Abstract
Expression and function of drug transporters and metabolic enzymes (DMEs) in the gastrointestinal tract are critical attributes of intestinal physiology that influence the absorption of orally administered compounds. The purpose of this study was to examine the effects of media composition and cell source on mRNA expression and function of pharmaceutically relevant transporters and DMEs from two different sources of Caco-2 cells. Briefly, cells were cultured in either Minimum Essential Medium Alpha (AMEM) or Dulbecco’s Modified Eagle’s Medium (DMEM). Total RNA was isolated from each experimental group, and mRNA expression was evaluated using Quantitative Reverse Transcriptase-Polymerase Chain Reaction (QRT-PCR) arrays. Principal component analysis (PCA) was used to analyze results, which indicated variable transporter and metabolic expression attributable to differences in media composition and cell source. In addition, transport properties of paracellular markers and proton-dependent oligopeptide transporter-mediated substrates across Caco-2 cell monolayers were assessed. Transport experiments demonstrated significant differences in both paracellular and transcellular permeation resultant from differences in media composition and cell source. These studies support previous findings that media composition and cell source may significantly impact expressional and functional characteristics of Caco-2 cells. Standardization of culture-related methodology may reduce variability associated with Caco-2 cells, enabling more meaningful intra- and inter-laboratory data comparisons.
Keywords: Caco-2 cells, Cell culture, Variability, Media composition, Active transport, Paracellular transport, Transporters, Gene expression, Permeability, Intestinal metabolism
Introduction
Increasing predictive efficiency in the lead candidate optimization and selection phases of research and development (R&D) and reducing clinical compound attrition are two of the most important challenges facing the pharmaceutical industry today.1 In order to help overcome these challenges, it is extremely important for preclinical drug candidate screening models to be well-controlled and characterized so that critical performance attributes within the model can be identified. The characterization of these models is of particular importance when considering High Throughput Screening (HTS) techniques, such as permeability assays, because of the large number of compounds that must be screened for optimal biopharmaceutical properties.2,3 Among the various models that are employed for HTS applications, the colorectal adenocarcinoma derived Caco-2 cell line4 is one of the most commonly utilized in vitro tools for determining the surrogate intestinal epithelial permeation of new drug candidates.5 Caco-2 cells possess multiple structural and functional characteristics that closely resemble cells of the human small intestine,6–8 enabling them to be marginally successful at predicting in vivo oral bioavailability from in vitro permeability data for certain compounds.9–11
The widespread utilization of Caco-2 cells throughout the pharmaceutical industry and their acceptance by regulatory agencies has also brought forth growing concerns regarding variability associated with cell culture systems.12,13 Inter-laboratory variability in cell line performance, including the Caco-2 model, has given rise to an increased interest in the need for greater molecular and functional characterization when they are used for drug screening applications.13,14 This is particularly true when conducting permeability experiments for compounds that may potentially be substrates for active transporters or drug metabolizing enzymes due to the highly variable transporter and/or metabolic expression across laboratories.15,16 This high degree of variability may result in poor inter-laboratory correlations and obfuscate human absorption predictions.17
Variability associated with transporter and metabolic expression can be attributed to a number of different factors including cell origin18, passage number15,19, seeding density20,21, addition of extracellular matrix proteins20, and culture media.20,22,23 This variability is further confounded by the lack of standardized culture and experimental conditions across laboratories (e.g., differences in culture media, CO2 concentration, time of trypsin incubation, etc.). For example, several studies have examined the effects that media composition can have on expressional and functional attributes of Caco-2 cells including the effects of single components of the media such as supplementation with fetal bovine serum (FBS)24–27, glutamine supplementation28,29, or glucose concentration.23,30,31 While single components such as those mentioned above appear to affect the expressional and functional properties of Caco-2 cells, studies examining the effects of different media composition on the expression of transporters and DMEs have been limited.17,32 Such studies would provide insight into the need for improved characterization of Caco-2 cells with respect to culture conditions in various drug screening laboratories.14 For example, recent data from our laboratory illustrated the potentially profound impact that media composition can have on genotypic, and selected phenotypic properties, of the human HT-29 adenocarcinoma cell line.33 Based on our work with the HT-29 cell line33 and the results presented in the current manuscript, it is our laboratory’s belief that a better understanding of induced transporter and DME expressional changes will allow pharmaceutical scientists to appreciate the limitations of permeability screening measurements and control for functional changes associated with seemingly minor differences in culture protocols across laboratories.
The aim of the present study was to characterize the expressional and functional differences in 84 pharmaceutically relevant transporters and 84 DMEs in Caco-2 cells that occur as a result of differences in culture media and cell source. Specifically, we investigated changes in mRNA expression that occurred in Caco-2 cells obtained from different sources (American Type Culture Collection (ATCC) and from another laboratory) that were cultured in either one of the two commonly utilized growth media: DMEM and AMEM. The ATCC recommends cell culture in AMEM, whereas many studies described in the literature reveal DMEM as the media of choice for Caco-2 cell culture.2,17,32 It is important to note that the composition of the two mediums is significantly different, as will be described below. All other culture-related conditions including lot numbers for media supplementation of FBS, glutamine, and penicillin/streptomycin were held constant throughout the studies to exclusively focus on the effects of media composition. Additionally, the permeability coefficients for the paracellular markers, mannitol and urea, and selected proton-dependent oligopeptide transporter (POT) substrates16,34 were determined to elucidate potential functional transport differences that occurred as a result of media compositional differences. We believe these results will help to illustrate the profound impact that media can have not only on the expression of certain transporters and DMEs, but also on the permeation properties of substrates across the cell monolayer.
Materials and Methods
Materials
Two sources of Caco-2 cells were obtained. Cell Source 1 was obtained from Dr. Thomas Cook’s laboratory at Rutgers University in 2000, while Cell Source 2 was purchased directly from the ATCC (Manassas, VA). AMEM and the nonessential amino acids were obtained from Mediatech, Inc. (Manassas, VA). The QRT-PCR drug transporter arrays (PAHS-070), drug metabolism arrays (PAHS-002), and RT2 First-Strand Kits (C-03) were obtained from SA Biosciences (Frederick, MD). The Absolutely RNA® isolation kits and Brilliant II SYBR Green QRT-PCR 2-Step Master Mix were obtained from Stratagene (La Jolla, CA). Radiolabeled carnosine (Car), GlySar, mannitol, urea, and valacyclovir (VACV) were obtained from Moravek Biochemicals (Brea, CA). Radiolabeled 5-aminolevulinic acid (5-ALA) and benzylpenicillin (Benz) were obtained from American Radiolabeled Chemicals (Saint Louis, MO). FBS, penicillin-streptomycin solution, trypsin-EDTA solution, Hank’s Balanced Salt Solution (HBSS), Phosphate Buffered Saline (PBS), culture flasks, and all other supplies were obtained from Sigma Chemical Company (St. Louis, MO).
Cell Culture (Media Supplementation and Cell Culture Protocol)
Two sources of Caco-2 cells were cultured in T-25 flasks at 37°C in 5% CO2 and 90% relative humidity. Each cell source was maintained in both AMEM and DMEM supplemented with 10% FBS, 1X nonessential amino acids, penicillin/streptomycin, and 2 mM glutamine. Media was changed every other day of culture and cells were passaged at 80–90% confluence at a split ratio of 1:3. Each cell source was passaged five times in each medium before use in experimentation. Both cell sources were within five passages of each other when studies were initiated and all studies were completed within twenty passages of the original passage number. All other culture conditions, including media supplementation lot numbers, were maintained so as to focus exclusively on the effects of media composition.
RNA Isolation
Total RNA was isolated using Absolutely RNA® isolation kits per the manufacturer’s instructions, as described previously by our laboratory.33,35 Briefly, Caco-2 cells were seeded in 6-well plates in triplicate at a density of 5×104 cells/cm2. Media was changed every other day (or daily if necessary), and total RNA was harvested on day 21 post-seeding. The RNA was quantified using ultraviolet absorbance at 260 nm and then the purity and integrity was confirmed using gel electrophoresis.
Drug Transporter and Drug Metabolism QRT-PCR Arrays
SA Biosciences QRT-PCR arrays were used to determine the mRNA expression levels of 84 pharmaceutically relevant transporters and 84 DMEs. The cDNA’s were prepared from the RNA isolated as described above using SA Biosciences’ RT2 First-Strand Kit per the manufacturer’s instructions. All arrays were performed in triplicate using Brilliant II SYBR Green Mastermix and a Stratagene Mx3000P Real-Time PCR system. Results were normalized to the average of five housekeeping genes; β-actin, β-2 microglobulin, glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine phosphoribosyltransferase 1, and ribosomal protein L13a. The limit of quantitation was set at a threshold cycle (Ct) of Ct ≥ 35, and genes expressed in this range were excluded and set as below the limit of quantitation (BLQ). Data analysis was performed using SABiosciences RT2 Profiler™ PCR Array Data Analysis software. ΔCt values were calculated using Equation 1, where CtGOI is the threshold cycle for the gene of interest, and CtHKG is the average threshold cycle for the five housekeeping genes. The normalized expression level (E) for each gene of interest to the average of the housekeeping genes is given by Equation 2, which is due to the doubling of the amount of each gene with each cycle and the inverse relationship between the threshold cycle and the initial gene expression level. The relative fold change in gene expression between an experimental (EXP) group and a control (CON) group is given by the ratio of the two normalized expression levels as illustrated in Equation 3, where ΔΔCt is equal to ΔCtEXP − ΔCtCON. Genes that are expressed at a higher level in the experimental test group compared to the control group will have fold changes greater than 1, while genes that are expressed at a lower level in the experimental test group compared to the control group will have fold changes less than 1. In order to represent fold change values that are less than 1 in a more meaningful way, fold regulation values were calculated and reported by taking the negative inverse of the fold change.
