Abstract
Glucose has been recognized as an energy source for a long time, but it has recently been suggested that the hexosamine biosynthesis pathway (HBP) and downstream protein O-GlcNAcylation have important functions in mouse preimplantation development. Thus, whether or not O-GlcNAcylation was present and what functions O-GlcNAcylation has in pig preimplantation development were investigated in the present study. The expressions of mRNA of glutaminefructose-6-phosphate aminotransferase (Gfpt), O-GlcNAc transferase (Ogt) and O-GlcNAcase (Oga), which are involved in the HBP and O-GlcNAc cycling, were examined in pig parthenogenetic diploids at each preimplantation developmental stage. Gfpt and Ogt were detected in diploids at all stages. Though Oga was detected at all stages except the 4-cell stage, OGA proteins were detected in diploids from the 2-cell to blastocyst stage. Furthermore, O-GlcNAcylated proteins in MII oocytes and diploids were also detected by immunofluorescence at every stage. Inhibition of OGT by 4.0 mM BADGP did not affect development up to the blastocyst stage, while inhibition of OGA by 300 µM PUGNAc decreased the proportion of diploids beyond the 4-cell stage. Four-cell diploids cultured with PUGNAc until 48 h developed to the blastocyst stage after culture in a PUGNAc-free medium until 144 h after electrostimulation. RNA polymerase II (Pol II) phosphorylation, which indicates the onset of mRNA transcription, was detected in nuclei of diploids in the control group at 48 h but not in the PUGNAc-treated group. These results indicate that HBP and O-GlcNAcylation have important functions in pig preimplantation development and that inhibition of OGA is fatal for development. It is also suggested that OGA inhibition disrupts normal Pol II regulation and may cause a zygotic gene activation error.
Keywords: O-GlcNAc modification, Parthenogenetic diploids, Pig, Preimplantation development, Zygotic gene activation
The roles of carbohydrates as energy substrates in preimplantation development of mammalian embryos have been studied for a long time. Embryos at the early cleaving stages predominantly use pyruvate and lactate through the metabolic pathway based on carboxylic acid as their main energy sources. As embryonic development progresses, glycolysis becomes the main pathway for energy production, and glucose uptake steadily increases from the zygotic stage to embryonic compaction in the mouse and pig [1, 2]. There have been so many reports concerned with the effects of glucose in culture medium on preimplantation development of embryos, and it has been shown that addition of glucose to the medium suppressed embryo development during the preimplantation period in the hamster [3] and before the 4-cell stage in the mouse [4]. It is also known that preimplantation development of 4-cell mouse embryos cannot progress to the blastocyst stage in a medium without glucose [5,6,7]. Although pig embryos develop to the blastocyst stage in the medium including only glucose as the major energy source [8], it has been reported that pig embryos developed in a medium without glucose [9, 10] and that addition of glucose to a medium that included a combination of pyruvate and lactate could not improve pig embryo development [11, 12]. Moreover, the suppressive effects of glucose addition have been observed in pig embryos before the 4-cell stage [12], and glucose supplementation has been shown to increase the viability of blastocysts [13]. These reports suggest that pig preimplantation embryos have two different phases of glucose requirement before and after the 4-cell stage.
The roles of glucose were considered only from the viewpoint of the energy source in these reports. Glucose is utilized not only as an energy source via glycolysis and the tricarboxylic acid cycle (TCA cycle) but also for ribose production via the pentose phosphate pathway and protein modification, for example, O-linked N-acetylglucosamine (O-GlcNAc) modification [14]. It has also been reported that glucose was consumed by pig embryos at all stages of preimplantation development [15] and partially metabolized via the pentose phosphate pathway before morula formation [2]. Recently, it was shown that glucose metabolized via the hexosamine biosynthetic pathway (HBP) was necessary for the expression of zygotic mRNAs and proteins for successful blastocyst formation in the mouse [16]. Although mouse embryos require glucose for the development to the blastocyst stage [5,6,7], pig embryos do not [9]. The differences in glucose requirement suggest that the role of the HBP and/or role of protein O-GlcNAc modification (O-GlcNAcylation) that is a protein posttranslational modification using a substrate produced via the HBP, differed between the mouse and pig.
The HBP branches from the glycolytic pathway and produces uridine diphosphate N-acetylglucosamine (UDP-GlcNAc). It is known that all eukaryotic cells have the HBP, and then synthesized UDP-GlcNAc was reversibly transferred to protein serine or threonine (Ser/Thr) residues [17, 18]. It has been reported that about 2 to 5% of glucose entering the cell was metabolized via the HBP in animal cells [19]. Fructose-6-phosphate (F6P) is an intermediate metabolite in the glycolytic pathway and also the first substrate for the HBP. F6P is metabolized by glutamine-fructose-6-phosphate aminotransferase (GFPT). The reaction is the rate-limiting step in the HBP (see http://rgd.mcw.edu/rgdweb/pathway/pathwayRecord.html?acc_id=PW:0000559&species= Mouse#annot).
O-GlcNAcylation is one of the posttranslational protein modifications. It has been shown that O-GlcNAcylation often competes with phosphorylation on the Ser/Thr residue and regulates a protein function [20]. O-GlcNAcylation is catalyzed by O-GlcNAc transferase (OGT) and removed by O-GlcNAcase (OGA). OGT transfers GlcNAc to proteins from UDP-GlcNAc, and OGA removes GlcNAc from O-GlcNAcylated proteins. The addition and removal of GlcNAc from modified proteins is called O-GlcNAc cycling. It has been reported that many proteins are O-GlcNAcylated, for example, RNA polymerase II (Pol II) [21, 22], transcription factors [23] and histone family proteins [24,25,26], and these reports suggested that O-GlcNAc cycling regulated gene expression, cell signaling, differentiation and so on. It was recently shown that the O-GlcNAcylation of the C-terminal domain (CTD) of the largest subunit of Pol II was involved with Pol II recruitment to the promoter region on DNA and that phosphorylation of the CTD is necessary to initiate mRNA transcription [22]. The authors of that study suggested that both O-GlcNAcylation and phosphorylation of the CTD also regulated mRNA expressions in embryo development, often called zygotic gene activation (ZGA).
Because of the high incidence of polyspermic fertilization in in vitro fertilized pig eggs, it is difficult to expect that the eggs will show high and stable developmental ability to the blastocyst stage. It was shown that electrostimulated pig diploids had high ability for developing to the blastocyst stage from the points of view of the total cell numbers and the durations not only until the first cell division but also until compaction and blastulation [27, 28]. All these abilities were comparable to those in in vivo fertilized eggs [29]. Furthermore, more than 50% of 4-cell diploids transferred into the oviducts of recipients 48 h post activation implanted, and beating hearts were observed in nearly all of the fetuses recovered on day 19 [30]. All this evidence indicates that pig electrostimulated diploids have a high ability to develop to the blastocyst stage and that their ability is comparable to that of in vivo fertilized eggs. Therefore, electrically activated diploids were employed instead of in vitro fertilized pig eggs with unknown ploidy in this experiment.
O-GlcNAcylation may have an important role in mammalian preimplantation development; however, there are no reports concerning the HBP and O-GlcNAcylation in pig preimplantation development. In the present study, the presence of O-GlcNAc modification and its functions during preimplantation development in the pig were investigated.
