Abstract
Replication protein A (RPA) is a highly conserved heterotrimeric single-stranded DNA-binding protein involved in DNA replication, recombination, and repair. In fission yeast, the Rpa1-D223Y mutation provokes telomere shortening. Here, we show that this mutation impairs lagging-strand telomere replication and leads to the accumulation of secondary structures and recruitment of the homologous recombination factor Rad52. The presence of these secondary DNA structures correlates with reduced association of shelterin subunits Pot1 and Ccq1 at telomeres. Strikingly, heterologous expression of the budding yeast Pif1 known to efficiently unwind G-quadruplex rescues all the telomeric defects of the D223Y cells. Furthermore, in vitro data show that the identical D to Y mutation in human RPA specifically affects its ability to bind G-quadruplex. We propose that RPA prevents the formation of G-quadruplex structures at lagging-strand telomeres to promote shelterin association and facilitate telomerase action at telomeres.
Keywords: G-quadruplex, replication, RPA, Schizosaccharomyces pombe, telomeres
Introduction
Telomeres are nucleoprotein structures that protect chromosome ends from degradation and ensure replication of the terminal DNA. In most eukaryotes, telomere length is maintained by telomerase that adds telomere repeats to the 3′ end of chromosomes (Pfeiffer & Lingner, 2013). Telomere elongation by telomerase is regulated by the shelterin complex that specifically binds to telomeric repeats (Palm & de Lange, 2008). In mammals, the shelterin complex consists of the telomeric-repeat-binding factors (TRF1 and TRF2), the TRF1-interacting protein 2 (TIN2), the transcriptional repressor/activator RAP1, the protection of telomere protein1 (POT1), and the POT1- and TIN2-organizing protein TPP1. Within the shelterin complex, TRF1 and TRF2 directly bind to telomeric duplex DNA, while POT1 binds to the 3′ single-stranded overhang, known as G-tail (Gilson & Géli, 2007; Verdun & Karlseder, 2007).
The discovery of the shelterin complex in fission yeast Schizosaccharomyces pombe highlighted evolutionarily conserved elements of telomere length regulation between fission yeast and mammalian cells (Miyoshi et al, 2008; Pfeiffer & Lingner, 2013). The fission yeast shelterin complex consists of Taz1 (TRF1/TRF2 ortholog) that specifically binds to duplex telomeric DNA, the G-tail-binding protein Pot1 and the four shelterin proteins Tpz1 (TPP1 ortholog), Rap1, Poz1, and Ccq1 that link Taz1 and Pot1 through a network of protein–protein interactions (Palm & de Lange, 2008; Moser & Nakamura, 2009; Dehé & Cooper, 2010). Poz1 is necessary to connect the Pot1-Tpz1 complex to the Taz1-Rap1 complex, and thus likely fulfills similar functional roles as mammalian TIN2, which connects TRF1 and TRF2 to the POT1-TPP1 complex (Gilson & Géli, 2007; Verdun & Karlseder, 2007; Miyoshi et al, 2008). Tpz1 also interacts with Ccq1, which is involved in both telomere capping and telomerase recruitment (Tomita & Cooper, 2008). Taz1 also interacts with Rif1, Rap1 and Poz1 that are all necessary for limiting telomere elongation by telomerase (Kanoh & Ishikawa, 2001).
The fission yeast telomerase complex contains the catalytic subunit Trt1TERT, the regulatory subunit Est1 and the TER1 RNA. Ccq1 initiates telomerase recruitment by interacting with the telomerase regulatory subunit at the S/G2-phase transition. ATRRad3- and ATMTel1-dependent phosphorylation of Ccq1 at Thr93 promotes late S/G2-phase recruitment of telomerase to telomeres (Moser et al, 2011; Yamazaki et al, 2012; Chang et al, 2013) by promoting Ccq1–Est1 interaction (Moser et al, 2011; Webb & Zakian, 2012). Recently, the OB-fold domain of Tpz1 was also shown to play a critical role in telomere elongation by telomerase (Armstrong et al, 2014). On the other hand, SUMOylation of Tpz1 by the SUMO ligase Pli1 in late S/G2-phase promotes association of the Stn1-Ten1 complex at telomeres and negatively regulates telomere association of telomerase (Garg et al, 2014; Miyagawa et al, 2014).
Replication Protein A (RPA) is a highly conserved heterotrimeric single-stranded DNA-binding protein involved in DNA replication, recombination, and repair (Wold, 1997). RPA was found to bind at telomeres in late S-phase, and several alleles of RPA result in a telomere shortening phenotype in budding and fission yeasts (Smith & Rothstein, 1995, 1999; Ono et al, 2003; Schramke et al, 2004; Moser et al, 2009). More recently, we showed that RPA facilitates telomerase action at telomeres (Luciano et al, 2012). In particular, in S. pombe, a mutation in the second OB-fold domain of the largest RPA subunit (Rpa1-D223Y) leads to substantial telomere shortening (Ono et al, 2003; Luciano et al, 2012). Furthermore, the rpa1-D223Y allele has a synergistic reduction in telomere length with rad3Δ and reduces the telomerase-dependent elongation of telomeres caused by the inactivation of poz1 or rap1 genes (Luciano et al, 2012). These data suggested that telomerase action is compromised in rpa1-D223Y cells; however, the mechanism by which RPA promotes telomerase action is not fully understood.
In this study, we show that the Rpa1-D223Y mutation increases the presence of DNA polymerase Polα (Polα), but not DNA polymerase ε (Polε), at telomeres during telomere replication. Our results indicate that the Rpa1-D223Y mutation provokes the accumulation at telomeres of secondary DNA structures and the recruitment of the homologous recombination factor Rad52. The presence of these secondary DNA structures at telomeres correlates with reduced amounts of Pot1 and Ccq1 at telomeres. Strikingly, we found that over-expression of Pif1 helicase family members was able to rescue the telomere phenotypes of the rpa1-D223Y mutant. Furthermore, we found that the corresponding mutation in human RPA (RPA1-D228Y) reduces the affinity for telomeric single-strand DNA (ssDNA) prone to form G-quadruplex (G4) and affects its binding mode. Taken together, our results suggest that RPA acts at the lagging telomere to prevent accumulation of G4 or G-rich structures thereby promoting telomere elongation by telomerase.
Results
rpa1-D223Y mutant causes dramatic replication defects at telomeres
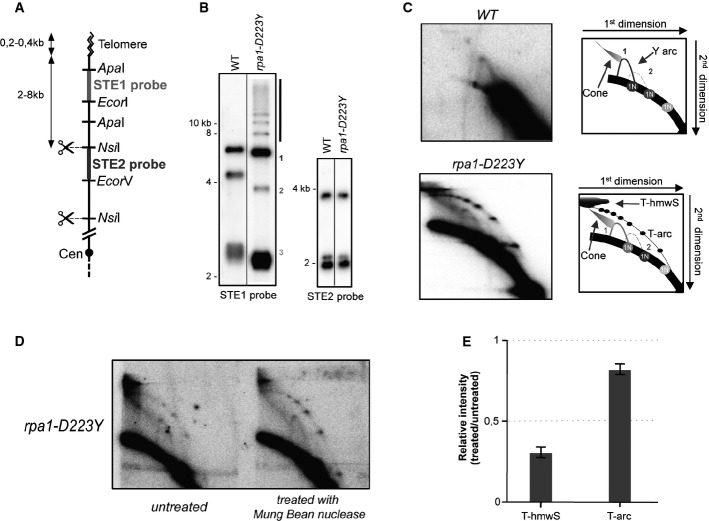
Previous work indicated that telomerase-mediated telomere elongation required the activity of Polα and Polδ (Diede & Gottschling, 1999). We thus decided to investigate how Rpa1-D223Y mutation (called D223Y below) impacts the DNA replication at telomeres by DNA two-dimensional gel electrophoresis analysis (2D-gel). As previously described (Miller et al, 2006), we digested DNA samples with NsiI to release telomere terminal fragments containing sub-telomeric sequences (Fig1A). We compared 2D-gel profiles between the parental WT and the D223Y mutant. In the first dimension, we observed three main bands after NsiI digestion revealed with either STE1 or STE2 subtelomeric probes (Fig1B). Telomere shortening in the D223Y mutant explains this size difference seen with the STE1 probe between WT and the D223Y mutant. The first dimension analysis indicates that subtelomeric regions were not rearranged in the D223Y mutant. However, we observed several additional higher bands (> 8 kb) in the D223Y mutant with the STE1 probe. In WT cells, 2D-gel analysis revealed that these telomere fragments were replicated as simple Y-arcs corresponding to replication forks progressing toward chromosome ends (Fig1C, upper panel). A cone-shaped signal extending from the crest of the Y-arcs that has been shown to represent four-way DNA junctions (double Y) can also be detected (Fachinetti et al, 2010). In the D223Y strain, the Y-arcs were also detected and the cone-signal was exacerbated (Fig1C, lower panel). Strikingly, we observed an additional telomeric-arc (T-arc) that migrates above the Y-arcs. The T-arc runs at the same position as bubble-arcs that are observed at replication origin loci (Sanchez et al, 1998; Segurado et al, 2002). Several structures of the same molecular weight appear to accumulate along the T-arc as evidenced by the 5–6 dots detected in the T-arc. Finally, at the apex of the T-arc and above the cone, we observed a massive accumulation of telomeric high-molecular weight structures (T-hmwS). To further gain insight into the nature of the structures that were generated during telomere replication, D223Y DNA sample was treated with the single-stranded-specific DNA Mung Bean nuclease prior 2D-gel analysis (Fig1D). Nuclease treatment significantly reduced the T-hmwS, but the T-arc remained largely intact (Fig1E). These data suggested that the T-hmwS contains ssDNA that is likely the product of recombination events. In contrast, T-arc appeared to contain little or no ssDNA, or alternatively, ssDNA is not accessible to the nuclease. Taken together, we concluded that the D223Y mutation causes dramatic replication defects at telomeres.