| (1) |
| (2) |
| (3) |
Permeability Studies
Transport experiments were conducted in triplicate in the apical to basolateral direction for 90 minutes as described previously by our laboratory.16,33–35 Briefly, cells were seeded onto 12 mm tissue culture treated, collagen-coated polyester membranes (0.4 μm pore size 3460 Transwells®) at a density of 5 × 104 cells/cm2. After seeding, the respective medium was changed every other day until the day of the study. All studies were performed on day 21 post-seeding. On the day of the study, the culture medium was removed and the cells were washed twice with transport buffer warmed to 37°C (Donor: 2-(N-morpholino)ethanesulfonic acid (MES) pH 5.5, Receiver: HBSS pH 7.4). The cells were then equilibrated in transport buffer for 15 minutes prior to each study. After this time, a working buffer solution was added to the cells for each respective study and the transport characteristics of [14C]mannitol, [14C]urea, [3H]VACV, [3H]GlySar, [3H]Car, [3H]Benz, and [3H]5-ALA were determined. Cells were kept on a rocker platform at 37°C and samples were taken at 15, 30, 45, 60 and 90 minutes for [14C]mannitol and radiolabeled POT substrates, while samples were taken at 10, 20, 30, 40, and 50 minutes for [14C] urea. Apparent permeability coefficients (PApp) were determined using Equation 4:
| (4) |
Where dQ/dt is the steady state appearance rate in the receiver compartment, C0 is the initial concentration in the donor compartment and A is the surface area of exposed membrane (cm2). Sink conditions were maintained throughout each study. The permeability coefficients through the cell monolayer (PM) were calculated from the apparent permeability coefficients by subtracting out the resistance of the aqueous boundary layer, collagen coat, and filter support (PABL/F/C) as illustrated in Equation 5. Additionally, the permeability coefficient of each POT substrate was normalized to the respective permeability coefficient of mannitol in order to focus on the transcellular permeation pathway.
| (5) |
Statistical Analysis
All QRT-PCR array studies were performed in triplicate and fold regulation between two test groups was compared using an unpaired, two-tailed student’s t-test at a 95% confidence level. PCA was performed using XLSTAT to analyze mRNA expressional correlations between all of the cell source and media composition studies performed. All transport studies were conducted in triplicate and permeability coefficients were calculated as described above. Permeability coefficients were compared using an unpaired, two-tailed student’s t-test at a 95% confidence level. Statistical significance is denoted in the tables and figures using the following notation: * for p ≤ 0.05, ** for p ≤ 0.01, and *** for p ≤ 0.001.
Results and Discussion
Recent studies from our laboratory demonstrated that differences in culture media composition may significantly alter mRNA expression of pharmaceutically relevant transporters and the permeation of model substrates across the human HT-29 adenocarcinoma cell line.33 Based on the results of those studies, we aimed to examine related media-dependent changes on the expression of 84 pharmaceutically relevant transporters and 84 DMEs, as well as the permeation of model substrates across two separate sources of Caco-2 cells.
Figure 1 illustrates the normalized average expression levels (log10(2−ΔCt)) of 84 DMEs at day 21 post-seeding in two sources of Caco-2 cells cultured in two different media compositions. [For a tabular version of the mRNA expression levels and the respective standard deviations for each gene, the reader is encouraged to visit the Supporting Information section of the publisher’s website.] Surprisingly, the cytochrome P450 (CYP) 3A4 mRNA primer set was not included by the vendor in the DME array. Since others have published that CYP3A4 is not detected or expressed at very low levels in Caco-2 cells,32,36 we did not perform an experiment to determine its expression. The upper and lower diagonal lines in each panel represent a ±2 fold regulation between the two different groups. Results above the upper diagonal line (green symbols) indicate DME mRNAs whose expression was up-regulated in the group on the vertical axis compared to the group on the horizontal axis. Conversely, the results below the lower diagonal line (red symbols) indicate the DME mRNAs whose expression was down-regulated in the group on the vertical axis compared to the group on the horizontal axis. Panels A and B demonstrate that multiple DME mRNAs were either up or down-regulated by more than two fold in each cell source when cultured with different media. For example, CYP17A1 was down-regulated approximately 21 fold in Cell Source 1 when cultured in AMEM comparative to DMEM (Figure 1A) and 9 fold in Cell Source 2 when cultured in AMEM comparative to DMEM (Figure 1B). Conversely, alcohol dehydrogenase isoform 1C (ADH1C) was up-regulated approximately 6 fold in Cell Source 1 cultured in AMEM when contrasted to DMEM (Figure 1A) and 12 fold in Cell Source 2 when cultured in AMEM compared to DMEM (Figure 1B).
Figure 1.
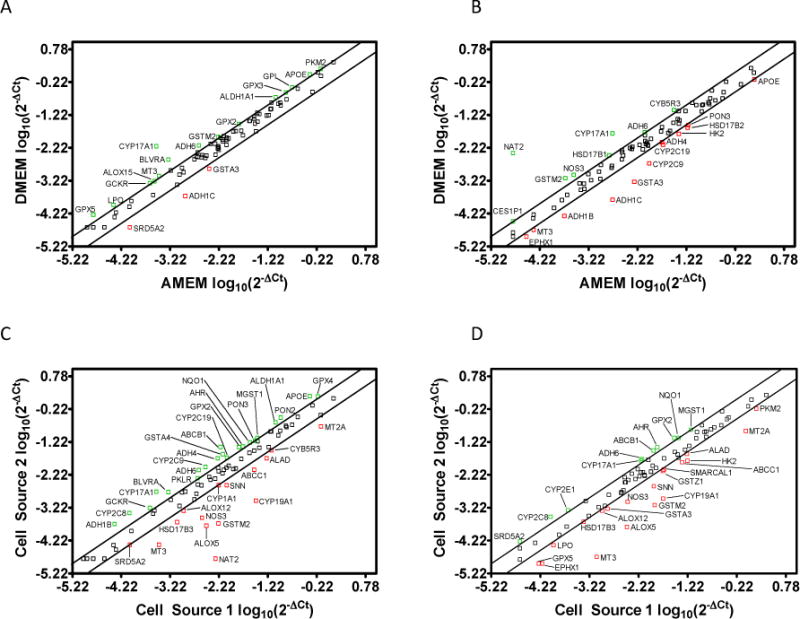
Scatter plots illustrating the normalized Drug Metabolizing Enzyme (DME) mRNA expression levels (log10(2−ΔCt)) at day 21 post-seeding for A) Cell Source 1 cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) vs. Minimum Essential Medium Alpha (AMEM); B) Cell Source 2 cultured in DMEM vs. AMEM; C) Cell Source 2 vs. Cell Source 1 cultured in AMEM; and D) Cell Source 2 vs. Cell Source 1 cultured in DMEM. The upper and lower diagonal lines indicate a two-fold up/down regulation, respectively.
DME mRNA expression differences due to cell source cultured in the same medium are revealed in Figure 1 (Panels C and D). For example, CYP2C8 displayed a 5 fold and 3 fold increase in expression in Cell Source 2 comparative to Cell Source 1 when cultured in AMEM (Figure 1C) and DMEM (Figure 1D), respectively. Furthermore, CYP19A1 expression was 35 fold lower in Cell Source 2 comparative to Cell Source 1 when cultured in AMEM (Figure 1C), but only 17 fold lower when cultured in DMEM (Figure 1D). Other notable DME mRNAs that exhibited significant expression level changes due to media differences or cell source include CYP2C9 (ibuprofen, warfarin, phenytoin metabolism), CYP2C19 (naproxen, propranolol, omeprazole metabolism), and NAT2 (furosemide, clonazepam, isoniazid metabolism). These results imply that the magnitude of the effects of media on DME mRNA expression can vary depending on the cell source and media composition.
In order to compare the relative expression levels of genes between each of the two groups presented in Figure 1, fold regulation values between each of the two groups were calculated as described above. Table 1 illustrates the average relative fold regulation (with statistical significance) between each of the test groups for the 84 DME mRNAs investigated. In total, 5 DME mRNAs in Cell Source 1 and 28 DME mRNAs in Cell Source 2 were statistically regulated when each cell source was cultured in differing media, suggesting that the effects of media on DME expression are dependent on cell source. When comparing the effects of cell source while holding media constant, a total of 43 DME mRNAs were statistically different when both sources were cultured in AMEM. However, only 15 DME mRNAs were significantly altered when both sources were cultured in DMEM. Therefore, it appears that DMEM may have the potential to reduce variability in the selected DME mRNA expressions between different sources of Caco-2 cells comparative to AMEM.
Table 1.