Materials and Methods
Collection, in vitro maturation and activation of oocytes
Pig ovaries were collected at local slaughterhouses and transported to our laboratory within 2 h. Ovaries were washed once with 0.2% (w/v) cetyltrimethylammonium bromide (CETAB; Wako Pure Chemical Industries, Osaka, Japan) and washed twice with Ca2+- and Mg2+-free Dulbecco’s phosphate buffered saline (PBS) containing 0.1% (w/v) polyvinyl alcohol (PVA; Sigma-Aldrich Chemical, St. Louis, MO, USA). Follicles that were 4–6 mm in diameter were cut out from ovaries in PBS-PVA using a pair of disposable surgical scalpels. Cumulus-oocyte-granulosa cell complexes (COGCs) were collected from follicles in tissue culture medium 199 (TCM-199) buffered with 25 mM 2-[4-(2-Hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES; Dojindo Molecular Technologies, Kumamoto, Japan), HEPES-199, and then washed with HEPES-199 followed by washing twice with the maturation-culture medium without human menopausal gonadotropin (hMG; ASKA Pharmaceutical, Tokyo, Japan). Several follicular shells were collected from healthy follicles 4–6 mm in diameter and then freed from the lining granulosa cells. Thirty to ninety COGCs were maturation cultured with a few follicular shells for 44–48 h in a 2.0 ml maturation-culture medium comprised of bicarbonate-buffered TCM-199 containing 10% (v/v) heat-treated fetal calf serum (FCS; MP Biomedicals, Santa Ana, CA, USA), 0.1 mg/ml sodium pyruvate, 0.08 mg/ml kanamycin sulfate, 2.2 mg/ml sodium bicarbonate and 0.1 IU/ml hMG in a CO2 incubator under a humidified atmosphere with 5% CO2 at 38.5 C. After maturation culture, 200 µl PBS-PVA containing 0.1% (w/v) hyaluronidase was added to the 2.0 ml maturation-culture medium. Then, oocytes were freed from cumulus cells in pig zygote medium 3 (PZM3) [9] by mechanical pipetting and washed three times in a field solution that consisted of 0.30 mM mannitol, 0.05 mM CaCl2, 0.10 mM MgSO4 and 0.01% (w/v) PVA. Washed oocytes were transferred into 100 µl of the field solution which was filled between parallel stainless steel electrodes in an electrofusion chamber (FTC-03; Shimadzu, Kyoto, Japan) and activated by a single squared pulse at 1,500 V/cm DC for 100 µsec. Electrostimulated oocytes were cultured in PZM3 containing 5 µg/ml cytochalasin B (CB, Sigma-Aldrich) for 4 h to inhibit ejection of the second polar body to produce presumptive parthenogenetic diploids.
Embryo culture and production of diploids at various stages of preimplantation development
CB-treated diploids were washed three times in PZM3. Groups of 8–12 presumptive diploids were cultured in a 10 µl droplet of PZM3 under mineral oil (Wako) until 144 h after electrostimulation in a CO2 incubator under a humidified atmosphere with 5% CO2 at 38.5 C. Cultured diploids were observed, and their developmental stages were morphologically determined every 24 h. Diploids generally developed to the 2-cell, early 4-cell, late 4-cell, morula, morula and blastocyst and blastocyst stages at 24, 48, 72, 96, 120 and 144 h, respectively, after electrostimulation. In all experiments, culture media had been preincubated more than 2 h before use for equilibration of the gas phase and temperature.
Detection of mRNA expression of glutamine fructose-6-phosphate aminotransferase (Gfpt), O-GlcNAc transferase (Ogt) and O-GlcNAcase (Oga) in oocytes and parthenogenetic diploids during preimplantation development
mRNAs of Gfpt, Ogt and Oga in diploids at each developmental stage during preimplantation were detected by reverse transcription PCR (RT-PCR). After in vitro culture, 48–50 embryos that developed to the most dominant stages in each culture period [12, 31] were pooled every 24 h during the culture-period. Therefore, diploids that developed to the 2-cell, early 4-cell, late 4-cell, morula, morula and blastocyst and blastocyst stages were collected at 24 h, 48 h, 72 h, 96 h, 120 h and 144 h after electrostimulation, respectively. Total RNA was extracted from embryos of each group using a GenEluteTM Mammalian Total RNA Miniprep Kit (Sigma-Aldrich) according to the manufacturer’s instructions. Then, RNA was isolated and was reverse transcribed into cDNA using SuperScript II Reverse Transcriptase (Life Technologies, Carlsbad, CA, USA) in a total volume of 20 µl. The first reaction mixture consisted of 1 µl of Oligo (dT), 8 µl of extracted RNA and 1 µl of 10 mM dNTP mix. This reaction was carried out for 5 min at 65 C, and then 2 µl of 10x RT buffer, 4 µl of 25 mM MgCl2, 2 µl of 0.1 M DTT, 0.25 µl SuperScript II reverse transcriptase with 0.75 µl water and 1 µl RNaseOUT recombinant RNase inhibitor were added to each tube. The RT reaction was carried out at 42 C for 50 min, followed by an inactivation step at 70 C for 15 min. The concentration of each synthesized cDNA was measured and diluted by 400 µg/ml. All RT products of the cDNAs were stored at −20 C until use.
Expressions of Gfpt, Ogt and Oga were analyzed by RT-PCR to determine whether parthenogenetic diploids were capable of O-GlcNAc cycling on any proteins during preimplantation development. PCR was run in a thermal cycler (C1000; Bio-Rad Laboratories, Hercules, CA, USA) using a program consisting of the first step at 94 C for 2 min followed by 45 repetitions of denaturation at 94 C for 30 sec, annealing at 56 C for 30 sec and extension at 72 C for 30 sec; any remaining single strand DNA was fully extended. All primer pairs were designed to detect exon domains. Sequences of the primers are listed in Table 1. Pig poly-ubiquitin mRNA (Ubc) was used as a positive control for the reaction [32].
Table 1. Sequences of primer pairs for detection of Gfpt, Ogt and Oga mRNAs.
| Gene | Sequence | Remarks | |
| Gfpt | Forward | ACTCGCTGTTTCCTGTGCC | Rate-limiting enzyme for HBP |
| Reverse | TCCAGTGCCTTAACTTTTCCC | ||
| Ogt | Forward | AGCGGGACTCAATTACCCTTT | Add O-GlcNAc to proteins |
| Reverse | ATTGTCTGGCTCTTGTCTCCA | ||
| Oga | Forward | AAAAGCATGATGGCTTGCCT | Remove O-GlcNAc from proteins |
| Reverse | CCACCCACCAGGTGAAGTTAA | ||
| Ubca | Forward | AAAAGCATGATGGCTTGCCT | Reference gene |
| Reverse | CCACCCACCAGGTGAAGTTAA | ||
a Kuijk et al., 2007.
Detection of O-GlcNAcase protein (OGA) by immunofluorescence in parthenogenetic diploids from the 2-cell to blastocyst stage
Expression of OGA was detected by immunofluorescent staining in diploids from the 2-cell to blastocyst stage. Diploids were collected as described for detection of RNA expression. Subsequently, diploids were fixed and permeabilized with PBS-PVA that contained 0.2% (v/v) Triton X-100 (Sigma-Aldrich) and 4% (w/v) paraformaldehyde (PFA, Sigma-Aldrich) for 25 min and washed twice in PBS-PVA. Fixed diploids were stored at 4 C in PBS-PVA containing 1% (w/v) bovine serum albumin (BSA; Wako) (BPP) before use. Immunofluorescence microscopy was carried out as described below. Diploids were treated with rabbit anti-human OGA antibody (1:100; Proteintech Group, Chicago, IL, USA) in BPP at 4 C overnight. Then they were washed three times in PBS-PVA and treated with Alexa Fluor 568 anti-rabbit IgG antibody (Life Technologies) in BPP for 1 h at room temperature. After washing in PBS-PVA, diploids were mounted in ProLong Gold Antifade Mountant (Life Technologies) containing 4',6-diamidino-2-phenylindole (DAPI) and observed under an epifluorescence microscope (BX53-FL; Olympus, Tokyo, Japan).
Detection of O-GlcNAcylated proteins by immunofluorescence in oocytes and parthenogenetic diploids during preimplantation development
O-GlcNAcylated proteins in metaphase II (MII) oocytes and preimplantation diploids from the 2-cell to blastocyst stage were detected by immunostaining with two different antibodies. CTD110.6 (Covance, Princeton, NJ, USA) and RL2 (Novus Biological, Littleton, CO, USA) are primed to synthetic peptides containing serine-O-GlcNAc, and cytoplasmic and intranuclear O-linked glycoproteins, respectively. When samples were treated with CTD110.6, oocytes and diploids were fixed with 4% PFA supplemented PBS-PVA at 4 C overnight and then permeabilized by PBS-PVA containing 0.2% Triton X-100 for 20 min. For the RL2 treatment, samples were fixed and permeabilized as described for OGA detection. Fixed oocytes and diploids were washed twice with PBS-PVA and stored at 4 C in BPP until use. Immunostaining was carried out as described above for OGA detection, except that CTD110.6 (1:100) or RL2 (1:100) was used as primary antibody. Alexa Fluor 546 labeled anti-mouse IgM and Alexa Fluor 488 labeled anti-mouse IgG (Life Technologies) were used as secondary antibodies for CTD110.6 and RL2, respectively. Mounting and observation were carried out as described above for OGA detection.
