Abstract
After nearly 30 years of effort, Ed Lewis published his 1978 landmark paper in which he described the analysis of a series of mutations that affect the identity of the segments that form along the anterior-posterior (AP) axis of the fly (Lewis 1978). The mutations behaved in a non-canonical fashion in complementation tests, forming what Ed Lewis called a “pseudo-allelic” series. Because of this, he never thought that the mutations represented segment-specific genes. As all of these mutations were grouped to a particular area of the Drosophila third chromosome, the locus became known of as the bithorax complex (BX-C). One of the key findings of Lewis’ article was that it revealed for the first time, to a wide scientific audience, that there was a remarkable correlation between the order of the segment-specific mutations along the chromosome and the order of the segments they affected along the AP axis. In Ed Lewis’ eyes, the mutants he discovered affected “segment-specific functions” that were sequentially activated along the chromosome as one moves from anterior to posterior along the body axis (the colinearity concept now cited in elementary biology textbooks). The nature of the “segment-specific functions” started to become clear when the BX-C was cloned through the pioneering chromosomal walk initiated in the mid 1980s by the Hogness and Bender laboratories (Bender et al. 1983a; Karch et al. 1985). Through this molecular biology effort, and along with genetic characterizations performed by Gines Morata’s group in Madrid (Sanchez-Herrero et al. 1985) and Robert Whittle’s in Sussex (Tiong et al. 1985), it soon became clear that the whole BX-C encoded only three protein-coding genes (Ubx, abd-A, and Abd-B). Later, immunostaining against the Ubx protein hinted that the segment-specific functions could, in fact, be cis-regulatory elements regulating the expression of the three protein-coding genes. In 1987, Peifer, Karch, and Bender proposed a comprehensive model of the functioning of the BX-C, in which the “segment-specific functions” appear as segment-specific enhancers regulating, Ubx, abd-A, or Abd-B (Peifer et al. 1987). Key to their model was that the segmental address of these enhancers was not an inherent ability of the enhancers themselves, but was determined by the chromosomal location in which they lay. In their view, the sequential activation of the segment-specific functions resulted from the sequential opening of chromatin domains along the chromosome as one moves from anterior to posterior. This model soon became known of as the open for business model. While the open for business model is quite easy to visualize at a conceptual level, molecular evidence to validate this model has been missing for almost 30 years. The recent publication describing the outstanding, joint effort from the Bender and Kingston laboratories now provides the missing proof to support this model (Bowman et al. 2014). The purpose of this article is to review the open for business model and take the reader through the genetic arguments that led to its elaboration.
Keywords: Chromatin domains, Boundaries, Insulators, Bithorax complex
Introduction
A quick overview of the model of Ed Lewis
Drosophila embryos and larvae harbor a head, three thoracic segments (T1–T3) and eight abdominal segments (A1–A8; see left panel of Fig. 1). At metamorphosis, the eighth abdominal segment gives rise to parts of the genital structures of the adult fly. When the whole BX-C is deleted, mutant embryos die before hatching, but at a stage where it is already possible to recognize the identities of the segments. Thus, it is possible to see that mutants lacking the BX-C have all posterior segments from T3 transformed into copies of T2 (to be precise, these transformations affect parasegments—see below—but Lewis talked in terms of segments). This finding led Ed Lewis to consider T2 as the ground state of development on which the activity of the BX-C built, thereby assigning identities to the more posterior segments (Lewis 1978) (Fig. 1).
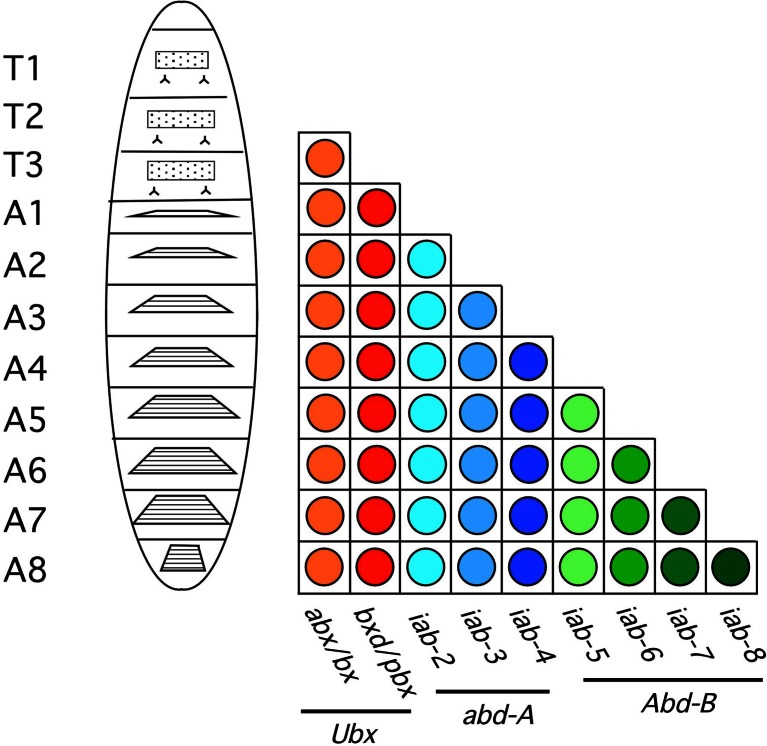
Fig. 1.
The model of Ed Lewis. Reproduced from Fig. 2 of Maeda and Karch (2006) with the permission of the Company of Biologists; DOI: 10.1242/dev.02323. A larva is drawn on the left with its three thoracic segments marked as T1, T2 and T3 as well as its eight abdominal segments marked A1 to A8. The diagram next to the larva represents the presence of absence of a segment-specific function that is required for the specification of each segment. As the segments-specific functions are aligned on the chromosome (in the x-axis) in the same order as the segments along the body axis, the diagram is represented in the form of a matrix. The fact that mutations in a given segment-specific function always transform that segment into the copy of the immediately adjacent segment anteriorly implies that the more anterior segments-specific functions are active in that segment. This led Ed Lewis to propose that segment-specific functions act in an additive fashion. The Ubx, abd-A, and Abd-B genes are indicated below the segment-specific functions, with the horizontal lines defining the mutations that are not complemented by the respective Ubx, abd-A, or Abd-B mutations
There are other mutations within the BX-C that primarily affect the identity of single segment under the control of the BX-C. Many of them allow survival to adulthood. These mutations define the nine “segment-specific functions”, abx/bx, bxd/pbx, and iab-2 through iab-8 that specify the identities of T3 and all eight abdominal segments (A1 through A8), respectively. Typically, loss-of-function mutations in the BX-C result in the transformation of a given segment into a copy of the segment directly anterior to it. The fact that mutations in individual “segment-specific functions” always cause transformations toward the segment immediately anterior to them and not toward the ground state (T2) indicates that everything required for the identity of the more anterior segments still functions in the more posterior segments. Thus, Ed Lewis proposed that the segment-specific functions act in an additive fashion: once they are turned on in the segment they specify, they remain active in the more posterior segments. Lewis synthesized these findings into two rules for BX-C regulation: “… a [segment-specific function] derepressed in one segment is derepressed in all segments posterior thereto…” and “the more posterior a segment… the greater the number of BX-C [segment-specific functions] that are in a derepressed state” (Lewis 1978). These rules are illustrated in the form of a matrix in which the anterior-posterior axis of the fly is represented along the y-axis and the activity state of the BX-C is represented along the x-axis (see Fig. 1).
The segment-specific functions are segment-specific enhancers
Three classes of mutations (Ubx, abd-A, and Abd-B) associated with embryonic lethality also exist within the BX-C. They cause the transformation of a group of segments into a more anterior segment (Lewis 1978; Sanchez-Herrero et al. 1985; Tiong et al. 1985). For example, Ubx (Ultrabithorax) mutant embryos have their T3 and A1 segments transformed into T2, as if both the abx/bx and bxd/pbx segment-specific functions were inactivated in Ubx alleles. In agreement with Ed Lewis’ observations, Ubx mutations fail to complement abx, bx, bxd, or pbx alleles. This lack of complementation is contrasted by the observation that heterozygous flies, with bx or abx mutations on one chromosome and bxd or pbx mutations on the other, looked normal. Ed Lewis proposed the term “pseudo-allelism” to describe these conflicting observations.
