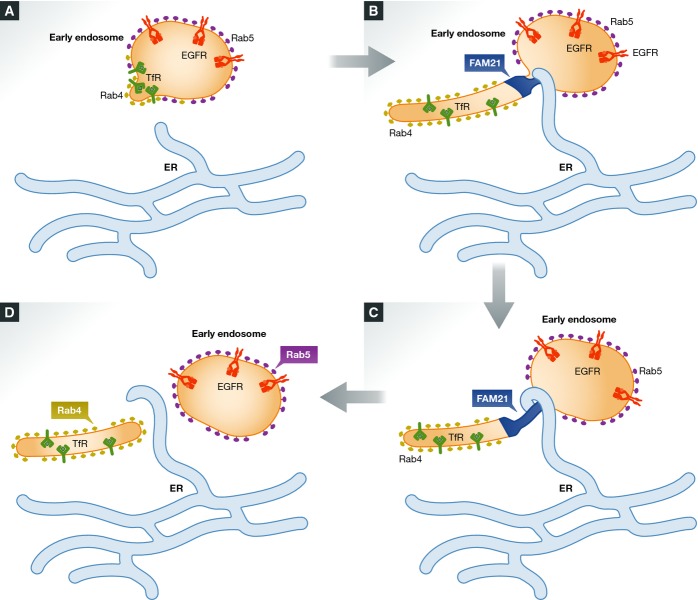
ER tubules define the position and timing of endosome fission
An unexpected function of ER tubules is to define sites for organelle fission. This was first described in the case of mitochondrial division (Friedman et al, 2011) but has recently been demonstrated for endosome fission as well (Rowland et al, 2014) (Fig6). Endosomal fission is frequently observed by live-cell microscopy and is thought to play an important role in endosome maturation as well as in formation of carriers destined for recycling of endocytosed cargo to the plasma membrane (Huotari & Helenius, 2011). Immediately prior to fission of EEs or LEs, contact sites are formed between the endosome and tubular ER elements on sites marked by the retromer-associated protein FAM-21, and overexpression of the ER-shaping protein reticulon 4 inhibits endosome fission. This suggests that ER–endosome contact sites define the position and timing of endosome fission (Rowland et al, 2014). Interestingly, the fission products of early endosomes that have been contacted by ER tubules typically consist of one part that contains the small GTPase Rab5, whereas the other part is enriched in another small GTPase, Rab4. Rab5 is involved in anterograde endocytic trafficking, whereas Rab4 is involved in recycling (Sonnichsen et al, 2000), suggesting that ER–endosome contact sites could indirectly mediate endosomal sorting. This, however, needs to be investigated.
Figure 6.
ER tubules define sites of endosome fission
(A) EEs contain internalized EGF receptors (EGFRs) and transferrin receptors (TfRs). TfRs accumulate in forming endosome tubules, whereas EGFRs are retained in the endosome body. (B) The retromer-associated protein FAM-21 localizes to the base of a forming tubule, which is contacted by a tubular ER element that defines the site of constriction. (C) The endosomal TfR containing endosomal tubule is constricted as ER folds around it. (D) Constriction is followed by fission into a Rab5-positive endosome that contains EGFR and a Rab4-positive endosome that contains TfR.
The molecular composition of ER–endosome contact sites involved in endosome fission is not known. It is possible that they might comprise some of the contact sites described above, but it is perhaps more likely that these contact sites have another composition. The finding that ER tubules define fission of both early and LEs raises the possibility that the endosomal moiety of the contact sites is a molecule found on both EEs and LEs. However, it is also possible that different types of contact sites might define fission of EEs and LEs. It is not known how ER–endosome contact sites function to define regions of endosome fission, but it is tempting to speculate that they serve to recruit components of the fission machinery (Rowland et al, 2014). Further studies will hopefully shed light on this.