Figure 1.
The rpa1-D223Y mutant exhibits severe replication defects at telomeres
- Relative position of the restriction sites in the telomeric and subtelomeric regions of chromosomes I and II. The subtelomeric probes (STE1 and STE2) used for 2D-gel hybridization are represented. Cen, centromere.
- Southern blot analysis of NsiI telomeric fragments (1st dimension) revealed by STE1 and STE2 probes.
- 2D-gel analysis of NsiI telomeric fragments. The Y-arc pattern is generated by the unidirectional movement of a replication fork across each telomeric fragment shown in the 1st dimension. The cone-shaped signal represents four-way DNA junctions (double Y). In the rpa1-D223Y strain (lower panel), additional telomeric high-molecular weight structures (T-hmwS) and an extra telomeric arc (T-arc) are observed.
- DNA of the rpa1-D223Y strain was treated with Mung Bean nuclease prior 2D-gel analysis.
- Quantification of T-hmwS and T-arc signal for Rpa1-D223Y sample with and without Mung Bean nuclease treatment. T-hmwS and T-arc signals are normalized to the 1N spot signal. Bars correspond to the mean of two independent experiments.
We also analyzed by 2D-gel the replication intermediates that are generated at the replication fork barrier (RFB) region of the rDNA known to be also difficult to replicate (Supplementary Fig S1A). In WT, we detected Y-arc and X-spike DNA structures as previously reported (Sanchez et al, 1998; Segurado et al, 2002). Although the presence of the D223Y mutation in Rpa1 leads to enhancement of fork pausing and X-shaped structures relative to WT, we neither detected the T-arc nor the T-hmwS at RFB site of the rDNA locus (Supplementary Fig S1B). Consistent with this observation, no obvious rDNA instability at chromosome III is detected by pulse-field gel electrophoresis in contrast to what has been shown in a rqh1Δ strain (Supplementary Fig S1C) (Coulon et al, 2004).
Dissociation of Polα, but not Polε, is delayed at telomeres of rpa1-D223Y cells
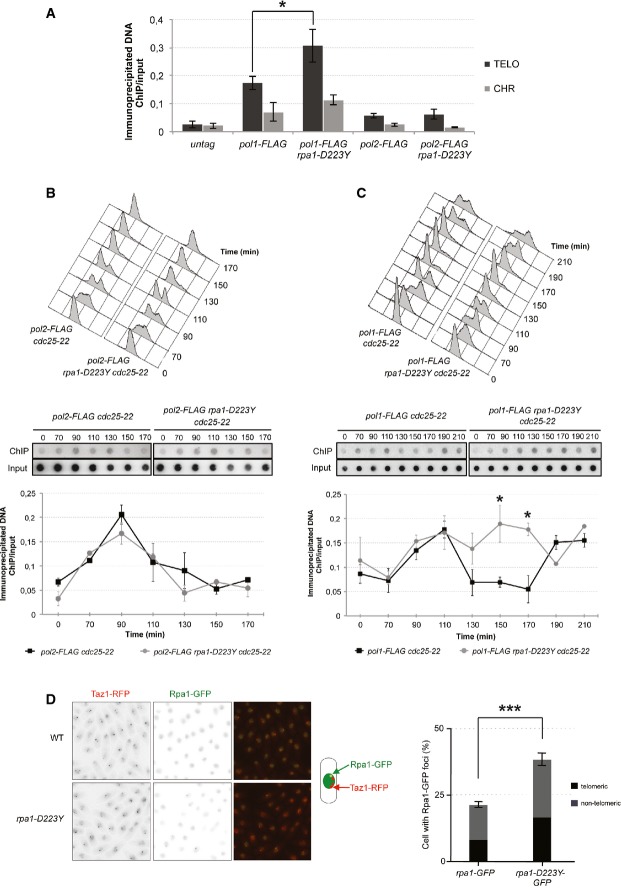
Because D223Y cells displayed a severe replication defect, we monitored the presence of Polα (Pol1) and Polε (Pol2) at telomeres (telomere length of strains used in this study is shown in Supplementary Fig S2). We first performed chromatin immunoprecipitation (ChIP) experiments of FLAG-tagged Polα and Polε in asynchronous WT and D223Y strains as previously described (Moser et al, 2009). The presence of Polε at telomeres was similar in WT and D223Y cells (Fig2A). In contrast to Polε, Polα presence at telomeres was markedly stronger particularly in the D223Y mutant for which a 2-fold increase in Polα at telomeres was observed compared to WT.
Figure 2.
Polα, but not Polε, is retained at telomeres in rpa1-D223Y cells
- ChIP experiments of FLAG-tagged Pol1 (Polα) and Pol2 (Polε) at telomeres in WT and rpa1-D223Y asynchronous cells. The immunoprecipitated DNA was analyzed by quantitative PCR with telomeric primers and chromosomic primers (listed in Supplementary Table S2). The ratio of bound DNA over input DNA is represented. Data are the mean of five and four independent experiments for Polα and Polε, respectively.
- FACS analysis of pol2-FLAG cdc25-22 and rap1-D223Y pol2-FLAG cdc25-22 strains. Cells were grown at 25°C until OD600nm = 0.4 and shifted to 36°C for 3.5 h to synchronize them. Cells were then released at 25°C from the G2/M block. Middle panel, Pol2-FLAG immunoprecipitated DNA were spotted onto Hybond-N+ membrane and hybridized with a telomeric probe. Signals were quantified with “Image Gauge” software. Lower panel, IP/input signals are plotted. Data are the mean of three independent experiments.
- Same as (B) for pol1-FLAG cdc25-22 and rap1-D223Y pol1-FLAG cdc25-22 strains.
- Number and telomeric localization of Rpa1-GFP and Rpa1-D223Y-GFP foci. Rpa1 foci were analyzed in cells expressing Taz1-RFP. Data are the mean of three independent experiments with n > 300 cells for each clone.
Data information: For all panels, error bars indicate standard deviation, and P-values are from the Fisher’s LSD test. *P < 0.05; ***P < 0.001.
We next monitored cell-cycle-regulated binding patterns of Polε and Polα with dot-blot ChIP assays by utilizing cdc25-22 synchronized cell cultures (Fig2B and C, top and middle panel). For Polε, temporal telomere association patterns were nearly identical for WT and D223Y cells, with a peak of binding at 90 min after release from cell-cycle arrest at G2/M boundary (Fig2B and C, bottom panel). For WT cells, binding of Polα at telomeres peaked at 110 min and dropped at 130 min consistent with the delayed replication of the lagging strand versus leading strand (Fig2C) (Moser et al, 2009). By contrast, for D223Y cells, while the initial timing of increase in Polα at telomeres was similar to WT cells, Polα strikingly showed persistent association at telomeres through G2 phase. These results were consistent with those obtained with asynchronous cells (Fig2A). Since both appearance of abnormal DNA structures and increased Polα association are consistent with incomplete telomere replication, we concluded that D223Y impaired lagging-strand synthesis at telomeres. Consistent with increased ssDNA accumulation at telomeres due to impaired lagging-strand synthesis, we also found an increased number of RPA foci that co-localize with Taz1 in the D223Y mutant although the proportion of telomeric Rpa1 foci versus non-telomeric Rpa1 foci was similar in WT and rpa1-D223Y cells (Fig2D and Supplementary Fig S3A).
Rad52 is recruited at telomeres and essential in D223Y cells
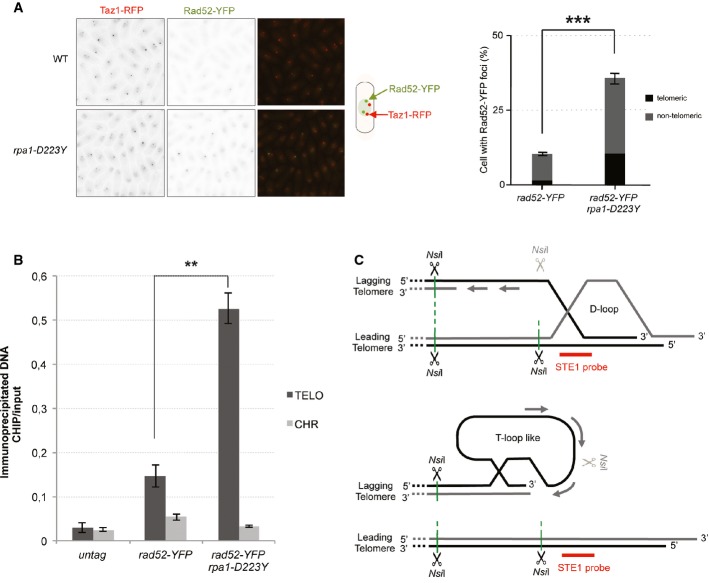
We first tested the genetic interaction of the D223Y allele with mutants of the homologous recombination pathway (HR). We were unable to obtain D223Y rad52Δ double mutant neither by tetrad dissection nor by random sporulation, whereas the double mutant D223Y rad51Δ was synthetically sick and exhibited severe sensitivity to HU (Supplementary Fig S3B). This indicated that HR sustained viability in D223Y cells. Consistent with this observation, we observe a 4-fold increase in Rad52-YFP foci in D223Y cells relative to WT (Fig3A and Supplementary Fig S3C). More specifically, co-localization of Rad52-GFP with Taz1-RFP in WT and in D223Y cells showed an increase in Rad52-GFP foci at telomeres. To determine whether Rad52 was present at telomeres, we performed ChIP experiments of Rad52-YFP in WT and D223Y asynchronous cells. We observed that association of Rad52 at telomeres was significantly increased in D223Y cells compared to WT (Fig3B).