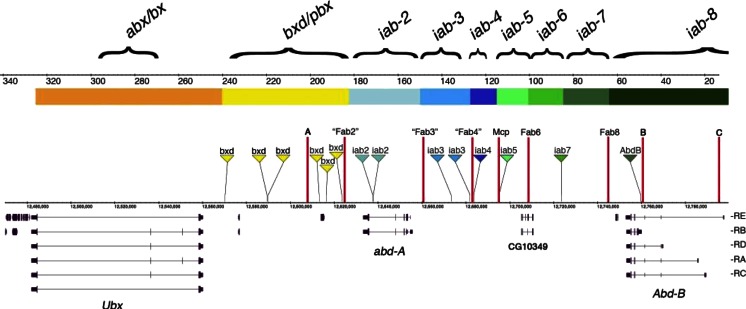
After the discovery that the BX-C encodes only three genes (Ubx, abd-A, and Abd-B), the phenomenon of pseudo-allelism was finally explained. In situ hybridization and antibodies generated against these proteins allowed the determination of their expression patterns (Akam 1983; Beachy et al. 1985; Bender et al. 1983a; Casanova et al. 1987; Celniker et al. 1990; Karch et al. 1990; Karch et al. 1985; Macias et al. 1990; Sanchez-Herrero 1991; White and Wilcox 1985). By staining various mutant embryos, it was finally understood that the “segment-specific functions” corresponded to cis-regulatory regions that regulate the expression of Ubx, abd-A, or Abd-B in a segment-specific fashion. The molecular organization of the BX-C is shown along the x-axis of Fig. 2, with the extent of each of the nine segment-specific function indicated by brackets above the DNA line. The Ubx, abd-A, and Abd-B transcription units are shown below. The regulatory interactions between the segment-specific functions and their respective target promoter follow a color code (Figs. 1 and 2). While the abx/bx and bxd/pbx regions regulate Ubx (as indicated in reddish color), iab-2 though iab-4 regulate abd-A (blueish). Finally, iab-5 through iab-8 regulate Abd-B (greenish; see (Maeda and Karch 2006) for review).
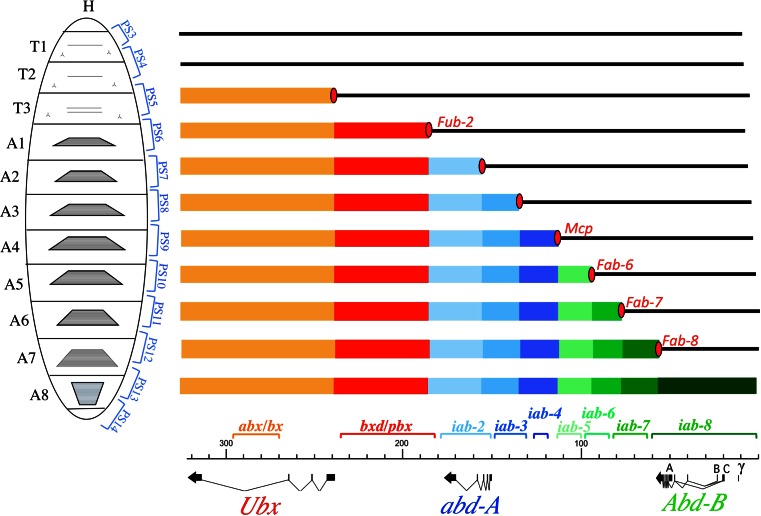
Fig. 2.
The open for business model. A larvae is represented on the left with the thoracic (T1–T3) and abdominal segmental boundaries (A1–A8) as well as the corresponding parasegmental boundaries (PS1–PS14; see text). The genomic map of the BX-C is drawn on the x-axis at the scale indicated in kilobases. The Ubx, abd-A, and Abd-B transcription units are drawn at scale below the genomic map. The extent occupied by the segments-specific functions are indicated by brackets above the DNA line. The sequential opening of the segment-specific regulatory domains is drawn for each parasegments. While colored rectangles indicate open for business, the solid black line represents closed chromatin (see text). Boundaries marking the borders between the open and closed domains are shown by red ovals. The boundaries that have been identified by mutational analysis are named. Note the similarity with the model of Ed Lewis where the dots shown in Fig. 1 are replaced by DNA domains
It should be noted that the embryonic expression patterns of Ubx, abd-A, and Abd-B (and some other homeotic genes) are made up of reiterated units along the AP axis. Each unit of expression is roughly equivalent to one segment in length, but slightly shifted relative to the morphological body segments that appear during mid embryogenesis. These units are known as parasegments (Martinez-Arias and Lawrence 1985). One parasegment (PS) is composed of the posterior compartment of one segment and the anterior compartment of the next segment. For example, PS5 corresponds to the posterior part of T2 and the anterior part of T3. For the most part, the parasegment-segment shift is rarely mentioned due to the fact that the visible adult cuticle is mostly made up of cells from the anterior half of each segment. Nevertheless, the correspondence between parasegments and segments are indicated in the figures of this paper.
Regulatory domains; the open for business model
The finding that the segment-specific functions are in fact cis-regulatory elements clarified the genetic schema that Ed Lewis had been working on for decades. Due to the size of the regulatory regions in question (from 10 to 60 kb), multiple enhancers were hypothesized to exist within each regulatory domain. This was supported by some of the early work using bxd mutations. There are many bxd mutations caused by chromosome breaks. These mutations make up an allelic series with differing strengths of transformation (lowering the Ubx expression in PS6). It turns out that mutations with breakpoints closer to the Ubx promoter cause stronger transformations, while mutations with breakpoints further away from the promoter cause weaker transformations (Bender et al. 1983b; Bender et al. 1985). The correlation between the loss of Ubx expression in PS6 and the amount of DNA from the bxd/pbx region that was separated from the Ubx target promoter [finally published 26 years later in Pease et al. (2013)] was taken as evidence for the existence of multiple enhancers.
Enhancers function from a variety of positions with respect to their target promoters and can often activate different promoters, depending on the circumstances. Given this promiscuity, clustering of the BX-C enhancers in discrete regions along the chromosome was puzzling. Peifer et al (1987) brought a plausible explanation to this question with the idea that parasegmental/segmental address may be conferred by the DNA domain in which the enhancers reside (Fig. 2). According to their view, each regulatory region should be a chromosomal domain that opens up in the appropriate parasegments during early embryogenesis, enabling the enhancers residing within the domain to perform their regulatory “business” with the target promoter (dubbed as the open for business model in Akam et al. 1988).
The idea that BX-C enhancers might be regulated coordinately through chromatin domains primarily came from the analysis of dominant gain-of-function (GOF) mutations, where a given segment develops like a copy of the segment that lies immediately posterior to it. Peifer et al (1987) focused on the dominant Cbx1 mutation to generate their model, but later work describing mutations that delete boundary elements separating regulatory domains also supported this idea. Two additional lines of evidence also pointed to coordination of enhancers within a chromatin domain. The first was the recovery of enhancer trap transposons within the BX-C that brought forward a visual argument to the segment-specific regulatory domain model. And secondly, experiments where special, early enhancers (initiators) were exchanged between different domains, were able to consolidate the model by entirely fulfilling the predictions made by the open for business model.
The Cbx1 mutation
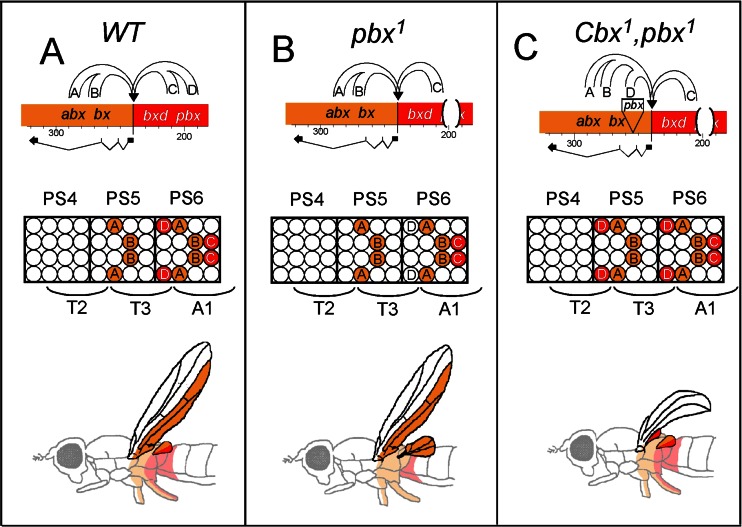
Cbx1 is a gain-of function mutation that transforms the posterior half of the wing (T2) into the posterior half of the haltere (T3). Fine structure mapping led Lewis to discover that the original Cbx1 chromosome contained two, separable mutations. One of them was associated with the dominant GOF phenotype and the other one was associated with a recessive phenotype. The recessive mutation was named postbithorax1 (pbx1) in accordance with the transformation of the posterior haltere into the posterior wing (Lewis 1954). Based on this phenotype, Lewis reasoned that the pbx+ function must be to “make” the posterior haltere. Given that the dominant GOF Cbx1 phenotype is to transform the posterior wing into the posterior haltere, it followed that Cbx1 must cause the expression of the pbx+ function one segment ahead, in T2. In 1983, the molecular lesions associated with the Cbx1 mutation were identified by Welcome Bender and confirmed Ed Lewis’ genetic predictions (Bender et al. 1983a). Bender found that in Cbx1, a 17-kb piece of DNA had been excised from the bxd/pbx region and transposed in reverse orientation 40 kb away within the second intron of Ubx (Fig. 3c). The deletion alone (pbx1) abolishes Ubx expression in the posterior half of the haltere imaginal disc (Fig. 3b), but its relocation 40 kb upstream activates Ubx expression in the posterior part of the wing imaginal disc (Cabrera et al. 1985; White and Akam 1985; White and Wilcox 1985). The loss of Ubx expression in the posterior compartment of the haltere disc in pbx1 mutants indicated that the 17-kb-long DNA element deleted enhancers responsible for Ubx expression in these cells (Fig. 3b). Given the positional flexibility of most enhancers, if these enhancers autonomously controlled their activity along the AP axis, then moving them from their endogenous location to the second intron of Ubx would not be expected to affect their function. And yet, moving these enhancers 40 kb changed the parasegment in which they activate Ubx. These observations suggested that position along the chromosome determines where BX-C enhancers are active along the AP axis. As the BX-C was first defined by segment-specific functions, a likely extension of the Cbx1 result would be that each segment-specific function derived from a region of the chromosome where enhancers were coordinately regulated along the AP axis. A model summarizing this idea is shown in Fig. 3 where a number of cell-type-specific enhancers from the abx/bx and bxd/pbx are depicted (A, B, C and D). In PS5, the abx/bx DNA domain opens up, enabling the A and B enhancers to activate Ubx in the A and B cells of PS5. As the domain remains open in more posterior parasegments (first rule of Ed Lewis model, see above), the A and B enhancers remain active as well in those more posterior parasegments (Fig. 3a). In the meantime, the bxd/pbx domain remains inactive in PS5 (see also Fig. 2). In PS6, the next adjacent domain (bxd/pbx) opens up, enabling the C and D enhancers to activate Ubx in different cell types (Fig. 3a). In Cbx1, the D enhancers are relocated in the domain that is active in PS5, enabling them to activate Ubx one parasegment ahead of their normal realm of activity (Fig. 3c). In this view, BX-C enhancers provide cell-type or tissue specificity and their location along the chromosome provides the segment/parasegmental information about where the enhancers should be activated along the AP axis.
