Inoue et al. report that CNOT3, a subunit of the CCR4–NOT deadenylase complex regulating mRNA decay and translational repression, controls Igh gene rearrangement and destabilizes the mRNA of the tumor suppressor p53. Loss of CNOT3 results in a block of pro- to pre–B cell transition.
Abstract
The CCR4–NOT deadenylase complex plays crucial roles in mRNA decay and translational repression induced by poly(A) tail shortening. Although the in vitro activities of each component of this complex have been well characterized, its in vivo role in immune cells remains unclear. Here we show that mice lacking the CNOT3 subunit of this complex, specifically in B cells, have a developmental block at the pro- to pre–B cell transition. CNOT3 regulated generation of germline transcripts in the VH region of the immunoglobulin heavy chain (Igh) locus, compaction of the locus, and subsequent Igh gene rearrangement and destabilized tumor suppressor p53 mRNA. The developmental defect in the absence of CNOT3 could be partially rescued by ablation of p53 or introduction of a pre-rearranged Igh transgene. Thus, our data suggest that the CCR4–NOT complex regulates B cell differentiation by controlling Igh rearrangement and destabilizing p53 mRNA.
B cell development is a complex process occurring in the fetal liver and then bone marrow. It begins with the proliferative expansion of progenitor cells that undergo sequential rearrangements of the Ig heavy chain (Igh) and Ig light chain (Igl) genes (Rajewsky, 1996; Meffre et al., 2000; Jung et al., 2006). Igh variable region exons are assembled from variable (VH), diversity (DH), and joining (JH) gene segments, a recombination process that must be tightly regulated to ensure lineage and stage specificity, as well as highly ordered; DH to JH joining occurs first in pre-pro–B cells, followed by VH to DHJH recombination in pro–B cells. Productive VHDHJH rearrangement results in the expression of a μ heavy chain that assembles with the surrogate light chains (λ5 and VpreB) to form a pre–BCR, which defines the pre–B cell differentiation stage. After further clonal expansion, pre–B cells undergo rearrangement of VL and JL elements in the Igl loci, resulting in transition to the immature B cell stage, marked by the cell surface expression of an IgM BCR. Ultimately, cells expressing functional, nonself-reactive BCRs are positively selected into the peripheral pool of long-lived mature B cells. These early B cell developmental steps are harmoniously regulated by transcriptional networks that integrate environmental cues to evoke gene expression programs appropriate to a particular developmental stage.
Emerging evidence has demonstrated that these transcriptional regulatory mechanisms on their own are not sufficient for proper B cell development and that posttranscriptional mechanisms are also required (Koralov et al., 2008). In regard to a general posttranscriptional regulator, attention has been recently paid to the CCR4–NOT multiprotein complex, which serves as one of the major deadenylases in eukaryotes (Collart and Panasenko, 2012; Miller and Reese, 2012). Deadenylation is the initial and often rate-limiting step in mRNA decay, resulting in the repression of translation (Decker and Parker, 1993). The CCR4–NOT complex consists of two major modules: the deadenylase module composed of two subunits with deadenylation enzymatic activity (CNOT6 or CNOT6L and CNOT7 or CNOT8) and the NOT module, which minimally consists of the CNOT1 scaffold protein, CNOT2, and CNOT3. Although the precise function of the NOT module remains largely elusive, a recent study indicates that it regulates the stability and activity of the deadenylase module and participates in recruitment of the CCR4–NOT complex to its specific target mRNAs (Wahle and Winkler, 2013). To ensure the target specificity, two targeting mechanisms have been proposed: first, sequence-specific RNA-binding proteins (RBPs) bring the CCR4–NOT complex to sequence elements in the 3′ untranslated region (3′-UTR) of the target mRNA, and second, instead of RBPs, the microRNA (miRNA) machinery recruits the CCR4–NOT complex to its target mRNA (Wahle and Winkler, 2013). In addition to its central role in specific mRNA degradation, the CCR4–NOT complex has also been implicated in transcription initiation and elongation and protein degradation (Collart and Panasenko, 2012; Miller and Reese, 2012).
The physiological significance of CCR4–NOT-mediated regulation in mammals has been addressed by using conventional knockout mice. CNOT7 deficiency leads to defects in spermatogenesis and anomalies in bone formation (Nakamura et al., 2004; Washio-Oikawa et al., 2007) and CNOT3 ablation halts embryogenesis, whereas its haploinsufficiency results in anomalies of heart function, bone formation, and energy metabolism (Neely et al., 2010; Morita et al., 2011; Watanabe et al., 2014). Although informative, the cellular and molecular bases of these severe phenotypes remain ill defined.
Here, we explored the role of CNOT3 in B cell development and activation and how, if at all, it participates in these processes. We first show that CNOT3 deficiency results in a developmental block at the pro- to pre–B cell transition. This developmental defect is attributable primarily to impaired Igh gene rearrangement in pro–B cells and increased apoptosis in pro- and pre–B cells. Notably, our data suggest that CNOT3 contributes to these biological phenomena both transcriptionally, by regulating initiation of germline transcription of the Igh locus, and posttranscriptionally, by deadenylating mRNA encoding the tumor suppressor p53.