Components of ER–endosome contact sites are affected in diseases
If we consider the various proteins involved in formation of ER–endosome contact sites, their dysfunctions have been implicated in several diseases. For instance, mutations in NPC1 and NPC2 are causative for the neurodegenerative Niemann–Pick disease, where cholesterol fails to reach the ER and accumulates in late endosomes (Mukherjee & Maxfield, 2004). Further, expression of ORP5 has been found to correlate with invasion and poor prognosis of pancreatic cancer (Koga et al, 2008), and expression of human ORP1L in macrophages enhances atherosclerotic lesion development in LDL receptor-deficient mice (Yan et al, 2007). Overexpression of STARD3 induces liver damage in mice (Tichauer et al, 2007), and increased expression of this protein in human tumours is associated with high-grade prostate cancer and HER2-positive breast cancer (Vinatzer et al, 2005; Stigliano et al, 2007). Single-nucleotide polymorphisms of STARD3NL are associated with bone mineral density (Rivadeneira et al, 2009). The phosphatase PTP1B has been characterized as both oncoprotein and tumour suppressor, depending on cancer type (Liu et al, 2015), and has also been implicated in metabolic disease (Elchebly et al, 1999). A polymorphism of protrudin is associated with hereditary spastic paraplegia, HSP (Mannan et al, 2006; Hashimoto et al, 2014), and mutations in several protrudin-binding proteins, including KIF5A, are causative of HSPs (Reid et al, 1999). Mutations in the associated protein Rab7 cause Charcot–Marie–Tooth disease type 2B (Verhoeven et al, 2003), and mutations in p150Glued cause motor neuron disease and the Parkinson-related Perry syndrome, depending on the localization of the mutation (Puls et al, 2003; Farrer et al, 2009). Even though diseases are associated with overexpression or mutations of ER–endosome contact site components, it remains to be investigated whether these can be related to contact sites as such or some other activities of the proteins involved. Given the well-documented roles of ER–endosome contact sites in crucial cellular functions, one would anyway expect them to play important roles in human health.
Conclusions and perspectives
Recent research has revealed the existence of multiple different ER–endosome contact sites with diverse functions. Although we cannot pinpoint any universal structural determinant that distinguishes such contact sites, some features can be identified that are common to several types of ER–endosome contact sites. Firstly, several of the contact sites contain the ER protein VAP-A, either as a structural component or as a targeting factor. Secondly, it is conspicuous that several contact sites contain cholesterol-binding proteins. This could reflect the involvement of such sites in cholesterol transport from ER to endosomes (as with ORP5) but also a regulatory role of cholesterol in contact site dynamics (as with ORP1L), or a role in maintaining a defined membrane lipid composition. In support of the latter, a well-characterized membrane contact site in budding yeast, the nucleus–vacuole junction, which promotes piecemeal autophagy of the nucleus, contains the sterol-binding protein Osh1. This protein, which binds the VAP-A homologue Scs2, has been proposed to control the membrane lipid composition of the nucleus–vacuole junction in order to establish a diffusion barrier (Dawaliby & Mayer, 2010). Thirdly, several ER–endosome contact sites contain components with confirmed or putative phosphoinositide-binding domains. While the FYVE domain of protrudin binds PtdIns3P on LEs, the possible lipid binding of the PH domains of ORP1L and ORP5 still needs to be established. In the case of protrudin, phosphoinositide binding is involved together with Rab7 in coincident detection of LE membranes, and a role of the ORP1L/ORP5 PH domains in membrane interactions is plausible. It is interesting to note that VAP-A, cholesterol-binding proteins and phosphoinositides are involved in contact sites between the ER and other membranes as well (Elbaz & Schuldiner, 2011; Stefan et al, 2011; Giordano et al, 2013; Helle et al, 2013; Mesmin et al, 2013), so the same principal mechanisms could be employed in several types of membrane contact sites.