Fig. 3.
Molecular genetics of Cbx 1. Modified from Fig. 4 of Peifer et al. 1987 and from Fig. 1.5 of Maeda and Karch (2009); DOI: 10.1016/S0070-2153(09)88001-0 with the permission of CSH Press and Elsevier, respectively. The abx/bx (orange) and bxd/pbx (red) regulatory regions, respectively active in PS5 and PS6 are represented on the top of each of the a, b, and c panels. A cartoon of the central nervous system in PS4, PS5, and PS6 is represented in the middle of each panel with the parasegmental borders on top and the corresponding segmental borders below. At the bottom of each panel, an adult thorax is shown with the PS5- and PS6-specific expression of Ubx drawn in orange and red, respectively. Note the PS5-PS6 boundary passing through the middle of the haltere. In panel a, enhancers A and B from the abx/bx regulatory domain turn on Ubx at a moderate level into the A and B cells of the CNS. These A and B enhancers remain active in the more posterior parasegments. Note that the C and D enhancers of the bxd/pbx regulatory region remain inactive in PS5. In PS6 however, these C and D enhancers activate Ubx at a higher level in the C and D cells. Panel b displays the pbx 1 mutation deleting the D enhancer. As a consequence, the D cells located in the posterior part of T3 do not express Ubx, leading in adults to the transformation of the posterior part of the haltere into posterior wing. In the Cbx 1 mutation (panel c), the D enhancer is relocated within the abx/bx regulatory, enabling them to function in PS5, as drawn in the cartoon of the CNS. This activity leads to the transformation of the posterior part of the wing into the posterior part of the haltere
The Mcp and Fab-7 boundary deletions
At the time of the proposal of the open for business model, Mcp1 (isolated by Lynn Crosby in Ed Lewis’ laboratory) was another GOF mutation that had been localized on the DNA map (Karch et al. 1985). For classical geneticists, dominant GOF mutations are enticing treats. To gain more insights into the mechanisms underlying a dominant mutation, the geneticist simply follows the tried-and-true method of inducing second site mutations that revert the dominant phenotype. For instance, Ed Lewis performed many screens to revert the Cbx1 phenotype. Nearly all revertants turned out to be chromosomal rearrangement breaks within the 70-kb-long Ubx transcription unit. This observation suggested that Cbx1 was causing misexpression of Ubx.
The Mcp1 mutation turned out to be a 3 kb deletion located near the region defined by mutational analysis as iab-5 (Fig. 2). However, while iab-5 mutations lead to an A5 to A4 transformation, Mcp1 causes the dominant transformation of A4 into A5. It was, therefore, thought that the deletion caused misexpression of iab-5 in A4, perhaps by removing a repressor involved in iab-5 repression in segments anterior to A5. The finding of Mcp revertants with rearrangement breakpoints in iab-5 confirmed the assumption that the deletion affected iab-5 regulation (Karch et al. 1985).
In the light of the open for business model, the location of Mcp1 at the border between the iab-4 and iab-5 regulatory domains inspired another interpretation. If the chromosomal domains were important for coordinately regulating BX-C enhancers in a parasegmentally controlled manner, then there must be some mechanism to limit the area of one domain from the area of the next. This interpretation would predict the presence of domain boundary elements. Accordingly, the Mcp1 deletion was thought to possibly be the deletion of a boundary element separating the iab-4 and iab-5 regulatory domains. In the context of the Mcp1 mutation, opening of the iab-4 domain in A4 would spread to iab-5, leading to the ectopic activation of iab-5 enhancers in A4.
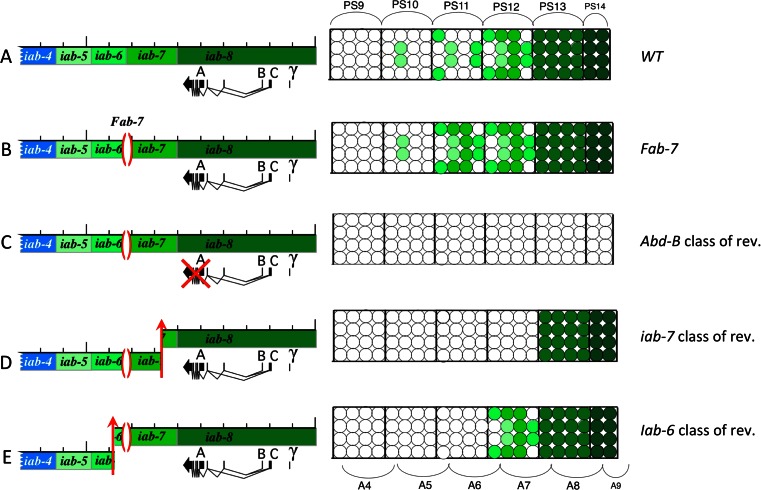
In 1985, the discovery of another GOF mutant, Fab-71 by Henrik Gyurkovics in Szeged, brought additional support to the concept of boundaries delimiting BX-C regulatory domains (Gyurkovics et al. 1990). In the case of the Fab-7 mutation, a 4.3-kb-long deletion occurred in the region delimiting iab-6 from iab-7 (Fig. 4b) and caused an A6 to A7 transformation. Following Ed Lewis’ model, the iab-7 function seems to be activated ectopically in A6. As for Mcp1, the simplest interpretation of Fab-7 consists in assuming that the deletion removes the binding site of a repressor/silencer complex that normally keeps iab-7 inactive in segments anterior to A7. But again the isolation and localization of revertants of Fab-71 make this simple interpretation unlikely. In this reversion screen, Fab-71 homozygotes were mutagenized with X-rays and crossed to WT females. The progeny of this cross would be expected to be heterozygous for the Fab-71 mutation and show the dominant transformation of A6 into A7 unless the X-ray treatment hit a region necessary for the manifestation of the GOF phenotype. Figure 4 summarizes the three classes of revertants that were recovered during this simple screen (Gyurkovics et al. 1990). The first class corresponded to Abd-B alleles (Fig. 4c). These chromosomes do not produce any Abd-B protein, confirming thereby that the Fab-71 mutation affects Abd-B regulation. The second class of revertants carry chromosomal rearrangements breakpoints within the iab-7 domain (Fig. 4d). In these mutants, the Fab-71 deletion along with iab-6 and iab-5 are separated away from the Abd-B target gene, causing the loss of Abd-B expression in A5/PS10 to A7/PS12. Homozygotes for such revertants are viable and have their A5/PS10 through A7/PS12 that develop like a copy of A4/PS9. This class of revertants confirms that iab-7 must be intact and in cis with both the Abd-B target gene and the Fab-71deletion to observe the GOF phenotype. Surprisingly, the third class of revertants disrupted iab-6 (Fig. 4e). This is the most interesting class regarding the open for business model, because it allowed for the ruling out of the simple hypothesis that the Fab-71 deletion removes binding sites for a repressive complex negatively regulating iab-7 in A6/PS11. If this had been the case, disruption of iab-6 should not interfere with this simple de-repression mechanism. Furthermore, the normal appearance of Abd-B expression in A7/PS11 in this iab-6 class of revertants clearly demonstrates that the Fab-71 deletion does not affect essential sequences for proper iab-7 activity. Based on this reversion experiment, it was concluded that in A6/PS11 of Fab-71 flies, the iab-6 and iab-7 domains are fused into a single functional unit with mixed characteristics: parasegmental/segmental address is provided by iab-6 and parasegmental/segmental identity is provided by iab-7. Revertants of Fab-71 mapping further to the left in iab-5 or iab-4 were never recovered, indicating that iab-6 worked in an autonomous fashion with respect to the Fab-71 phenotype.
Fig. 4.