Figure 3.
Rad52 is recruited at telomeres in rpa1-D223Y cells
- The number of Rad52-YFP foci was analyzed in cells expressing Taz1-RFP. Data are the mean of three independent experiments with n > 300 cells.
- Enrichment of Rad52-YFP at telomeres in WT and rpa1-D223Y asynchronous cells. ChIPs were performed with an anti-GFP antibody. The immunoprecipitated DNA was analyzed by quantitative PCR with telomeric primers and chromosome-specific primers (listed in Supplementary Table S2). The ratio of bound DNA over input DNA is represented. Data are the mean of three independent experiments.
- Model representing Rad52-dependent DNA structures that may arise at telomeres as a consequence of lagging-strand replication defects. NsiI sites within ssDNA might become insensitive to the NsiI cut thereby generating larger DNA fragments that result from a cut at a distal NsiI site.
Data information: For all panels, error bars indicate standard deviation and P-values are from the Fisher’s LSD test. **P < 0.01; ***P < 0.001.
Taken together, results of Figs1, 2 and 3 indicated that replication is impaired at the G-rich lagging-strand telomeres, and suggested that HR factors are recruited to resolve these structures. At this stage, we imagined that incomplete replication of the lagging telomere could generate ssDNA and that this ssDNA would be prone to recombine either with a leading telomere and initiate the formation of circular structures like a D-loop, or with itself to form a T-loop-like structure (Fig3C). The question arises about the structure that could prevent completion of replication at the lagging telomere and the generation of unreplicated ssDNA.
Overexpression of Pfh1Pif1 rescues rpa1-D223Y short telomere phenotype and replication defects
Among secondary structures that could be generated at telomeres, G4 structures have been proposed to be formed at S. pombe telomeres (Sabouri et al, 2014) although one does not have the formal proof of their formation. Interestingly, at telomeres, G4 could only be formed in the G-rich strand at the lagging telomere where Polα is retained in the D223Y cells. To test the presence of secondary structures, we overexpressed spDna2, Rqh1 (RecQ helicase), and Pfh1Pif1 helicases. Homologs of these helicases in Saccharomyces cerevisiae (scDna2, Sgs1RecQ, and scPif1) are known to unwind G4 with different efficiencies (Sun et al, 1999; Masuda-Sasa et al, 2008; Paeschke et al, 2011, 2013; Zhou et al, 2014). Moreover, we found that the D223Y mutant exhibits negative genetic interaction with each of these three helicases. The D223Y allele is synthetically lethal with dna2-C2 ts mutant and synthetically sick with rqh1Δ, rqh1-hd (helicase dead) and pfh1-mt* strains (Pinter et al, 2008; Supplementary Fig S4).
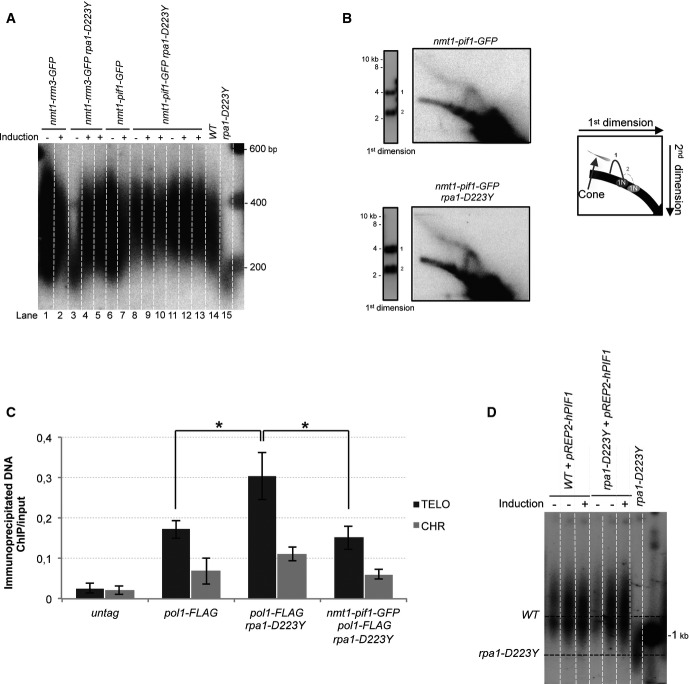
D223Y cells were transformed with pREP1 plasmid expressing spDna2, Rqh1, or Pfh1Pif1 DNA helicases under the control of the nmt1 promoter. We checked that these plasmids were able to rescue the growth defect or HU sensitivity of the corresponding dna2-C2, pfh1-mt*, and rqh1Δ strains (Supplementary Fig S5). Interestingly, among these three candidates, only overexpression of Pfh1Pif1 was able to restore WT telomere length of D223Y cells (Fig4A, lanes 8, 9). Noteworthy, a milder overexpression of Pfh1Pif1 with pREP41 did not restore telomere length of D223Y cells (Supplementary Fig S6). Consistent with a recent study, we observed that overexpression of Pfh1Pif1 by itself slightly lengthened the telomeres of a WT strain (Fig4A, lanes 6, 7) (McDonald et al, 2014). Thus, among the tested helicases, only Pfh1Pif1 was able to rescue the telomere defect of the D223Y mutant. The ability of Pfh1Pif1 to unwind G4 has never been tested in vitro. However, scPif1 has been shown to be a potent G4 unwinder, more efficient than Sgs1RecQ (Paeschke et al, 2013). Our observation that Pfh1Pif1, but not Rqh1, is able to restore telomere length reinforces the hypothesis that accumulation of G4 could be the cause of the telomeric defect in D223Y cells.
Figure 4.
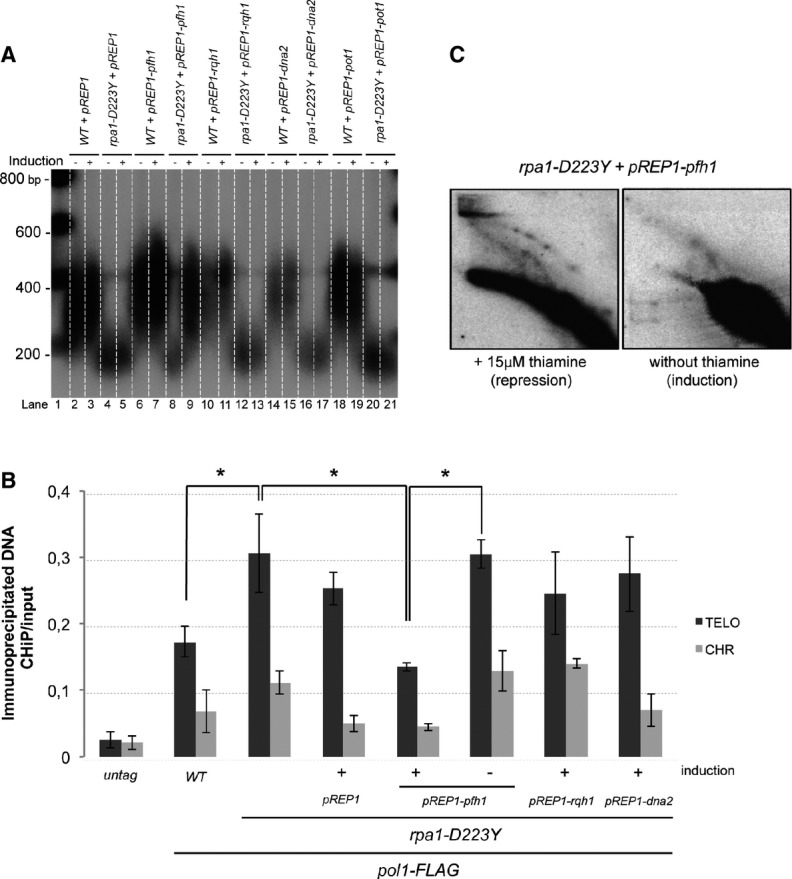
Overexpression of Pfh1Pif1 rescues the rpa1-D223Y short telomere phenotype
- Telomere length of strains overexpressing Pot1, Rqh1, spDNA2, or Pfh1 was analyzed by Southern hybridization. The three helicase genes are under the control of the nmt1 promoter in pREP1 plasmid. Cells were grown in minimum medium ± 15 μM of thiamine. Genomic DNAs were digested by ApaI and hybridized with a telomeric probe (see Materials and Methods).
- ChIP experiments of FLAG-tagged Pol1 (Polα) in asynchronous cells overexpressing Rqh1, Dna2, or Pfh1Pif1 helicases. Immunoprecipitated DNAs were analyzed by quantitative PCR with either telomeric or chromosomic primers (see Supplementary Table S2). The ratio of bound DNA over input DNA is represented. Data are the mean of three independent experiments. Error bars indicate standard deviation. P-values are from the Fisher’s LSD test. *P < 0.05.
- 2D-gel analysis of NsiI telomeric fragments of rpa1-D223Y strain containing the pREP1-pfh1 plasmid in the presence (repression) or absence (induction) of thiamine. The heterologous expression of the Pfh1 helicase suppresses the formation of the T-arc and the T-hmwS.
Then, we wondered whether overexpression of Pot1 rescues the telomere length defect of D223Y mutant since both proteins compete for ssDNA binding at telomeres and overexpression of Pot1 rescues the telomere loss of D223Y taz1Δ cells (Kibe et al, 2007). Importantly, overexpression of Pot1 under the same conditions as Pfh1Pif1 did not rescue the telomere shortening of the D223Y mutant (Fig4A, lanes 20, 21). This suggests that RPA has a telomeric function that is not shared by Pot1 in addition to the protection of telomeric ssDNA. However, we mitigated this conclusion since Pot1 access to telomeres may be limited under these conditions.