The known functions of ER–endosome contact sites include endosome positioning, cholesterol transfer, receptor dephosphorylation, endosome fission, and negative control of endosome fusion, and there are also good arguments for a role of ER–endosome contact sites in Ca2+ transfer (van der Kant & Neefjes, 2014). The functional repertoire of ER–endosome contact sites is likely to expand as new contact sites are identified (Table1). At the same time, it will be important to characterize the compositions of known contact sites in more detail. With the exception of contact sites that define endosome fission, the core components of known ER–endosome contact sites have been identified, but this does not exclude the possibility that further molecules are involved and we do not know the exact structural organizations of the various contact sites. We also know very little about the dynamics of ER–endosome contact site formation and turnover and how such dynamics are regulated. In the case of PTP1B-containing contact sites, it is reasonable to assume that contact sites are negatively regulated by receptor dephosphorylation since a substrate-trap mutant of PTP1B causes excessive contact site formation (Eden et al, 2010). Likewise, protrudin-containing contact sites are likely to be regulated by the GTPase activity of Rab7 since protrudin only binds to the GTP-bound form of Rab7 and since expression of a GTPase-defective mutant of Rab7 increases the number of contact sites (Raiborg et al, 2015). Phosphoinositide-dependent contact sites might also be regulated by phosphoinositide phosphorylation or dephosphorylation.
Table 1.
Compositions and functions of ER–endosome contact sites
| Contact site | Function | References | |
|---|---|---|---|
| ER | Endosome | ||
| PTP1B | EGFR | Receptor dephosphorylation in-trans | Eden et al (2010) |
| PTP1B | G-CSFR | Receptor dephosphorylation in-trans | Palande et al (2011) |
| VAP-A | STARD3/MLN64 | Endosome positioning? | Alpy et al (2013); van der Kant et al (2013b) |
| VAP-A | STARD3NL/MENTHO | Control of endosome tubulation? | Alpy et al (2013) |
| VAP-A | ORP1L | Negative regulation of dynein association with LEs (when cholesterol concentration is low) | Rocha et al (2009) |
| ORP5 | NPC1 | Cholesterol transport between LEs and ER? | Du et al (2011); Du & Yang (2013) |
| Protrudin (+VAP-A) | Rab7-GTP + PtdIns3P | LE translocation to cell periphery mediated by transfer of kinesin-1 from ER to LEs | Raiborg et al (2015) |
| ? | FAM21 ? | Definition of sites for endosome fission | Rowland et al (2014) |
The moieties associated with ER and endosome membranes are indicated.
We have witnessed the opening of an exciting new field in cell biology, and many major questions remain to be addressed. Do the handful of ER–endosome contact sites identified to date represent the full complement of such contact sites, or are they just the tip of the iceberg? Are there additional components involved in the identified ER–endosome contact sites? Which are their structural organizations? Are there additional functions of ER–endosome contact sites to be discovered? How are their dynamics regulated, and is there crosstalk between different contact sites? Are defective ER–endosome contact sites causative of diseases? Addressing these questions will require combined approaches in molecular cell biology, cellular imaging, biochemistry, structural biology, and molecular medicine. The fast progress made in this young research field during a relatively narrow time span suggests that we can anticipate new insight in the near future.
Acknowledgments
CR and EMW are senior research fellows of the Norwegian Cancer Society and the South-Eastern Norway Regional Health Authority, respectively. HS has been supported by the Norwegian Cancer Society, the South-Eastern Norway Regional Health Authority, and the European Research Council. This work was partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 179571.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Alpy F, Latchumanan VK, Kedinger V, Janoshazi A, Thiele C, Wendling C, Rio MC, Tomasetto C. Functional characterization of the MENTAL domain. J Biol Chem. 2005;280:17945–17952. doi: 10.1074/jbc.M500723200. [DOI] [PubMed] [Google Scholar]