RESULTS
CNOT3 is essential for early B cell development
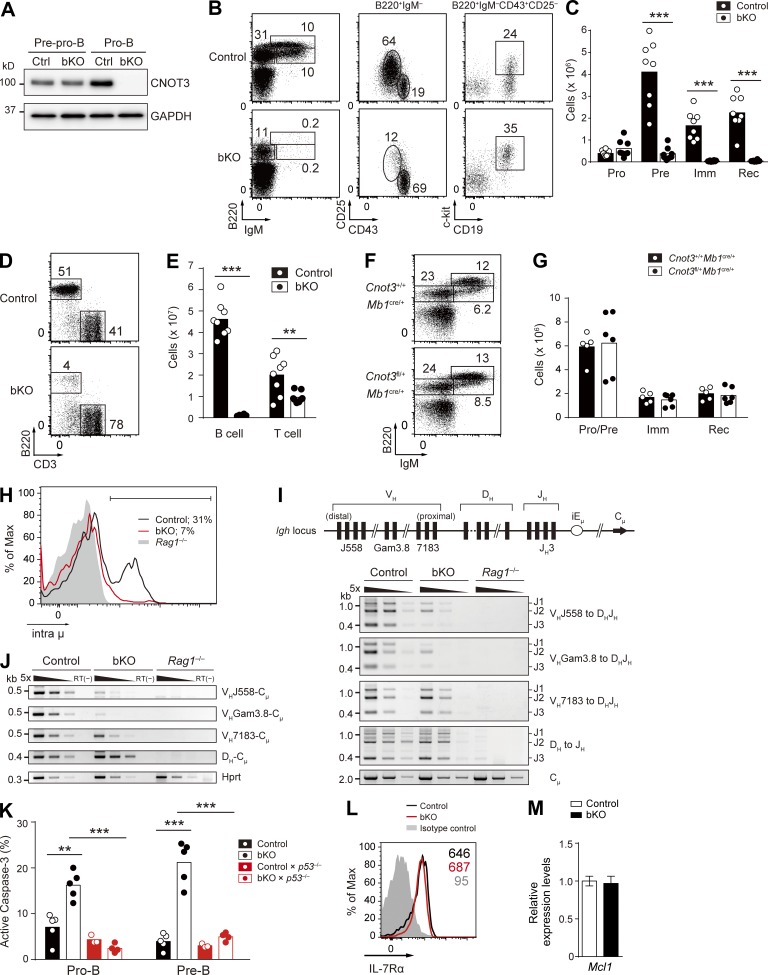
Because the CNOT3 subunit has been thought to be a key component for exerting the biological functions of the CCR4–NOT complex (Collart et al., 2013), we decided to first focus on clarifying the function of CNOT3. We confirmed that CNOT3 and other subunits of the CCR4–NOT complex are expressed in the bone marrow during early B cell development and that CNOT3 protein is up-regulated in pro-B and pre–B cells (Fig. 1 A and not depicted). To determine its possible function at these developmental stages, we conditionally deleted Cnot3 in B lineage cells by crossing with the mb1-cre deleter strain (Cnot3fl/flMb1cre/+; indicated as bKO hereafter; Fig. 1, B–D; Hobeika et al., 2006). In bKO mice, floxed exons of the Cnot3 allele and CNOT3 protein were efficiently deleted at the pro–B cell stage (Figs. 1 E and 2 A). In the absence of CNOT3, other subunits of the complex were still expressed, although at somewhat decreased levels (Fig. 1 F). In bKO mice, the number of pro–B cells was increased to some extent, whereas pre–B cells were greatly reduced compared with control mice (Cnot3+/+Mb1cre/+). Immature and recirculating B cells in the bone marrow and B cells in the spleen were barely detectable (Fig. 2, B–E). Thus, CNOT3 is essential for the development of B lymphocytes and plays its critical role during the differentiation of pro-B to pre–B cells. A single Cnot3 allele was sufficient to support normal B cell development (Fig. 2, F and G).
Figure 1.
Generation of the Cnot3 conditional knockout mice. (A) Total lysates from 1.5 × 105 sorted pre-pro-B (Lin−B220+IgM−CD43+CD25−CD19−) and pro–B cells (Lin−B220+IgM−CD43+CD25−CD19+) and 3 × 105 pre-B (Lin−B220+IgM−CD43−CD25+), immature B (Lin−B220intIgM+), and recirculating B (Lin−B220hiIgM+) cells were separated by SDS-PAGE and immunoblotted with anti-CNOT3 antibody. Almost equal protein loading was confirmed by GAPDH. (B) Structure of the targeted Cnot3 allele. Open triangles indicate loxP sequences. Numbered boxes represent exons in the Cnot3 gene. Negative (HSV-TK) and positive (PGK-neo) selection markers are indicated by open rectangles (TK, thymidine kinase; neo, neomycin). The loxP-flanked neo-cassette was removed by transient transfection of the pIC-Cre vector into ES clones of Cnot3neo/+ mice and PCR screening. Arrowheads indicate the position of the primers used in C and D. (C and D) PCR genotyping of genomic DNA isolated from Cnot3+/+, Cnot3neo/+, Cnot3null/+, and Cnot3flox/+ mice using primers a–c (C) and primers d–f (D). (E) RNA-seq–mapped reads at the Cnot3 locus from control and bKO pro–B cells. (F) Expression of subunits of the CCR4–NOT complex in control and bKO pro–B cells. Total lysates from 1.5 × 105 sorted pro–B cells of control and bKO mice were separated by SDS-PAGE and immunoblotted with antibodies against the CCR4–NOT subunits. GAPDH, loading control. (A and F) Data are representative of two independent experiments.
Figure 2.
CNOT3 is essential for early B cell development. (A) CNOT3 protein expression in sorted pre-pro–B (Lin−B220+IgM−CD43+CD25−CD19−) and pro–B (Lin−B220+IgM−CD43+CD25−CD19+) cells in the bone marrow from control and bKO mice was analyzed by Western blotting. GAPDH, loading control. (B and C) Flow cytometry (B) and absolute number of B cell subsets (C) of the bone marrow cells from control (n = 8) and bKO (n = 8) mice; B220+IgM−CD43+CD25−CD19+c-kit+ pro–B (Pro), B220+IgM−CD43−CD25+ pre–B (Pre), B220intIgM+ immature B (Imm), and B220hiIgM+ recirculating B (Rec) cells. (D and E) Flow cytometry (D) and absolute number (E) of splenic B cells (B220+CD3−) and T cells (B220−CD3+) from control (n = 8) and bKO (n = 8) mice. (F and G) Flow cytometry (F) and absolute number of B cell subsets (G) of bone marrow from Cnot3+/+Mb1cre/+ (n = 5) and Cnot3fl/+Mb1cre/+ (n = 6) mice; B220+IgM− pro–B and pre-B (Pro/Pre), B220intIgM+ immature B, and B220hiIgM+ recirculating B cells. Numbers in B, D, and F indicate the percentage of cells. (H) Flow cytometry of intracellular Ig μ chain (intra μ) expression in pro–B cells from control, bKO, and Rag1−/− mice. Percentages indicate the frequency of intra μ–positive cells (bracketed line). (I, top) Schematic drawing of the germline Igh locus with the relative location of the different VH family members. (bottom) PCR analysis of DH-JH and VH-DHJH rearrangements with fivefold serial dilutions of genomic DNA from sorted pro–B cells of control, bKO, and Rag1−/− mice. Cμ, loading control. (J) RT-PCR analysis of the expression of the rearranged DH-Cμ and VH-Cμ transcripts with fivefold serial dilutions of cDNA prepared from sorted pro–B cells of control, bKO, and Rag1−/− mice. Hprt, loading control; RT(−), no reverse transcription. (K) Frequency of apoptotic pro- and pre–B cells from mice of the indicated genotypes (n = 3–5), as assessed by positive staining of active caspase-3. (L) Flow cytometry analysis of surface expression of IL-7Rα on pro–B cells from control and bKO mice. Gray histogram, isotype control signal in control mice. Numbers indicate the mean fluorescence intensity of each population. (M) Relative mRNA expression levels of Mcl1 in bKO pro–B cells compared with control pro–B cells, as determined by real-time qPCR. Error bar represents SD. n = 3 biological replicates. (C, E, G, and K) Each symbol represents a single mouse, and bars indicate the mean. (A–M) Data are representative of three (A, B, D, F, H, I, and J) or two (L and M) independent experiments or are pooled from three (C and E) or two (G and K) experiments. **, P < 0.01; ***, P < 0.001; Student’s t test.