Treatment of diploids with an OGT inhbitor (BADGP) or an OGA inhibitor (PUGNAc)
Parthenogenetically activated diploids were treated with BADGP or PUGNAc to determine the roles of O-GlcNAc cycling in pig preimplantation development. To examine the effects of OGT inhibition, diploids were cultured in PZM3 with 0, 1.0, 2.0 and 4.0 mM BADGP and DMSO at the same concentration of 4.0 mM BADGP containing group (vehicle control) from immediately after CB-treatment to 144 h after electrostimulation. Diploids were observed every 24 h with an inverted microscope, and their developmental stages were recorded as described above. The effects of OGA inhibition on pig preimplantation development were also examined. In this case, diploids were cultured in PZM3 with 0, 75, 150 and 300 µM PUGNAc and DMSO at the same concentration of 300 µM PUGNAc containing group (vehicle control), and their development was observed every 24 h as described above.
As it was shown that the addition of 300 µM PUGNAc led to developmental cessation of diploids around the 4-cell stage in these experiments, the period in which OGA activity was critical for development beyond the 4-cell stage was examined. Diploids were cultured in PZM3 with 300 µM PUGNAc until 48 h, and then only those at the 4-cell stage were selected and divided into two groups and cultured in PZM3 with 0 or 300 µM PUGNAc to 144 h. Their development was observed every 24 h as described above.
Detection of Pol II and phosphorylated Pol II (pPol II)
It has been reported that Pol II phosphorylation is important for mRNA transcription [22] and that pPol II is detected when mRNA transcription becomes active in mouse embryos [33]. Therefore, the Pol II expression and its phosphorylation were examined in the pig 4-cell diploids.
Cross-reactions were examined between primary antibodies, anti-RNA Pol II monoclonal mouse antibody (8WG16, immunoglobulin isotype, IgG2a; Covance) and anti-pPol II monoclonal antibody (H14, immunoglobulin isotype, IgM; Covance), and secondary antibodies, Alexa Fluor 564 labeled anti-mouse IgM antibody and Alexa Fluor 488 labeled anti-mouse IgG antibody, before double immunofluorescence staining. Blastocysts cultured in PZM3 until 144 h were collected and fixed in 4% PFA at 4 C overnight. Fixed blastocysts were permeabilized with PBS-PVA containing 0.2% Triton X-100 for 20 min, washed twice in PBS-PVA and then stored at 4 C in BPP until staining. These embryos were treated with 8WG16 (1:100) or H14 (1:100) followed by treatment with both the Alexa Fluor 488 labeled anti-mouse IgG antibody and Alexa Fluor 564 labeled anti-mouse IgM antibody. Embryos were mounted and observed under an epifluorescence microscope.
Four-cell diploids cultured in PZM3 with 0 (control) or 300 µM PUGNAc were collected 48 h after electrostimulation. Fixation and immunofluorescence staining were carried out as described above. Diploids were mounted and observed under an epifluorescence microscope and a confocal laser fluorescence microscope (FV-1000; Olympus).
Detection of RNA synthesis in diploids cultured in PZM3 with or without PUGNAc
RNA synthesis in diploids cultured in PZM3 with 0 (control) and 300 μM PUGNAc was detected to examine the decrease in RNA synthesis caused by PUGNAc treatment. Synthesized RNA was visualized using a Click-iT RNA Alexa Fluor 594 Imaging Kit (Life Technologies). Detection was carried out according to the manufacture’s instructions. Diploids were cultured in each medium until 96 h as described above, 10 µl of 20 mM 5-ethynyl uridine (EU) were added to 10 µl medium, and then the diploids were further cultured for 1 h. Diploids that incorporated EU were fixed and permeabilized with PBS-PVA containing 0.2% Triton X-100 for 20 min. Fixed diploids were washed twice in PBS-PVA and then placed in a reaction cocktail which consisting of 43 µl reaction buffer, 5 µl reaction buffer additive, 2 µl CuSO4 solution and 0.2 µl Alexa Fluor azide. EU incorporated by diploids to synthesize RNA was reacted with Alexa Fluor azide for 1 h at 37.0 C. Reacted diploids were washed three times and counterstained with DAPI. Diploids were mounted and observed under an epifluorescence microscope.
Detection of Claudin-4 expression in diploids treated with PUGNAc
Localization of Claudin-4, a tight junction sealing protein, was assessed in diploids cultured from 4 h in PZM3 with 0 (control) or 300 μM PUGNAc to examine the effects of PUGNAc treatment on the formation of tight junctions. After culture of diploids for 72 to 120 h in the control group and for 120 h in the PUGNAc-treated group, the surviving diploids were collected and subjected to immunofluorescent staining. These diploids were treated with a mouse anti-Claudin-4 antibody (1:100, Life Technologies) and then Alexa Fluor 488 labeled anti-mouse IgG antibody (1:500, Life Technologies). Procedures for fixation and immunostaining were carried out as described for detection of OGA.
Statistical analysis
Values for proportions of embryonic development were subjected to arcsine transformation in each replication, and then transformed values were analyzed using one-way ANOVA. If the ANOVA revealed a significant effect, the treatments were compared with the Tukey-Kramer method.
Results
The RNA expressions of Gfpt, Ogt and Oga mRNAs detected by RT-PCR are shown in Fig. 1. Gfpt and Ogt were expressed in diploids at all stages of preimplantation development. On the other hand, Oga was expressed in diploids at every stage except for the 4-cell stage.
Fig. 1.
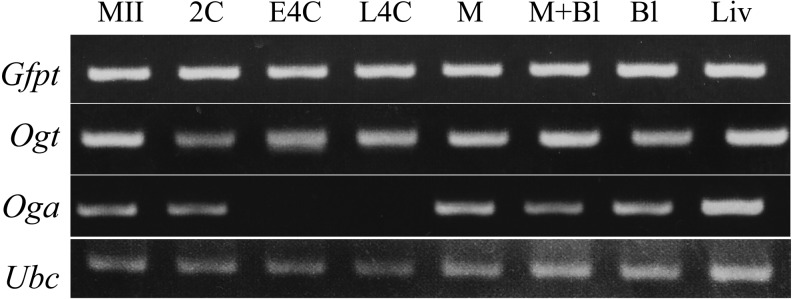
mRNA expression of Gfpt, Ogt and Oga involved in the hexosamine biosynthesis pathway (HBP) and O-GlcNAcylation in pig oocytes at the second metaphase stage and parthenogenetically activated diploids during preimplantation development. Liver and Ubc were used as controls for expression and concentration, respectively. MII, second metaphase; 2C, 2-cell; E4C, early 4-cell; L4C, late 4-cell; M, morula; M+Bl, morula and blastocyst; Bl, blastocyst; and Liv, liver.
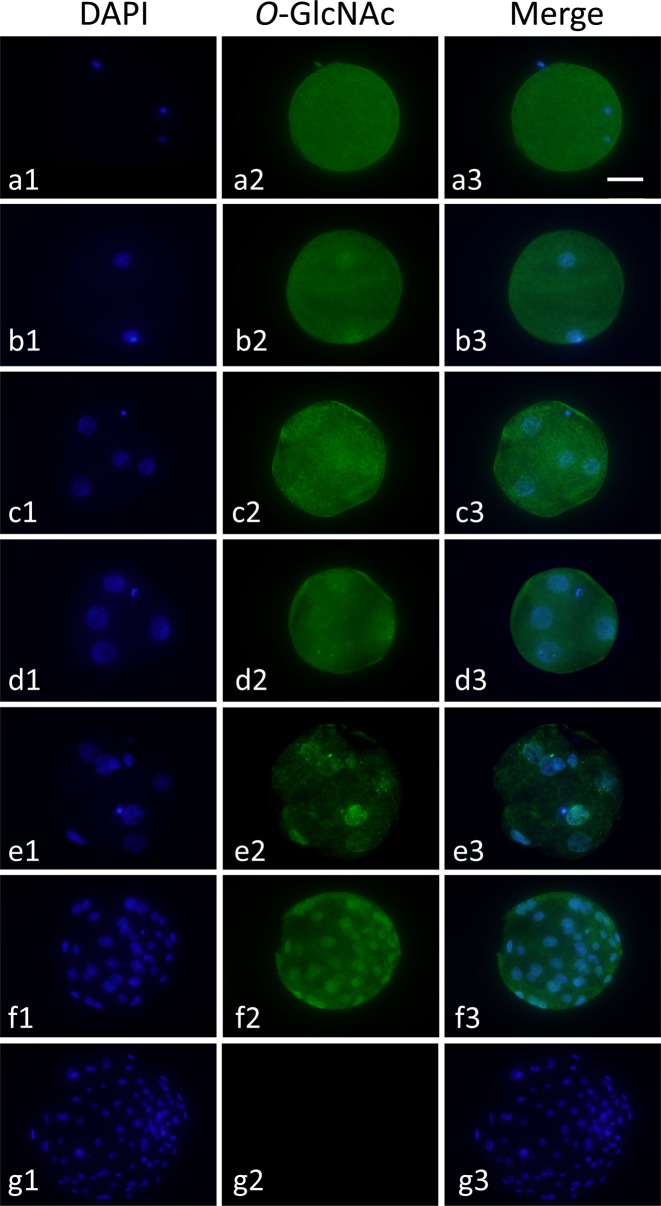
Protein modification with O-GlcNAc (O-GlcNAcylation) in metaphase II (MII) oocytes and diploids at the 2-cell to blastocyst stage was investigated by immunofluorescence using two kinds of primary antibodies against O-GlcNAcylated proteins, CTD110.6 and RL2. Only typical results of O-GlcNAcylated protein localization using RL2 are shown in Fig. 2; because the results of staining with CTD 110.6 were very similar to those of RL2. O-GlcNAcylated proteins were detected in the cytoplasm of MII oocytes and in the cytoplasm and nucleoplasm of diploids at each developmental stage. O-GlcNAcylated proteins were localized at the same level in the cytoplasm and nucleoplasm in diploids before the morula stage. On the other hand, O-GlcNAcylation showed rather nucleoplasmic localization in diploids at the morula and blastocyst stages (Fig. 2, e2 and f2).