Fab-7 mutation and revertants. The Abd-B transcription unit and associated regulatory domains iab-5 through iab-8 are drawn on the left of each panel. The parasegment-specific expression pattern of Abd-B are represented on the right in the form of cartoon of the central nervous system (with parasegmental and segmental borders indicated respectively above and below). In WT, panel a, Abd-B is expressed at a low level in a few cells in PS10. This expression is controlled by the iab-5 regulatory domain. In PS11, a few more cells express Abd-B at a slightly higher level, under the control of iab-6. In PS12 additional cells express Abd-B at a higher level, under the control of the iab-7 domain. Finally, in PS13, Abd-B appears in all cells at a higher level, under the control of iab-8. It should be noticed that in PS14, a truncated form of Abd-B is expressed from alternate promoters (B, C, and γ) at even a higher level. The enhancers controlling Abd-B expression in PS14 are not known. In panel b, the Fab-7 1deletion located between iab-6 and iab-7 is drawn on the genomic map. This deletion leads to the ectopic activation of iab-7 in PS11, resulting in the appearance of the PS12-specific Abd-B expression pattern in PS11. Panel c represents the first class of Fab-7 1 revertants that inactivate Abd-B. This class confirmed that Fab-7 1 affects Abd-B regulation. Panel d represents the second class of Fab-7 1 mutations that map within the iab-7 region. As the rearrangement breakpoints separate iab-5, iab-6, and part of iab-7 from their Abd-B target promoter, Abd-B expression is lost in PS10, PS11, and PS12. This class of revertants confirmed that Fab-7 1 is misregulating iab-7. Finally, the 3rd class of Fab-7 1 revertants in which a rearrangement breakpoint occurred in iab-6 is represented in panel e. Overall, this analysis established that the Fab-7 1 GOF phenotype appears only if the whole region from iab-6 to the Abd-B transcription is intact in cis
Additional boundary mutations
Mcp1 and Fab-71 were discovered as spontaneous mutations probably because the identities of the affected abdominal segments are easily recognized in the adult fly. Although the open-for-business model predicts the existence of boundary elements flanking each of the nine regulatory domains, additional boundary mutations did not appear in traditional, non-directed, genetic screens. This is probably because the remaining abdominal segments look very similar, making homeotic transformations difficult to identify. Nevertheless, there are three additional boundaries in the abdominal region of the BX-C that are genetically characterized. The Fab-8 boundary demarcates the iab-7 from the iab-8 regulatory domain (Barges et al. 2000). It was isolated by imprecise P-element excision using a P-element insertion line that was recovered on the basis of its sterility phenotype (Spradling et al. 1999). Fortuitously, this P-element inserted within Fab-8. Imprecise excision of this P-element showed that deletion of the region between iab-7 and iab-8 induces a partial transformation of A7 into A8 (Barges et al. 2000), again supporting the idea of a boundary element between iab-7 and iab-8. The case of Fab-6, separating iab-5 from iab-6 is less straightforward. It was first functionally inferred on the DNA map by the differences in phenotype between two internal deficiencies sharing the same distal breakpoint (toward Abd-B) but differing at their proximal breakpoints (toward abd-A; (Mihaly et al. 2006)). Later, this region was cleanly deleted and flies mutant for Fab-6 displayed a weak but consistent boundary phenotype (Iampietro et al. 2010). Finally, the Fub boundary marks the border between the bxd/pbx domain specifying A1 and the iab-2 domain that specifies A2 (Bender and Lucas 2013). Fub mutations were recovered by targeted mutagenesis following a hypothesis-driven experiment (see below).
Painting DNA domains of the BX-C with enhancer trap lines
From 1982 (Rubin and Spradling 1982) to 2006 (Groth et al. 2004), transgenesis in Drosophila was accomplished using P-element transposons as vectors. Insertion of the transgenic constructs was more or less random. Because of the promiscuous nature of enhancers and other chromatin regulatory elements (such as Polycomb-Response-Elements and heterochromatin), expression from the transgenes was often influenced by the neighboring chromosomal environment, a phenomenon known as position-effect (PE). The phenomenon of PE inspired Cahir O’Kane and Walter Gehring to engineer a lacZ-based reporter transposon aimed at trapping the activity of regulatory elements in the vicinity of the insertion site of a transposon (O’Kane and Gehring 1987). Using this P-element, O’Kane and Gehring, discovered that about 1/3 of the insertion lines gave rise to a lacZ expression pattern that was spatially and/or temporally restricted. This breakthrough observation opened up new avenues for identifying genes based on their expression pattern. Of the thousands of lines that have been generated in Drosophila laboratories across the world, only few landed in the BX-C.
The use of P-elements with lacZ reporter genes to study enhancers and other chromosomal regulatory elements led to the astonishing discovery of a phenomenon called homing, in which a DNA fragment can direct its insertion to the vicinity of the site from which it originates. Homing is rare and was first discovered with a fragment from the regulatory region of the engrailed gene (Hama et al. 1990; Kassis et al. 1992). Another such homing fragment is a 7-kb-long DNA fragment derived from the region between bxd/pbx and iab-2. In this case, 18 % of the P-element constructs carrying this homing fragment inserted into the BX-C (Bender and Hudson 2000). While the mechanisms behind homing remain elusive, it is worthwhile mentioning that the homing pigeon fragment spans the Fub boundary that separates the bxd/pbx regulatory domain from the iab-2 domain (see above; Bender and Lucas 2013). The idea of boundaries mediating homing is further substantiated by a more recent case of homing discovered at the eve locus by Fujioka and Jaynes (Fujioka et al. 2009). In this case, the homing fragment spans the homie boundary that insulates the eve locus from the next adjacent gene TER94 (Fujioka et al. 2013).
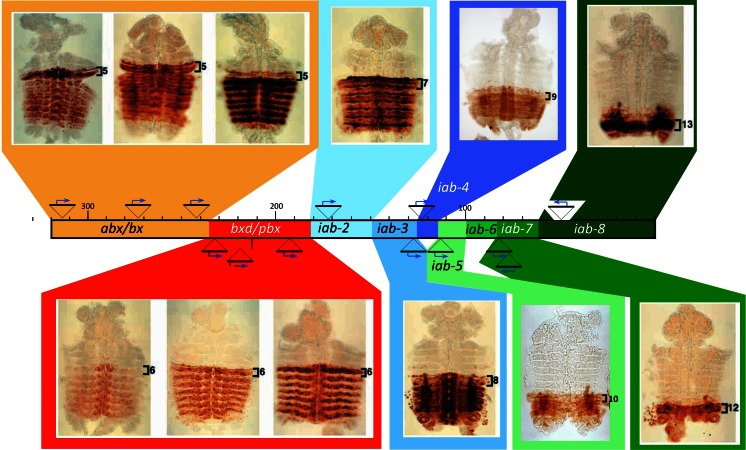
With the help of the homing pigeon fragment, the lab of Welcome Bender generated numerous new enhancers trap lines spread throughout the BX-C. Figure 5 shows some of these lines. The colored lines in this figure correspond to the DNA domains that are depicted in Fig. 2. If we focus on the three transposons inserted within the 75 kb region colored in orange, we find that the anterior border of expression of the lacZ reporter genes marks precisely PS5. This region comprises the sites of the abx/bx mutations that activate Ubx expression in PS5 and more posterior parasegments. Obviously, the promoters of the lacZ reporter genes in these three lines are trapping different sets of enhancers, as revealed by their different tissue specificities of expression. Nevertheless, all three enhancer trap lines share the same anterior border of expression in PS5. Meanwhile, the anterior border of expression of the next three enhancer trap lines (within the region colored in red) is shifted one parasegment posterior, in PS6. These three insertions map to the region previously assigned to the bxd/pbx region that controls Ubx expression in PS6. Once again, the tissue distribution and intensities of lacZ expression varies between the three lines but the anterior border of each starts at PS6.
Fig. 5.
Painting the regulatory domains with enhancer trap lines. Reproduced from Fig. 5 of Maeda and Karch (2006); DOI: 10.1242/dev.02323 with the permission of the Company of Biologists. The 300-kb-long genomic DNA of the bithorax are represented as a long rectangle in the middle of the figures, with the insertion sites of the various enhancer trap P[lacZ] transposons indicated by triangles above it. Embryos stained with antibodies directed against ß-galactosidase are shown above and below the DNA lines. They were cut along the dorsal midlines and flattened on a microscope slide. The anterior parasegmental boundary of lacZ expression is indicated in each embryo. Note that this anterior border of expression moves by increment of 1 parasegment when the insertion site of the P[lacZ] transposon moves from left to right on the DNA map. The extent of each regulatory domain was determined by integrating the insertion sites of the P[lacZ] transposons with the locations of various rearrangement breakpoints associated with iab mutations and with the locations of the Mcp, Fab-7, Fab-8 mutations
By examining a large number of enhancer trap lines in the BX-C, Bender and Hudson (2000) made three major observations. First, enhancer trap lines that are spread over large distances often produce the same expression pattern, whereas other located just a few kb away produce a different pattern. This, for example, is the case for the rightmost transposon in the orange domain and the leftmost transposon in the red domain. These two transposons are located only a few kilobases apart but nevertheless express in different parasegments (PS5 and PS6 respectively; Fig. 5). Second, the anterior border of lacZ expression always progress toward the posterior by increment of one parasegment. And third, once an enhancer trap line is activated in a given parasegment, it remains active in the more posterior parasegments, following the first rule of Ed Lewis (see above). Taken together, the enhancer trap experiments provide additional visual evidence that there are distinct, and precisely definable domains of coordinated activity within the BX-C. As the known boundary elements mapped to the transition zones between domains, these experiments also helped to validate the idea that boundary elements limit the extent of domain activity.