Successful VHDHJH recombination and the resultant production of the Ig μ chain are essential for the pro-B to pre–B transition (Kitamura et al., 1991; Jung and Alt, 2004). We found that intracellular Ig μ protein levels were greatly reduced in bKO pro–B cells (Fig. 2 H). To directly address whether the impaired μ chain expression resulted from inefficient VHDHJH recombination, the status of the Igh locus was examined in bKO pro–B cells. PCR analyses of genomic DNA isolated from pro–B cells clearly demonstrated that although DH to JH and the DH-proximal VH7183 to DHJH rearrangements occur normally in bKO mice, the more distal VHGam3.8 to DHJH and the most distal VHJ558 to DHJH rearrangements were greatly impaired (Fig. 2 I). Consistently, the corresponding transcript levels correlated with the genomic DNA recombination frequency (Fig. 2 J). Collectively, these findings suggest the selective involvement of CNOT3 in recombination of the IgH distal VH gene segments. However, the observed defect in the distal-specific VH-DHJH recombination could also be explained by a survival defect of CNOT3-deficient pro–B cells, as B cells using the distal VHJ558 family members expand at later stages of B cell development compared with those using the proximal VH7183 family (Malynn et al., 1990). In fact, staining of active caspase-3 revealed an approximately two- and fivefold higher frequency of apoptotic pro- and pre–B cells, respectively, in bKO mice (Fig. 2 K). Because IL-7 signaling is essential for survival of pro–B cells (Malin et al., 2010), we examined its status, demonstrating that IL-7Rα surface expression as well as a downstream event, the mRNA expression of Mcl1, is unaffected by the loss of CNOT3 (Fig. 2, L and M). Thus, apparently normal IL-7 signaling takes place in the absence of CNOT3.
CNOT3 regulates pro–B cell survival by destabilizing p53 mRNA
To clarify the molecular basis of the survival defect in CNOT3-deficient pro–B cells, we performed comprehensive gene expression profiling of control and bKO pro–B cells. Deep sequencing of mRNA and subsequent statistical analysis led to the identification of 79 up-regulated and 107 down-regulated genes in mutant cells (Fig. 3 A and Table S1). Bioinformatic clustering with DAVID resources and KEGG pathway analysis (Huang et al., 2009) revealed a significant enrichment of genes involved in the p53 signaling pathway among the differentially expressed genes (Fig. 3, B and C). The mRNA levels of molecules known to be important for early B cell development were not significantly affected by the loss of CNOT3 (Fig. 3 D), except for Cd79a (∼2-fold decrease) and Irf4 (∼2.5-fold increase). In addition to the p53 signaling pathway, among factors related to the cell survival, Pim1 (∼2-fold increase), Mef2a (∼2-fold decrease), and Tgfbr1 (∼2-fold decrease; Fortunel et al., 2000; McKinsey et al., 2002; Bednarski et al., 2012) were differentially expressed (Table S1), which might play a role in the survival defect of bKO pro–B cells.
Figure 3.
Transcriptome analysis of control and bKO pro–B cells. (A) A scatter plot of gene expression differences between control and bKO pro–B cells. RNA-seq was performed for sorted pro–B cells (Lin−B220+IgM−CD43+CD25−CD19+) for each genotype. Differentially expressed genes (FDR <0.05) are highlighted in red (up-regulated in bKO) or blue (down-regulated in bKO). As our pro–B cell preparation for the RNA-seq comprises both c-kit+ pro–B cells and c-kit− large pre–B cells, which are missing in bKO mice, these genes may also include those that are differentially regulated as a result of the analysis of a cell mixture. n = 4 biological replicates. (B) KEGG pathway enrichment analysis performed on differentially expressed genes between control and bKO pro–B cells using DAVID resources version 6.7. The most enriched 10 pathways based on their p-values are shown. (C) FPKM values of selected genes in the p53 signaling pathway in control and bKO pro–B cells. (D) Relative expression levels of selected molecules involved in early B cell development. FPKM values in bKO pro–B cells were normalized to those in control pro–B cells. (C and D) Error bars represent 95% confidence intervals. *, q < 0.05; **, q < 0.01.
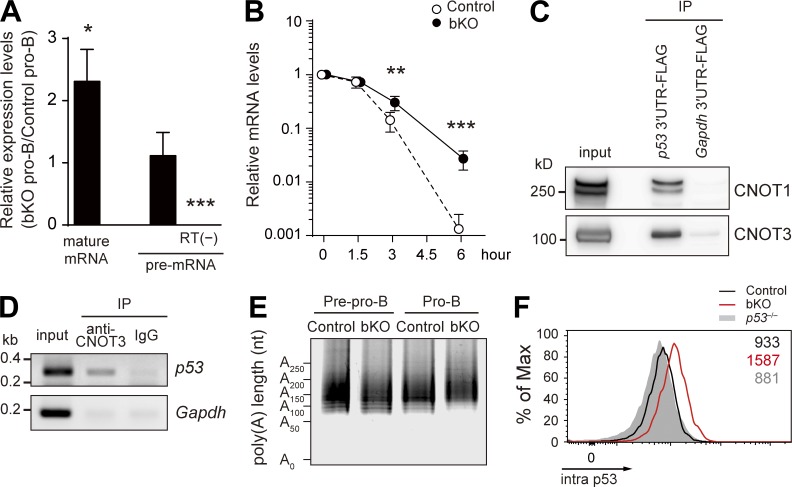
In this study, we focused on the p53 pathway because it is known to be involved in survival of early B lymphocytes (Lu et al., 1999). Real-time quantitative PCR (qPCR) analysis revealed the significant elevation of p53 mature mRNA in bKO pro–B cells (Fig. 4 A). We next determined whether this increase was caused by transcriptional and/or posttranscriptional regulation. There was no significant difference of the level of the p53 unspliced transcript (pre-mRNA) between mutant and control pro–B cells (Fig. 4 A), indicating that CNOT3 likely down-modulates the level of mature p53 mRNA in a posttranscriptional manner. This conclusion was further substantiated by the measurement of p53 mature mRNA half-life, which was significantly longer in the absence of CNOT3 (Fig. 4 B). We next examined whether p53 mRNA is directly targeted for deadenylation by the CCR4–NOT complex. A FLAG-tagged 3′-UTR of p53 mRNA, but not of control Gapdh mRNA, associated with the CCR4–NOT complex in pro–B cell line lysates in vitro (Fig. 4 C); conversely, anti-CNOT3 antibody coimmunoprecipitated p53 mRNA in wild-type pro–B cells (Fig. 4 D). These results indicate that physical association of the CCR4–NOT complex with the 3′-UTR of p53 mRNA occurs in pro–B cells, although our data do not distinguish whether this association is direct or indirect. The targeting of the CCR4–NOT complex to p53 mRNA resulted in shortening of the poly(A) tail; in the absence of CNOT3, the poly(A) tail length of the p53 transcripts was significantly elongated, as revealed by the ligase-mediated poly(A) test (LM-PAT) assay (Fig. 4 E; Sallés et al., 1999). A similar effect of the CNOT3 deficiency on p53 mRNA was also seen in pre–B cells (Fig. 5, A and B).
Figure 4.