Average fold regulation of Drug Metabolizing Enzyme (DME) mRNA transcripts at day 21 post-seeding in each cell source cultured in each medium.
| Gene Symbol | Average Fold Regulation for Cell Source 1 Cultured in DMEM Compared to AMEM | Average Fold Regulation for Cell Source 2 Cultured in DMEM Compared to AMEM | Average Fold Regulation for Cell Source 2 Compared to Cell Source 1 Cultured in AMEM | Average Fold Regulation for Cell Source 2 Compared to Cell Source 1 Cultured in DMEM |
|---|---|---|---|---|
| ABP1 | 1.68 | 1.06 | 1.04 | −1.52 |
| ADH1B | 1.07 | −3.79 | 4.39 | 1.08 |
| ADH1C | −5.96** | −11.64** | 1.51 | −1.29 |
| ADH4 | 1.02 | −2.14** | 3.38*** | 1.55 |
| ADH5 | 1.23 | −1.18 | 1.17 | −1.25 |
| ADH6 | 3.04 | 2.12** | 3.62* | 2.52* |
| AHR | −1.29 | −1.06 | 2.34** | 2.86** |
| ALAD | 1.02 | 1.35 | −2.97* | −2.24 |
| ALDH1A1 | 2.42 | −1.42 | 2.74** | −1.25 |
| ALOX12 | −1.05 | 1.01 | −2.35 | −2.23 |
| ALOX15 | 2.04 | −1.46 | 1.73* | −1.72 |
| ALOX5 | 1.05 | −1.10 | −19.45*** | −22.39* |
| APOE | 2.36 | −2.05* | 3.41* | −1.42 |
| ARNT | 1.05 | −1.43* | 1.31** | −1.15 |
| ASNA1 | 1.60 | 1.13 | −1.00 | −1.42 |
| BLVRA | 4.77 | −1.07 | 3.25* | −1.57 |
| BLVRB | 1.32 | −1.06 | 1.22 | −1.15 |
| CES2 | 1.79 | 1.12 | 1.14 | −1.41 |
| CES1P1 | 1.51 | 2.09 | 1.08 | 1.50 |
| CHST1 | 1.89 | −1.44 | 1.36 | −2.01 |
| COMT | 1.31 | −1.59 | 1.82 | −1.15 |
| CYP11B2 | BLQ | −1.06 | 1.65* | −1.47 |
| CYP17A1 | 21.49* | 8.95*** | 5.89*** | 2.45** |
| CYP19A1 | −1.80 | 1.16 | −34.99*** | −16.77* |
| CYP1A1 | −1.14 | 1.52* | −2.03* | −1.16 |
| CYP2B6 | 1.41 | −1.20 | 1.68 | −1.00 |
| CYP2C19 | 1.22 | −2.67*** | 2.29** | −1.42 |
| CYP2C8 | 1.10 | −1.31 | 4.73* | 3.28* |
| CYP2C9 | −1.08 | −5.17* | 3.29* | −1.45 |
| CYP2D6 | 1.43 | 1.68 | −1.76 | −1.49 |
| CYP2E1 | −1.23 | 1.26 | 1.46 | 2.26** |
| CYP2F1 | 1.50 | −1.32 | 1.23 | −1.61 |
| CYP2J2 | 1.17 | 1.74* | 1.09 | 1.62 |
| CYP3A5 | 1.16 | −1.41 | 1.72 | 1.05 |
| CYB5R3 | 1.99* | 2.52* | −2.14 | −1.69 |
| EPHX1 | 1.36 | −2.77* | −1.47 | −5.55* |
| FAAH | 1.59 | 1.20 | −1.09 | −1.44 |
| FBP1 | 1.56 | −1.41 | 1.11 | −1.99 |
| GAD1 | 1.97 | 1.41 | 1.05 | −1.33 |
| GCKR | 2.18 | 1.61 | 2.56* | 1.89 |
| GGT1 | 1.22 | −1.01 | 1.20 | −1.03 |
| GPI | 2.19 | 1.67* | 1.06 | −1.24 |
| GPX1 | 1.35 | 1.25 | −1.02 | −1.11 |
| GPX2 | 2.08 | 1.88* | 2.73** | 2.47* |
| GPX3 | 2.05 | 1.04 | 1.31* | −1.51 |
| GPX4 | 1.90 | −1.24 | 2.29** | −1.03 |
| GPX5 | 3.54 | −1.44 | 1.07 | −4.79 |
| GSR | 1.41 | 1.15 | 1.37 | 1.11 |
| GSTA3 | −2.73** | −9.23** | 1.36 | −2.49 |
| GSTA4 | 1.31 | −1.51 | 3.54** | 1.79 |
| GSTM2 | 2.20 | 3.59* | −29.15** | −17.85 |
| GSTM3 | 1.99 | −1.53 | 1.69 | −1.81 |
| GSTM5 | BLQ | −1.44 | 1.65* | −2.01 |
| GSTP1 | 1.82 | 1.27 | −1.04 | −1.50 |
| GSTT1 | BLQ | −1.44 | 1.65* | −2.01 |
| GSTZ1 | 1.59 | −1.14 | −1.32 | −2.38 |
| HK2 | −1.05 | −2.63 | −1.49 | −3.72 |
| HSD17B1 | 1.57 | 2.21* | −1.31 | 1.08 |
| HSD17B2 | 1.01 | −2.55*** | 1.71*** | −1.51 |
| HSD17B3 | −1.83 | 1.02 | −3.81*** | −2.03 |
| LPO | 2.61 | −1.02 | 1.03 | −2.58 |
| MARCKS | −1.63 | −1.52* | 1.59* | 1.71 |
| MGST1 | 1.91 | 1.76* | 2.18** | 2.01 |
| MGST2 | 1.32 | −1.05 | 1.80** | 1.30 |
| MGST3 | 1.54 | −1.17 | 1.42* | −1.28 |
| MPO | −1.15 | −1.44 | −1.60 | −2.01 |
| MT2A | 1.26 | −1.39* | −4.13*** | −7.22* |
| MT3 | 2.35 | −2.36 | −7.94* | −44.06* |
| MTHFR | 1.51 | 1.11 | −1.07 | −1.46 |
| NAT1 | −1.16 | −1.37* | 1.16 | −1.02 |
| NAT2 | −1.37 | 248.31*** | −303.45*** | 1.12 |
| NOS3 | 1.33 | 3.06** | −9.12** | −3.97* |
| NQO1 | 1.52 | 1.37 | 2.23** | 2.02* |
| PKLR | 1.99 | 1.37 | 2.16* | 1.48 |
| PKM2 | 2.07 | 1.57* | −1.85 | −2.44 |
| PON1 | −1.01 | −1.09 | −1.37 | −1.48 |
| PON2 | 1.06 | −1.71* | 2.99** | 1.65 |
| PON3 | −1.00 | −2.20* | 2.00* | −1.10 |
| SMARCAL1 | 1.47 | −1.09 | −1.41* | −2.26 |
| SNN | 1.41 | −1.07 | −2.99* | −4.53* |
| SRD5A1 | 1.16 | 1.03 | 1.07 | −1.05 |
| SRD5A2 | −3.85* | 1.25 | −2.01 | 2.39 |
Significance is denoted using the following notation
p < 0.05
p < 0.01
p < 0.001.
It is interesting to note that a greater number of DME mRNAs exhibited statistically significant differences in expression levels due to cell source than due to media composition. This observation may be attributable to the heterogeneity of the cell line and the fact that differing culture conditions may induce selective pressure which results in the preferential growth of a particular subpopulation of cells.37 The heterogeneity of the cell line may also explain why only 5 genes were statistically different in Cell Source 1 when contrasting AMEM vs. DMEM. However, 28 genes were statistically different in Cell Source 2 when comparing AMEM vs. DMEM, which may be resultant from obtaining an earlier, heterogeneous lineage of the Caco-2 cells from the ATCC. For example, it may be possible that Cell Source 2 has a larger subpopulation of cells that are more sensitive to changes in media composition than Cell Source 1, which have been cultured for several years and been subjected to selective pressure. Further studies would need to be conducted in order to confirm this hypothesis.
In an attempt to identify correlations of mRNA expression with cell source and media composition, PCA was performed on the data set. PCA correlation results are illustrated in Figure 2 (Panel A) and reveal a high correlation (> .9) in DME mRNA expression for a given cell source cultured in differing media (e.g., Cell Source 1 AMEM vs. Cell Source 1 DMEM) despite the fact that a considerable number of DMEs were up- or down-regulated by more than two fold when cultured in different media. Furthermore, a weaker correlation was found to exist when comparing different cell sources cultured in the same media (e.g. Cell Source 1 AMEM vs. Cell Source 2 AMEM). It is interesting to note that the PCA revealed a weaker correlation between the different cell sources when cultured in AMEM (0.799) comparative to culturing in DMEM (0.932). This further supports our previous statement that DMEM may have the potential to reduce variability in DME mRNA expression between different cell sources. Collectively, these results imply that cell source may have a greater impact on DME mRNA expression comparative to media composition, even though media composition does influence expression for a number of DMEs.
Figure 2.
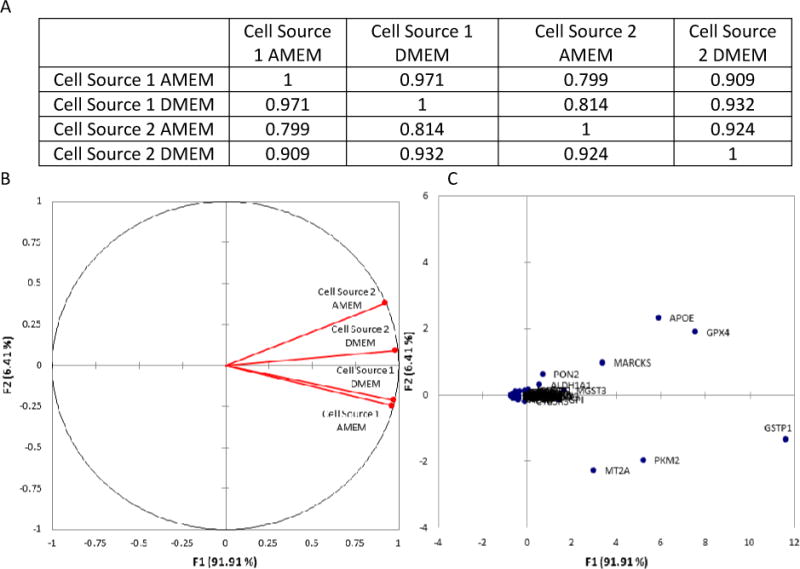
Principal Component Analysis (PCA) results for the normalized Drug Metabolizing Enzyme (DME) mRNA expression levels at day 21 post-seeding. A) Illustrates the Pearson Correlation Matrix for each of the four test groups; B) Illustrates the correlations circle which is useful for interpreting the meaning of the factors used in the PCA; and C) Illustrates a scatter plot of the factor scores for each of the 84 metabolic genes studied.