Fig. 2.
Localization of O-GlcNAcylated proteins in a pig metaphase II (MII) oocytes and parthenogenetically activated diploids during preimplantation development. MII oocytes (a1–a3) and diploids at the 2-cell (b1–b3), early 4-cell (c1–c3), late 4-cell (d1–d3), morula (e1–e3) and blastocyst stages (f1–f3) were treated with mouse anti-O-GlcNAcylated protein antibody (RL2) and then stained with anti-mouse IgG antibody labeled with Alexa Fluor 488 (green, center column). In the negative control (g1–g3), blastocysts were treated with only the secondary antibody. All diploids were counterstained with DAPI (blue, left column). Merged images are shown in the right column. Oocytes and diploids were observed under an epifluorescence microscope. Scale bar = 50 µm.
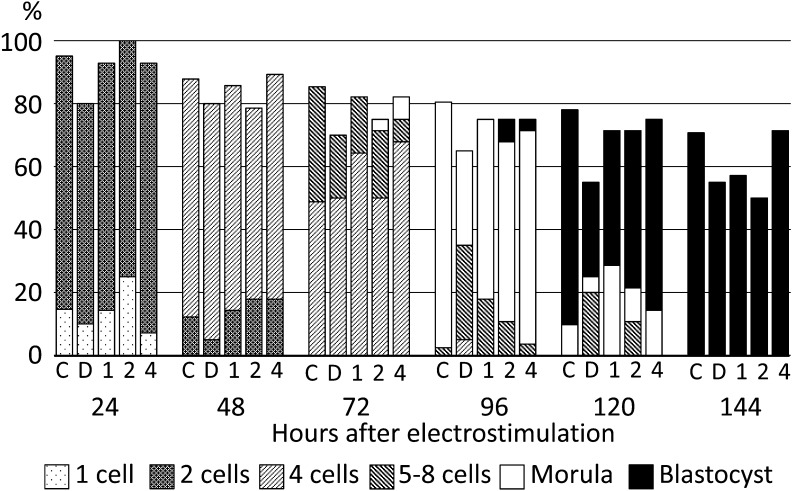
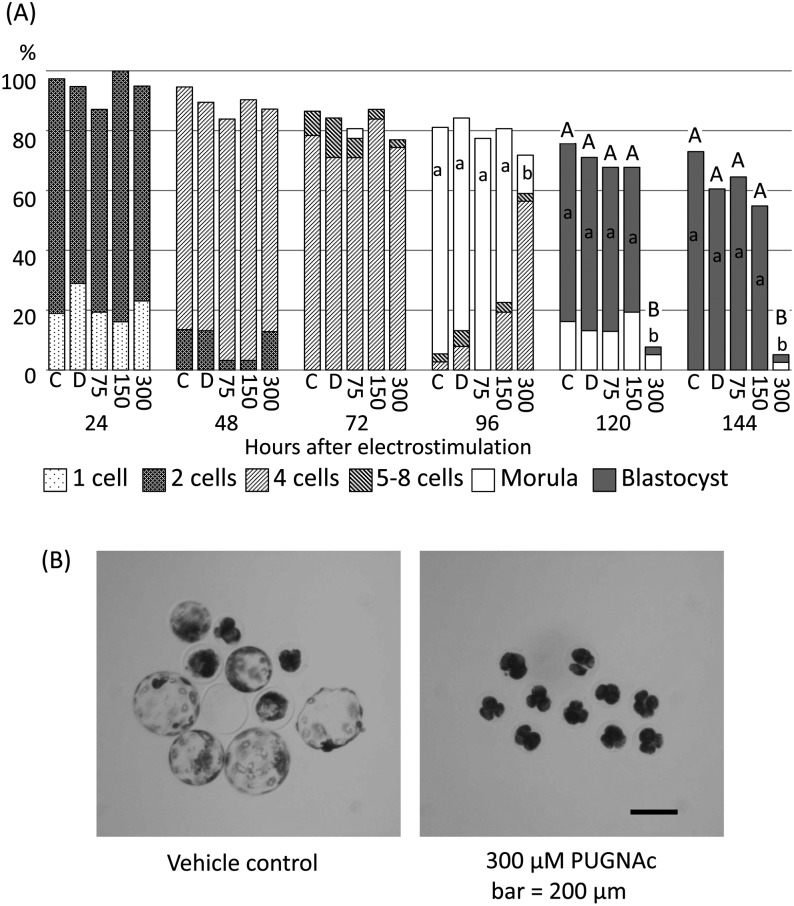
As the results described above showed the existence of O-GlcNAcylation in pig oocytes and diploids during preimplantation development, diploids were cultured under the presence of BADGP or PUGNAc. When diploids were cultured with 1.0 to 4.0 mM BADGP, there were no significant differences in the proportion of diploids that normally developed and reached the most advanced developmental stages among the control, vehicle control and experimental groups at each observation point (Fig. 3). When diploids were cultured with PUGNAc from the beginning of culture, no differences were observed in embryonic development at each stage among each experimental group until 72 h after electrostimulation (Fig. 4A). On the other hand, the proportions of morulae and blastocysts were significantly lower in the 300 µM PUGNAc group than in the others after 96 h (P < 0.05, Fig. 4A). The proportions of normally developed diploids were also significantly lower in the 300 µM PUGNAc group than in the others (P < 0.05, Fig. 4A). Typical results of diploids cultured in the vehicle control and 300 µM PUGNAc groups at 144 h are shown in Fig. 4B. The diploids cultured with 300 µM PUGNAc arrested at the 4-cell stage, while 70% (7/10) of diploids in the vehicle control developed to expanded blastocysts normally (Fig. 4B).
Fig. 3.
Effects of various concentrations of BADGP on preimplantation development of pig parthenogenetically activated diploids. Diploids were cultured in PZM3 with 0 (C, control; D, DMSO as vehicle control), 1.0, 2.0 and 4.0 mM BADGP. Trials were repeated at least three times in each experimental group, and the total numbers of cultured diploids for the analysis were 48, 28, 28, 28 and 27 for the control, vehicle control and 1.0, 2.0 and 4.0 mM BADGP groups, respectively. There were no significant differences in the proportion of diploids that developed to the most advanced stage or in the proportions of diploids that developed normally at each observation point among the experimental groups.
Fig. 4.