- Alpy F, Tomasetto C. MLN64 and MENTHO, two mediators of endosomal cholesterol transport. Biochem Soc Trans. 2006;34:343–345. doi: 10.1042/BST0340343. [DOI] [PubMed] [Google Scholar]
- Alpy F, Rousseau A, Schwab Y, Legueux F, Stoll I, Wendling C, Spiegelhalter C, Kessler P, Mathelin C, Rio MC, Levine TP, Tomasetto C. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J Cell Sci. 2013;126:5500–5512. doi: 10.1242/jcs.139295. [DOI] [PubMed] [Google Scholar]
- Chang J, Lee S, Blackstone C. Protrudin binds atlastins and endoplasmic reticulum-shaping proteins and regulates network formation. Proc Natl Acad Sci USA. 2013;110:14954–14959. doi: 10.1073/pnas.1307391110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu BB, Liao YC, Qi W, Xie C, Du X, Wang J, Yang H, Miao HH, Li BL, Song BL. Cholesterol Transport through Lysosome-Peroxisome Membrane Contacts. Cell. 2015;161:291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- D’Angelo G, Polishchuk E, Di TG, Santoro M, Di CA, Godi A, West G, Bielawski J, Chuang CC, van der Spoel AC, Platt FM, Hannun YA, Polishchuk R, Mattjus P, De Matteis MA. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- Dawaliby R, Mayer A. Microautophagy of the nucleus coincides with a vacuolar diffusion barrier at nuclear-vacuolar junctions. Mol Biol Cell. 2010;21:4173–4183. doi: 10.1091/mbc.E09-09-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell. 2009;17:712–723. doi: 10.1016/j.devcel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- Du X, Kumar J, Ferguson C, Schulz TA, Ong YS, Hong W, Prinz WA, Parton RG, Brown AJ, Yang H. A role for oxysterol-binding protein-related protein 5 in endosomal cholesterol trafficking. J Cell Biol. 2011;192:121–135. doi: 10.1083/jcb.201004142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Yang H. Endosomal cholesterol trafficking: protein factors at a glance. Acta Biochim Biophys Sin (Shanghai) 2013;45:11–17. doi: 10.1093/abbs/gms095. [DOI] [PubMed] [Google Scholar]
- Eden ER, White IJ, Tsapara A, Futter CE. Membrane contacts between endosomes and ER provide sites for PTP1B-epidermal growth factor receptor interaction. Nat Cell Biol. 2010;12:267–272. doi: 10.1038/ncb2026. [DOI] [PubMed] [Google Scholar]
- Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan CC, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, Dachsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, Tsuboi Y, Yamada T, Gutmann L, Elibol B, Bhatia KP, Wider C, Vilarino-Guell C, Ross OA, Brown LA, Castanedes-Casey M, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldhammer M, Uetani N, Miranda-Saavedra D, Tremblay ML. PTP1B: a simple enzyme for a complex world. Crit Rev Biochem Mol Biol. 2013;48:430–445. doi: 10.3109/10409238.2013.819830. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, Dibenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Dibenedetto JR, West M, Rowland AA, Voeltz GK. Endoplasmic reticulum-endosome contact increases as endosomes traffic and mature. Mol Biol Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De CP. PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell. 2013;153:1494–1509. doi: 10.1016/j.cell.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, Amano A, Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Shirane M, Matsuzaki F, Saita S, Ohnishi T, Nakayama KI. Protrudin regulates endoplasmic reticulum morphology and function associated with the pathogenesis of hereditary spastic paraplegia. J Biol Chem. 2014;989:12946–12961. doi: 10.1074/jbc.M113.528687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helle SC, Kanfer G, Kolar K, Lang A, Michel AH, Kornmann B. Organization and function of membrane contact sites. Biochim Biophys Acta. 2013;1833:2526–2541. doi: 10.1016/j.bbamcr.2013.01.028. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol Rev. 2008;88:1089–1118. doi: 10.1152/physrev.00023.2007. [DOI] [PubMed] [Google Scholar]