PCA was also utilized to identify specific DME mRNAs whose expression levels were the most sensitive to changes in culture media and cell source. Figure 2 (Panel B) illustrates the projection of the initial variables in the factor space F1, F2. It can be observed from Panel B that factor F2 is related to cell source (Cell Source 1 and 2 on opposite sides of the center line), while factor F1 is related to media. These two factors were selected to analyze results because they collectively account for more than 98% of the variability in the data set. This information is useful when analyzing the observations plot in Panel C, because it allows for identification of DME mRNAs which have shown trends to be influenced by either media composition or cell source. These genes include APOE (apolipoprotein E), GPX4 (glutathione peroxidase 4), MARCKS (myristoylated alanine-rich C-kinase substrate), PON2 (paraoxonase 2), GSTP1 (glutathione S transferase P1), PKM2 (pyruvate kinase M2), and MT2A (metallothionein 2A). Moreover, all of these genes have relatively high expression levels comparative to the housekeeping genes (Supporting Information, Table 1), although the functional relevance of these results would require further studies beyond the scope of this manuscript. However, it is interesting to note that many of these genes play a role in stress response.38–43 Therefore, it may be possible that the cells are up-regulating these DME mRNAs in response to some form of stress induced by the system (e.g. competing cell subpopulations or media component differences).
In addition to the DME isoforms discussed above, QRT-PCR arrays were also utilized to evaluate the mRNA expression of pharmaceutically relevant transporters. Figure 3 illustrates the normalized mRNA expression levels (log10(2−ΔCt)) of 84 transporters at day 21 post-seeding in two sources of Caco-2 cells cultured in two different media compositions. [A tabular version of the mRNA expression levels for each gene is available in the Supporting Information section of the publisher’s website]. Figure 3 (Panels A and B) illustrates the selected transporter mRNA expression level differences attributable to media composition for each cell source (e.g., Cell Source 1 AMEM vs. DMEM), while Panels C and D illustrate expression differences attributable to cell source (e.g., Cell Source 1 vs. Cell Source 2 AMEM). The expression levels of several ATP Binding Cassette (ABC) efflux transporters were altered by changes in media composition or cell source including P-glycoprotein (P-gp; ABCB1), sister to P-gp (ABCB11), and multidrug resistance associated protein (MRP) isoforms 1, 3, and 5 (ABCC1, ABCC3, ABCC5). For example, P-gp was expressed approximately 7 fold higher in Cell Source 2 compared to Cell Source 1 cultured in AMEM and approximately 3 fold higher in Cell Source 2 compared to Cell Source 1 cultured in DMEM. It is interesting to note that the difference in expression levels of P-gp between Cell Source 2 and Cell Source 1 was decreased when both sources were cultured in DMEM compared to when both sources were cultured in AMEM. Further work to assess the impact of the stress response may be warranted. However, it is clear that the findings may be of particular importance for laboratories that are screening for P-gp substrates and do not have standardized culture protocols. Similarly, sister to P-gp demonstrated a 13 fold reduction in expression in Cell Source 2 comparative to Cell Source 1 cultured in AMEM and a 4 fold reduction in Cell Source 2 compared to Cell Source 1 cultured in DMEM.
Figure 3.
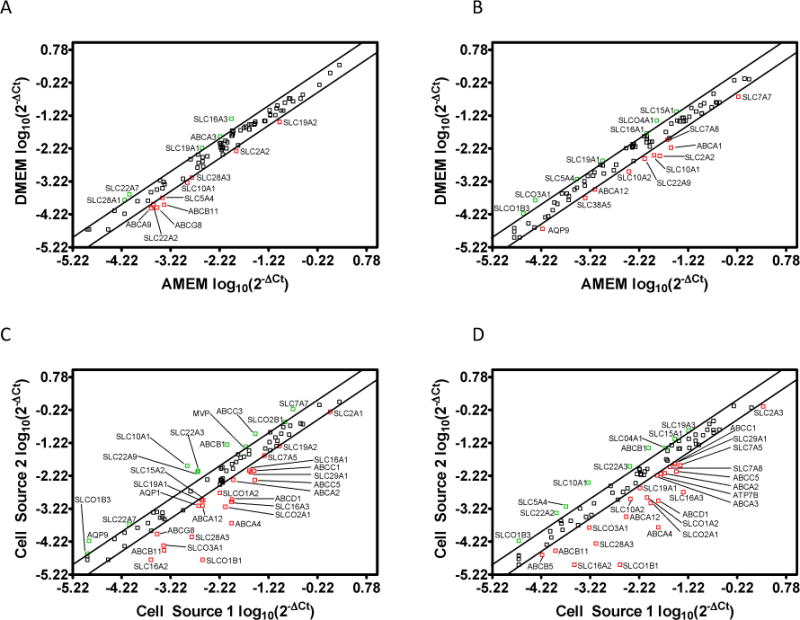
Scatter plots illustrating the normalized drug transporter mRNA expression level (log10(2−ΔCt)) at day 21 post-seeding for A) Cell Source 1 cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) vs. Minimum Essential Medium Alpha (AMEM); B) Cell Source 2 cultured in DMEM vs. AMEM; C) Cell Source 2 vs. Cell Source 1 cultured in AMEM; and D) Cell Source 2 vs. Cell Source 1 cultured in DMEM. The upper and lower diagonal lines indicate a two-fold up/down regulation, respectively.
In addition to the ABC efflux transporters discussed above, the expression levels of several solute carrier (SLC) transporter members were also affected by media and cell source. For example, the organic cation transporter SLC22A3 displayed a 4 fold increase in expression in Cell Source 2 comparative to Cell Source 1 cultured in AMEM and a 3 fold increase in Cell Source 2 comparative to Cell Source 1 cultured in DMEM. Likewise, the monocarboxylate transporter SLC16A2 had a 14 and 24 fold reduction in expression in Cell Source 2 compared to Cell Source 1 when cultured in AMEM and DMEM, respectively.
Table 2 displays the transporter mRNA fold regulation values and statistical significance between each of the test groups. A total of 39 transporters were statistically regulated in Cell Source 1 as a result of media differences compared to 33 transporters in Cell Source 2. Interestingly, only 19 of these transporters were consistently regulated in both cell sources when cultured with different mediums: 3 ABC members, aquaporin 9, and 15 SLC members. For example, the folate transporter (SLC19A1) was down-regulated in both cell sources when cultured in AMEM comparative to DMEM. This observation may be resultant of the fact that AMEM has a 4 fold lower concentration of folic acid comparative to DMEM (Table 3). Table 3 provides a comprehensive listing of compositional differences in the media that may provide some rationale for the changes observed in these studies. Conversely, the thiamine transporter (SLC19A2) was up-regulated in both cell sources cultured in AMEM comparative to DMEM despite the fact that AMEM also has a 4 fold lower concentration of thiamine comparative to DMEM (Table 3). Therefore, transporter mRNA expression appears to be differentially regulated by media composition. Other notable transporters that displayed consistent trends across both cell sources cultured in the different mediums include the facilitated glucose transporters (SLC2A2 and SLC2A3), pedptide transporter isoforms 1 and 2 (PepT1 and PepT2) (SLC15A1 and SLC15A2), and the cationic amino acid transporter (SLC7A8).
Table 2.
Average fold regulation of drug transporter mRNA transcripts at day 21 post-seeding in each cell source cultured in each medium.