Additional boundaries in the BX-C
In Fig. 5, we took into account the positions of the enhancers trap insertion sites and the sites of mutations causing iab phenotypes to draw the extent of the regulatory domains. If, as mentioned above, boundary elements limit the extent of each domain, then we can infer the position of other boundary elements using this figure. P-elements in close proximity but expressed in different parasegments, give the most precise information for mapping boundaries. This is the case, for example, for the boundary separating the abx/bx domain (orange) from the bxd/pbx domain (red). This region contains the Ubx promoter. Similarly, the putative boundary (Fab-3) separating iab-4 from iab-5 can be localized accurately between the 2 transposons inserted on each side of it and that are expressed in PS9 and PS10, respectively.
In 2007, the laboratory of Rob White performed a whole genome search for chromatin sites associated with the CTCF insulator factor (Holohan et al. 2007). As BX-C boundaries have been shown to behave as insulators in ectopic contexts, the White lab spent some part of their analysis on the distribution of CTCF sites within the BX-C. Using a figure based on Hudson and Bender’s mapping data, they described an almost perfect match between the boundaries as shown in Fig. 5 and the presence of CTCF sites. It appears then that Fub, Fab-2, Fab-3, Fab-4, Mcp, and Fab-8 are all highlighted by the presence of CTCF binding sites (Fig. 6). Surprisingly, the best characterized boundary Fab-7, represents a conspicuous exception to this rule.
Fig. 6.
Position of CTCF sites in the BX-C as determined by chip’n chips. Picture taken from Holohan et al. (2007); DOI: 10.1371/journal.pgen.0030112 and reproduced with the permission of PLoS Genetics. The representation of the BX-C genomic region is taken from our review article published in 2006 in Development (Maeda and Karch 2006). The corresponding genomic region as described in Flybase (http://flybase.org/) is reproduced below the painted BX-C genomic region. Note the presence of CTCF binding in all the boundaries to the exception of Fab-7 (see text). It should be noticed that boundary deletions of Fab-2 have been since then recovered and named Fub (Bender and Lucas 2013) in reference to the Ultra-abdominal (Uab) alleles initially identified by Lewis (1978)
Initiator elements function as “domain control regions”
Using transgenic approaches with lacZ reporter genes, several laboratories searched the BX-C regulatory regions for new and important regulatory elements. Among the elements identified were early embryonic enhancers (initiators), cell-type-specific enhancers, silencers and insulators (Simon et al. 1990) (Muller and Bienz 1992) (Busturia and Bienz 1993) (Zhou et al. 1996) (Hagstrom et al. 1996) (Fritsch et al. 1999) (Zhou et al. 1999) (Barges et al. 2000) (Horard et al. 2000) (Shimell et al. 2000) (Gruzdeva et al. 2005) (Mihaly et al. 2006). What was surprising from these analyses was that there were very few elements discovered that were restricted along the A-P axis. For example, an individual cell-type-specific enhancer might drive expression only in neuroblasts, but this expression was not restricted along the A-P axis. Likewise, the silencers and insulators discovered would perform their activity irrespective of A-P position. In fact, when any domain was dissected, one could expect to find only one or two elements within each domain that were limited along the A-P axis. These rare elements, now called initiators, had the ability to turn on reporter gene expression from the specific parasegment controlled by the iab domain from which it was isolated, and in more posterior parasegments (Busturia and Bienz 1993; Mihaly et al. 2006; Simon et al. 1990). From the transgenic analysis alone, a mystery developed on how a group of non-AP restricted enhancers could be used to determine an AP restricted event (i.e., the creation of a specific segment). However, the transgenic analysis fit perfectly with predictions of the open for business model. According to the open for business model, most BX-C enhancers should be naïve to AP position, relying instead upon some spatial cue to come from the domain in which it resides. The only thing missing from this model was the identity of that cue and how the whole domain perceived its AP position. As the only elements found in BX-C that autonomously respond to an AP position, the early embryonic enhancer/initiators were proposed to read a parasegmental address and to communicate this knowledge to the rest of the elements within a domain.
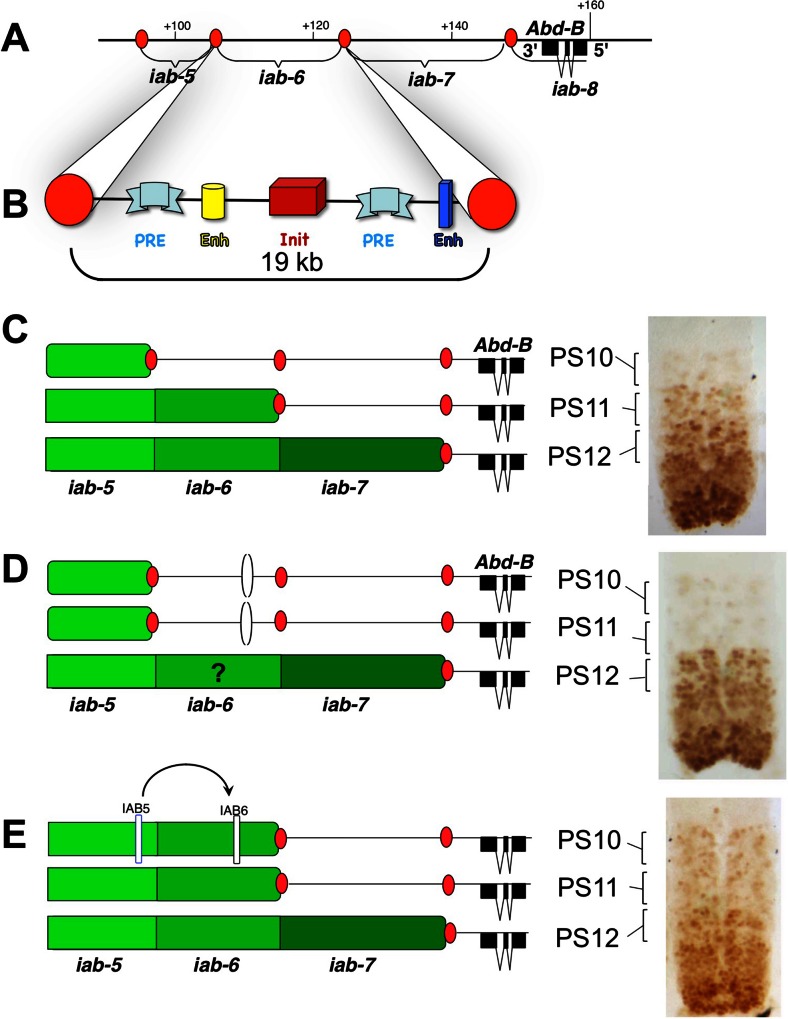
If initiator elements truly perform this function, then there are certain predictions that can be made. First, the removal of an initiator from a domain should completely abolish the activity of the whole domain. And second, switching an initiator from one domain for the initiator of a more anterior domain should cause activation of the more posterior domain in the parasegment specified by the more anterior initiator. We have directly addressed both of these predictions using a technique that coupled homologous recombination and ΦC31 site-specific integration to target the iab-6 regulatory domain for mutagenesis (Iampietro et al. 2010). Using this method, we showed that removal of the iab-6 initiator (a 927 bp fragment) abolishes iab-6 function, even though 18 kb of iab-6 sequences remains. This results in an A6 to A5 transformation, as the iab-5 domain is active in parasegments posterior to A5/PS10 (Fig. 7d). Next, we showed that switching the iab-6 initiator with that of iab-5 caused the enhancers present in iab-6 to become active one parasegment too anterior, in PS10(A5). This caused a A5 to A6 transformation, as seen in Fig. 7e. Thus, our study proved that initiators function as a “domain control regions” to read A-P positional information and accordingly, coordinate the various enhancers (which pattern the parasegment) within a domain. How initiators accomplish this feat remains to be discovered, but clearly, our data suggests a hierarchical nature to the regulatory elements in the BX-C consistent with the predictions of the open for business model.
Fig. 7.
Initiators function as domain control regions. Figure reproduced from the review article of Maeda and Karch (2011); DOI: 10.1016/j.gde.2011.01.021, with the permission of Elsevier. This article was reviewing work published by our laboratory in 2010 (Iampietro et al. 2010). The Abd-B genes and associated regulatory regions iab-5, iab-6, iab-7, and iab-8 regulatory regions is drawn in panel a with a central nervous system (CNS) dissected out of an embryo stained with antibodies against Abd-B (see also legend of Fig. 4 for the parasegmental expression of Abd-B in the CNS.) In panel b, a magnification of the 19-kb-long iab-6 domain is drawn in the form of a cartoon. Ovals indicate boundaries. PREs, cell type-specific enhancer (Enh) and initiator elements (Init) are drawn. Panel c shows the sequential opening of the iab-5, iab-6, and iab-7 regulatory domains in PS10, PS11, and PS12, respectively. Panel d shows the consequence of deleting the iab-6 initator alone (a 927-bp-long deletion). Despite the fact that 18.1 kb of iab-6 remains intact, the whole domain seems inactive as revealed by the Abd-B expression pattern in PS11 which is a reiteration of the expression observed in PS10. In agreement with this embryonic phenotype, the adult flies emerge with a complete transformation of A6 into A5. The initiator swapping experiment is shown in panel e. In this strain, the iab-6 initiator was removed and replaced by the initiator of iab-5. Note the PS10 Abd-B expression pattern that is similar to the pattern normally present in PS11, indicating that the iab-6 domain is opened in PS10. In agreement with this effect in embryos, adult flies emerge with a transformation of A5 into A6. In these initiator swapping flies, the parasegmental address is provided by the iab-5 initiator and the segmental identity is provided by the cell-type-specific enhancers of iab-6
H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex
Since its conception, the open-for-business model always proposed that enhancer domains somehow open in parasegments where they should be active. Thus far, we have described the evidence supporting a parasegment-specific activation of chromosomal domains and how these domains sense a parasegmental address. What we have not described is what it means to be active or open. To do this, we must first introduce the last group of players in this puzzle: the Polycomb-Group (Pc-G) and trithorax-Group (trx-G) genes.