CNOT3-mediated posttranscriptional regulation of p53 mRNA. (A) Relative expression levels of spliced p53 mature mRNA and unspliced pre-mRNA in bKO pro–B cells compared with control pro–B cells, as determined by real-time qPCR. Error bars represent SD. n = 4 biological replicates. RT(−), no reverse transcription. (B) Stability of p53 mature mRNA. Pro–B cells from control and bKO mice were treated with actinomycin D for the indicated periods of time, and the relative mRNA levels compared with time 0 were determined by real-time qPCR. Data are shown as mean ± SD of n = 3 biological replicates. (C) Association of CNOT1 and CNOT3 with the 3′-UTR of p53 mRNA. FLAG-tagged 3′-UTRs of p53 and Gapdh mRNA were incubated with lysates of an Abelson virus–transformed pro–B cell line and immunoprecipitated with anti-FLAG antibody, followed by Western blotting. (D) Association of CNOT3 with p53 mRNA by RIP assay. Wild-type pro–B cell lysates were immunoprecipitated with anti-CNOT3 antibody or control IgG, and the immunoprecipitates were analyzed by RT-PCR with primers specific to p53 and Gapdh mRNA. (E) Poly(A) tail length of p53 mRNA in pre-pro–B and pro–B cells of control and bKO mice was determined by the LM-PAT assay. (F) Flow cytometry of intracellular p53 protein expression in pro–B cells from control, bKO, and p53−/− mice. Numbers indicate the mean fluorescence intensity of each population. Data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test.
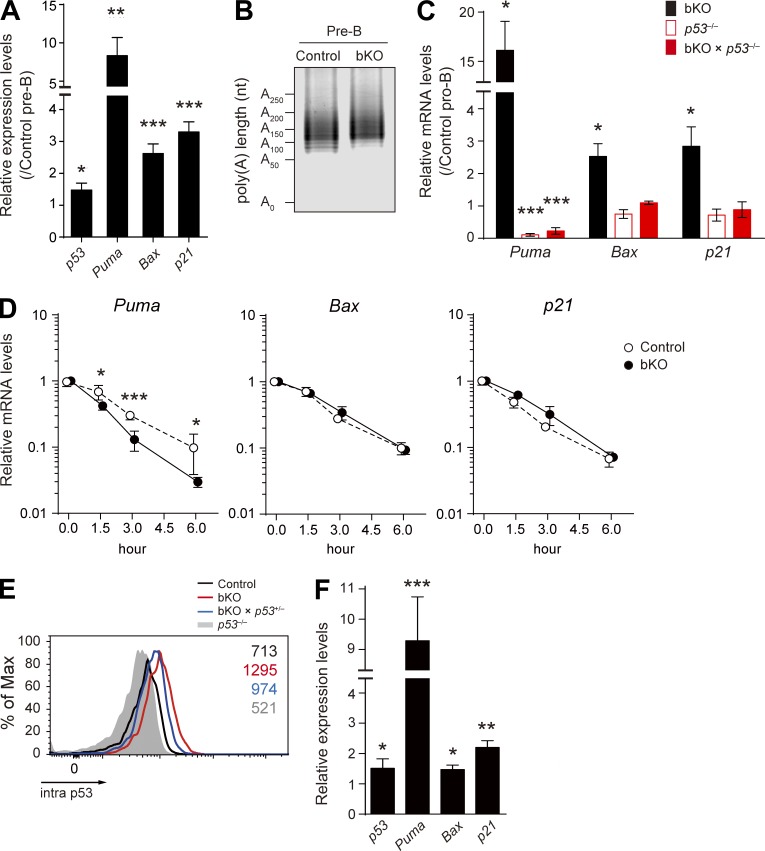
Figure 5.
Analysis of p53 pathway genes in pro-B and pre–B cells. (A) Real-time qPCR analysis of the relative mRNA expression levels of p53, Puma, Bax, and p21 in pre–B cells of bKO mice, normalized by control pre–B cells. (B) Poly(A) tail length of p53 mRNA in pre–B cells of control and bKO mice was determined by the LM-PAT assay. (C) Real-time qPCR to analyze the relative mRNA expression levels of Puma, Bax, and p21 in pro–B cells from the indicated mice compared with those in control pro–B cells. (D) Stability of Puma, Bax, and p21 mRNAs was analyzed by real-time qPCR as described in Fig. 4 B. Data are shown as mean ± SD of n = 3 biological replicates. (E) Flow cytometry of intracellular p53 protein expression in pro–B cells from control, bKO, bKO × p53+/−, and p53−/− mice. Numbers indicate the mean fluorescence intensity of each population. (F) Real-time qPCR to analyze the relative mRNA expression levels of p53, Puma, Bax, and p21 in pro–B cells from bKO × p53+/− mice compared with control pro–B cells. (A, C, and F) Error bars represent SD. n = 3 biological replicates. Data are representative of two independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test.
The increase in p53 mRNA in bKO pro–B cells was about twofold (Fig. 4 A), whereas the increase in p53 protein was more dramatic (Fig. 4 F). This difference could be the result of enhanced translational efficiency caused by the elongated poly(A) tail and/or by release of poly(A) tail–independent translational repression imposed by the CCR4–NOT complex (Cooke et al., 2010).
We also confirmed the up-regulation of p53 target mRNAs encoding proapoptotic factors, Puma and Bax, and a cell cycle regulator, p21, in bKO pro–B and pre–B cells (Fig. 5, A and C). However, in contrast to the case of p53 (Fig. 4 B), these mRNAs were not stabilized in mutant pro–B cells (Fig. 5 D); the Puma mRNA even appeared to be somewhat destabilized in the absence of CNOT3. Because the elevated Puma, Bax, and p21 mRNA levels in mutant cells were normalized by additional deletion of the p53 gene (Fig. 5 C), their up-regulation appears to be primarily caused by p53-mediated transcriptional regulation.
Genetic ablation of p53 rescues the survival defect but not the impaired Igh gene rearrangement
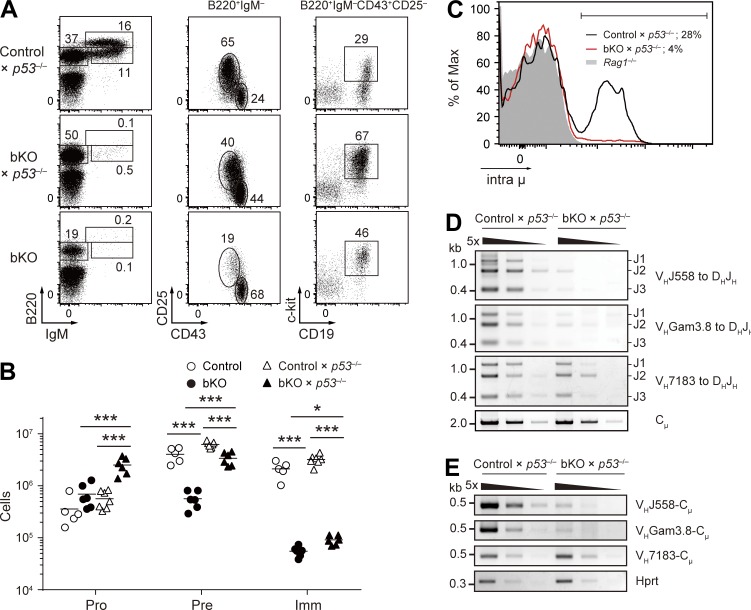
Having demonstrated the activated p53 pathway in CNOT3-deficient pro–B cells, we wished to test whether this indeed results in a survival defect, causing the distal VH to DHJH recombination anomaly. The ideal way to do this would be to restore the p53 level to the near normal, but not null, level in the CNOT3-knockout background. For this purpose, we considered using bKO × p53+/− mice; however, their pro–B cells were found to express higher levels of p53 protein and its target mRNAs such as Puma compared with control pro–B cells (Fig. 5, E and F). Therefore, instead, bKO × p53−/− mice were used to address the putative cause–effect relationship. As shown in Fig. 2 K, the proportion of apoptotic cells in bKO pro–B/pre–B cells was restored to the level of control cells by genetic ablation of p53. Moreover, the reduced pre–B cell numbers in bKO mice were significantly restored, albeit incompletely, upon loss of p53 (Fig. 6, A and B). Importantly, we found that the decreased percentage of intra μ–positive cells among bKO pro–B cells could not be complemented by p53 deletion (Fig. 6 C). Consistent with this result, rearrangement of distal VHJ558 and VHGam3.8 to DHJH and the resultant VHCμ transcripts could hardly be detected in pro–B cells deficient in both CNOT3 and p53 (Fig. 6, D and E). Based on these data, we conclude that CNOT3 is a novel regulator of IgH distal VH to DHJH recombination.