| Gene Symbol | Average Fold Regulation for Cell Source 1 Cultured in DMEM Compared to AMEM | Average Fold Regulation for Cell Source 2 Cultured in DMEM Compared to AMEM | Average Fold Regulation for Cell Source 2 Compared to Cell Source 1 Cultured in AMEM | Average Fold Regulation for Cell Source 2 Compared to Cell Source 1 Cultured in DMEM |
|---|---|---|---|---|
| ABCA1 | −1.91* | −4.20** | 1.88* | −1.17 |
| ABCA12 | 1.22 | −2.18 | −3.58* | −9.47*** |
| ABCA13 | −1.52* | 1.04 | −1.86 | −1.18 |
| ABCA2 | 1.74* | 1.59 | −2.54 | −2.78*** |
| ABCA3 | 2.29** | 1.89** | −1.87 | −2.27*** |
| ABCA4 | 1.40 | −1.34 | −48.10** | −90.47*** |
| ABCA9 | −2.61* | −1.04 | −1.87* | 1.34 |
| ABCB1 | 1.88 | −1.28 | 6.56*** | 2.72* |
| ABCB11 | −3.77** | −1.02 | −13.53** | −3.65* |
| ABCB4 | −1.34 | −1.03 | 1.06 | 1.38 |
| ABCB5 | −1.10 | −1.82 | −1.56 | −2.59* |
| ABCB6 | 1.38 | −1.15 | 1.01 | −1.57** |
| ABCC1 | 1.47 | 1.67* | −3.67*** | −3.21 |
| ABCC10 | 1.83 | −1.05 | 1.33 | −1.44 |
| ABCC11 | 1.20** | 1.62 | 1.26 | 1.71 |
| ABCC12 | 1.79* | −1.47 | 1.67 | −1.58 |
| ABCC2 | −1.40 | −1.70* | 1.52 | 1.25 |
| ABCC3 | 1.21* | −1.64* | 3.50** | 1.77** |
| ABCC4 | −1.12 | −1.13 | 1.61** | 1.60** |
| ABCC5 | 1.10 | 1.81** | −7.05* | −4.27*** |
| ABCC6 | 1.37 | −1.02 | 1.55 | 1.10 |
| ABCD1 | 1.36 | −1.09 | −9.50** | −14.12*** |
| ABCD3 | −1.26 | −1.01 | −1.27 | −1.01 |
| ABCD4 | −1.50* | 1.63 | −2.00* | 1.22 |
| ABCF1 | −1.02 | 1.18 | −1.80* | −1.49** |
| ABCG2 | −1.22 | 1.58* | −1.54* | 1.25 |
| ABCG8 | −3.28 | −1.27 | −3.11** | −1.21 |
| AQP1 | −1.02 | 1.52* | −3.03** | −1.96** |
| AQP7 | 1.04 | −1.84* | 1.38 | −1.38 |
| AQP9 | 1.60* | −2.95** | 4.84*** | 1.03 |
| ATP6V0C | 1.27 | 1.14 | −1.47 | −1.64** |
| ATP7A | −1.35 | 1.10 | 1.03 | 1.52** |
| ATP7B | 1.48 | −1.43 | −1.14 | −2.40*** |
| MVP | 1.35* | 1.12 | 2.21** | 1.84** |
| SLC10A1 | −2.36 | −3.25** | 9.22*** | 6.71** |
| SLC10A2 | −1.75* | −3.12** | −1.86* | −3.33** |
| SLC15A1 | 1.33* | 2.28** | 1.45** | 2.49** |
| SLC15A2 | −1.63*** | 1.68** | −2.56*** | 1.07 |
| SLC16A1 | −1.07 | 2.03** | −2.86** | −1.31** |
| SLC16A2 | 1.20 | −1.46 | −13.78** | −24.13* |
| SLC16A3 | 4.57*** | 1.97** | −10.76* | −25.04*** |
| SLC19A1 | 2.38** | 2.38* | −2.44* | −2.44*** |
| SLC19A2 | −2.63*** | −1.22* | −2.07** | 1.04 |
| SLC19A3 | −1.07 | 1.11 | 1.95** | 2.31*** |
| SLC22A1 | 1.09 | 1.18 | −1.58 | −1.46 |
| SLC22A2 | −2.18* | −1.04 | 1.74* | 3.65** |
| SLC22A3 | 1.79** | 1.35 | 3.95** | 3.00*** |
| SLC22A6 | 1.73* | 1.31 | 1.41 | 1.06 |
| SLC22A7 | 2.80** | 1.30 | 2.30* | 1.07 |
| SLC22A8 | 1.73* | −1.85 | 1.78 | −1.79* |
| SLC22A9 | −1.35* | −2.64*** | 3.68*** | 1.88** |
| SLC28A1 | 2.40** | 1.23 | 1.63 | −1.20 |
| SLC28A2 | −1.10 | 1.34 | 1.12 | 1.64 |
| SLC28A3 | −2.07 | −1.60 | −19.40** | −14.99** |
| SLC29A1 | 1.25* | 1.85* | −3.43*** | −2.32*** |
| SLC29A2 | 1.34 | −1.10 | 1.31 | −1.13* |
| SLC2A1 | 1.08 | 1.35 | −2.07** | −1.66** |
| SLC2A2 | −2.60*** | −4.42** | 1.20 | −1.42* |
| SLC2A3 | 1.22* | −1.38* | −1.60** | −2.69*** |
| SLC31A1 | 1.33** | 1.47* | −1.31* | −1.19 |
| SLC38A2 | −1.15 | −1.49* | −1.34 | −1.72*** |
| SLC38A5 | 1.08 | −2.52* | 1.51 | −1.79 |
| SLC3A1 | −1.28 | −1.61* | 1.05 | −1.20 |
| SLC3A2 | −1.37* | −1.58 | −1.16 | −1.34 |
| SLC5A1 | 1.02 | 1.50** | −1.46 | 1.01 |
| SLC5A4 | −2.13* | 2.17** | −1.26 | 3.66*** |
| SLC25A13 | 1.14 | 1.09 | −1.82* | −1.91*** |
| SLC7A11 | −1.06 | 1.36 | −1.40 | 1.03 |
| SLC7A5 | −1.19 | −2.00 | −2.00* | −3.35*** |
| SLC7A6 | −1.34* | −1.10 | 1.05 | 1.28* |
| SLC7A7 | −1.17 | −2.83** | 3.34** | 1.39* |
| SLC7A8 | 1.36* | −2.04* | 1.22 | −2.29*** |
| SLC7A9 | −1.05 | 1.27 | 1.13 | 1.51 |
| SLCO1A2 | 1.43* | −1.39* | −3.33** | −6.65*** |
| SLCO1B1 | −1.11 | −1.46 | −155.92*** | −205.93*** |
| SLCO1B3 | 1.73* | 2.43 | 2.14 | 3.01 |
| SLCO2A1 | 1.31 | 1.31 | −11.48** | −11.47*** |
| SLCO2B1 | 1.15 | −1.73 | 2.01 | 1.01 |
| SLCO3A1 | 1.33 | 3.54 | −9.46** | −3.56* |
| SLCO4A1 | 1.93* | 3.11*** | 1.27 | 2.05*** |
| TAP1 | 1.73** | 1.20 | −1.14 | −1.65** |
| TAP2 | 1.21 | 1.75** | −1.39 | 1.05 |
| VDAC1 | 1.19* | 1.40 | 1.20 | 1.41* |
| VDAC2 | −1.28*** | −1.07 | 1.29 | 1.54** |
Significance is denoted using the following notation
p < 0.05
p <0.01
p < 0.001.
Table 3.
Comparison of media compositions for Minimum Essential Medium Alpha (AMEM) and Dulbecco’s Modified Eagle’s Medium (DMEM).
| Component | AMEM (mg/L) | DMEM (mg/L) |
|---|---|---|
| Inorganic Salts | ||
| CaCl2 | 200 | 200 |
| Fe(NO3)3 • 9H2O | — | 0.1 |
| KCl | 400 | 400 |
| MgSO4 | 97.7 | 97.7 |
| NaCl | 6800 | 6400 |
| NaHCO3 | 2200 | 3700 |
| NaH2PO4 | 121.7 | 109 |
| Amino Acids | ||
| L-Alanine | 25 | — |
| L-Arginine • HCl | 126.4 | 84 |
| L-Asparagine • H2O | 50 | — |
| L-Aspartic Acid | 30 | — |
| L-Cysteine • HCl • H2O | 100 | — |
| L-Cysteine • 2HCl | 31.2 | 62.6 |
| L-Glutamic Acid | 75 | — |
| L-Glutamine | — | 584 |
| Glycine | 50 | 30 |
| L-Histidine • HCl • H2O | 41.9 | 42 |
| L-Isoleucine | 52.5 | 105 |
| L-Leucine | 52.5 | 105 |
| L-Lysine HCl | 72.5 | 146 |
| L-Methionine | 15 | 30 |
| L-Phenylalanine | 32.5 | 66 |
| L-Proline | 40 | — |
| L-Serine | 25 | 42 |
| L-Threonine | 47.6 | 95 |
| L-Tryptophan | 10 | 16 |
| L-Tyrosine • 2Na+ • 2H2O | 51.9 | 103.8 |
| L-Valine | 46.8 | 94 |
| Vitamins | ||
| Ascorbic Acid | 50 | — |
| Biotin | 0.1 | — |
| D-Ca2+ Pantothenate | 1 | 4 |
| Choline Chloride | 1 | 4 |
| Folic Acid | 1 | 4 |
| i-Inositol | 2 | 7.2 |
| Niacinamide | — | 4 |
| Nicotinamide | 1 | — |
| Pyridoxine • HCl | 1 | 4 |
| Riboflavin | 0.1 | 0.4 |
| Thiamine • HCl | 1 | 4 |
| Vitamin B | 1.36 | — |
| Other | ||
| D-Glucose | 1000 | 4500 |
| Lipoic Acid | 0.2 | — |
| Phenol Red, Na+ | 10 | 15.9 |
| Sodium Pyruvate | 110 | 110 |
With respect to cell source, 44 transporters were statistically different between the two cell sources cultured in AMEM, contrasted to 52 transporters when each source was cultured in DMEM (Table 2). A total of 33 transporters displayed consistent trends between the cell sources when cultured in each medium: 9 ABC members, aquaporin 1, major vault protein, and 22 SLC members. It is interesting to note that a larger fraction of ABC transporters was affected by cell source (9/33, 27%) than by media composition (3/19, 16%). In contrast, a smaller fraction of SLC transporters was affected by cell source (22/33, 67%) than media composition (15/19, 79%). This may be resultant of the functional role that many of the SLC members play in nutrient uptake and homeostasis comparative to the ABC members.
PCA was also performed on the transporter data set to identify transporters whose mRNA expression is the most sensitive to changes in media composition and cell source. PCA results indicate overall transporter mRNA expression trends similar to those observed for the DME mRNAs. Figure 4 (Panel A) reveals a strong correlation (> .9) in transporter expression for a given cell source cultured in differing media. Moreover, Cell Source 1 may be more homogeneous (or at least comprised of a different population of cells) than Cell Source 2 because its transporter expression appears to be less susceptible to changes in media composition. Additionally, weaker correlations were found to exist between different cell sources cultured in the same media. These results imply that cell source may have a greater impact on transporter mRNA expression than culture media, which was also observed for DME mRNA expression. Because of this variability associated with cell sourcing, it is important for researchers to assay cell lines with respect to their laboratory’s expression of mRNAs of interest (relative to the application) throughout different stages of experimentation.
Figure 4.