During embryogenesis, the bithorax complex is thought to go through two phases of regulation: an early, initiation phase and a later, maintenance phase (see Maeda and Karch 2006 for review). Due to the timing of bithorax gene expression, the initiation phase is thought to be under the control of the transcription factors encoded by the maternal, gap and pair-rule genes that are responsible for the subdivision of the early embryos into 14 parasegments (for reviews, see for example Ingham, 1988; Hoch and Jackle, 1993; Kornberg and Tabata, 1993; DiNardo et al., 1994). These transcription factors are thought to interact with the initiators of each cis-regulatory domain to determine their ultimate expression pattern along the AP axis (Casares and Sanchez-Herrero 1995; Irish et al. 1989; Shimell et al. 1994; White and Lehmann 1986). For example, the combination of gap and pair-rule gene products present in PS11 are thought to bind to the iab-6 initiator to allow the iab-6 cis-regulatory region to control Abd-B expression in PS11/A6, while at the same time preventing the iab-7 cis-regulatory region from becoming active. Supporting this view, initiator elements do seem to contain numerous binding sites for these early transcription factors and in a few cases, have been shown to be dependent upon the activity of these transcription factors (Busturia and Bienz 1993; Ho et al. 2009; Qian et al. 1991; Shimell et al. 1994; Starr et al. 2011).
However, because the gap and pair-rule genes are only transiently expressed in the early embryo, and the activity states of the segment-specific cis-regulatory regions seem to be fixed for the life of the fly, a system to maintain homeotic gene expression is required within each cis-regulatory domain (Struhl and Akam 1985). The maintenance of homeotic gene expression has been shown to require the products of the Pc-G and trx-G genes. While the Pc-G products are thought to function as negative regulators, maintaining the inactive state of the cis-regulatory regions not in use, the trx-G products function as positive regulators, maintaining the active state of active regulatory regions (Kennison 1993; Paro 1990; Pirrotta 1997; Simon 1995). Both the Pc-G and trx-G products are known to bind within the parasegment-specific cis-regulatory domains to specific elements called Polycomb/Trithorax Response Elements (PREs/TREs) and are thought to maintain the active or inactive state of each domain by modifying its chromatin structure (Brown and Kassis 2013; Muller and Kassis 2006; Schwartz and Pirrotta 2008; Simon and Kingston 2009).
Pc-G proteins have been shown to form distinct chromatin repressive complexes (PRC1 and PRC2) with distinct chromatin modifying activities. While the PRC1 complex seems to ubiquitylate histone H2A (de Napoles et al. 2004), (Scheuermann et al. 2010) PRC2 seems to primarily methylate histone H3 on lysine K27 (Czermin et al. 2002; Muller et al. 2002; Ng et al. 2000). Although the molecular details on how these epigenetic changes may result in chromatin compaction and lowering gene expression remain poorly understood, it is known that both of these chromatin marks correlate with repressive chromatin environments.
Association of the Polycomb protein with the chromatin of the BX-C was first shown in 1993 by chromatin immuno-precipitations (ChIP) experiments performed with Drosophila tissue culture cells (Orlando and Paro 1993). Similar ChIP experiments performed in Drosophila Kc cells showed that the mark of PRC2, H3K27me3, was also present in the BX-C. Interestingly, this chromatin mark covered all of the BX-C, suggesting that in Kc cells, the whole BX-C was repressed. Later, when ChIP data was analyzed from a different Drosophila cell line, the SF4 cells, only the Ubx and abd-A portion of the BX-C was covered by H3K27me3 marks. The Abd-B gene and its associated iab-5 through iab-8 regulatory domains was completely devoid of H3K27me3, and conversely was associated with hyperacetylation of histone H4 (H4Ac), a mark associated with active genes (Beisel et al. 2007; Schwartz et al. 2006). This epigenetic signature fit well with the expression profil of the BX-C Hox genes in these two cell lines. While Kc cells do not express any BX-C homeotic genes, Sf4 cells express exclusively Abd-B. Later work, comparing tissue from the wing and haltere discs also supported a correlation between BX-C homeotic gene expression, in this case Ubx, and the lack of the H3K27me3 mark (Papp and Muller 2006). However, there was one problem. Based on the open for business model, one would expect that the H3K27me3 marks should be progressively stripped off from the chromatin by an increment of one domain at a time as one moves from anterior to posterior along the AP axis. The work from the cell lines seemed to slightly contradict this prediction, as only the Abd-B region seemed to lack H3K27me3 and show H4Ac. The work in the discs did not resolve this discrepancy as they only examined the Ubx region of the BX-C in anterior tissues where no other BX-C gene should be open. Therefore, a real test for domain opening of the open for business was still needed. Unfortunately, to truly test the open for business model, experiments would have to be done using purified populations of cells derived from different parasegments of the embryo spanning domains of expression of more than one BX-C homeotic gene. Until recently, this task seemed impossible due to the difficulty of separating and sorting Drosophila embryonic cells.
In 2010, Deal and Henikoff developed a system called INTACT to bypass this problem for ChIP by sorting specific populations of Drosophila embryonic nuclei. This method uses a nuclear envelope protein expressed under the control of a cell-type-specific promoter to anchor an mCherry marker with the biotin ligase recognition peptide (BLRP). After tissue disruption, specific nuclei are sorted by FACS or by affinity purification on streptavidin columns (Deal and Henikoff 2010). While in Drosophila, the vast number of Gal4 driver lines allows the INTACT marker to be expressed in almost any cell-type or tissue, it is often a problem to limit this expression to exclude cells that are not of interest. This was the challenge in the BX-C: to express the marker in all of the various cell-tyes in a parasegment, but to exclude the cells of neighboring parasegments. Lessons from the lacZ enhancer trap lines showed that transgenes inserted into the BX-C could drive expression of a marker in a particular parasegment. However, as the enhancer trap lines also revealed, these drivers would remain active in all the posterior parasegments (as predicted by Ed Lewis’ first rules).
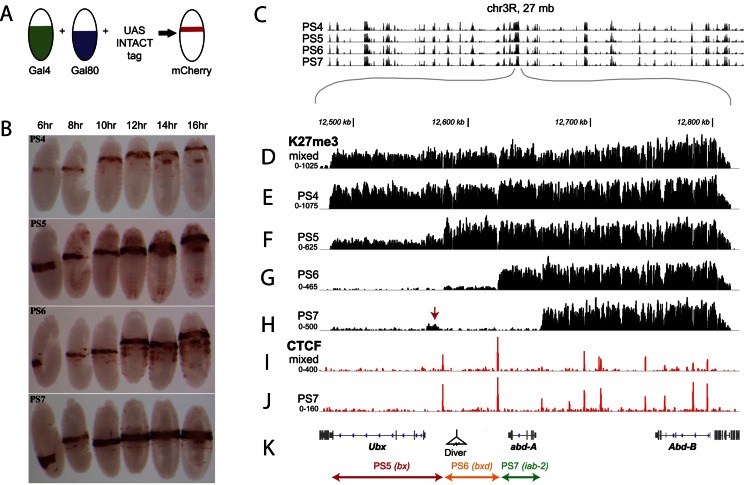
Through the work of the Bender and Kingston lab, this problem was finally solved (Bowman et al. 2014). As mentioned above, reporters inserted into the BX-C express according to the activity of the domain in which they are inserted. As such, the reporter starts its expression in one parasegment and continues throughout all of the parasegments more posterior. Thus, if a Gal4 reporter was inserted into the abx/bx domain, it would express the Gal4 activator from PS5 until the posterior end of the embryo. Meanwhile, a Gal4 inserted into the bxd/pbx domain would express from PS6 until the posterior end of the embryo. What Bender’s group did, was to create double insert lines where a Gal4 transgene was inserted into one cis-regulatory domain and a Gal80 expressing transgene (an inhibitor of Gal4 activity) was inserted into the next more posterior domain (Fig. 8a). In this way, they were able to express Gal4 in a broad region of the embryo, but have it only active in one parasegment (Bowman et al. 2014) (Fig.8b). Of course, this was much harder to do than it seems on paper, as transgenes inserted into the BX-C often trap different tissue-specific enhancers, depending upon where in the domain the transgenes inserted. Still, through the tenacious fine tuning of the Bender group, a set of lines having Gal4 activity exclusively in PS4, PS5, PS6 or PS7 was finally created.