Next, we performed ChIP experiments of FLAG-tagged Polα in asynchronous D223Y cells overexpressing spDna2, Rqh1, and Pfh1Pif1. In agreement with telomere length rescue, only the overexpression of Pfh1Pif1 reduced Polα association at telomeres to the level comparable to WT cells (Fig4B). Overexpression of Pfh1Pif1 also prevented the accumulation of T-arc and T-hmwS when tested by 2D-gel (Fig4C). Thus, we concluded that the short telomere length of D223Y cells was linked to the impediment of telomere replication.
Overexpression of scPif1 restores rpa1-D223Y telomere replication defects and telomere shortening
Because Pfh1Pif1 has two homologs in S. cerevisiae, Rrm3 and scPif1, we decided to overexpress scPif1 and Rrm3 in D223Y cells. We took advantage of S. pombe strains that have been engineered in V. Zakian’s laboratory to overexpress Rrm3-GFP and scPif1-GFP fusion proteins with nmt1 promoter at leu1 genomic locus (Pinter et al, 2008). We first tested the effect of the overexpression of Rrm3 and scPif1 on telomere length in D223Y cells. Expression of both helicases was able to rescue the telomere shortening phenotype of the D223Y mutant (Fig5A, lanes 4–5, 9–10 and 12–13). However, in non-induced conditions, the basal expression of scPif1-GFP driven by the nmt1 promoter was sufficient by itself to restore a WT telomere length of the D223Y cells (lanes 8 and 11). In contrast, suppression of the short telomere length of the D223Y mutant required the full induction of the Rrm3-GFP (compare lanes 3–8 and 11). Quantification of GFP fluorescence signals indicated that basal expressions of scPif1 and Rrm3 were similar, while after induction, scPif1 and Rrm3 expression level increased by 2.5- and 1.8-fold, respectively (Supplementary Fig S7). These results are consistent with data from Paeschke et al (2013) showing that Rrm3 was able to suppress G4-induced genome stability when the level of scPif1 was low (Paeschke et al, 2013). Because scPif1 is a much more potent G4 unwinder than Rrm3, we concluded that an important function of RPA (defective in the D223Y mutant) could be to prevent accumulation of G4 occurring at the lagging telomere. Noteworthy, scPif1 has been recently described to unwind tetramolecular G4 (Byrd & Raney, 2015) raising the possibility that this type of G4 may be formed in telomeric or subtelomeric regions of S. pombe. Another possibility would be that the D-loop branch migration function described for scPif1 is responsible for the restoration of the telomere length defect of D223Y cells (Saini et al, 2013; Wilson et al, 2013).
Figure 5.
Overexpression of scPif1 restores rpa1-D223Y telomere replication defects and telomere shortening
- Telomere length of strains overexpressing Rrm3 and scPif1. Genomic DNAs digested by ApaI were analyzed by Southern hybridization. RRM3 and scPIF1 under the control of nmt1 promoter were induced by removing thiamine from the minimum medium.
- 2D-gel analysis of NsiI telomeric fragments in a strain expressing scPif1 at basal level (see legend of Fig1). nmt1-pif1-GFP and nmt1-pif1-GFP rpa1-D223Y cells were grown in YES medium under conditions in which the nmt1 promoter is not induced.
- ChiP experiments of FLAG-tagged Pol1 (Polα) at telomeres in asynchronous cells expressing scPif1 at basal level. The immunoprecipitated DNA was analyzed by quantitative PCR with telomeric and chromosomic primers and represented as in Fig2A. Data are the mean of three independent experiments. Error bars indicate standard deviation. P-values are from the Fisher’s LSD test. *P < 0.05.
- Telomere length of strains overexpressing hPIF1. hPIF1 is under the control of nmt1 promoter in pREP2 plasmid. Cells were grown in minimum medium ± 15 μM of thiamine. Genomic DNAs were digested by EcoRI and analyzed by Southern hybridization.
We further performed 2D-gel analysis and ChIP of Polα in D223Y cells expressing basal levels of scPif1-GFP. Remarkably, Fig5B shows that the T-hmwS and T-arc that we previously observed in D223Y cells disappeared when scPif1-GFP was expressed at a basal level in the D223Y mutant. Thus, we hypothesized that the D223Y mutant accumulated G4 structures at the lagging telomere that were removed by scPif1. This assumption was further strengthened by ChIP experiments of Polα in the D223Y mutant expressing a basal level of scPif1-GFP showing that the accumulation Polα at telomeres was also rescued by the expression of scPif1-GFP (Fig5C). Importantly, neither the sensitivity to hydroxyurea (HU) exhibited by the D223Y mutant, nor the D223Y rad52Δ synthetic lethality, nor the D223Y rad51Δ synthetic sickness were rescued by scPif1-GFP (data not shown).
The human PIF1 (hPIF1) has also been described to unwind G4 (Sanders, 2010; Paeschke et al, 2013). Therefore, we wondered whether hPIF1 was also able to restore the telomere length defect of D223Y cells. hPIF1 cDNA was cloned in pREP1 plasmid and introduced in WT and D223Y cells. The Fig5D shows that hPIF1 restores the telomere shortening of D223Y cells under conditions where hPIF1 is overexpressed or expressed at its basal level. Our results suggest that the ability of scPif1 and hPIF1 to restore telomere length defect of D223Y cells stems from their property to unwind G4.
Taken together, our results suggest that the G4-unwinding function of scPif1 is responsible for the rescue of the telomere phenotypes of the D223Y mutant.
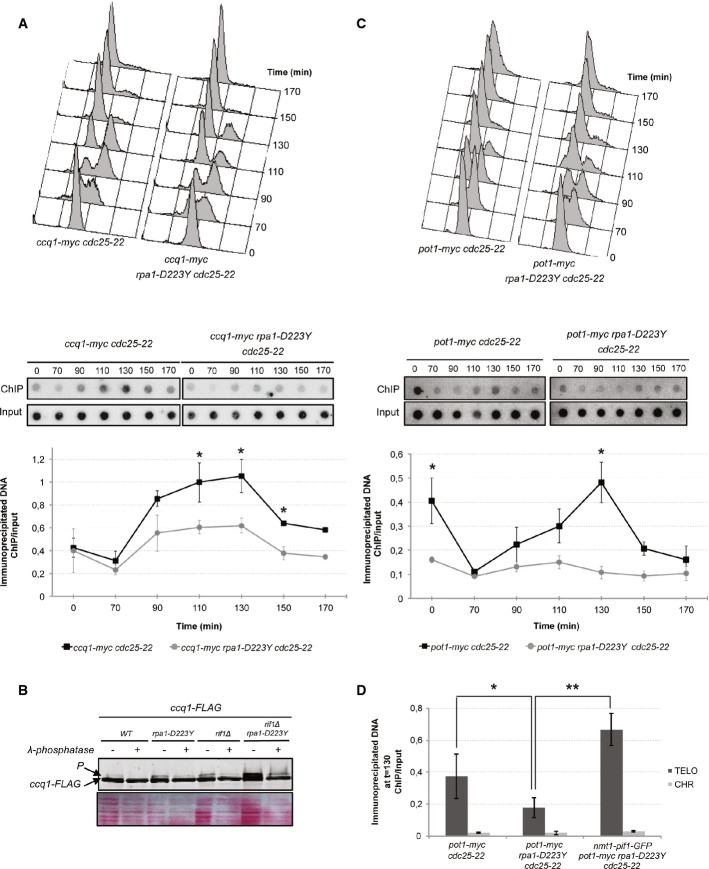
Recruitment of Ccq1 and Pot1 are impaired in rpa1-D223Y cells
To test whether the structures generated at telomeres in the D223Y mutant (Fig1C) might affect association of shelterin at telomeres, we monitored the binding of Ccq1 and Pot1 in D223Y cells. We performed ChIP experiments of Ccq1-myc and Pot1-myc in synchronized cultures. In WT cells, Ccq1 peaks at telomeres between 90 and 130 min and then drops to its initial level (Fig6A). The binding of Ccq1 in the D223Y mutant follows the same profile as the WT, but its level is reduced by 1.7-fold compared to the WT. In addition, we monitored the phosphorylation of Ccq1-FLAG in WT and D223Y in asynchronous cells in the presence or absence of Rif1 (to enhance Ccq1 phosphorylation) (Fig6B). In the presence or absence of Rif1, we observed that Ccq1 phosphorylation was stronger in D223Y cells than in WT. This could be explained by the fact the PT93-Ccq1/Est1 interaction is stimulated because telomeres are short in D223Y cells.
Figure 6.
Recruitment of Ccq1 and Pot1 are impaired in rpa1-D223Y cells
- Upper panel, FACS analysis of ccq1-myc cdc25-22 and rap1-D223Y ccq1-myc cdc25-22 strains. Cells were grown as previously described in Fig2B. Lower panel, ChIP of Ccq1-myc was performed with anti-myc 9E10 antibodies. Immunoprecipitated DNA was spotted onto Hybond-N+ membrane and hybridized with a telomeric probe. Signals were quantified with “Image Gauge” software. ChIP over input signals are plotted.
- Levels of FLAG-tagged Ccq1 in rpa1-WT and rpa1-D223Y cells in WT or rif1Δ genetic background. Cell lysates were treated with phosphatase inhibitors or with λ-phosphatase as indicated. Ponceau-S staining of the membrane was used as a loading control.
- Same as (A) for pot1-myc cdc25-22 and rap1-D223Y pot1-myc cdc25-22 strains.
- ChIP of Pot1-myc in synchronized cells at 130 min when the binding of Pot1 at telomeres reaches its maximum. The expression of scPif1 at its basal level restores the defect of Pot1 binding in rpa1-D223Y mutant. Immunoprecipitated DNAs were analyzed by quantitative PCR with either telomeric or chromosomic primers. The ratio of bound DNA over input DNA is represented.