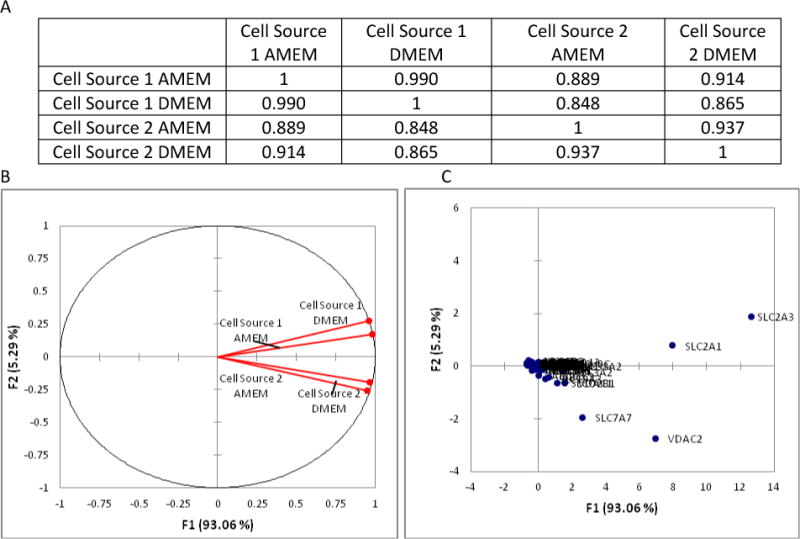
Principal Component Analysis (PCA) results for the normalized drug transporter gene expression levels at day 21 post-seeding. A) Illustrates the Pearson Correlation Matrix for each of the four test groups; B) Illustrates the correlations circle which is useful for interpreting the meaning of the factors used in the PCA; and C) Illustrates a scatter plot of the factor scores for each of the 84 transporters studied.
Figure 4 (Panel B) illustrates the correlation circle in the factor space F1, F2, which accounts for more than 98% of the variability. Similar to the DME isoform PCA, factor F2 is closely related to cell source and factor F1 is related to culture media. These factors are useful for identifying which transporters have shown trends to be the most sensitive to changes in media and cell source, which include SLC2A1, SLC2A3, SLC7A7, and VDAC (Panel C). Given the large differences in glucose and amino acid concentrations between the different mediums (Table 3), it is not surprising that the facilitative glucose transporters (SLC2A1 and SLC2A3) and cationic amino acid transporter (SLC7A7) displayed significant trends for large degrees of transporter expression.
The results presented above indicate that differences in media composition and cell source can have potentially significant consequences on mRNA expression of pharmaceutically relevant transporters and DMEs. However, it is important to acknowledge that transcriptional differences in mRNA expression do not necessarily result in a linear relationship with protein translation or posttranslational modifications to functional proteins. As a result, further studies would be required to ascertain potential relationships between genotypic changes observed in these studies and subsequent phenotypic changes in functional protein. To establish these relationships, members of the POT family were selected for further analysis in an attempt to correlate mRNA expressional changes with some measure of functional activity based on our laboratory’s primary research interests. Additionally, mannitol and urea (established paracellular markers) were selected to determine the effects of media composition and cell source on monolayer integrity.44,45
Figure 5 (Panels A and B) illustrates statistically significant differences in the permeability coefficients for mannitol and urea as a result of media composition and cell source. Specifically, cells cultured in AMEM resulted in greater permeability coefficients for mannitol and urea than cells cultured in DMEM, indicating that media composition can influence the paracellular permeation pathway. In addition, Cell Source 2 resulted in lower permeability coefficients than Cell Source 1 for both paracellular markers, which may be resultant of cell line heterogeneity or different cell source homogeneity due to sub-culturing pressure.37 It is important to note that similar trends were observed for both mannitol and urea, suggesting that media-dependent changes in paracellular permeability are affected consistently.
Figure 5.
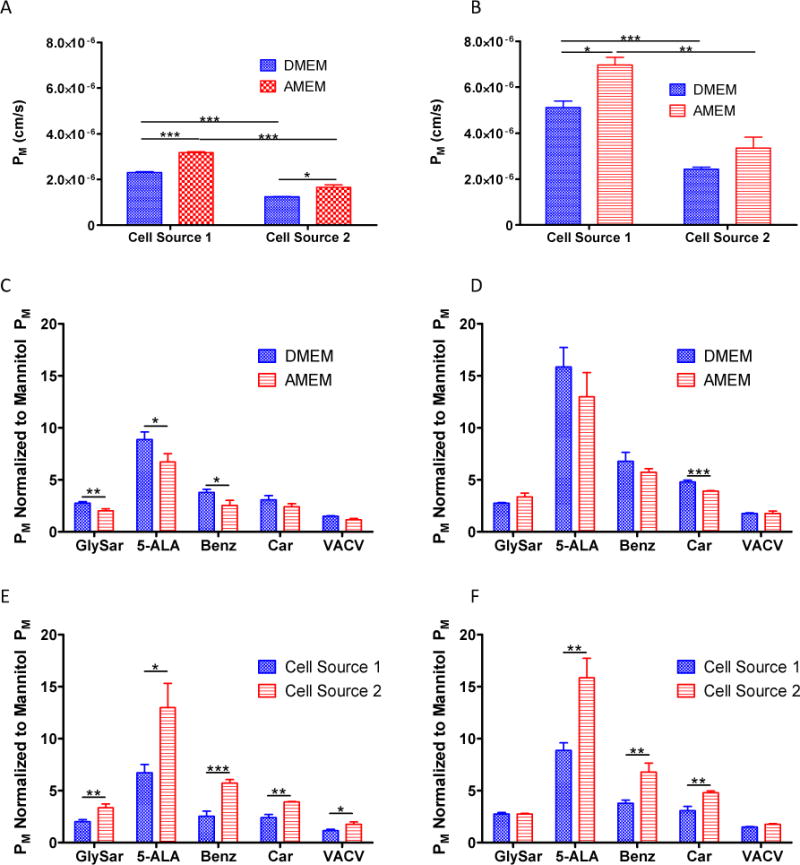
Illustrations of the paracellular permeability coefficients (PM, Apical to Basolateral) for A) mannitol and B) urea for each cell source cultured in each medium. Mannitol normalized monolayer permeability coefficients (PM, Apical to Basolateral) for selected proton dependent oligopeptide transporter (POT) substrates in C) Cell Source 1 cultured in Minimum Essential Medium Alpha (AMEM) vs. Dulbecco’s Modified Eagle’s Medium (DMEM); D) Cell Source 2 cultured in AMEM vs. DMEM; E) Cell Source 1 vs. Cell Source 2 cultured in AMEM; and F) Cell Source 1 vs. Cell Source 2 cultured in DMEM. Results are presented as the mean ± standard deviation for three replicates. *p < 0.05; **p < 0.01; ***p < 0.001.
In addition to the paracellular permeability markers, we also evaluated five POT substrates (GlySar, 5-ALA, Benz, Car, and VACV) to determine media-dependent effects on transporter-mediated permeability. As discussed previously, permeability coefficients for POT substrates (PM) have been normalized to each group’s mannitol permeability coefficient in order to normalize for paracellular confounders and to better focus the transport kinetics on the transcellular permeation pathway. Figure 5 (Panels C and D) illustrates media-dependent changes in permeability for each cell source. In general, DMEM gave rise to apparently higher permeability coefficients for 8 out of the ten possible comparisons of the five POT substrates examined in the two cell sources, although statistical significance was not established for all. This finding was in agreement with mRNA expression changes in which PepT1 (SLC15A1) was down-regulated in AMEM when contrasted to DMEM in both cell sources (Table 2). Statistically significant media-dependent differences in permeability were observed for GlySar, 5-ALA, and Benz in Cell Source 1 (Panel C), and for Car in Cell Source 2 (Panel D).
When comparing different cell sources cultured in the same medium (Panels E and F), increased permeability coefficients were observed for Cell Source 2 compared to Cell Source 1. This finding was also in agreement with mRNA expression changes in which PepT1 revealed increased expression in Cell Source 2 compared to Cell Source 1 (Table 2). Statistically significant cell-dependent differences in permeability were observed for all five POT substrates when cultured in AMEM (Panel E), and for all substrates except GlySar and VACV when cultured in DMEM (Panel F). Collectively, these results suggest that mRNA expression changes for PepT1 correlate with functional transport differences for POT substrates. However, other transporters with overlapping affinity for these substrates may potentially confound the results and alter the interpretation for PepT1. For example, PepT2 possesses similar substrate specificity to PepT1 for a number of compounds.46 While the mRNA expression of PepT2 in the current study is relatively low comparative to PepT1 (Supporting Information, Table 2), the effects of PepT2 may potentially obfuscate the permeability results. Therefore, further experimentation is needed in order to delineate whether the transport differences are directly correlated with PepT1 mRNA expression, or whether they are a result of competition with confounding transporters.
Conclusions
Culturing induced differences in mRNA expression and function of DMEs and pharmaceutically relevant transporters may ultimately lead to intra- and inter-laboratory variability associated with cell-based screening systems.13,14 The results presented here expand upon current knowledge about media-induced variation in Caco-2 cells by examining a more comprehensive set of pharmaceutically relevant transporters and DMEs. Our results indicated that Caco-2 cells from the same source exhibited significant variation with respect to transporter and DME mRNA expression when cultured with two different media compositions that are commonly used throughout our field. For example, cells cultured in DMEM demonstrated decreased variability with respect to DME expression compared to cells cultured in AMEM. Interestingly, this trend was not observed for transporter expression implying that mRNA expression may be differentially regulated by media composition.
Because media-induced variability may affect various cell lines in differing manners, it is important for researchers to assess the effects of media and culture conditions on the expression and function of genes of interest in cell lines utilized in their respective laboratories throughout the course of experimentation. Based on our results, it appears that one technique for performing this characterization would be to utilize a similar QRT-PCR approach coupled with PCA to identify potential confounding transporters or DMEs that may affect experimental results. Such characterization may provide valuable information that could potentially explain discrepancies in data when making comparisons for studies conducted with the same substrates in different laboratories.