Fig. 8.
H3K27 modifications define segment-specific regulatory domains. The figure compiles Figs. 2 and 3 of Bowman et al. (2014); DOI 10.7554/eLife.02833 reproduced with the permission of eLife. Panel a shows the strategy to obtain strains expressing active yeast Gal4 activator in single parasegments. The homing fragment was used to attract transposons harboring either the Gal4 activator or the Gal80 repressor in the regulatory domains (see detailed procedure in figure supplement 1 of Bowman et al. 2014). Drivers for the Gal4 activator or the Gal80 repressors each with different anterior limit of expressions are combined by simple crosses (see also text). Panel b shows the resulting expression pattern for strains expressing active Gal4 in PS4, PS5, PS6, and PS7, respectively. Note the existence of weak leakiness in anterior parasegments in the PS5-specific combination (see remark below). These Gal4 strains active in single parasegments are then crossed to INTACT construct (Deal and Henikoff 2010) to purify nuclei from single parasegment and perform ChIP-seq experiments with antibodies recognizing H3K27me3 modification. Panel c is a control experiment revealing that the overall H3K27me3 profile over a region of 27 Mb centered around the BX-C is invariant in the nuclei isolated from PS4, PS5, PS6, and PS7. Panels d through h show the H3K27me3 profile over the entire BX-C. The H3K27me3 profile from whole embryo (panel d) does not differ from the profile obtained from PS4 nuclei (panel e) where the entire BX-C is repressed. In PS5 however (panel f), the H3K27me3 profile is greatly reduced over the PS5-specific regulatory domain (as indicated in k). The fact that the H3K27me3 profile does not reach the background levels seen in the more posterior domains (panel g and h) probably stems from the leakiness of the PS5 specific driver in anterior parasegments, suggesting that the preparation is contaminated with nuclei originating from anterior inactive regions. Note the progressive loss of H3K27me3 modifications in nuclei derived from PS6 and PS7 (panel g and h, respectively). Panels i and j show that the CTCF binding profiles do not differ in nuclei isolated from PS7 and mixed nuclei isolated from whole embryos, suggesting that this boundary factor is bound in a constitutive fashion, regardless of the state of activity of the regulatory domans of the BX-C
Using these lines, nuclei were isolated from individual parasegments for ChIP-seq experiments. H3K27me3 ChIP on these samples confirmed the remarkable domain opening, predicted by the domain model (Fig. 8c through h). Nuclei derived from PS4 had the whole BX-C covered with H3K27me3 (Fig. 8e). Nuclei derived from PS5 had H3K27me3 retracting from the area of the chromosome attributed to the PS5 controlling abx/bx domain (Fig. 8f). Nuclei derived from PS6 had H3K27me3 retracting from the area attributed to both the abx/bx domain and the bxd/pbx domain (controlling Ubx in PS6; Fig. 8g). And finally, nuclei derived from PS7 had H3K27me3 retracting from the area from spanning the abx/bx domain until the iab-2 domain (Fig. 8h) (Bowman et al. 2014).
With regards to the open for business model, the work of Bowman et al. confirmed a number of important details. First was the precision in which a domain was activated. In their experiment, Bowman et al. found that the retracting H3K27me3 signal essential went from one point on the chromosome to another, without any sloping intermediate zones. ChIP experiments directed against the boundary protein CTCF confirmed that these places of abrupt transition coincided with expected boundary elements. Next, in the absence of H3K27me3, H3K27 acetylation marks were found. As H3K27Ac is a mark associated with active chromatin, it seems like domains that are not silenced become active, or open. Lastly, these experiments showed that even in parasegments where a given homeotic gene is not the primary segment-determining gene, domains controlling more anterior homeotic genes are still active. This was seen in the PS7 ChIP experiments where the abx/bx and bxd/pbx domains remained active, even though it is iab-2 controlling abd-A expression that is the primary determinant of PS7 identity.
“Sure enough, I was [we were] right.”
To spatially regulate the activation of homeotic genes along the AP axis, the open for business model proposes that enhancer containing domains open sequentially along the chromosome as one follows the anterior-posterior axis of the fly (Fig. 2). At first glance, this model seems quite simple to comprehend. Yet, within this model lie numerous implications, implications that have taken over 25 years to validate. Through the experiments that have been described in this review, we can now see many of the hidden details inherent in the open for business model. First, each domain contains a core set of regulatory elements that function in a hierarchical manner. At the bottom of this hierarchy seems to be the cell-type-specific enhancers that turn on single homeotic genes in cells appropriate for a specific parasegment. Controlling these enhancers are the PRE silencers and TREs that either compact the enhancers into a heterochromatin-like structure in parasegments where they are not needed or open the domain in parasegments where they should be active. To keep the domains separate are domain boundary elements that prevent the spreading of active or inactive chromatin from one domain to another. And lastly, there are the initiators that somehow instruct the PREs/TREs where they should or should not be active.
Although many questions still remain about the mechanisms by which each of the elements perform their function, the open for business model has been, for the most part, validated. As part of the community of researchers who contributed to the creation and validation of this model, this has been quite comforting. In our lab, we have a term for the experience of making a startling prediction that proves to be true. We call such instances, “Walter Gehring moments”, in honor of the numerous instances where Walter Gehring and his colleagues made incredible predictions that, through the elegant fusion of genetics and molecular biology, were proven to be true. For this reason, it seems quite fitting that this review to be placed in a series of articles dedicated to his memory.
Acknowledgments
We are indebted to Welcome Bender, Henrik Gyurkovics, Lazslo Sipos, Jozsef Mihaly, Mark Peifer, Martin Müller and Paul Schedl for numerous, insightful and inspiring discussions. Special thanks go also to the State of Geneva and the Swiss national Fund for Research for their constant support over the last 28 years.
Compliance with ethical standards
This research was funded by grants from the State of Geneva given to F.K. and by the Swiss National Fund for Research (grant number 310003A_149634).
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Footnotes
This article is dedicated to the memory of Walter Gehring.
References
- Akam ME. The location of Ultrabithorax transcripts in Drosophila tissue sections. EMBO J. 1983;2:2075–2084. doi: 10.1002/j.1460-2075.1983.tb01703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M, Dawson I, Tear G. Homeotic genes and the control of segment diversity. Development. 1988;104:123–133. [Google Scholar]
- Barges S, Mihaly J, Galloni M, Hagstrom K, Müller M, Shanower G, Schedl P, Gyurkovics H, Karch F. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development. 2000;127:779–790. doi: 10.1242/dev.127.4.779. [DOI] [PubMed] [Google Scholar]
- Beachy PA, Helfand SL, Hogness DS. Segmental distribution of bithorax complex proteins during Drosophila development. Nature. 1985;313:545–551. doi: 10.1038/313545a0. [DOI] [PubMed] [Google Scholar]
- Beisel C, Buness A, Roustan-Espinosa IM, Koch B, Schmitt S, Haas SA, Hild M, Katsuyama T, Paro R. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci U S A. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Hudson A. P element homing to the Drosophila bithorax complex. Development. 2000;127:3981–3992. doi: 10.1242/dev.127.18.3981. [DOI] [PubMed] [Google Scholar]
- Bender W, Lucas M. The border between the ultrabithorax and abdominal—a regulatory domains in the Drosophila bithorax complex. Genetics. 2013;193:1135–1147. doi: 10.1534/genetics.112.146340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Akam M, Karch F, Beachy PA, Peifer M, Spierer P, Lewis EB, Hogness DS. Molecular genetics of the bithorax complex in Drosophila melanogaster. Science. 1983;221:23–29. doi: 10.1126/science.221.4605.23. [DOI] [PubMed] [Google Scholar]
- Bender W, Spierer P, Hogness DS. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983;168:17–33. doi: 10.1016/S0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Bender W, Weiffenbach B, Karch F, Peifer M. Domains of cis-interaction in the bithorax complex. Cold Spring Harb Symp Quant Biol. 1985;50:173–180. doi: 10.1101/SQB.1985.050.01.023. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Deaton AM, Domingues H, Wang PI, Sadreyev RI, Kingston RE, Bender W. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. Elife. 2014;3:e02833. doi: 10.7554/eLife.02833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Kassis JA. Architectural and functional diversity of polycomb group response elements in Drosophila. Genetics. 2013;195:407–419. doi: 10.1534/genetics.113.153247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busturia A, Bienz M. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 1993;12:1415–1425. doi: 10.1002/j.1460-2075.1993.tb05785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera C, Botas J, Garcia-Bellido A. Distribution of Ultrabithorax proteins in mutants of Drosophila bithorax complex and its trans-regulatory genes. Nature. 1985;318:569–571. doi: 10.1038/318569a0. [DOI] [Google Scholar]