Data information: In (A, C, D), the represented data are the mean of three independent experiments. Error bars indicate standard deviation, and P-values are from the Fisher’s LSD test. *P < 0.05; **P < 0.01.
For its part, binding of Pot1 in WT cells at telomeres was maximum at 130 min, in agreement with previous results (Fig6C) (Moser et al, 2009). We found that binding of Pot1 was severely affected in D223Y synchronized cells, suggesting that telomeric ssDNA was less accessible to Pot1 in the D223Y mutant. Strikingly, expression of scPif1 was able to rescue the binding of Pot1 at telomeres (Fig6D).
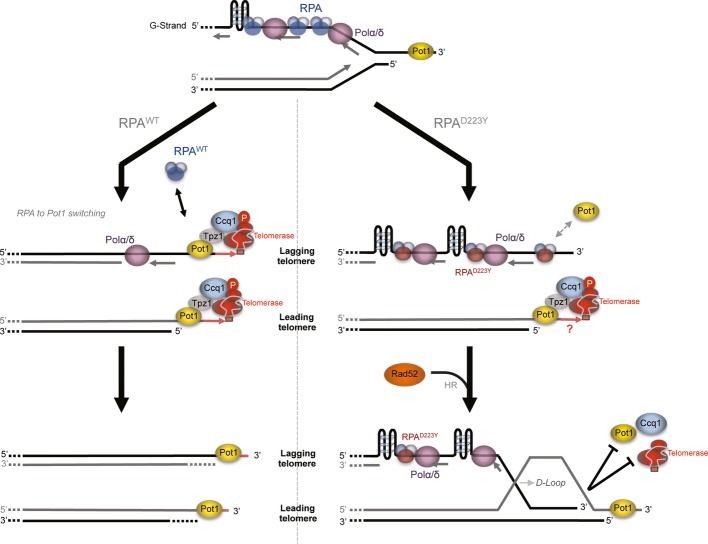
Taken together, these data suggest that the replication defect of D223Y allele impedes replication at telomeres and the correct recruitment of Pot1 and Ccq1 shelterin components that are necessary for telomerase action (see Fig 8 and Discussion).
Figure 8.
Model of RPA preventing secondary structures accumulations at the lagging telomere in fission yeast
RPA is recruited at telomeres with the replication machinery. Differential arrival of leading and lagging DNA polymerases generates ssDNA at the G-rich strand telomere and may lead to G4 formation. RPAWT ensures correct replication of the lagging telomere by preventing G4 accumulation. RPA-to-Pot1 switch occurs normally. Shelterin complex formation allows efficient recruitment of telomerase. In rpa1-D223Y cells, the accumulation of G4 alters the proper replication of the lagging telomere. Exposed ssDNA is processed by HR pathway. RPA-to-Pot1 switch and shelterin complex formation are hindered, which may lead to less efficient telomere extension by telomerase (see Discussion).
The D228Y mutation in human RPA1 affects its DNA-binding activity to telomeric DNA
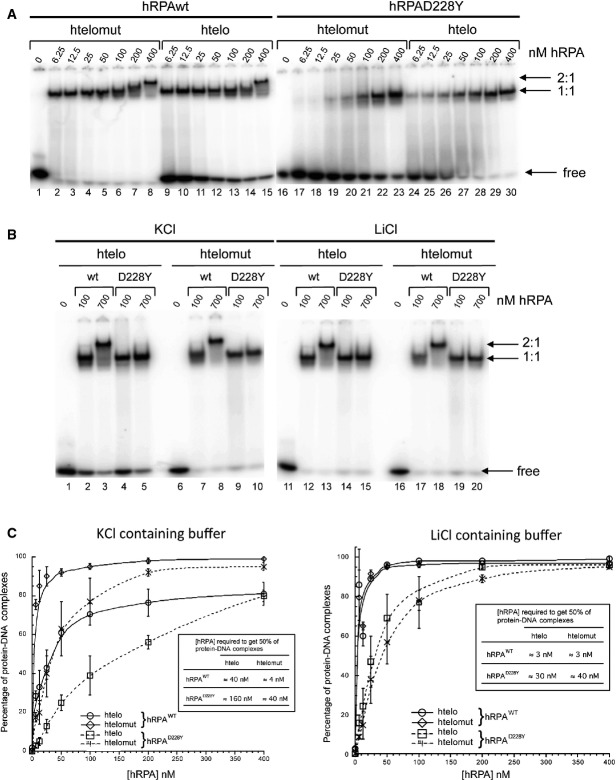
The aspartic acid residue of Rpa1 at position 223 in S. pombe and 228 in human is highly conserved in eukaryotes. This residue lies within the loop region opposite the DNA-contacting surface of the DNA-binding domain A (DBD-A) in the crystal structure of human RPA1 (Bochkarev et al, 1997). We choose to purify the hRPAD228Y because the budding yeast RPAD228Y turned out to be very difficult to purify to homogeneity (our unpublished results) (Deng et al, 2014). The mechanism by which hRPA binds to ssDNA has been previously described (Salas et al, 2006) (Supplementary Fig S8A). Briefly, hRPA has two modes of binding the 1:1 and 2:1 hRPA:DNA complexes. In 1:1 binding mode, one hRPA binds to ssDNA through the RPA1 and RPA2 subunits. In this mode, hRPA binds to 13–22 nucleotides of DNA with contacts to DBD-A, DBD-B, and DBD-C of RPA1 and the DBD-D of RPA2. In the 2:1 binding mode, two hRPA are bound solely through their RPA1 subunit, suggesting a compact 8–10-nt binding mode where only the DBD-A and DBD-B of RPA1 are bound along 8–10 nucleotides preventing RPA2 to contact DNA. This mode of binding is particularly important when the ssDNA sequences are able to form G4 structures (Salas et al, 2006; Safa et al, 2014). Indeed, G4 structures are in a dynamic equilibrium with unfolded and partially folded states. hRPA will bind first with 1:1 mode and then switch to 2:1 mode to unfold G4 (Supplementary Fig S8B).
In order to assess whether the hRPA:DNA interaction was affected by the presence of the D228Y mutation, we carried out binding experiments with two different DNA sequences: the 21-nt-long oligonucleotide that contains the human telomeric repeat sequence (htelo) and the 21-nt-long oligonucleotide (htelomut) in which the four guanines of the htelo oligonucleotide have been replaced by four cytosines. Notably, the htelo oligonucleotide mimics the physiological telomeric G-rich ssDNA and forms G4 in vitro (Safa et al, 2014), while the htelomut oligonucleotide does not. We purified recombinant hRPAWT and hRPAD228Y (Supplementary Fig S8C) and assayed their association with htelo and htelomut oligonucleotides by electrophoretic mobility shift assays (EMSA) under conditions where htelo oligonucleotide forms G4 (20°C, 100 nM K+) or does not (20°C, 100 mM Li+) (Fig7A). For hRPA, slow-migrating species corresponding to 1:1 and 2:1 hRPA:DNA complexes appeared for htelo and htelomut as the protein concentration increased (Fig7A). In contrast, the hRPAD228Y mutant was impeded in its ability to form the 2:1 complexes for both htelo and htelomut even at 700 nM of hRPAD228Y (Fig7B). Quantification of the EMSA clearly revealed that RPA displayed a lower affinity for htelo than htelomut in K+ while displaying similar affinities for both substrates in Li+ (Fig7C). The affinity of the mutant hRPAD228Y to both oligonucleotides was markedly decreased compared to hRPA under both conditions (K+ or Li+) (Fig7C). In K+, the affinity of hRPAD228Y to htelo was dramatically reduced (Fig7C). Taken together, these EMSA data indicate first that the mutant hRPAD228Y has a lower affinity for ssDNA than hRPA and second that hRPAD228Y is not able to switch from 1:1 to 2:1 binding mode and therefore may unfold less efficiently G4.
Figure 7.
The D228Y mutation affects hRPA DNA-binding activity
- Radiolabeled htelo [(GGGTTA)3GGG] and htelomut (GGCTTACGGTTAGCGTTACGG) (2 nM) were incubated with various amounts of hRPA or hRPAD228Y and the mixture separated by electrophoresis on a 5% acrylamide gel.
- 700 nM of hRPAD228Y did not allow the formation of 2:1 hRPA:DNA complexes, independently of the nature of the ssDNA (htelo or htelomut) and the monovalent cation (K+ or Li+).
- Quantification of the binding of hRPA or hRPAD228Y to ssDNA in KCl (left panel) and LiCl (right panel) containing buffer. The percentage of hRPA:DNA complexes corresponds to the ratio of bound DNA/total DNA (free and bound DNA). Error bars correspond to the standard deviation from three independent experiments. Insert table: concentration of hRPA required to form 50% of hRPA:DNA complex.
Discussion
Telomeres from S. pombe are about 300 bp long with unusual heterogeneous sequence G2-8TTAC(A) that are likely to form intra- or intermolecular G4 (Sabouri et al, 2014; Byrd & Raney, 2015). Previous results have demonstrated a differential arrival of leading- and lagging-strand DNA polymerases at telomeres in fission yeast. This uncoupling creates extended telomeric ssDNA at the lagging telomere until ssDNA is converted to duplex telomeric DNA by the lagging-strand polymerases (Moser et al, 2009; Chang et al, 2013). In agreement with these observations, RPA binding to telomeres in fission yeast increases as Polε arrives and then decreases as Polα and Polδ replicate the lagging telomeres (Luciano et al, 2012; Chang et al, 2013). Consistent with these results, specific enrichment of RPA at lagging strand versus leading strand has been recently confirmed in budding yeast (Yu et al, 2014).