- Holtta-Vuori M, Alpy F, Tanhuanpaa K, Jokitalo E, Mutka AL, Ikonen E. MLN64 is involved in actin-mediated dynamics of late endocytic organelles. Mol Biol Cell. 2005;16:3873–3886. doi: 10.1091/mbc.E04-12-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honscher C, Ungermann C. A close-up view of membrane contact sites between the endoplasmic reticulum and the endolysosomal system: from yeast to man. Crit Rev Biochem Mol Biol. 2014;49:262–268. doi: 10.3109/10409238.2013.875512. [DOI] [PubMed] [Google Scholar]
- Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat Rev Mol Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- Kallen CB, Billheimer JT, Summers SA, Stayrook SE, Lewis M, Strauss JF., III Steroidogenic acute regulatory protein (StAR) is a sterol transfer protein. J Biol Chem. 1998;273:26285–26288. doi: 10.1074/jbc.273.41.26285. [DOI] [PubMed] [Google Scholar]
- van der Kant R, Fish A, Janssen L, Janssen H, Krom S, Ho N, Brummelkamp T, Carette J, Rocha N, Neefjes J. Late endosomal transport and tethering are coupled processes controlled by RILP and the cholesterol sensor ORP1L. J Cell Sci. 2013a;126:3462–3474. doi: 10.1242/jcs.129270. [DOI] [PubMed] [Google Scholar]
- van der Kant R, Neefjes J. Small regulators, major consequences - Ca2+ and cholesterol at the endosome-ER interface. J Cell Sci. 2014;127:929–938. doi: 10.1242/jcs.137539. [DOI] [PubMed] [Google Scholar]
- van der Kant R, Zondervan I, Janssen L, Neefjes J. Cholesterol-binding molecules MLN64 and ORP1L mark distinct late endosomes with transporters ABCA3 and NPC1. J Lipid Res. 2013b;54:2153–2165. doi: 10.1194/jlr.M037325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga Y, Ishikawa S, Nakamura T, Masuda T, Nagai Y, Takamori H, Hirota M, Kanemitsu K, Baba Y, Baba H. Oxysterol binding protein-related protein-5 is related to invasion and poor prognosis in pancreatic cancer. Cancer Sci. 2008;99:2387–2394. doi: 10.1111/j.1349-7006.2008.00987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Liu H, Wu Y, Zhu S, Liang W, Wang Z, Wang Y, Lv T, Yao Y, Yuan D, Song Y. PTP1B promotes cell proliferation and metastasis through activating src and ERK1/2 in non-small cell lung cancer. Cancer Lett. 2015;359:218–225. doi: 10.1016/j.canlet.2015.01.020. [DOI] [PubMed] [Google Scholar]
- Maeda K, Anand K, Chiapparino A, Kumar A, Poletto M, Kaksonen M, Gavin AC. Interactome map uncovers phosphatidylserine transport by oxysterol-binding proteins. Nature. 2013;501:257–261. doi: 10.1038/nature12430. [DOI] [PubMed] [Google Scholar]
- Mannan AU, Krawen P, Sauter SM, Boehm J, Chronowska A, Paulus W, Neesen J, Engel W. ZFYVE27 (SPG33), a novel spastin-binding protein, is mutated in hereditary spastic paraplegia. Am J Hum Genet. 2006;79:351–357. doi: 10.1086/504927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, von Moser FJ, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Maxfield FR. Lipid and cholesterol trafficking in NPC. Biochim Biophys Acta. 2004;1685:28–37. doi: 10.1016/j.bbalip.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Neufeld EB, Cooney AM, Pitha J, Dawidowicz EA, Dwyer NK, Pentchev PG, Blanchette-Mackie EJ. Intracellular trafficking of cholesterol monitored with a cyclodextrin. J Biol Chem. 1996;271:21604–21613. doi: 10.1074/jbc.271.35.21604. [DOI] [PubMed] [Google Scholar]
- Olkkonen VM, Li S. Oxysterol-binding proteins: sterol and phosphoinositide sensors coordinating transport, signaling and metabolism. Prog Lipid Res. 2013;52:529–538. doi: 10.1016/j.plipres.2013.06.004. [DOI] [PubMed] [Google Scholar]
- Ostman A, Bohmer FD. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001;11:258–266. doi: 10.1016/s0962-8924(01)01990-0. [DOI] [PubMed] [Google Scholar]