Furthermore, our results demonstrated that Caco-2 cells obtained from two different sources also displayed significant variation in their transporter and DME expression profiles when cultured under identical conditions. These results may be due to the inherent heterogeneity of the cell line and the possibility that different culture conditions may allow for preferential growth of a particular subpopulation of cells. Due to expression differences in transporters and DMEs and the inherent heterogeneity of the Caco-2 cell line, some researchers are utilizing clonal cells derived from parental Caco-2 cells such as the Caco-2BBe.47 While these cell lines may have reduced variability, their utilization in drug screening laboratories has not replaced the widespread use of Caco-2 cells. Therefore, it is important to understand which genes in parental Caco-2 cells are most sensitive to changes in culture conditions and cell source so that intra- and inter-laboratory differences may potentially be accounted for.
As noted above, our laboratory’s interest has been in elucidating peptide permeation pathways. To address our interests, these results imply that mRNA expression changes for PepT1 correlate with functional differences observed for POT substrates. Determining whether mRNA expressional changes in other transporter or DME isoforms correlate with functional differences for other substrates is beyond the scope of this current study and would require many additional studies. The results presented here, along with results from our previously published work33, are intended to raise awareness and provide insights into the importance of characterizing cell lines with respect to media and culture conditions. If the data presented here are indicative of functional differences in other cell lines, then a substantial amount of variability may potentially be reduced through the use of standardized culture media. Such standardization could potentially improve the quality of drug screening approaches and enable more meaningful intra- and inter-laboratory data comparisons. It is our laboratory’s belief that a better understanding of induced transporter and DME expressional changes will allow pharmaceutical scientists to appreciate the limitations of permeability screening measurements and control for functional changes associated with seemingly minor differences in culture protocols across laboratories.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the financial support received from the National Institute of General Medical Sciences (RO1-GM65448). The authors would also like to acknowledge the assistance of Dr. Stephanie Mowery who aided us with the principal component analysis.
Abbreviations
- 5-ALA
5-aminolevulinic acid
- ABC
ATP-Binding Cassette
- AMEM
Minimum Essential Medium Alpha
- APOE
Apolipoprotein E
- ATCC
American Type Culture Collection
- Benz
Benzylpenicillin
- BLQ
Below the Limit of Quantitation
- Car
Carnosine
- Ct
Threshold Cycle
- CYP
Cytochrome P450
- DME
Drug Metabolizing Enzymes
- DMEM
Dulbecco’s Modified Eagle’s Media
- FBS
Fetal Bovine Serum
- GI
Gastrointestinal
- GPX4
Glutathione Peroxidase 4
- GSTP1
Glutathione S transferase Protein 1
- HBSS
Hank’s Balanced Salt Solution
- HTS
High Throughput Screening
- MARCKS
Myristoylated Alanine-Rich C-Kinase Substrate
- MES
2-(N-morpholino)ethanesulfonic acid
- MRP
Multidrug Resistance Associated Protein
- MT2A
Metallothionein 2A
- QRT-PCR
Quantitative Reverse Transcriptase-Polymerase Chain Reaction
- PABL/F/C
Permeability Coefficient through Aqueous Boundary Layer, Filter Support, Collagen
- PApp
Apparent Permeability Coefficient
- PBS
Phosphate Buffered Saline
- PCA
Principal Component Analysis
- PepT1
Peptide Transporter 1
- PepT2
Peptide Transporter 2
- P-gp
P-glycoprotein (MDR1)
- PM
Cell Monolayer Permeability Coefficient
- PON2
Paraoxonase 2
- POT
Proton-dependent Oligopeptide Transporter
- PKM2
Pyruvate Kinase M2
- R&D
Research and Development
- SLC
Solute Carrier Transporter
- TEER
Transepithelial Electrical Resistance
- VACV
Valacycolvir
Footnotes
Supporting Information
Electronic supporting information is available via the internet at http://onlinelibrary.wiley.com. This information includes the average normalized gene expression levels (2−ΔCt) and standard deviations for each of the 84 drug metabolizing enzymes (DMEs) and transporters evaluated. Results with threshold cycle (Ct) values greater than 35 were considered below the limit of quantitation (BLQ).
References
- 1.Paul SM, Mytelka DS, Dunwiddie CT, Persinger CC, Munos BH, Lindborg SR, Schacht AL. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Reviews Drug Discovery. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 2.Balimane PV, Patel K, Marino A, Chong SH. Utility of 96 well Caco-2 cell system for increased throughput of P-gp screening in drug discovery. European Journal of Pharmaceutics and Biopharmaceutics. 2004;58(1):99–105. doi: 10.1016/j.ejpb.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 3.Balani SK, Miwa GT, Gan LS, Wu JT, Lee FW. Strategy of utilizing in vitro and in vivo ADME tools for lead optimization and drug candidate selection. Current Topics in Medicinal Chemistry. 2005;5(11):1033–1038. doi: 10.2174/156802605774297038. [DOI] [PubMed] [Google Scholar]
- 4.Fogh J, Fogh JM, Orfeo T. 127 Cultured Human Tumor-Cell Lines Producing Tumors in Nude Mice. J Natl Cancer I. 1977;59(1):221–226. doi: 10.1093/jnci/59.1.221. [DOI] [PubMed] [Google Scholar]
- 5.Artursson P, Palm K, Luthman K. Caco-2 monolayers in experimental and theoretical predictions of drug transport. Advanced Drug Delivery Reviews. 2001;46(1–3):27–43. doi: 10.1016/s0169-409x(00)00128-9. [DOI] [PubMed] [Google Scholar]
- 6.Pinto M, Robineleon S, Appay MD, Kedinger M, Triadou N, Dussaulx E, Lacroix B, Simonassmann P, Haffen K, Fogh J, Zweibaum A. Enterocyte-Like Differentiation and Polarization of the Human-Colon Carcinoma Cell-Line Caco-2 in Culture. Biology of the Cell. 1983;47(3):323–330. [Google Scholar]
- 7.Artursson P. Epithelial Transport of Drugs in Cell-Culture. 1. A Model for Studying the Passive Diffusion of Drugs over Intestinal Absorptive (Caco-2) Cells. Journal of Pharmaceutical Sciences. 1990;79(6):476–482. doi: 10.1002/jps.2600790604. [DOI] [PubMed] [Google Scholar]
- 8.Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the Human-Colon Carcinoma Cell-Line (Caco-2) as a Model System for Intestinal Epithelial Permeability. Gastroenterology. 1989;96(3):736–749. [PubMed] [Google Scholar]
- 9.Artursson P, Karlsson J. Correlation between Oral-Drug Absorption in Humans and Apparent Drug Permeability Coefficients in Human Intestinal Epithelial (Caco-2) Cells. Biochemical and Biophysical Research Communications. 1991;175(3):880–885. doi: 10.1016/0006-291x(91)91647-u. [DOI] [PubMed] [Google Scholar]
- 10.Rubas W, Cromwell MEM, Shahrokh Z, Villagran J, Nguyen TN, Wellton M, Nguyen TH, Mrsny RJ. Flux measurements across Caco-2 monolayers may predict transport in human large intestinal tissue. Journal of Pharmaceutical Sciences. 1996;85(2):165–169. doi: 10.1021/js950267+. [DOI] [PubMed] [Google Scholar]
- 11.Yee SY. In vitro permeability across Caco3 cells (colonic) can predict in vivo (small intestinal) absorption in man – Fact or myth. Pharmaceutical Research. 1997;14(6):763–766. doi: 10.1023/a:1012102522787. [DOI] [PubMed] [Google Scholar]
- 12.Volpe DA. Application of method suitability for drug permeability classification. AAPS J. 12(4):670–678. doi: 10.1208/s12248-010-9227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volpe DA. Variability in Caco-2 and MDCK cell-based intestinal permeability assays. Journal of Pharmaceutical Sciences. 2008;97(2):712–725. doi: 10.1002/jps.21010. [DOI] [PubMed] [Google Scholar]
- 14.Sambuy Y, Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biology and Toxicology. 2005;21(1):1–26. doi: 10.1007/s10565-005-0085-6. [DOI] [PubMed] [Google Scholar]
- 15.Anderle P, Niederer E, Rubas W, Hilgendorf C, Spahn-Langguth H, Wunderli-Allenspach H, Merkle HP, Langguth P. P-glycoprotein (P-gp) mediated efflux in Caco-2 cell monolayers: The influence of culturing conditions and drug exposure on P-gp expression levels. Journal of Pharmaceutical Sciences. 1998;87(6):757–762. doi: 10.1021/js970372e. [DOI] [PubMed] [Google Scholar]
- 16.Herrera-Ruiz D, Faria TN, Bhardwaj RK, Timoszyk J, Gudmundsson OS, Moench P, Wall DA, Smith RL, Knipp GT. A novel hPepT1 stably transfected cell line: Establishing a correlation between expression and function. Molecular Pharmaceutics. 2004;1(2):136–144. doi: 10.1021/mp034011l. [DOI] [PubMed] [Google Scholar]