In this study, we show that the D223Y mutation located in the second OB fold of the largest subunit of RPA (DBD-A) provokes the accumulation of secondary structures during telomere replication and the recruitment of Rad52 at chromosome ends. Our results indicate that these aberrant telomeric structures originate from lagging telomere replication and require the HR pathway to be processed. The presence of these secondary DNA structures correlates with reduced amounts of Pot1 and Ccq1 at telomeres that are both involved in the recruitment of telomerase. Strikingly, overexpression of members of the Pif1 helicase family (Pfh1Pif1, Rrm3, scPif1, and hPIF1) rescues telomere length of D223Y cells. From these results, we propose a model in which RPA prevents the formation of G4 structures during the transient accumulation of telomeric ssDNA that occurs at the lagging telomere (Fig8). These results are consistent with the previously described role of RPA in preventing formation of hairpin DNA in S. cerevisiae (Chen et al, 2013; Deng et al, 2014).
This model is strengthened by in vitro results that we obtained with recombinant human RPA complex. Here, we show that the D228Y mutation overall decreases the affinity of RPA for human single-stranded telomeric DNA able to form G4 structures in vitro. The DNA-binding mode of RPA to ssDNA is also affected by this mutation as shown by the fact that the RPAD228Y heterotrimer loses its capacity to form 2:1 complexes which was shown to be crucial to unfold G4 (Salas et al, 2006; Safa et al, 2014). Overall, these in vitro data suggest that besides decreasing the affinity for DNA, the mutation D228Y impedes the ability of hRPA to unfold G4 (Salas et al, 2006; Safa et al, 2014). These data may explain why the Rpa1-D223Y mutation leads to the accumulation of G4 in vivo in S. pombe.
In a recent study, McDonald and collaborators have proposed that Pfh1Pif1 is a positive regulator of telomere length (McDonald et al, 2014). They proposed that Pfh1Pif1 has a RPA-dependent role in telomere lengthening because overexpression of Pfh1Pif1 (driven by a plasmid in which Pfh1Pif1 is under the control of its own promoter) does not rescue the short telomere phenotype of D223Y cells. In contrast, we observed a rescue of the short telomere phenotype of D223Y cells when Pfh1Pif1 is overexpressed under a strong promoter (nmt1). The strength of the promoter certainly explains this apparent discrepancy. Supporting this idea, telomeres of D223Y cells remain short when Pfh1Pif1 is overexpressed under the control of nmt41 promoter (roughly 100 times weaker than nmt1) (Supplementary Fig S6).
Here, we also show that the telomere defects of D223Y cells are rescued not only by overexpression of Pfh1Pif1 but also by scPif1, Rrm3, and hPIF1 that all share the ability to unwind G4 in vivo (Paeschke et al, 2013). Strikingly, basal heterologous expression of scPif1 rescues all the telomeric defects of D223Y cells that we have tested so far, including short telomeres, accumulation of secondary structures at telomeres (monitored by 2D-gel) and extended presence of Polα at telomeres (Fig5). Similar to scPif1, the basal heterologous expression hPIF1 rescued the telomere shortening of D223Y cells. Given that one of the main function of scPif1 and hPIF1 is to remove G4 (Paeschke et al, 2011, 2013; Zhou et al, 2014), these results are consistent with our assumption that the D223Y mutation leads to accumulation of G4 at lagging telomeres during telomere replication either by preventing the formation of G4 or by unwinding them once they are formed. Like at telomeres, G4 could arise at the rDNA locus (Capra et al, 2010) which is known to be hard to replicate because of the presence of multiple pausing sites (Krings & Bastia, 2004). Although the rDNA is not unstable in D223Y cells, its replication was hindered in the D223Y mutant (Supplementary Fig S1). However, the replication defects observed at the rDNA locus were not comparable with the ones observed at telomeres suggesting that at telomeres, the effect of the D223Y mutation is exacerbated because of extensive exposed ssDNA and high G4 levels.
In D223Y cells, we observe in 2D-gel analysis the appearance of an additional T-arc and a massive accumulation of T-hmwS. Nuclease treatment significantly reduced the T-hmwS, but the T-arc remains intact suggesting that T-hmwS contains ssDNA. The T-arc which migrates at the location of circular DNA could be the result of the emergence of multiple bubble-arcs at each telomere that come from the firing of cryptic origins within NsiI fragments. Indeed, subtelomeric sequences are AT-rich regions that may contain cryptic origins (Segurado et al, 2003). Another explanation would be that the T-arc represents the generation of D-loop structures within subtelomeric regions. In this scenario, accumulation of G4 at the lagging telomere in the D223Y mutant would prevent replication of the lagging-strand telomere allowing unreplicated telomeric ssDNA to invade the duplex DNA of the leading telomere (Fig8). However, one could expect that the D-loop structures are sensitive to Mung Bean nuclease treatment. Further investigations will be necessary to clearly determine the exact DNA structures of the T-arc. The T-hmwS could be explained by the fact that ssDNA is generated in subtelomeric regions. NsiI sites with ssDNA would become then insensitive to the NsiI cut and generate larger DNA fragments with a cut at a distal NsiI site (Fig3C). Because HR is required in D223Y cells, entangled telomeres or T-loop-like structures may generate these T-hmwS.
In fission yeast, rpa1-D223Y allele along with mutation in Rad3ATR or Rad26ATRIP shows extensive shortening of telomeres, whereas mutations in Tel1ATM or MRN complex have very little effect on telomere length (Nakamura et al, 2002; Ono et al, 2003). In contrast to fission yeast, mutations in Tel1ATM and MRXMRN lead to telomere shortening, while the rfa1-D228Y allele and mutants of Mec1ATR or Ddc2ATRIP have minor effect on telomere length in budding yeast (Smith et al, 2000; Craven et al, 2002). We have shown previously that telomere elongation by telomerase in budding yeast is likely to occur mainly at the leading telomere (Faure et al, 2010). In fission yeast, our data are rather consistent with the hypothesis that fission yeast primarily regulates telomere length by controlling telomerase activity on the lagging telomere through Rad3ATR-Rad26ATRIP (Moser et al, 2009). Indeed, we show that defects in the replication of the lagging telomeres are very strongly correlated with telomere shortening. Further studies are clearly needed to test this hypothesis.
In mammals, G4 formation during telomere replication also represents a threat to fulfill complete chromosome replication (Arnoult et al, 2009). Two types of proteins have been shown to unwind G4 in vitro: the DNA helicases including BLM, WRN, PIF1, DNA2, (Paeschke et al, 2013) and the ssDNA-binding proteins RPA and POT1 (Salas et al, 2006; Fan et al, 2009; Prakash et al, 2011; Hwang et al, 2012; Ray et al, 2013). Recently, Zimmermann et al (2014) have proposed that the BLM helicase is recruited at telomeres by TRF1 to facilitate lagging telomere DNA synthesis by removing secondary structures like G4 (Zimmermann et al, 2014). In this study, we show that in fission yeast RPA plays a major role in this process. One can imagine that several factors act sequentially to prevent G4 formation during lagging telomeres synthesis in human cells. Likely, RPA may come first to the lagging telomere with the replication fork. Then, RPA-to-POT1 switch might be operated to prevent ATR activation and to preserve telomere stability (Flynn et al, 2011). TRF1-dependent BLM recruitment or WRN might be necessary during this step to unwind the G4 that might be formed (Arnoult et al, 2009; Zimmermann et al, 2014). Further studies will be necessary to understand the precise mechanisms that cells have evolved to preserve telomere stability during replication.
Materials and Methods
Strains, constructs, and media
Schizosaccharomyces pombe methods and media have been described previously (Coulon et al, 2004). Strains used in this study are listed in Supplementary Table S1. Cells were grown in YES (yeast extract with supplements) or EMM (Edinburgh minimal medium) supplemented with amino acids as required. To construct pREP1-pfh1, pREP1-dna2, pREP1-rqh1, pREP1-pot1, and pREP41-pfh1 plasmids, genomic DNA was amplified by PCR (Q5-polymerase, NEB), sub-cloned with TA-Cloning® kit (Invitrogen), and integrated in pREP1. To construct pREP2-hPIF1, cDNA of hPIF1 was extracted from the plasmid pRS414(Paeschke et al, 2013) and integrated in pREP2 (for details: Supplementary Table S3). For strains carrying pREP plasmids, cells were grown at 32°C in the presence (or not) of 15 μM of thiamine in EMM with appropriate amino acids. Induction of genes under the control of the nmt1 promoter occurs in the absence of thiamine.
Cell-cycle synchronization
Temperature-sensitive allele cdc25-22 was used to synchronize cells at the G2/M transition. To induce the arrest, cells were incubated at 36°C for 4 h and then shifted to 25°C to induce synchronous cell cycle reentry. Cell cycle progression was monitored by FACS analysis.
Chromatin immunoprecipitation
Cells (100 ml at OD600nm = 0.5) were processed as previously described (Moser et al, 2009), with minor modifications. Antibodies (M2 mouse monoclonal anti-FLAG antibody (Sigma-Aldrich), anti-GFP monoclonal antibody (JL-8, Clontech) or anti-myc mouse monoclonal 9E10 (Santa Cruz)) was added to whole-cell extracts and incubated 3 h at 4°C on a rotator wheel, and then magnetic Dynabeads (Invitrogen) were added for 3 h at 4°C. Recovered DNA was analyzed either by SYBR Green-based real-time PCR (Takara) using telomeric (Telo) or chromosomic (Chr) primers that are listed in Supplementary Table S2 or by dot-blot hybridization. For dot-blot assay, recovered DNA was denatured in 1.5 N NaOH, 3 M NaCl for 10 min at room temperature, spotted directly onto Hybond-N+ membrane (GE healthcare), and cross-linked with UV (1,200 J/m2). The membrane was then hybridized with a telomeric probe. Quantification of ChIP signal by “Image Gauge” software was performed by subtraction of background signal. The ratios of ChIP/Input signal are presented. Dot-blot hybridization was used as a complementary approach to qPCR to analyze the presence of target protein in WT and D223Y cells that exhibit difference in telomere size.