Effects of various concentrations of PUGNAc on preimplantation development of pig parthenogenetically activated diploids. Diploids were cultured in PZM3 with 0 (C, control; D, DMSO as vehicle control), 75, 150 and 300 µM PUGNAc. Trials were repeated at least three times in each experimental group, and the total numbers of cultured diploids for the analysis were 37, 38, 31, 31 and 39 for the control, vehicle control and 75, 150 and 300 µM PUGNAc groups, respectively. (A) Proportions of diploids at normally developing stages at each observation period. The different small letters (a, b) indicate significant differences in the proportions of diploids that developed to the most advanced stage at each observation period among the experimental groups (P < 0.05). The different capital letters (A, B) indicate significant differences in proportions of normally developing diploids at each observation period among the experimental groups (P < 0.05). (B) Typical images of diploids cultured in the vehicle control group and 300 µM PUGNAc group from 4 to 144 h after electrostimulation. Seven out of ten (70%) diploids developed further than the expanded blastocyst stage in the vehicle control, and all 10 diploids cultured with 300 µM PUGNAc cased development at the 4-cell stage. Scale bar = 200 µm.
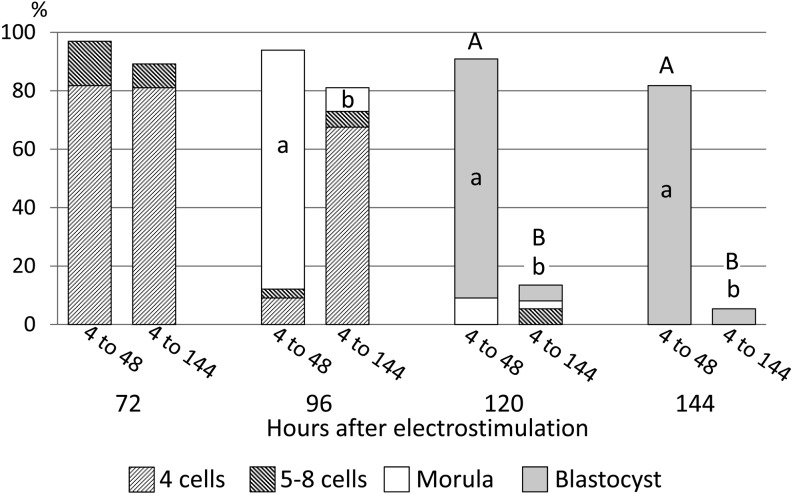
As diploids treated with 300 µM PUGNAc ceased development at the 4-cell stage, diploids were cultured with 300 µM PUGNAc until 48 h, and then 4-cell diploids were selected. The proportion of 4-cell diploids used in this experiment at 48 h was 70.0 ± 4.2% (average ± SE). They were divided into two groups that were cultured in PZM3 with (4 to 144) or without 300 µM PUGNAc (4 to 48) until 144 h to show which period of PUGNAc treatment, before 48 h (4 to 48) or after 48 h (4 to 144), was critical for developmental cessation at the 4-cell stage. The proportion of morulae was significantly lower in diploids cultured with PUGNAc from 4 to 144 h than in those cultured from 4 to 48 h (Fig. 5, P < 0.05), and most diploids cultured with PUGNAc from 4 to 144 h ceased development at the 4-cell stage. The lower proportion of morulae reflected the lower proportions of blastocysts caused by successive treatment with PUGNAc at 120 and 144 h (P < 0.05; Fig. 5). Thus successive treatment of diploids with PUGNAc resulted in very low blastocyst formation, whereas treatment of diploids with PUGNAc until 48 h did not affect the potency of their development to the blastocyst stage.
Fig. 5.
Stage-specific effects of PUGNAc on preimplantation development of pig parthenogenetically activated diploids. Diploids were cultured in PZM3 with 300 µM PUGNAc from 4 to 48 h followed by culture in PZM3 with 0 (4 to 48) or 300 µM PUGNAc (4 to 144) from 48 to 144 h after electrostimulation. Trials were repeated at least three times in each experimental group, and the total numbers of diploids for the analysis were 33 and 37 in the 4 to 48 and 4 to 144 groups, respectively. Different small letters (a, b) indicate significant differences in the proportions of diploids that developed to the most advanced stage at each observation period between the experimental groups (P < 0.05). Different capital letters (A, B) indicate significant differences in proportions of normally developing diploids at each observation period between the two experimental groups (P < 0.05).
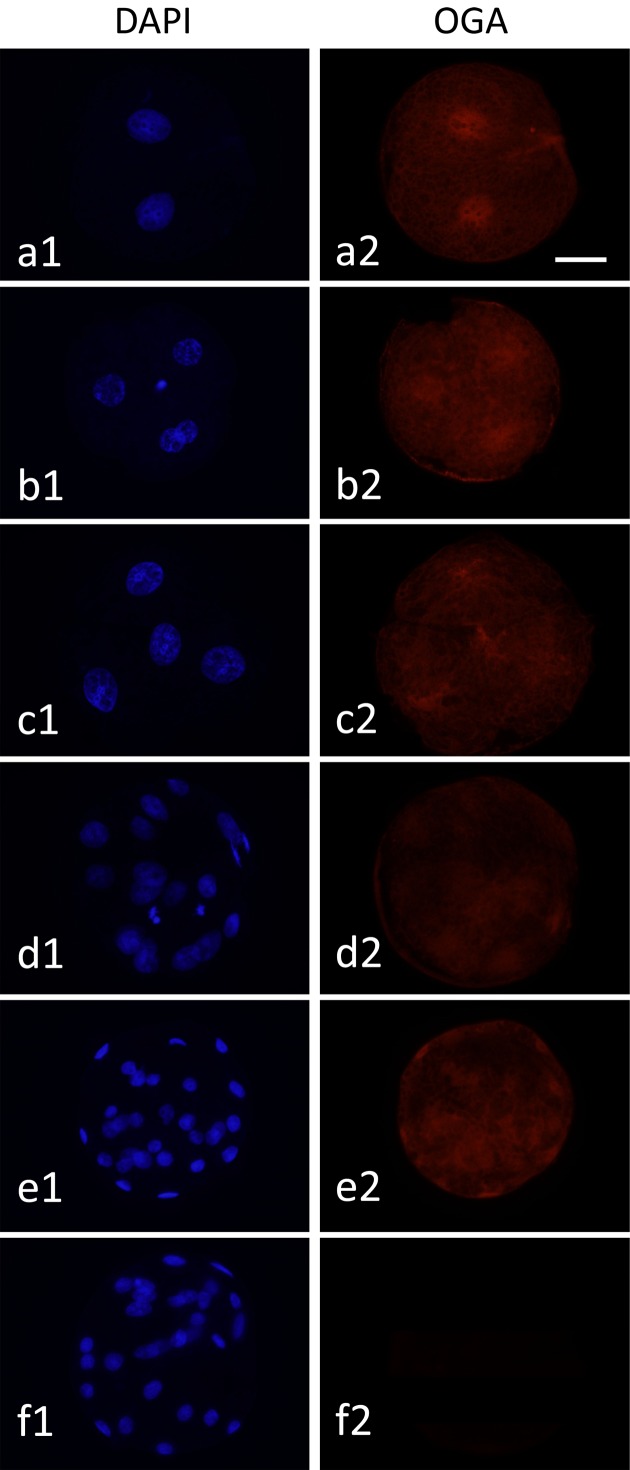
As inhibition of OGA clearly suppressed the development of diploids beyond the 4-cell stage, the expression of OGA in diploids during preimplantation development was examined by the immunofluorescence microscopy. As shown in Fig. 6, OGA was detected in diploids at all stages from the 2-cell to blastocyst stage (Fig. 6).
Fig. 6.
Localization of OGA proteins in pig parthenogenetically activated diploids during preimplantation development. Diploids at the 2-cell (a1 and a2), early 4-cell (b1 and b2), late 4-cell (c1 and c2), morula (d1 and d2) and blastocyst stages (e1 and e2) were treated with mouse anti-OGA antibody and then stained with anti-mouse IgM antibody labeled with Alexa Fluor 564 (red). In the negative control (f1 and f2), blastocysts were treated with only the secondary antibody. All diploids were counterstained with DAPI (blue). Diploids were observed under an epifluorescence microscope. Scale bar = 50 µm.