Figure 6.
Analysis of Cnot3 and p53 double-deficient mice. (A and B) Flow cytometry (A) and absolute number of B cell subsets (B) in the bone marrow cells from the indicated mice (n = 5–7) using the same gating strategy as in Fig. 2 (B and C). (B) Each symbol represents a single mouse, and bars indicate the mean. *, P < 0.05; ***, P < 0.001; Student’s t test. (C) Flow cytometry of intra μ expression in pro–B cells from control × p53−/−, bKO × p53−/−, and Rag1−/− mice. Percentages indicate the frequency of intra μ–positive cells (bracketed line). (D) PCR analysis of VH-DHJH rearrangements with fivefold serial dilutions of genomic DNA from sorted pro–B cells of control × p53−/− and bKO × p53−/− mice. (E) RT-PCR analysis of the expression of the rearranged VH-Cμ transcripts with fivefold serial dilutions of cDNA prepared from sorted pro–B cells of control × p53−/− and bKO × p53−/− mice. (A–E) Data are representative of three independent experiments (A and C–E) or are pooled from three experiments (B).
CNOT3 is required for the efficient Igh distal VH to DHJH rearrangement
Distal VH to DHJH rearrangement is known to be selectively affected by deficiencies in several molecules such as in the B cell commitment factor Pax5 (Fuxa et al., 2004), the general transcription factor YY1 (Liu et al., 2007), and the histone modifier Ezh2 (Su et al., 2003); however, all of these genes were expressed at comparable levels in bKO and control pro–B cells (Fig. 3 D). Given that proximal VH to DHJH rearrangement occurred normally in bKO pro–B cells, the diminished distal VH to DHJH recombination in CNOT3-deficient pro–B cells seemed likely caused by limited accessibility of these particular VH loci to the recombination machinery.
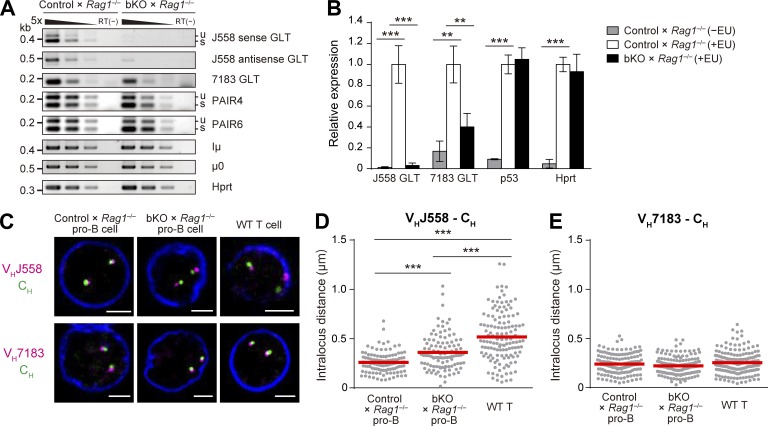
According to a recently proposed model (Verma-Gaur et al., 2012), germline transcription across the Igh locus (Yancopoulos and Alt, 1985) plays a key role in facilitating locus compaction, which juxtaposes the distant VH genes and DHJH segments, allowing access to these distal genes for efficient recombination (Kosak et al., 2002; Sayegh et al., 2005; Jhunjhunwala et al., 2008). We found that the germline transcripts (GLTs) that initiate from the heavy chain intronic enhancer (Iμ) and from the most 3′ D segment (DQ52) promoter (μ0) were comparably abundant in control and bKO pro–B cells on a Rag1−/− background (Fig. 7 A). The levels of intergenic antisense transcripts from Pax5-associated intergenic repeat (PAIR) elements, PAIR4 and PAIR6, which are the major intergenic antisense transcripts induced by Pax5 and are thought to be involved in the regulation of distal VH-DHJH recombination (Ebert et al., 2011; Verma-Gaur et al., 2012), were also unaffected by the absence of CNOT3. In contrast, the GLTs in the proximal VH7183 gene segments were somewhat decreased in bKO × Rag1−/− pro–B cells. Strikingly, both sense and antisense GLTs in the distal VHJ558 gene segments were greatly reduced in these cells (Fig. 7 A). To determine whether this decrease is caused by transcriptional or posttranscriptional events, expression levels of newly transcribed nascent RNA were quantified by pulse labeling of pro–B cells with a uridine analogue, 5-ethynyl uridine (EU), which is efficiently incorporated into the nascent RNA. Quantification of the EU-incorporated RNAs revealed that nascent VH7183 GLTs were reduced ∼60% and nascent VHJ558 GLTs were reduced more than ∼95% in bKO × Rag1−/− pro–B cells (Fig. 7 B). Thus, CNOT3 is highly suggested to be required for the germline transcription of the Igh V genes, particularly in the distal VHJ558 family. In contrast, nascent p53 mRNA levels were comparable between control and CNOT3-deficient cells, consistent with the unaltered p53 pre-mRNA levels in bKO pro–B cells described above (Fig. 4 A).
Figure 7.