Two-dimensional (2D) gel electrophoresis
2D-gel electrophoresis was performed as previously described (Noguchi et al, 2003) with minor modifications. For analysis of telomeres, 10 μg of DNA was digested with 60 U of NsiI. Precipitated DNAs were run first on a 0.4% agarose gel for 12 h at room temperature. Large bands were incised from first dimension agarose gel (from 1 kb to the top of the gel) in order to detect all the high-molecular weight structures. For the second dimension, samples were run in 1% agarose gel containing 0.33 μg/ml of ethidium bromide for 8 h at 4°C. Gels were transferred to Hybond-XL membranes (GE healthcare), cross-linked with UV and probed with a subtelomeric probe (STE). Genomic DNA was incubated with 1 U of Mung Bean nuclease (NEB) per μg of DNA and precipitated prior 2D-gel electrophoresis.
Telomere length analysis (Southern Hybridization)
Genomic DNA was prepared from 15 ml of cells at OD600nm = 1 and digested either with ApaI or EcoRI. The digested DNA was resolved in a 1.2% agarose gel and blotted onto a Hybond-XL membrane. After transfer, the membrane was cross-linked with UV and hybridized with a telomeric probe. Telomere length of strains used in this study is shown in Supplementary Fig S2. 32P labeling of DNA probes was performed with Rediprime II DNA-Labeling System kit (Amersham). The telomeric and subtelomeric STE DNA probes were extracted by digestion of pIRT2-Telo plasmid by SacI/PstI and EcorI-ApaI, respectively (Rog et al, 2010).
Microscopy
Live cell analysis of telomeric foci was performed in an imaging chamber (CoverWell PCI-2.5, Grace Bio-Labs, Bend, OR) filled with 1 ml of 1% agarose in minimal medium and sealed with a 22 × 22-mm glass coverslip. Images of Z stacks (maximum five stacks of 0.3–0.4-μm steps, to avoid photobleaching) were taken at 32°C, and images were acquired from an inverted wide-field microscope (Nikon Eclipse TI) equipped with a Neo 5.5 sCMOS camera (Andor Technology Ltd), a LED light source (Lumencor Spectra), and a 100× objective (1.45 NA). Images were recorded using the MetaMorph software package (Molecular Devices France, St. Gregoire, France). Relative distance distribution analysis between foci was achieved using image J software and Excel for display.
Purification of hRPAWT and hRPAD228Y
We purified the hRPAD228Y because the budding yeast RPAD228Y turns out to be very difficult to purify to homogeneity. The D228Y mutation was introduced by site directed mutagenesis in p11d-tRPA (Henricksen et al, 1994). The p11d-tRPAWT or p11d-tRPAD228Y plasmid (sequence verified by sequencing from ATGC) was used to transform BL21(DE3) cells. Cells freshly transformed with either p11d-tRPAWT or p11d-tRPAD228Y were used to inoculate 2 or 6 l of TB media (1.2% tryptone, 2.4% yeast extract, 0.4% glycerol, 100 μg/ml ampicillin), respectively, and were incubated at 34°C overnight with slow shaking (150 rpm). The next morning, cells were left growing (170 rpm) at 34°C until they reached an OD600nm of 0.4. Protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.3 mM. After 2 h of induction at 34°C under slow shaking (170 rpm), the cells were harvested by centrifugation (5,000 g, 4°C, 30 min) and kept at −80°C until used. hRPA and hRPAD228Y were purified according to the procedure developed by Dupaigne et al (2008) for the yeast RPA. Briefly, 9 g (for hRPAWT) or 20 g (for hRPAD228Y) of cell paste were resuspended in 45 or 70 ml of lysis buffer, respectively (25 mM Tris pH 7.5, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol), supplemented with a cocktail of protease inhibitors (Roche), stirred on ice for 30 or 60 min, respectively, and sonicated (30 cycles of 1 min with bursts of 30 s/min). The crude extract was centrifuged (25,000 g, 4°C, 1 h), and the soluble material was loaded on a 5-ml ssDNA cellulose column (1.5 cm diameter, 1 ml/min) pre-equilibrated in lysis buffer. The column was next washed with 25 ml of W1 buffer (25 mM Tris pH 7.5, 500 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). In the case of the hRPAWT protein, the column was also washed with 25 ml of W2 buffer (25 mM Tris pH 7.5, 750 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol) and the hRPAWT protein was eluted with 40 ml of E buffer (25 mM Tris pH 7.5, 1.5 M NaCl, 50% ethylene glycol, 1 mM EDTA, 1 mM DTT, 10% glycerol). For hRPAD228Y, the mutation changed the binding property of the protein onto the ssDNA cellulose resin (Amersham Biosciences) and the protein was almost completely eluted in the W2 buffer. Fractions containing either the WT or mutant protein were pooled and dialyzed overnight against 2 l of B buffer (25 mM Tris pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). The dialyzed sample was loaded on a 1-ml MacroPrep HighQ column (Bio-Rad, 1 cm diameter, 0.7 ml/min) pre-equilibrated with B buffer. The column was next washed with 7 ml of B buffer and the protein was eluted with a 17 ml gradient of NaCl (from 100 mM to 500 mM NaCl) followed by a 20-ml step of C buffer (25 mM Tris pH 7.5, 1 M NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). Fractions containing the protein were pooled, dialyzed overnight against 2 l of storage buffer (25 mM Tris pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol). Aliquots of 20–100 μl were stored at −80°C. After purification, the proteins were estimated to be between 80–90% pure by staining of the protein gel with SYPRO (Invitrogen) (Supplementary Fig S8). The concentration of the protein was measured by UV spectroscopy using an extinction coefficient of 88,085 M−1cm−1.
Electrophoretic mobility shift assay
hRPA:DNA-binding assays were realized according to the procedure developed by Safa et al (2014). Briefly, oligonucleotides were labeled with γ[32P]ATP (3,000 Ci/mmol) using PNK (NEB). 32P-labeled oligonucleotides were purified on a denaturing 15% acrylamide gel. For all EMSA experiments, hRPA was diluted and pre-incubated (20 min at 4°C) in buffer containing 50 mM Tris–HCl pH 7.5, 1 mM DTT, 10% glycerol, 0.2 mg/ml BSA, 0.1 mM EDTA, and 100 mM KCl or 100 mM LiCl. In a standard reaction, radiolabeled oligonucleotide (2 nM) was incubated with various amounts of protein in 10 μl reaction buffer (5 mM HEPES pH 7.9, 0.1 mg/ml BSA, 100 mM KCl, and 2% glycerol) for 20 min at 20°C. Samples were then loaded on a native 5% acrylamide gel (acrylamide:bisacrylamide mass ratio = 29:1) made in 0.5× TBE buffer. Electrophoresis was performed for 40 min at 20 V/cm at room temperature. After electrophoresis, the gel was exposed on a phosphorimager screen for at least 10 h. The screen was scanned with the Phosphorimager TYPHOON instrument (Molecular Dynamics). The samples in the gel were quantified using ImageQuant version 5.1.
Pulse-field gel electrophoresis
Pulsed-field gel electrophoresis samples were prepared as described previously (Nakamura et al, 2002). To observe whole chromosomes, a 1% agarose gel was subjected to electrophoresis with constant circulation of TBE 0.5× buffer at 14°C for 48 h (2 V/cm, included angle 106°, initial and final switch time 30 min) and stained with ethidium bromide.
Acknowledgments
We thank M. Ueno, F. Ishikawa, J. Cooper, PM. Dehé, S. Lambert, P. Russell, M. Feirrera, E. Noguchi, and V. Zakian for sharing yeast strains and plasmids. We are very grateful to K. Paeschke, M. Bochman, and V. Zakian, for the gift of Pif1 constructs. We thank Christophe Machu for technical assistance. VG laboratory is supported by the «Ligue Nationale contre le Cancer» (LNCC) (équipe labellisée). JA has been supported by the LNCC and the Association pour la Recherche sur le Cancer (ARC). TMN is supported by the NIH grant GM078253.