- 17.Seithel A, Karlsson J, Hilgendorf C, Bjorquist A, Ungell AL. Variability in mRNA expression of ABC- and SLC-transporters in human intestinal cells: Comparison between human segments and Caco-2 cells. European Journal of Pharmaceutical Sciences. 2006;28(4):291–299. doi: 10.1016/j.ejps.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Behrens I, Kamm W, Dantzig AH, Kissel T. Variation of peptide transporter (PepT1 expression in Caco-2 cells as a function and HPT1) of cell origin. Journal of Pharmaceutical Sciences. 2004;93(7):1743–1754. doi: 10.1002/jps.20062. [DOI] [PubMed] [Google Scholar]
- 19.Chantret I, Rodolosse A, Barbat A, Dussaulx E, Brotlaroche E, Zweibaum A, Rousset M. Differential Expression of Sucrase-Isomaltase in Clones Isolated from Early and Late Passages of the Cell-Line Caco-2 – Evidence for Glucose-Dependent Negative Regulation. Journal of Cell Science. 1994;107:213–225. doi: 10.1242/jcs.107.1.213. [DOI] [PubMed] [Google Scholar]
- 20.Behrens I, Kissel T. Do cell culture conditions influence the carrier-mediated transport of peptides in Caco-2 cell monolayers? European Journal of Pharmaceutical Sciences. 2003;19(5):433–442. doi: 10.1016/s0928-0987(03)00146-5. [DOI] [PubMed] [Google Scholar]
- 21.Natoli M, Leoni BD, D’Agnano I, D’Onofrio M, Brandi R, Arisi I, Zucco F, Felsani A. Cell Growing Density Affects the Structural and Functional Properties of Caco-2 Differentiated Monolayer. Journal of Cellular Physiology. 2011;226(6):1531–1543. doi: 10.1002/jcp.22487. [DOI] [PubMed] [Google Scholar]
- 22.SchmiedlinRen P, Thummel KE, Fisher JM, Paine MF, Lown KS, Watkins PB. Expression of enzymatically active CYP3A4 by Caco-2 cells grown on extracellular matrix-coated permeable supports in the presence of 1 alpha,25-dihydroxyvitamin D-3. Molecular Pharmacology. 1997;51(5):741–754. doi: 10.1124/mol.51.5.741. [DOI] [PubMed] [Google Scholar]
- 23.D’Souza VM, Buckley DJ, Buckley AR, Pauletti GM. Extracellular glucose concentration alters functional activity of the intestinal oligopeptide transporter (PepT-1) in Caco-2 cells. Journal of Pharmaceutical Sciences. 2003;92(3):594–603. doi: 10.1002/jps.10325. [DOI] [PubMed] [Google Scholar]
- 24.Jumarie C, Malo C. Caco-2 Cells Cultured in Serum-Free Medium as a Model for the Study of Enterocytic Differentiation Invitro. Journal of Cellular Physiology. 1991;149(1):24–33. doi: 10.1002/jcp.1041490105. [DOI] [PubMed] [Google Scholar]
- 25.Gangloff MB, Lai CD, VanCampen DR, Miller DD, Norvell WA, Glahn RP. Ferrous iron uptake but not transfer is down-regulated in Caco-2 cells grown in high iron serum-free medium. Journal of Nutrition. 1996;126(12):3118–3127. doi: 10.1093/jn/126.12.3118. [DOI] [PubMed] [Google Scholar]
- 26.Ranaldi G, Consalvo R, Sambuy Y, Scarino ML. Permeability characteristics of parental and clonal human intestinal Caco-2 cell lines differentiated in serum-supplemented and serum-free media. Toxicology in Vitro. 2003;17(5–6):761–767. doi: 10.1016/s0887-2333(03)00095-x. [DOI] [PubMed] [Google Scholar]
- 27.Zucco F, Batto AF, Bises G, Chambaz J, Chiusolo A, Consalvo R, Cross H, Dal Negro G, de Angelis I, Fabre G, Guillou F, Hoffman S, Laplanche L, Morel E, Pincon-Raymond M, Prieto P, Turco L, Ranaldi G, Rousset M, Sambuy Y, Scarino ML, Torreilles F, Stammati A. An inter-laboratory study to evaluate the effects of medium composition on the differentiation and barrier function of Caco-2 cell lines. Atla-Alternatives to Laboratory Animals. 2005;33(6):603–618. doi: 10.1177/026119290503300618. [DOI] [PubMed] [Google Scholar]
- 28.Le Bacquer O, Nazih H, Blottiere H, Meynial-Denis D, Laboisse C, Darmaun D. Effects of glutamine deprivation on protein synthesis in a model of human enterocytes in culture. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2001;281(6):G1340–G1347. doi: 10.1152/ajpgi.2001.281.6.G1340. [DOI] [PubMed] [Google Scholar]
- 29.Li N, Lewis P, Samuelson D, Liboni K, Neu J. Glutamine regulates Caco-2 cell tight junction proteins. American Journal of Physiology-Gastrointestinal and Liver Physiology. 2004;287(3):G726–G733. doi: 10.1152/ajpgi.00012.2004. [DOI] [PubMed] [Google Scholar]
- 30.D’Souza VM, Shertzer HG, Menon AG, Pauletti GM. High glucose concentration in isotonic media alters Caco-2 cell permeability. Aaps Pharmsci. 2003;5(3) doi: 10.1208/ps050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Q, Sai Y, Kato Y, Tamai I, Tsuji A. Influence of drugs and nutrients on transporter gene expression levels in Caco-2 and LS180 intestinal epithelial cell lines. Pharmaceutical Research. 2003;20(8):1119–1124. doi: 10.1023/a:1025076326061. [DOI] [PubMed] [Google Scholar]
- 32.Hayeshi R, Hilgendorf C, Artursson P, Augustijns P, Brodin B, Dehertogh P, Fisher K, Fossati L, Hovenkamp E, Korjamo T, Masungi C, Maubon N, Mols R, Mullertz A, Monkkonen J, O’Driscoll C, Oppers-Tiemissen HM, Ragnarsson EG, Rooseboom M, Ungell AL. Comparison of drug transporter gene expression and functionality in Caco-2 cells from 10 different laboratories. Eur J Pharm Sci. 2008;35(5):383–396. doi: 10.1016/j.ejps.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Lindley DJ, Roth WJ, Carl SM, Knipp GT. The effects of media on pharmaceutically relevant transporters in the human HT-29 adenocarcinoma cell line: Does culture media need to be controlled? J Pharm Sci. 101(4):1616–1630. doi: 10.1002/jps.23036. [DOI] [PubMed] [Google Scholar]
- 34.Bhardwaj RK, Herrera-Ruiz D, Sinko PJ, Gudmundsson OS, Knipp G. Delineation of human peptide transporter 1 (hPepT1)-mediated uptake and transport of substrates with varying transporter affinities utilizing stably transfected hPepT1/Madin-Darby canine kidney clones and caco-2 cells. Journal of Pharmacology and Experimental Therapeutics. 2005;314(3):1093–1100. doi: 10.1124/jpet.105.087148. [DOI] [PubMed] [Google Scholar]
- 35.Carl SM, Lindley DJ, Couraud PO, Weksler BB, Romero I, Mowery SA, Knipp GT. ABC and SLC Transporter Expression and Pot Substrate Characterization across the Human CMEC/D3 Blood-Brain Barrier Cell Line. Molecular Pharmaceutics. 2010;7(4):1057–1068. doi: 10.1021/mp900178j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taipalensuu J, Tornblom H, Lindberg G, Einarsson C, Sjoqvist F, Melhus H, Garberg P, Sjostrom B, Lundgren B, Artursson P. Correlation of gene expression of ten drug efflux proteins of the ATP-binding cassette transporter family in normal human jejunum and in human intestinal epithelial Caco-2 cell monolayers. Journal of Pharmacology and Experimental Therapeutics. 2001;299(1):164–170. [PubMed] [Google Scholar]
- 37.Walter E, Kissel T. Heterogeneity in the Human Intestinal-Cell Line Caco-2 Leads to Differences in Transepithelial Transport. European Journal of Pharmaceutical Sciences. 1995;3(4):215–230. [Google Scholar]
- 38.Jofre-Monseny L, Minihane AM, Rimbach G. Impact of apoE genotype on oxidative stress, inflammation and disease risk. Mol Nutr Food Res. 2008;52(1):131–145. doi: 10.1002/mnfr.200700322. [DOI] [PubMed] [Google Scholar]
- 39.Efferth T, Oesch F. Oxidative stress response of tumor cells: microarray-based comparison between artemisinins and anthracyclines. Biochem Pharmacol. 2004;68(1):3–10. doi: 10.1016/j.bcp.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Levy E, Trudel K, Bendayan M, Seidman E, Delvin E, Elchebly M, Lavoie JC, Precourt LP, Amre D, Sinnett D. Biological role, protein expression, subcellular localization, and oxidative stress response of paraoxonase 2 in the intestine of humans and rats. Am J Physiol Gastrointest Liver Physiol. 2007;293(6):G1252–1261. doi: 10.1152/ajpgi.00369.2007. [DOI] [PubMed] [Google Scholar]
- 41.Ranganathan PN, Whalen R, Boyer TD. Characterization of the molecular forms of glutathione S-transferase P1 in human gastric cancer cells (Kato III) and in normal human erythrocytes. Biochem J. 2005;386(Pt 3):525–533. doi: 10.1042/BJ20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 334(6060):1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59(1):95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- 44.Adson A, Burton PS, Raub TJ, Barsuhn CL, Audus KL, Ho NFH. Passive Diffusion of Weak Organic Electrolytes across Caco-2 Cell Monolayers – Uncoupling the Contributions of Hydrodynamic, Transcellular, and Paracellular Barriers. J Pharm Sci. 1995;84(10):1197–1204. doi: 10.1002/jps.2600841011. [DOI] [PubMed] [Google Scholar]
- 45.Knipp GT, Ho NFH, Barsuhn CL, Borchardt RT. Paracellular diffusion in Caco-2 cell monolayers: Effect of perturbation on the transport of hydrophilic compounds that vary in charge and size. J Pharm Sci. 1997;86(10):1105–1110. doi: 10.1021/js9700309. [DOI] [PubMed] [Google Scholar]
- 46.Brandsch M, Knutter I, Bosse-Doenecke E. Pharmaceutical and pharmacological importance of peptide transporters. J Pharm Pharmacol. 2008;60(5):543–585. doi: 10.1211/jpp.60.5.0002. [DOI] [PubMed] [Google Scholar]
- 47.Peterson MD, Mooseker MS. Characterization of the Enterocyte-Like Brush-Border Cytoskeleton of the C2bbe Clones of the Human Intestinal-Cell Line, Caco-2. J Cell Sci. 1992;102:581–600. doi: 10.1242/jcs.102.3.581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