Involvement of CNOT3 in the VH germline transcription and the Igh locus contraction. (A) RT-PCR analysis of the Igh GLTs, strand-specific VHJ558 GLTs, VH7183 GLTs (including sense and antisense strands), PAIR4, PAIR6, Iμ, and μ0, with fivefold serial dilutions of cDNA prepared from sorted pro–B cells (B220+CD19+c-kit+) of control × Rag1−/− and bKO × Rag1−/− mice. u and s, unspliced and spliced transcripts, respectively. Hprt, loading control; RT(−), no reverse transcription. (B) Quantification of newly transcribed nascent RNA levels of VHJ558 GLTs (including sense and antisense strands), VH7183 GLTs, p53 mRNA, and Hprt mRNA. Pro–B cells were pulse-labeled with vehicle (−EU) or EU (+EU) for 30 min, and the EU-labeled nascent RNAs were quantified by real-time qPCR and normalized to the control × Rag1−/− (+EU) sample. Error bars represent SD. n = 3 biological replicates. **, P < 0.01; ***, P < 0.001; Student’s t test. (C–E) 3D DNA-FISH analysis of the Igh locus of control × Rag1−/− pro–B, bKO × Rag1−/− pro–B, and wild-type splenic CD4 T cells. (C) Confocal images of Igh segments and nuclear envelope (anti–Lamin B1, blue). Bars, 2 µm. (D and E) Quantification of the intralocus distances between VHJ558 (D) or VH7183 (E) and CH signals. Red lines indicate the mean distance of at least 100 alleles per each cell type. ***, P < 0.001; unpaired Student’s t test. Loss of CNOT3 did not largely affect the nuclear periphery-to-center repositioning of the Igh locus. Percentages of VHJ558 probes at the nuclear periphery, defined as a distance of <0.5 µm between VH probes and nuclear envelope (Lamin B1), are 11.6% (control × Rag1−/− pro–B), 13.0% (bKO × Rag1−/− pro–B), and 25.5% (wild-type splenic CD4 T). (A–E) Data are representative of three (A) or two (B–E) independent experiments.
Next, we measured the distances between VH probes (VHJ558 or VH7183) and the constant region probe (CH) on the same Igh allele using three-dimensional DNA fluorescence in situ hybridization (3D DNA-FISH). The distribution of intralocus distances between VHJ558 and CH probes in control × Rag1−/− pro–B cells are more constrained compared with those in splenic CD4 T cells (Fig. 7, C and D), as reported previously (Sayegh et al., 2005). We found that the absence of CNOT3 resulted in a significant increase in the spatial distances between the VHJ558 and CH loci compared with control pro–B cells, although the increase was not as extensive as seen in T cells (Fig. 7, C and D). In regard to the less extensive spatial distances compared with T cells, our data could not completely exclude the following possibility. Only a small amount of CNOT3 might be remaining, probably because of the late onset of Cnot3 deletion by mb1-cre, thereby causing the less efficient decontraction of the Igh locus in bKO × Rag1−/− pro–B cells compared with T cells. In this regard, our deletion detection system might not have sufficed to detect such small changes. We also analyzed the distances between proximal VH7183 and CH probes, showing they were not significantly affected in the absence of CNOT3 (Fig. 7, C and E). These results suggest that CNOT3 is required for the regulation of efficient compaction of the Igh locus in pro–B cells.
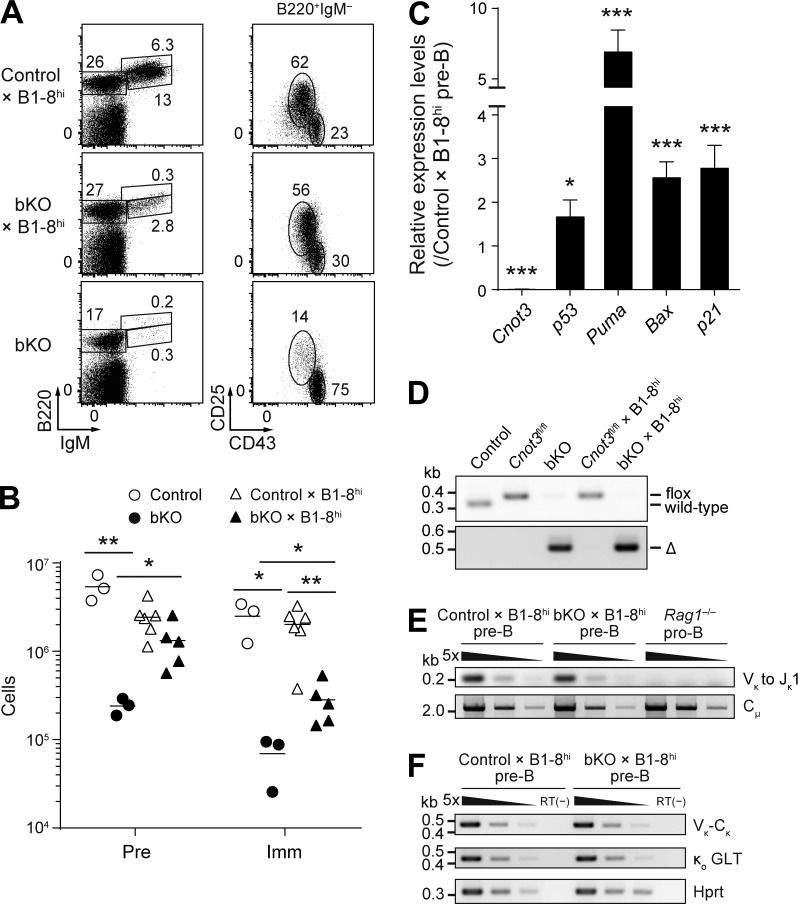
To test whether the inability of CNOT3-deficient pro–B cells to perform distal VH-DHJH rearrangements contributes to the developmental block observed in vivo, we made use of the B1-8hi mouse line, which carries a pre-rearranged VHDHJH segment in the endogenous Igh locus (Shih et al., 2002). As shown in Fig. 8 (A and B), compared with bKO mice, bKO × B1-8hi mice displayed increased numbers of pre–B and immature B cells in the bone marrow, indicating that the developmental block imposed by CNOT3 ablation was partially alleviated by the introduction of the pre-rearranged VHDHJH segment. Note that Cnot3fl alleles were efficiently deleted in bKO and bKO × B1-8hi pre–B cells (Fig. 8, C and D). To address whether the only partial restoration of the pre–B cell population in bKO × B1-8hi mice is caused by the inefficient Igl VJ rearrangements, we analyzed the Vκ to Jκ rearrangements at the Igκ locus in CNOT3-deficient pre–B cells. Genomic PCR and RT-PCR analyses revealed the VκJκ rearrangements and κo germline transcription were unaffected by the absence of CNOT3 (Fig. 8, E and F). Hence, rather, the still-augmented p53 pathway in bKO × B1-8hi mice (Fig. 8 C) is likely to be one of the reasons for the partial restoration of the pre–B cell population in bKO × B1-8hi mice.
Figure 8.
Partial rescue of the differentiation defect in bKO mice by the introduction of pre-rearranged VHDHJH segment. (A and B) Flow cytometry (A) and absolute number of B cell subsets (B) of the bone marrow cells from the indicated mice (n = 3–6) using the same gating strategy as in Fig. 2 (B and C). (B) Each symbol represents a single mouse, and bars indicate the mean. (C) Real-time qPCR to analyze the relative mRNA expression levels of Cnot3, p53, Puma, Bax, and p21 in bKO × B1-8hi pre–B cells, normalized by control × B1-8hi pre–B cells. Cnot3 primers are designed in the floxed exons to confirm efficient deletion of Cnot3. Error bars represent SD. n = 3 biological replicates. (D) PCR genotyping of pre–B cells of the indicated mice. The PCR fragments corresponding to wild-type, floxed, or deleted (Δ) alleles are indicated to the right. (E) PCR analysis of Vκ-Jκ1 rearrangements with fivefold serial dilutions of genomic DNA from sorted control × B1-8hi pre–B cells, bKO × B1-8hi pre–B cells, and Rag1−/− pro–B cells. Cμ, loading control. (F) RT-PCR analysis of the expression of the rearranged Vκ-Cκ transcripts and κo GLTs with fivefold serial dilutions of cDNA prepared from sorted pre–B cells of control × B1-8hi and bKO × B1-8hi mice. Hprt, loading control; RT(−), no reverse transcription. (A–F) Data are representative of three (A) or two (C–F) independent experiments or are pooled from two experiments (B). *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student’s t test.