- Palande K, Roovers O, Gits J, Verwijmeren C, Iuchi Y, Fujii J, Neel BG, Karisch R, Tavernier J, Touw IP. Peroxiredoxin-controlled G-CSF signalling at the endoplasmic reticulum-early endosome interface. J Cell Sci. 2011;124:3695–3705. doi: 10.1242/jcs.089656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur EL, Tokito M, Mann E, Floeter MK, Bidus K, Drayna D, Oh SJ, Brown RH, Jr, Ludlow CL, Fischbeck KH. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–456. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Wenzel EM, Pedersen NM, Olsvik H, Schink KO, Schultz SW, Vietri M, Nisi V, Bucci C, Brech A, Johansen T, Stenmark H. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC. A new locus for autosomal dominant “pure” hereditary spastic paraplegia mapping to chromosome 12q13, and evidence for further genetic heterogeneity. Am J Hum Genet. 1999;65:757–763. doi: 10.1086/302555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, et al. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N, Kuijl C, van der Kant R, Janssen L, Houben D, Janssen H, Zwart W, Neefjes J. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J Cell Biol. 2009;185:1209–1225. doi: 10.1083/jcb.200811005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13:607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Chitwood PJ, Phillips MJ, Voeltz G. ER contact sites define the position and timing of endosome fission. Cell. 2014;159:1027–1041. doi: 10.1016/j.cell.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Shirane M, Nakayama KI. Protrudin induces neurite formation by directional membrane trafficking. Science. 2006;314:818–821. doi: 10.1126/science.1134027. [DOI] [PubMed] [Google Scholar]
- Skjeldal FM, Strunze S, Bergeland T, Walseng E, Gregers TF, Bakke O. The fusion of early endosomes induces molecular-motor-driven tubule formation and fission. J Cell Sci. 2012;125:1910–1919. doi: 10.1242/jcs.092569. [DOI] [PubMed] [Google Scholar]
- Sonnichsen B, De RS, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkin A, von Zastrow M. Endocytosis and signalling: intertwining molecular networks. Nat Rev Mol Cell Biol. 2009;10:609–622. doi: 10.1038/nrm2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stigliano A, Gandini O, Cerquetti L, Gazzaniga P, Misiti S, Monti S, Gradilone A, Falasca P, Poggi M, Brunetti E, Agliano AM, Toscano V. Increased metastatic lymph node 64 and CYP17 expression are associated with high stage prostate cancer. J Endocrinol. 2007;194:55–61. doi: 10.1677/JOE-07-0131. [DOI] [PubMed] [Google Scholar]
- Tichauer JE, Morales MG, Amigo L, Galdames L, Klein A, Quinones V, Ferrada C, Alvarez AR, Rio MC, Miquel JF, Rigotti A, Zanlungo S. Overexpression of the cholesterol-binding protein MLN64 induces liver damage in the mouse. World J Gastroenterol. 2007;13:3071–3079. doi: 10.3748/wjg.v13.i22.3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven K, De Jonghe P, Coen K, Verpoorten N, Auer-Grumbach M, Kwon JM, FitzPatrick D, Schmedding E, De Vriendt E, Jacobs A, Van Gerwen V, Wagner K, Hartung HP, Timmerman V. Mutations in the small GTP-ase late endosomal protein RAB7 cause Charcot-Marie-Tooth type 2B neuropathy. Am J Hum Genet. 2003;72:722–727. doi: 10.1086/367847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer U, Dampier B, Streubel B, Pacher M, Seewald MJ, Stratowa C, Kaserer K, Schreiber M. Expression of HER2 and the coamplified genes GRB7 and MLN64 in human breast cancer: quantitative real-time reverse transcription-PCR as a diagnostic alternative to immunohistochemistry and fluorescence in situ hybridization. Clin Cancer Res. 2005;11:8348–8357. doi: 10.1158/1078-0432.CCR-05-0841. [DOI] [PubMed] [Google Scholar]
- Wang CW, Miao YH, Chang YS. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci. 2014;127:1214–1228. doi: 10.1242/jcs.137737. [DOI] [PubMed] [Google Scholar]
- Yan D, Jauhiainen M, Hildebrand RB, van Willems DK, Van Berkel TJ, Ehnholm C, Van EM, Olkkonen VM. Expression of human OSBP-related protein 1L in macrophages enhances atherosclerotic lesion development in LDL receptor-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:1618–1624. doi: 10.1161/ATVBAHA.107.144121. [DOI] [PubMed] [Google Scholar]