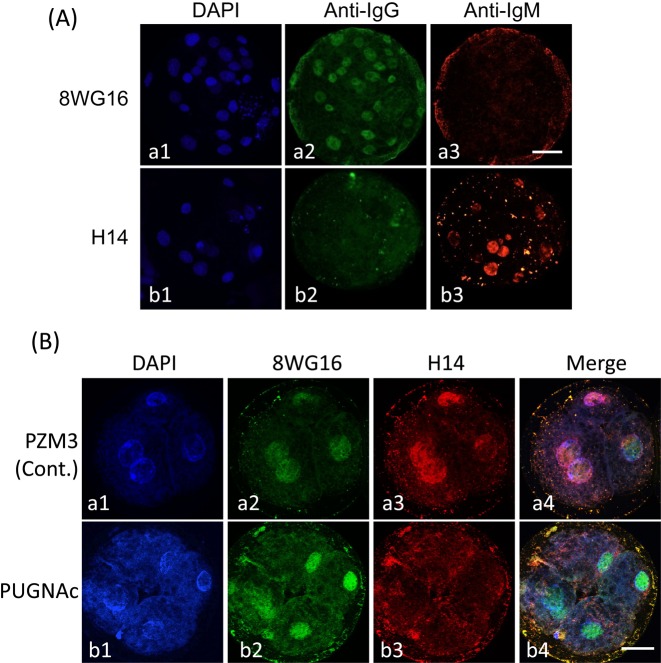
Then it was confirmed whether the treatment of diploids with 300 µM PUGNAc caused the decrease in phosphorylation of Pol II. Before the experiment, cross-reactions between primary antibodies, 8WG16 and H14, and secondary antibodies, anti-mouse IgG and IgM antibodies, were tested (Fig. 7A). The 8WG16 antibody did not react with the anti-IgM antibody (Fig. 7A, a3) but did react with the anti-IgG antibody (Fig. 7A, a2). The H14 antibody did not react with the anti-IgG antibody (Fig. 7A, b2) but did react with the anti-IgM antibody (Fig. 7A, b3). Pol II was detected in almost all nuclei (Fig. 7A, a2), and pPol II was detected in only some nuclei (Fig. 7A, b3) of a blastocyst.
Fig. 7.
Localization of Pol II and phosphorylated Pol II (pPol II) in pig parthenogenetically activated diploids. (A) Cross-reactivity test between primary antibodies (8WG16 and H14) and secondary antibodies (anti-mouse IgG antibody labeled with Alexa Fluor 488 and anti-mouse IgM antibody labeled with Alexa Fluor 564). Blastocysts after culture of diploids for 144 h and treatment with 8WG16, which reacted with Pol II and phosphorylated Pol II (pPol II), and H14, which reacted with pPol II alone. Both primary antibodies were derived from primed mice; the immunoglobulin isotypes of 8WG16 and H14 were IgG and IgM, respectively. Therefore, it had to be confirmed that only pPol II was detected by H14 when diploids were subjected to double immunofluorostaining using these primary and secondary antibodies. The anti-mouse IgG antibody labeled with Alexa Fluor 488 reacted with 8WG16 (a2); but not H14 (b2), while the anti-mouse IgM labeled with Alexa Fluor 564 reacted with H14 (b3); but not 8WG16 (a3). Diploids were observed under an epifluorescence microscope. (B) Localization of Pol II and pPol II in pig parthenogenetically activated diploids exposed to PUGNAc. Diploids were cultured in PZM3 containing 0 (control) and 300 µM PUGNAc from 4 to 48 h after electrostimulation. Four-cell diploids were treated with 8WG16 and H14 and then stained with Alexa Fluor 488 and Alexa Fluor 564 labeled secondary antibodies. Pol II is shown by green (a2 and b2), and pPol II is shown by red (a3 and b3). All diploids were counterstained with DAPI (blue; a1 and b1). Merged images are shown in the right column (a4 and b4). Pol II was detected in nuclei in both groups (a2 and b2), whereas pPol II was only detected in nuclei in the control group (a3). Diploids were observed under a confocal laser microscope. Scale bar = 50 µm.
Fig. 7B shows the results of double immunostaining using 8WG16 and H14 in diploids cultured with or without PUGNAc until 48 h. RNA transcription occurs at the 4-cell stage in pig embryos, so the Pol II phosphorylation was determined at 48 h. Pol II, which was detected as nonphosphorylated and phosphorylated Pol II, was observed in almost all nuclei irrespective of PUGNAc treatment (Fig. 7B, a2 and b2). On the other hand, pPol II was observed in nuclei of diploids cultured without PUGNAc (Fig. 7B, a3 and a4) but not in those with PUGNAc (Fig. 7B, b3 and b4).
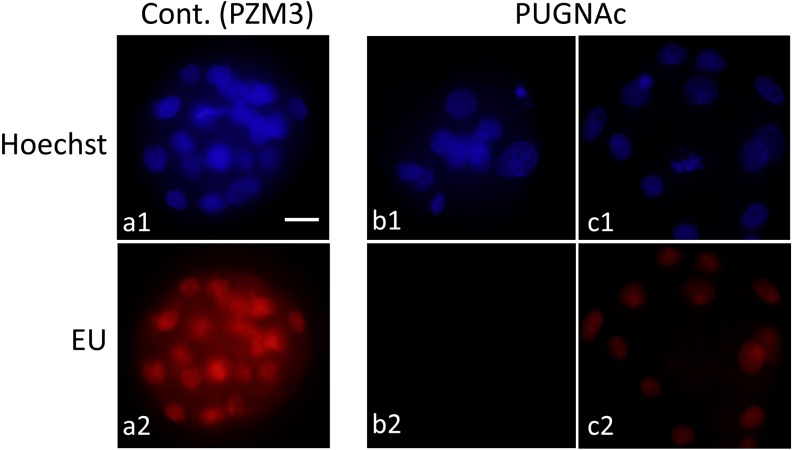
Diploids cultured with 0 or 300 µM PUGNAc were subjected to additional culture with EU 96 h after electrostimulation to detect translation of RNAs. The incorporation of EU was detected in nuclei of diploids that developed to the morula stage, irrespective of PUGNAc treatment (Fig. 8, a2 and c2), but not in most of the diploids that did not show compaction caused by PUGNAc treatment (Fig. 8, b2). An EU signal was not detected before 96 h in diploids cultured even without PUGNAc.
Fig. 8.
RNA transcription in pig parthenogenetically activated diploids under inhibition of OGA activity. Diploids were cultured in PZM3 with 0 (control; a1 and a2) and 300 µM PUGNAc (b1, b2, c1 and c2) until 96 h. Then they were cultured in PZM3 with 5-ethyl uridine (EU) for 1 h, and nascent RNAs were detected by Click-iT reaction (red). Diploids treated with PUGNAc typically showed no signal of EU (b2), though all blastomeres showed the signal in the control (a2). In addition, some diploids that showed compaction irrespective of PUGNAc treatment also showed the signal (c2). Diploids were observed under an epifluorescence microscope. Scale bar = 50 µm.
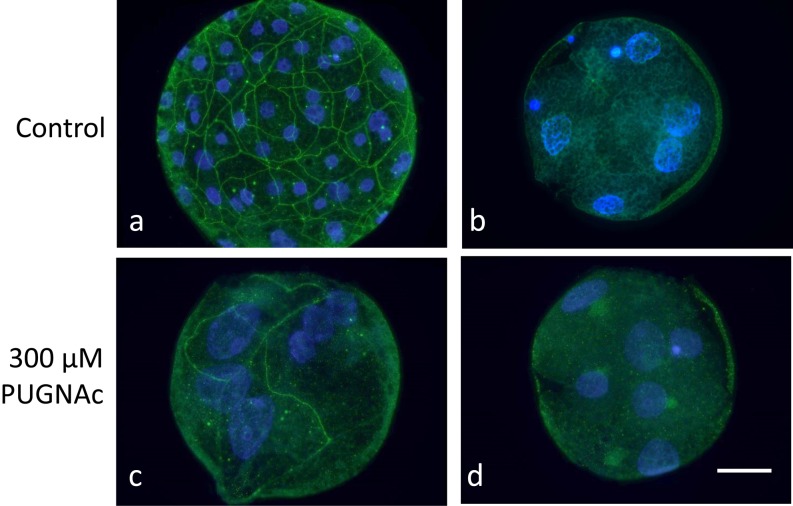
As the RNAs translation was detected in diploids at the morula stage irrespective of PUGNAc treatment, expression of Claudin-4, which is one of the tight junction proteins, was examined. Claudin-4 was observed on the cell boundary in blastocysts of control diploids at 120 h (Fig. 9, a). The Claudin-4 partially expressed on the cell boundary in diploids that developed to the morula stage irrespective of PUGNAc treatment at 120 h (Fig. 9, c), whereas boundary expression was not detected in diploids before compaction at 120 h in the PUGNAc-treated group (Fig. 9, d), and the localization pattern was very similar to that in normally developed 5-cell diploids in the control group at 72 h (Fig. 9, b).