DISCUSSION
In the present work, we addressed the physiological function of CNOT3 and its action mechanisms by B cell–specific deletion of the Cnot3 gene and by identifying functionally critical CNOT3 targets in the in vivo context. One of the causes of impaired B cell development in the absence of CNOT3 lies in reduced rearrangement of distal VH genes, including the VHJ558 family, as a result of defective VH locus contraction. The fact that the VHJ558 family is the largest VH gene family that occupies the 5′ distal half of the VH region explains the major reduction of intracellular μ chain–positive pro–B cells in the absence of CNOT3.
The mechanism by which each of the ∼200 VH gene segments scattered over a 2.5-Mbp region has an equal opportunity to establish contact and recombine with the DHJH element was a mystery until it was realized that large-scale locus contraction could occur through chromatin looping (Kosak et al., 2002; Fuxa et al., 2004; Sayegh et al., 2005; Jhunjhunwala et al., 2008). Recent chromosome conformation capture sequencing experiments (4C-seq) have revealed the existence of two layers of chromatin contraction, local and large scale (Medvedovic et al., 2013). The local chromatin loops, ranging in length from 0.5 to 1.3 Mb, as well as VH GLTs are observed even in the absence of Pax5 (Hesslein et al., 2003). Subsequently, Pax5 and YY1 activate PAIRs, particularly PAIR4 and PAIR6, by inducing noncoding intergenic antisense transcripts, thereby promoting PAIRs to form the base of large-scale chromatin loops. Our data suggest that, similar to noncoding intergenic antisense transcripts, the VHJ558 sense and antisense GLTs are likely to be required for the local chromatin looping, which, in turn, could be a prerequisite for subsequent large-scale looping mediated by Pax5 and YY1.
The mechanism by which CNOT3 induces VHJ558 germline transcription remains elusive. The presence of defective nascent transcripts in the mutant pro–B cells makes the involvement of CNOT3 in the transcription processes by itself likely, although its participation in destabilization of mRNAs encoding suppressor-type transcription factors could not be completely excluded. In support of the transcriptional regulation model, recent genome-wide RNAi screening experiments identified CNOT3 as one of the molecules that form a unique module in the transcription network required for mouse embryonic stem (ES) cell self-renewal (Hu et al., 2009). Assuming that CNOT3 participates in transcription processes, three possibilities could be envisaged: (1) regulating transcription initiation by its interaction with transcription factors (Badarinarayana et al., 2000; Lemaire and Collart, 2000; Deluen et al., 2002), (2) controlling transcription elongation by its binding to Pol II elongation factor Paf1c (Kruk et al., 2011), and (3) controlling histone modifications (Laribee et al., 2007; Mulder et al., 2007; Neely et al., 2010).
The CCR4–NOT complex acts as one component for the nonsense-mediated mRNA decay (NMD) mechanism; aberrant mRNAs containing premature translation termination codons are detected by the NMD mechanism, thereby being degraded by the action of the CCR4–NOT complex (Loh et al., 2013). For instance, in early B lymphocytes, nonsense Igh mRNA from a nonproductively rearranged allele is rapidly degraded by this mechanism (Li and Wilkinson, 1998). Given the recent evidence for the importance of the NMD mechanism in development of early lymphocytes (Frischmeyer-Guerrerio et al., 2011; Lutz et al., 2011), it is possible that the NMD pathway is perturbed in the CNOT3-deficient pro–B cells, which may, at least to some extent, contribute to the defective pro– to pre–B cell transition.
p53 contributes to the suppression of pro–B cell expansion during the checkpoint associated with Igh locus recombination and the subsequent formation of the pre–BCR (Lu et al., 1999). This process requires the creation of DNA double-strand breaks and is thought to activate the p53 response, which initiates transcription of genes that arrest cell cycle and induce apoptosis (Khanna and Jackson, 2001). Therefore, a reasonable speculation is that CNOT3-mediated suppression of p53 expression may allow pro–B cells to survive the physiological genomic stress required for proper recombination at the Igh locus and to subsequently facilitate the expansion of successfully recombined cells. Up-regulation of some subunits of the CCR4–NOT complex, including CNOT3, in pro– and pre–B cell stages may support this speculation.
Because miRNAs are involved in the recruitment of the CCR4–NOT complex to the target mRNAs (Wahle and Winkler, 2013), we anticipated that the phenotypes of CNOT3- and Dicer-deficient mice might have some overlap. However, key biological targets differ in these two mutant mice. Bim expression was dramatically increased in Dicer-deficient pro–B cells (Koralov et al., 2008) but was normal in the absence of CNOT3 (Fig. 3 D and not depicted). Given that CNOT1 interacts with the miRNA machinery–associated proteins GW182/TNRC6 (Braun et al., 2011), one straightforward explanation among many is that even in the absence of the CNOT3 subunit, the CCR4–NOT complex, including the CNOT1 subunit, is recruited to the miRNA machinery, thereby functioning as a posttranscriptional regulator. In contrast, our data clearly demonstrate that CNOT3 is required for targeting the CCR4–NOT complex to p53 mRNA. Considering the previous evidence that NOT3, a Drosophila melanogaster homologue of CNOT3, directly interacts with Bic-C, an RBP, thereby recruiting the CCR4–NOT complex to target mRNAs (Chicoine et al., 2007), we would propose that the interaction between RBPs and CNOT3 for recruitment of the CCR4–NOT complex to p53 mRNA is a possible mechanism operating in pro- and pre–B cells.
Such CNOT3-associated RBPs are thought to function by destabilizing on p53 mRNA. Counteracting this action are stabilizing RBPs, for example HuR, which was initially identified as an AU-rich element-binding protein with a potential to stabilize mRNA (Barreau et al., 2005). By using Lck-Cre–dependent deletion of HuR, one study demonstrated that the transition through the β-selection checkpoint during T cell development (the TCRβ locus recombination stage) was promoted and that protein expression of p53 was reduced in HuR-deficient thymocytes (Papadaki et al., 2009), suggesting that HuR functions as a positive regulator of p53 mRNA stability in pro– and pre–T cell stages. Hence, it is reasonable to anticipate that p53 expression is regulated posttranscriptionally, mediated by the balance of opposing RBPs, thereby contributing significantly to early lymphocyte development.
MATERIALS AND METHODS
Mice.