- Casanova J, Sanchez-Herrero E, Busturia A, Morata G. Double and triple mutant combinations of the bithorax complex of Drosophila. EMBO J. 1987;6:3103–3109. doi: 10.1002/j.1460-2075.1987.tb02619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casares F, Sanchez Herrero E. Regulation of the infraabdominal regions of the bithorax complex of Drosophila by gap genes. Development. 1995;121:1855–1866. doi: 10.1242/dev.121.6.1855. [DOI] [PubMed] [Google Scholar]
- Celniker SE, Sharma S, Keelan DJ, Lewis EB. The molecular genetics of the bithorax complex of Drosophila: cis- regulation in the Abdominal-B domain. EMBO J. 1990;9:4277–4286. doi: 10.1002/j.1460-2075.1990.tb07876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/S0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinardo S, Heemskerk J, Dougan S, O'Farrell PH (1994) The making of a maggot: patterning the Drosophila embryonic epidermis. Curr Opin Genet Dev 4, 529–534 [DOI] [PMC free article] [PubMed]
- Fritsch C, Brown JL, Kassis JA, Muller J. The DNA-binding Polycomb group protein Pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development. 1999;126:3905–3913. doi: 10.1242/dev.126.17.3905. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Wu X, Jaynes JB. A chromatin insulator mediates transgene homing and very long-range enhancer-promoter communication. Development. 2009;136:3077–3087. doi: 10.1242/dev.036467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka M, Sun G, Jaynes JB. The Drosophila eve insulator Homie promotes eve expression and protects the adjacent gene from repression by polycomb spreading. PLoS Genet. 2013;9:e1003883. doi: 10.1371/journal.pgen.1003883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruzdeva N, Kyrchanova O, Parshikov A, Kullyev A, Georgiev P. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol Cell Biol. 2005;25:3682–3689. doi: 10.1128/MCB.25.9.3682-3689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyurkovics H, Gausz J, Kummer J, Karch F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 1990;9:2579–2585. doi: 10.1002/j.1460-2075.1990.tb07439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- Hama C, Ali Z, Kornberg TB. Region-specific recombination and expression are directed by portions of the Drosophila engrailed promoter. Genes Dev. 1990;4:1079–1093. doi: 10.1101/gad.4.7.1079. [DOI] [PubMed] [Google Scholar]
- Ho MC, Johnsen H, Goetz SE, Schiller BJ, Bae E, Tran DA, Shur AS, Allen JM, Rau C, Bender W, et al. Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet. 2009;5:e1000709. doi: 10.1371/journal.pgen.1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch M, Jackle H (1993) Transcriptional regulation and spatial patterning in Drosophila. Curr Opin Genet Dev 3:566–573 [DOI] [PubMed]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF genomic binding sites in Drosophila and the organisation of the bithorax complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horard B, Tatout C, Poux S, Pirrotta V. Structure of a polycomb response element and in vitro binding of polycomb group complexes containing GAGA factor. Mol Cell Biol. 2000;20:3187–3197. doi: 10.1128/MCB.20.9.3187-3197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iampietro C, Gummalla M, Mutero A, Karch F, Maeda RK. Initiator elements function to determine the activity state of BX-C enhancers. PLoS Genet. 2010;6:e1001260. doi: 10.1371/journal.pgen.1001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW (1988) The molecular genetics of embryonic pattern formation in Drosophila. Nature 335:25–34 [DOI] [PubMed]
- Irish VF, Martinez-Arias A, Akam M. Spatial regulation of the Antennapedia and Ultrabithorax homeotic genes during Drosophila early development. EMBO J. 1989;8:1527–1537. doi: 10.1002/j.1460-2075.1989.tb03537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch F, Weiffenbach B, Peifer M, Bender W, Duncan I, Celniker S, Crosby M, Lewis EB. The abdominal region of the bithorax complex. Cell. 1985;43:81–96. doi: 10.1016/0092-8674(85)90014-5. [DOI] [PubMed] [Google Scholar]
- Karch F, Bender W, Weiffenbach B. abdA expression in Drosophila embryos. Genes Dev. 1990;4:1573–1587. doi: 10.1101/gad.4.9.1573. [DOI] [PubMed] [Google Scholar]
- Kassis JA, Noll E, VanSickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. Proc Natl Acad Sci U S A. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. Transcriptional activation of Drosophila homeotic genes from distant regulatory elements. Trends Genet. 1993;9:75–78. doi: 10.1016/0168-9525(93)90227-9. [DOI] [PubMed] [Google Scholar]
- Kornberg TB, Tabata T (1993) Segmentation of the Drosophila embryo. Curr Opin Genet Dev 3:585–594 [DOI] [PubMed]
- Lewis EB (1954) Pseudoallelism and the gene concept. Proc IX Int Congr Genet Caryol Suppl 100–105
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Macias A, Casanova J, Morata G. Expression and regulation of the abd-A gene of Drosophila. Development. 1990;110:1197–1207. doi: 10.1242/dev.110.4.1197. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The ABC of the BX-C: the bithorax complex explained. Development. 2006;133:1413–1422. doi: 10.1242/dev.02323. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. The bithorax complex of Drosophila an exceptional Hox cluster. Curr Top Dev Biol. 2009;88:1–33. doi: 10.1016/S0070-2153(09)88001-0. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Gene expression in time and space: additive vs hierarchical organization of cis-regulatory regions. Curr Opin Genet Dev. 2011;21:187–193. doi: 10.1016/j.gde.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Martinez-Arias A, Lawrence P. Parasegments and compartments in the Drosophila embryo. Nature. 1985;313:639–642. doi: 10.1038/313639a0. [DOI] [PubMed] [Google Scholar]
- Mihaly J, Barges S, Sipos L, Maeda R, Cleard F, Hogga I, Bender W, Gyurkovics H, Karch F. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development. 2006;133:2983–2993. doi: 10.1242/dev.02451. [DOI] [PubMed] [Google Scholar]
- Muller J, Bienz M. Sharp anterior boundary of homeotic gene expression conferred by the fushi tarazu protein. EMBO J. 1992;11:3653–3661. doi: 10.1002/j.1460-2075.1992.tb05450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/S0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- Ng J, Hart CM, Morgan K, Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/MCB.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V, Paro R. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-N. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–421. doi: 10.1016/0168-9525(90)90303-N. [DOI] [PubMed] [Google Scholar]
- Pease B, Borges AC, Bender W. Noncoding RNAs of the Ultrabithorax domain of the Drosophila bithorax complex. Genetics. 2013;195:1253–1264. doi: 10.1534/genetics.113.155036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peifer M, Karch F, Bender W. The bithorax complex: control of segmental identity. Genes Dev. 1987;1:891–898. doi: 10.1101/gad.1.9.891. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 1997;13:314–318. doi: 10.1016/S0168-9525(97)01178-5. [DOI] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E. Control of the expression of the bithorax complex genes abdominal-A and abdominal-B by cis-regulatory regions in Drosophila embryos. Development. 1991;111:437–449. doi: 10.1242/dev.111.2.437. [DOI] [PubMed] [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G. Genetic organization of Drosophila bithorax complex. Nature. 1985;313:108–113. doi: 10.1038/313108a0. [DOI] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Muller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb complexes and epigenetic states. Curr Opin Cell Biol. 2008;20:266–273. doi: 10.1016/j.ceb.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Simon J, Bender W, O’Connor MB. Enhancer point mutation results in a homeotic transformation in Drosophila. Science. 1994;264:968–971. doi: 10.1126/science.7909957. [DOI] [PubMed] [Google Scholar]
- Shimell MJ, Peterson AJ, Burr J, Simon JA, O’Connor MB. Functional analysis of repressor binding sites in the iab-2 regulatory region of the abdominal-A homeotic gene. Dev Biol. 2000;218:38–52. doi: 10.1006/dbio.1999.9576. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-X. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrn2731. [DOI] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O’Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Stern D, Beaton A, Rhem EJ, Laverty T, Mozden N, Misra S, Rubin GM. The Berkeley Drosophila Genome project gene disruption project: SINGLE P-element insertions mutating 25 % of vital Drosophila genes. Genetics. 1999;153:135–177. doi: 10.1093/genetics/153.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr MO, Ho MC, Gunther EJ, Tu YK, Shur AS, Goetz SE, Borok MJ, Kang V, Drewell RA. Molecular dissection of cis-regulatory modules at the Drosophila bithorax complex reveals critical transcription factor signature motifs. Dev Biol. 2011;359:290–302. doi: 10.1016/j.ydbio.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Akam M. Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J. 1985;4:3259–3264. doi: 10.1002/j.1460-2075.1985.tb04075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong S, Bone LM, Whittle JR. Recessive lethal mutations within the bithorax-complex in Drosophila. Mol Gen Genet. 1985;200:335–342. doi: 10.1007/BF00425445. [DOI] [PubMed] [Google Scholar]
- White RA, Akam M. Contrabithorax mutations cause inappropriate expression of Ultrabithorax products in Drosophila. Nature. 1985;318:567–569. doi: 10.1038/318567a0. [DOI] [Google Scholar]
- White RA, Lehmann R. A gap gene, hunchback, regulates the spatial expression of Ultrabithorax. Cell. 1986;47:311–321. doi: 10.1016/0092-8674(86)90453-8. [DOI] [PubMed] [Google Scholar]
- White RAH, Wilcox M (1985) Regulation of the distribution of Ultrabithorax proteins in Drosophila. Nature 318 [DOI] [PMC free article] [PubMed]
- Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]
- Zhou J, Ashe H, Burks C, Levine M. Characterization of the transvection mediating region of the abdominal- B locus in Drosophila. Development. 1999;126:3057–3065. doi: 10.1242/dev.126.14.3057. [DOI] [PubMed] [Google Scholar]