Fig. 9.
Expression of a tight junction sealing protein, Claudin-4, in pig parthenogenetically activated diploids under inhibition of OGA activity. Diploids were cultured in PZM3 with 0 (a) and 300 µM PUGNAc from 4 to 120 h after electrostimulation (c and d). The control showed normal development to the morula and blastocyst stages, while most of the PUGNAc-treated diploids showed developmental delay and cessation at 120 h. Diploids with normal development to the 5–8 cell stage in the control group at 72 h were also analyzed to compare the Claudin-4 expression pattern with that of diploids that showed developmental delay caused by PUGNAc treatment (b). Blastocysts showed clear cell-boundary localization of Caludin-4 (a, green) in the control group. A small number of morulae in the PUGNAc-treated group also showed the cell-boundary localization of Claudin-4 (c; green), but the signal was generally partial or weak. Diploids before compaction did not show any boundary localization of Claudin-4 in the PUGNAc-treated group (d), and the expression pattern was similar to that of normally developed 5-cell diploids before compaction in the control group. All diploids were counterstained with DAPI (blue). Diploids were observed under an epifluorescence microscope. Scale bar = 50 µm.
Discussion
The presence and roles of O-GlcNAcylation, one of the functional protein modifications in mammalian cells, were examined in pig parthenogenetically activated diploids during preimplantation development. The mRNA expressions of three enzymes involved with O-GlcNAcylation and localization of O-GlcNAcylated proteins were examined to confirm the presence of O-GlcNAc modification in pig embryos. Two enzymes related to protein O-GlcNAcylation, O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA), were inhibited to examine the roles of O-GlcNAcylation in preimplantation development.
The mRNA of Gfpt, which is the rate-limiting enzyme in the HBP, and Ogt were expressed in embryos throughout the preimplantation stages. The mRNA of Oga was also detected at all preimplantation stages except for the 4-cell stage, but OGA proteins were also detected in 4-cell diploids. These results suggest that glucose is metabolized via the HBP and used for O-GlcNAc cycling in pig embryos. Since it has been reported that all eukaryotic cells had O-GlcNAc modification and both addition and removeal of O-GlcNAc [20], it seems that MII oocytes accumulated maternal OGA proteins and mRNA for embryo development before the completion of oocyte maturation. It has been supposed that the expression of RNAs that contribute to embryo development switches from maternal to zygotic at the 4-cell stage in pig embryos [34]; thus it is thought that Oga mRNA was not transcribed during the 4-cell stage and was re-expressed from the morula stage to support embryo development. The resumption of Oga expression beginning at the morula stage suggests that OGA has an important role in pig preimplantation development beyond the 4-cell stage.
O-GlcNAcylated proteins were detected in the cytoplasm and nucleoplasm of oocytes and embryos at all stages of preimplantation development, but O-GlcNAcylated proteins were observed mainly in the nucleoplasm beginning in the morula stage. This suggests that the roles of O-GlcNAcylation in pig preimplantation development change around the morula and blastocyst stages. Since it is unclear whether O-GlcNAcylated proteins in cytoplasm were moved into the nucleoplasm or de-O-GlcNAcylated in this study, it is necessary to identify which proteins were O-GlcNAcylated in morulae and blastocysts. The difference in localization may suggest that O-GlcNAcylation has a relationship with differentiation, because it has been reported that many transcription factors are the target of O-GlcNAcylation, for example, Oct4 and Sox2 [23], and it has been reported that the first differentiation starts in the morula to blastocyst stage in mammalian preimplantation development [35, 36].
Although embryo development beyond the 4-cell stage was severely impaired by the addition of 300 µM PUGNAc, embryos that developed to the blastocyst stage were not affected by the treatment until 48 h. On the other hand, the addition of an OGT inhibitor, BADGP, to the medium did not affect embryo development to the blastocyst stage. Furthermore, inhibition of GFPT with azaserine did not lead to developmental suppression in a preliminarily experiment (data not shown). These results indicate that removal of O-GlcNAcylation is essential for development beyond the 4-cell stage and that developmental failure is caused by treatment with PUGNAc beginning at the 4-cell stage. These results in the pig seem to be different from those reported previously in the mouse [16]. The removal of O-GlcNAcylation is important for embryo development in the pig, while the addition of O-GlcNAcylation may be important for it in the mouse.
Phosphorylation of Pol II in the CTD (Pol II-CTD) was detected in control 4-cell diploids; but not in 4-cell diploids cultured with 300 µM PUGNAc. Moreover, RNAs synthesis in diploids cultured with PUGNAc was also suppressed. Both these results suggest that exposure to the OGA inhibitor caused failure or delay of ZGA, since Pol II-CTD was phosphorylated prior to ZGA [33].
ZGA, which refer to the start of RNA expression using zygotic genomes [37], occurs during the 4-cell stage in pig embryos [38]. Previous studies reported that the metabolic characteristics [12, 27, 28] and RNA transcription pattern [34] change during 4-cell stage. Although developmental arrest at the 4-cell stage, which was caused by interference with Pol II-CTD phosphorylation and successive suppression of RNA transcription by PUGNAc treatment, indicates that inhibition of OGA results in ZGA failure in pig preimplantation embryos, it is unclear which proteins are O-GlcNAcylated, i.e., which are critical factors for pig preimplantation development.
Recently, it was suggested that O-GlcNAcylated Pol II-CTD was necessary to form preinitiation complex in the promoter region [22]. If Pol II-CTD de-O-GlcNAcylation was inhibited directly, abnormally O-GlcNAcylated Pol II may be detectable in the promoter region of the nuclei of 4-cell pig embryos treated with PUGNAc. It was also possible that O-GlcNAcylation was stayed in other proteins. A decrease in maternal proteins is also important for ZGA [37], and it has been shown that OGA inhibition causes a decrease in proteasome activity [39] and that O-GlcNAcylated proteins are often degraded slowly [40, 41]. Degradation of the maternal proteins might be delayed by PUGNAc treatment, and then abnormal retention of maternal proteins might result in failure of embryo development. Since histone family members are also target proteins of O-GlcNAcylation [24,25,26], derangement of chromatin remodeling might be induced in PUGNAc-treated embryos.
As RNA translation occurred in diploids at the morula stage irrespective of PUGNAc treatment, expression of Claudin-4, which is a tight junction sealing protein with important roles for the first mammalian embryonic differentiation [42], was examined. Claudin-4 protein was expressed on the cell boundary in control diploids at the blastocyst stage 120 h after electrostimulation, but it was not expressed or was partially expressed in PUGNAc-treated diploids. The unnatural Claudin expression pattern may be caused by OGA inhibition.
The electrostimulated pig diploids used in this study were similar to in vivo fertilized embryos in many previous reports in terms of morphological and physiological characteristics during preimplantation development [12, 27, 28]. Therefore, it is suggested that there are not so many differences between diploids and embryos in terms of RNA and protein expression pattern and the sequence of preimplantation development. The present results caused by dysfunction of O-GlcNAc cycling in parthenogenetic diploids may reflect those of fertilized embryos.
In conclusion, O-GlcNAcylated proteins are present in MII oocytes and preimplantation embryos, and the cycling of O-GlcNAcylation is active in embryos during preimplantation development in the pig. Furthermore, pig embryos show developmental arrest at the 4-cell stage as a result of inhibition of OGA, perhaps via inhibition of Pol II-CTD phosphorylation and successive suppression of RNA transcription.
Acknowledgments
This study was supported by JSPS Grant-in-Aid for Scientific Research (B), Grant Number 25292160.