Author contributions
JA generated the data shown in Figs1, 2A–C, 3B and C, 4, 5, 6, 8, Supplementary Figs S2, S3B, S4, S5, S6 and S7. LM generated the data shown in Supplementary Figs S2, S4, S5, S6 and S7. ED and CS generated the data shown in Fig7 and Supplementary Fig S8. TG and YG generated the data shown in Figs2D and 3A, Supplementary Fig S3A and C. SC generated the data shown in Figs1, 6B and 8 Supplementary Figs S1, S2 and S4. JA, LM, ED, TG, TMN, YG, CS, VG and SC participated in the design and interpretations of the results; VG and SC wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figures, Supplementary Tables
Review Process File
References
- Armstrong CA, Pearson SR, Amelina H, Moiseeva V, Tomita K. Telomerase activation after recruitment in fission yeast. Curr Biol. 2014;24:2006–2011. doi: 10.1016/j.cub.2014.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnoult N, Saintomé C, Ourliac-Garnier I, Riou J-F, Londoño-Vallejo A. Human POT1 is required for efficient telomere C-rich strand replication in the absence of WRN. Genes Dev. 2009;23:2915–2924. doi: 10.1101/gad.544009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]
- Byrd AK, Raney KD. A parallel quadruplex DNA is bound tightly but unfolded slowly by Pif1 helicase. J Biol Chem. 2015;290:6482–6494. doi: 10.1074/jbc.M114.630749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra JA, Paeschke K, Singh M, Zakian VA. G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comp Biol. 2010;6:e1000861. doi: 10.1371/journal.pcbi.1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-T, Moser BA, Nakamura TM. Fission yeast shelterin regulates DNA polymerases and Rad3ATR kinase to limit telomere extension. PLoS Genet. 2013;9:e1003936. doi: 10.1371/journal.pgen.1003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lisby M, Symington LS. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol Cell. 2013;50:589–600. doi: 10.1016/j.molcel.2013.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon S, Gaillard P-HL, Chahwan C, McDonald WH, Yates JR, Russell P. Slx1-Slx4 are subunits of a structure-specific endonuclease that maintains ribosomal DNA in fission yeast. Mol Biol Cell. 2004;15:71–80. doi: 10.1091/mbc.E03-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RJ, Greenwell PW, Dominska M, Petes TD. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics. 2002;161:493–507. doi: 10.1093/genetics/161.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P-M, Cooper JP. Fission yeast telomeres forecast the end of the crisis. FEBS Lett. 2010;584:3725–3733. doi: 10.1016/j.febslet.2010.07.045. [DOI] [PubMed] [Google Scholar]
- Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat Struct Mol Biol. 2014;21:405–412. doi: 10.1038/nsmb.2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–254. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell. 2010;39:595–605. doi: 10.1016/j.molcel.2010.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J-H, Bochkareva E, Bochkarev A, Gray DM. Circular dichroism spectra and electrophoretic mobility shift assays show that human replication protein A binds and melts intramolecular G-quadruplex structures †,‡. Biochemistry. 2009;48:1099–1111. doi: 10.1021/bi801538h. [DOI] [PubMed] [Google Scholar]
- Faure V, Coulon S, Hardy J, Géli V. Cdc13 and telomerase bind through different mechanisms at the lagging- and leading-strand telomeres. Mol Cell. 2010;38:842–852. doi: 10.1016/j.molcel.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg M, Gurung RL, Mansoubi S, Ahmed JO, Dave A, Watts FZ, Bianchi A. Tpz1TPP1 SUMOylation reveals evolutionary conservation of SUMO-dependent Stn1 telomere association. EMBO Rep. 2014;15:871–877. doi: 10.15252/embr.201438919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Géli V. How telomeres are replicated. Nat Rev Mol Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Hwang H, Buncher N, Opresko PL, Myong S. POT1-TPP1 regulates telomeric overhang structural dynamics. Structure. 2012;20:1872–1880. doi: 10.1016/j.str.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Kibe T, Ono Y, Sato K, Ueno M. Fission yeast Taz1 and RPA are synergistically required to prevent rapid telomere loss. Mol Biol Cell. 2007;18:2378–2387. doi: 10.1091/mbc.E06-12-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2004;101:14085–14090. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano P, Coulon S, Faure V, Corda Y, Bos J, Brill SJ, Gilson E, Simon MN, Géli V. RPA facilitates telomerase activity at chromosome ends in budding and fission yeasts. EMBO J. 2012;31:2034–2046. doi: 10.1038/emboj.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda-Sasa T, Polaczek P, Peng XP, Chen L, Campbell JL. Processing of G4 DNA by Dna2 helicase/nuclease and replication protein A (RPA) provides insights into the mechanism of DNA2/RPA substrate recognition. J Biol Chem. 2008;283:24359–24373. doi: 10.1074/jbc.M802244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KR, Sabouri N, Webb CJ, Zakian VA. The Pif1 family helicase Pfh1 facilitates telomere replication and has an RPA-dependent role during telomere lengthening. DNA Repair. 2014;24:80–86. doi: 10.1016/j.dnarep.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Miyagawa K, Low RS, Santosa V, Tsuji H, Moser BA, Fujisawa S, Harland JL, Raguimova ON, Go A, Ueno M, Matsuyama A, Yoshida M, Nakamura TM, Tanaka K. SUMOylation regulates telomere length by targeting the shelterin subunit Tpz1(Tpp1) to modulate shelterin-Stn1 interaction in fission yeast. Proc Natl Acad Sci USA. 2014;111:5950–5955. doi: 10.1073/pnas.1401359111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- Moser BA, Nakamura TM. Protection and replication of telomeres in fission yeast. Biochem Cell Biol. 2009;87:747–758. doi: 10.1139/o09-037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Subramanian L, Chang Y-T, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser BA, Chang Y-T, Kosti J, Nakamura TM. Tel1ATM and Rad3ATR kinases promote Ccq1-Est1 interaction to maintain telomeres in fission yeast. Nat Struct Mol Biol. 2011;18:1408–1413. doi: 10.1038/nsmb.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Moser BA, Russell P. Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics. 2002;161:1437–1452. doi: 10.1093/genetics/161.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–7874. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono Y, Tomita K, Matsuura A, Nakagawa T, Masukata H, Uritani M, Ushimaru T, Ueno M. A novel allele of fission yeast rad11 that causes defects in DNA repair and telomere length regulation. Nucleic Acids Res. 2003;31:7141. doi: 10.1093/nar/gkg917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paeschke K, Bochman ML, Garcia PD, Cejka P, Friedman KL, Kowalczykowski SC, Zakian VA. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013;497:458–462. doi: 10.1038/nature12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Pfeiffer V, Lingner J. Replication of telomeres and the regulation of telomerase. Cold Spring Harb Perspect Biol. 2013;5:a010405. doi: 10.1101/cshperspect.a010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter SF, Aubert SD, Zakian VA. The Schizosaccharomyces pombe Pfh1p DNA helicase is essential for the maintenance of nuclear and mitochondrial DNA. Mol Cell Biol. 2008;28:6594–6608. doi: 10.1128/MCB.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash A, Natarajan A, Marky LA, Ouellette MM, Borgstahl GEO. Identification of the DNA-binding domains of human replication protein A that recognize G-quadruplex DNA. J Nucleic Acids. 2011;2011:1–14. doi: 10.4061/2011/896947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Qureshi MH, Malcolm DW, Budhathoki JB, Çelik U, Balci H. RPA-mediated unfolding of systematically varying G-quadruplex structures. Biophys J. 2013;104:2235–2245. doi: 10.1016/j.bpj.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2010;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014;12:101. doi: 10.1186/s12915-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safa L, Delagoutte E, Petruseva I, Alberti P, Lavrik O, Riou J-F, Saintomé C. Binding polarity of RPA to telomeric sequences and influence of G-quadruplex stability. Biochimie. 2014;103:80–88. doi: 10.1016/j.biochi.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Saini N, Ramakrishnan S, Elango R, Ayyar S, Zhang Y, Deem A, Ira G, Haber JE, Lobachev KS, Malkova A. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–392. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas TR, Petruseva I, Lavrik O, Bourdoncle A, Mergny J-L, Favre A, Saintomé C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006;34:4857–4865. doi: 10.1093/nar/gkl564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez JA, Kim SM, Huberman JA. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp Cell Res. 1998;238:220–230. doi: 10.1006/excr.1997.3835. [DOI] [PubMed] [Google Scholar]
- Sanders CM. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem J. 2010;430:119–128. doi: 10.1042/BJ20100612. [DOI] [PubMed] [Google Scholar]
- Schramke V, Luciano P, Brevet V, Guillot S, Corda Y, Longhese MP, Gilson E, Géli V. RPA regulates telomerase action by providing Est1p access to chromosome ends. Nat Genet. 2004;36:46–54. doi: 10.1038/ng1284. [DOI] [PubMed] [Google Scholar]
- Segurado M, Gómez M, Antequera F. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol Cell. 2002;10:907–916. doi: 10.1016/s1097-2765(02)00684-6. [DOI] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F. Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep. 2003;4:1048–1053. doi: 10.1038/sj.embor.7400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Rothstein R. A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol Cell Biol. 1995;15:1632–1641. doi: 10.1128/mcb.15.3.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Rothstein R. An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics. 1999;151:447–458. doi: 10.1093/genetics/151.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Zou H, Rothstein R. Characterization of genetic interactions with RFA1: the role of RPA in DNA replication and telomere maintenance. Biochimie. 2000;82:71–78. doi: 10.1016/s0300-9084(00)00183-8. [DOI] [PubMed] [Google Scholar]
- Sun H, Bennett RJ, Maizels N. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 1999;27:1978–1984. doi: 10.1093/nar/27.9.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Cooper JP. Fission yeast Ccq1 is telomerase recruiter and local checkpoint controller. Genes Dev. 2008;22:3461–3474. doi: 10.1101/gad.498608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- Webb CJ, Zakian VA. Schizosaccharomyces pombe Ccq1 and TER1 bind the 14-3-3-like domain of Est1, which promotes and stabilizes telomerase-telomere association. Genes Dev. 2012;26:82–91. doi: 10.1101/gad.181826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, Kwon Y, Xu Y, Chung W-H, Chi P, Niu H, Mayle R, Chen X, Malkova A, Sung P, Ira G. Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration. Nature. 2013;502:393–396. doi: 10.1038/nature12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Tarumoto Y, Ishikawa F. Tel1(ATM) and Rad3(ATR) phosphorylate the telomere protein Ccq1 to recruit telomerase and elongate telomeres in fission yeast. Genes Dev. 2012;26:241–246. doi: 10.1101/gad.177873.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou Z-X, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z. Strand-specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell. 2014;56:551–563. doi: 10.1016/j.molcel.2014.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Zhang J, Bochman ML, Zakian VA, Ha T. Periodic DNA patrolling underlies diverse functions of Pif1 on R-loops and G-rich DNA. eLife. 2014;3:e02190. doi: 10.7554/eLife.02190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Kibe T, Kabir S, de Lange T. TRF1 negotiates TTAGGG repeat-associated replication problems by recruiting the BLM helicase and the TPP1/POT1 repressor of ATR signaling. Genes Dev. 2014;28:2477–2491. doi: 10.1101/gad.251611.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, Supplementary Tables
Review Process File