Reference
- 1.Leese HJ, Barton AM. Pyruvate and glucose uptake by mouse ova and preimplantation embryos. J Reprod Fertil 1984; 72: 9–13. [DOI] [PubMed] [Google Scholar]
- 2.Flood MR, Wiebold JL. Glucose metabolism by preimplantation pig embryos. J Reprod Fertil 1988; 84: 7–12. [DOI] [PubMed] [Google Scholar]
- 3.Seshagiri PB, Bavister BD. Glucose inhibits development of hamster 8-cell embryos in vitro. Biol Reprod 1989; 40: 599–606. [DOI] [PubMed] [Google Scholar]
- 4.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86: 679–688. [DOI] [PubMed] [Google Scholar]
- 5.Chatot CL, Lewis JL, Torres I, Ziomek CA. Development of 1-cell embryos from different strains of mice in CZB medium. Biol Reprod 1990; 42: 432–440. [DOI] [PubMed] [Google Scholar]
- 6.Chatot CL, Lewis-Williams J, Torres I, Ziomek CA. One-minute exposure of 4-cell mouse embryos to glucose overcomes morula block in CZB medium. Mol Reprod Dev 1994; 37: 407–412. [DOI] [PubMed] [Google Scholar]
- 7.Amarnath D, Wakayama S, Zhu J, Moawad AR, Wakayama T, Campbell KH. The novel use of modified pig zygotic medium for the efficient culture of the preimplantation mouse embryos. Theriogenology 2011; 76: 1639–1646. [DOI] [PubMed] [Google Scholar]
- 8.Petters RM, Wells KD. Culture of pig embryos. J Reprod Fertil Suppl 1993; 48: 61–73. [PubMed] [Google Scholar]
- 9.Yoshioka K, Suzuki C, Tanaka A, Anas IM, Iwamura S. Birth of piglets derived from porcine zygotes cultured in a chemically defined medium. Biol Reprod 2002; 66: 112–119. [DOI] [PubMed] [Google Scholar]
- 10.Karja NW, Medvedev S, Onishi A, Fuchimoto D, Iwamoto M, Otoi T, Nagai T. Effect of replacement of pyruvate/lactate in culture medium with glucose on preimplantation development of porcine embryos in vitro. J Reprod Dev 2004; 50: 587–592. [DOI] [PubMed] [Google Scholar]
- 11.Park Y, Hong J, Yong H, Lim J, Lee E. Effect of exogenous carbohydrates in a serum-free culture medium on the development of in vitro matured and fertilized porcine embryos. Zygote 2005; 13: 269–275. [DOI] [PubMed] [Google Scholar]
- 12.Shibutani M, Lee J, Miyano T, Miyake M. Demands for carbohydrates as major energy substrates depend on the preimplantation developmental stage in pig embryos: Differential use of fructose by parthenogenetic diploids before and after the 4-cell stage in the pig. J Reprod Dev 2015; 61: 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mito T, Yoshioka K, Yamashita S, Suzuki C, Noguchi M, Hoshi H. Glucose and glycine synergistically enhance the in vitro development of porcine blastocysts in a chemically defined medium. Reprod Fertil Dev 2012; 24: 443–450. [DOI] [PubMed] [Google Scholar]
- 14.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem 1984; 259: 3308–3317. [PubMed] [Google Scholar]
- 15.Sturmey RG, Leese HJ. Energy metabolism in pig oocytes and early embryos. Reproduction 2003; 126: 197–204. [DOI] [PubMed] [Google Scholar]
- 16.Pantaleon M, Scott J, Kaye PL. Nutrient sensing by the early mouse embryo: hexosamine biosynthesis and glucose signaling during preimplantation development. Biol Reprod 2008; 78: 595–600. [DOI] [PubMed] [Google Scholar]
- 17.Holt GD, Snow CM, Senior A, Haltiwanger RS, Gerace L, Hart GW. Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J Cell Biol 1987; 104: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanover JA, Cohen CK, Willingham MC, Park MK. O-linked N-acetylglucosamine is attached to proteins of the nuclear pore. Evidence for cytoplasmic and nucleoplasmic glycoproteins. J Biol Chem 1987; 262: 9887–9894. [PubMed] [Google Scholar]
- 19.Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem 1991; 266: 4706–4712. [PubMed] [Google Scholar]
- 20.Hanover JA, Krause MW, Love DC. Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat Rev Mol Cell Biol 2012; 13: 312–321. [DOI] [PubMed] [Google Scholar]
- 21.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 2001; 40: 7845–7852. [DOI] [PubMed] [Google Scholar]
- 22.Ranuncolo SM, Ghosh S, Hanover JA, Hart GW, Lewis BA. Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J Biol Chem 2012; 287: 23549–23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell 2012; 11: 62–74. [DOI] [PubMed] [Google Scholar]
- 24.Fujiki R, Hashiba W, Sekine H, Yokoyama A, Chikanishi T, Ito S, Imai Y, Kim J, He HH, Igarashi K, Kanno J, Ohtake F, Kitagawa H, Roeder RG, Brown M, Kato S. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 2011; 480: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakabe K, Wang Z, Hart GW. Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc Natl Acad Sci USA 2010; 107: 19915–19920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong JJ, Nguyen BL, Bridger R, Medrano EE, Wells L, Pan S, Sifers RN. β-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J Biol Chem 2012; 287: 12195–12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Thuan N, Harayama H, Miyake M. Characteristics of preimplantational development of porcine parthenogenetic diploids relative to the existence of amino acids in vitro. Biol Reprod 2002; 67: 1688–1698. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen VT, Kure-bayashi S, Harayama H, Nagai T, Miyake M. Stage-specific effects of the osmolarity of a culture medium on the development of parthenogenetic diploids in the pig. Theriogenology 2003; 59: 719–734. [DOI] [PubMed] [Google Scholar]
- 29.Papaioannou VE, Ebert KM. The preimplantation pig embryo: cell number and allocation to trophectoderm and inner cell mass of the blastocyst in vivo and in vitro. Development 1988; 102: 793–803. [DOI] [PubMed] [Google Scholar]
- 30.Kure-bayashi S, Miyake M, Okada K, Kato S. Successful implantation of in vitro-matured, electro-activated oocytes in the pig. Theriogenology 2000; 53: 1105–1119. [DOI] [PubMed] [Google Scholar]
- 31.Kure-Bayashi S, Miyake M, Katayama M, Miyano T, Kato S. Development of porcine blastocysts from in vitro- matured and activated haploid and diploid oocytes. Theriogenology 1996; 46: 1027–1036. [DOI] [PubMed] [Google Scholar]
- 32.Kuijk EW, du Puy L, van Tol HT, Haagsman HP, Colenbrander B, Roelen BA. Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and preimplantation embryos. BMC Dev Biol 2007; 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokoro M, Shin SW, Nishikawa S, Lee HH, Hatanaka Y, Amano T, Mitani T, Kato H, Anzai M, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Deposition of acetylated histones by RNAP II promoter clearance may occur at onset of zygotic gene activation in preimplantation mouse embryos. J Reprod Dev 2010; 56: 607–615. [DOI] [PubMed] [Google Scholar]
- 34.Cao S, Han J, Wu J, Li Q, Liu S, Zhang W, Pei Y, Ruan X, Liu Z, Wang X, Lim B, Li N. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 2014; 15: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo G, Huss M, Tong GQ, Wang C, Li Sun L, Clarke ND, Robson P. Resolution of cell fate decisions revealed by single-cell gene expression analysis from zygote to blastocyst. Dev Cell 2010; 18: 675–685. [DOI] [PubMed] [Google Scholar]
- 36.Fujii T, Sakurai N, Osaki T, Iwagami G, Hirayama H, Minamihashi A, Hashizume T, Sawai K. Changes in the expression patterns of the genes involved in the segregation and function of inner cell mass and trophectoderm lineages during porcine preimplantation development. J Reprod Dev 2013; 59: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin SW, Tokoro M, Nishikawa S, Lee HH, Hatanaka Y, Nishihara T, Amano T, Anzai M, Kato H, Mitani T, Kishigami S, Saeki K, Hosoi Y, Iritani A, Matsumoto K. Inhibition of the ubiquitin-proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev 2010; 56: 655–663. [DOI] [PubMed] [Google Scholar]
- 38.Jarrell VL, Day BN, Prather RS. The transition from maternal to zygotic control of development occurs during the 4-cell stage in the domestic pig, Sus scrofa: quantitative and qualitative aspects of protein synthesis. Biol Reprod 1991; 44: 62–68. [DOI] [PubMed] [Google Scholar]
- 39.Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 2003; 115: 715–725. [DOI] [PubMed] [Google Scholar]
- 40.Park SY, Kim HS, Kim NH, Ji S, Cha SY, Kang JG, Ota I, Shimada K, Konishi N, Nam HW, Hong SW, Yang WH, Roth J, Yook JI, Cho JW. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J 2010; 29: 3787–3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardivillé S, Hoedt E, Mariller C, Benaïssa M, Pierce A. O-GlcNAcylation/phosphorylation cycling at Ser10 controls both transcriptional activity and stability of delta-lactoferrin. J Biol Chem 2010; 285: 19205–19218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol 2007; 312: 509–522. [DOI] [PubMed] [Google Scholar]