To generate Cnot3fl/+ mice, ES clones of Cnot3neo/+ mice (Morita et al., 2011) were transiently transfected with a pIC-Cre vector and screened by PCR and Southern blot analyses to select the clones in which the loxP flanked neo-cassette was correctly removed (Fig. 1, B–D). Positive ES clones were used for microinjection to obtain chimeric mice. These chimeric mice were then crossed with C57BL/6 mice to obtain animals with germline transmission of the targeted allele. Cnot3fl/+ F1 mice were backcrossed to the C57BL/6 background for at least 10 generations. Homozygous mutant (Cnot3fl/fl) mice produced by crossing Cnot3fl/+ heterozygotes were born at Mendelian ratios, were fertile, and had no obvious aberrant phenotype. Mb1-cre mice (provided by E. Hobeika and M. Reth, Max Planck Institute of Immunology and Epigenetics; Hobeika et al., 2006), p53−/− mice (Gondo et al., 1994), B1-8hi IgH knock-in mice (provided by M.C. Nussenzweig, The Rockefeller University, New York, NY; Shih et al., 2002), and Rag1−/− mice (Jax 002216; JAX Mice database) are described elsewhere and were maintained on a C57BL/6 background. C57BL/6 mice were purchased from CLEA Japan. All mice were bred and maintained under specific pathogen–free conditions, and all animal experiments were performed under institutional guidelines of the RIKEN Yokohama Research Institute and Osaka University.
Flow cytometry analysis.
7–12-wk-old experimental and control mice were used for cell type analyses. Single-cell suspensions of splenocytes and bone marrow cells from two tibiae and femurs lysed of red blood cells were stained with the fluorochrome-conjugated antibodies and analyzed using FACSCanto II (BD), and data were evaluated with FlowJo software. Anti-CD11b (M1/70), CD25 (PC61.5), CD43 (R2/60), CD117 (2B8), CD127 (A7R34), B220 (RA-6B2), IgM (II/41), F4/80 (BM8), Gr-1 (RB6-8C5), and TER-119 (TER-119) antibodies were from eBioscience. Anti-CD19 (1D3) antibody was from BD. Alexa Fluor 647–anti-p53 (1C12) antibody was from Cell Signaling Technology. Intracellular Ig μ staining was performed as described previously (Reichlin et al., 2001). In brief, cells were first stained for surface antigens and then fixed, permeabilized, and stained with anti-IgM using an Intracellular Fixation & Permeabilization Buffer Set (eBioscience). Intracellular p53 staining was performed by using a Foxp3 staining kit (eBioscience) according to the manufacturer’s instructions. For purification of the bone marrow cell populations, cells were first stained with a mixture of biotin-conjugated antibodies (anti-CD11b, F4/80, Gr-1, TER-119, and CD3ε), incubated with Streptavidin Microbeads (Miltenyi Biotec), and then enriched by depletion of labeled cells using autoMACS system (Miltenyi Biotec) before surface labeling for cell sorting (FACSAria II; BD). For active caspase-3 staining, equal numbers of the bone marrow cells were incubated in culture medium (IMDM supplemented with 10% FBS, nonessential amino acids, sodium pyruvate, 2-mercaptethanol, l-glutamine, and antibiotics) for 3 h at 37°C. After labeling with the appropriate antibodies, cells were stained with FITC-DEVD-FMK to detect active caspase-3 (CaspGLOW Fluorescein Active Caspase-3 Staining kit; eBioscience) according to the manufacturer’s instructions.
RNA analysis.
Total RNA was extracted from purified cell populations using TRIzol reagent (Invitrogen). For RT-PCR, DNase I (Invitrogen)–treated RNA was reverse transcribed with a mixture of random primers and oligo(dT) using a High Capacity RNA-to-cDNA kit (Applied Biosystems). Semi–qRT-PCR was performed using the primers listed below, and the PCR products were separated on agarose gels and visualized by ethidium bromide staining. Most primer sequences were described previously (Schlissel et al., 1991; Wang et al., 2002; Fuxa et al., 2004; Quong et al., 2004; Liu et al., 2007; Reynaud et al., 2008; Ebert et al., 2011). To measure strand-specific VHJ558 GLTs, 50 ng of total RNA was reverse transcribed using strand-specific RT primers and Superscript II reverse transcription (Invitrogen), followed by semi–qRT-PCR as described previously (Bolland et al., 2004). Real-time qPCR was performed using Power SYBR Green PCR Master Mix and StepOnePlus Real-Time PCR systems (Applied Biosystems). All qPCR experiments were run with three technical replicates in 96-well plates. Gapdh mRNA levels were used for normalization. Primers used for qPCR analysis are listed below. For the analysis of mRNA stability, sorted pro–B cells were cultured in the presence of 5 µg/ml actinomycin D. Total RNA extracted at the indicated time points was subjected to real-time qPCR analysis. RNA-immunoprecipitation (RIP) assay was performed using the RiboCluster Profiler RIP-assay kit (MBL) according to the manufacturer’s instructions using anti-CNOT3 monoclonal antibody, which has been generated by using bacterially expressed full-length CNOT3 as an immunogen (Bio Matrix Research Inc.) and control mouse IgG. Eluted RNA was analyzed by RT-PCR. To quantify newly transcribed nascent RNA levels, MACS-purified CD19+ pro–B cells (95–98% purity) were incubated with 0.5 mM EU (Invitrogen) for 30 min at 37°C. EU-labeled RNA was biotinylated and purified on Dynabeads MyOne Streptavidin T1 using Click-iT Nascent RNA Capture kit (Invitrogen) according to the manufacturer’s instructions. The captured RNA on beads was reverse transcribed into cDNA, which was used as the template for real-time qRT-PCR analysis.
The following primers were used for RT-PCR analysis: VHJ558-Cμ forward, 5′-CGAGCTCTCCARCACAGCCTWCATGCARCTCARC-3′; VHJ558-Cμ reverse, 5′-ATGCAGATCTCTGTTTTTGCCTCC-3′; VHGam3.8-Cμ forward, 5′-CAAGGGACGGTTTGCCTTCTCTTTGGAA-3′; VHGam3.8-Cμ reverse, 5′-ATGCAGATCTCTGTTTTTGCCTCC-3′; VH7183-Cμ forward, 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′; VH7183-Cμ reverse, 5′-ATGCAGATCTCTGTTTTTGCCTCC-3′; DH-Cμ forward, 5′-TTCAAAGCACAATGCCTGGCT-3′; DH-Cμ reverse, 5′-ATGCAGATCTCTGTTTTTGCCTCC-3′; Hprt forward, 5′-GGGGGCTATAAGTTCTTTGCTGACC-3′; Hprt reverse, 5′-TCCAACACTTCGAGAGGTCCTTTTCAC-3′; VHJ558 sense GLT RT primer, 5′-GAGCTTGCTGCACCTCCA-3′; VHJ558 sense GLT forward, 5′-ATGGGATGGAGCTGGATCTT-3′; VHJ558 sense GLT reverse, 5′-GACACACTCAGGATGTGTTTGTAG-3′; VHJ558 antisense GLT RT primer, 5′-CACTGAACACACTGACTCTAACCA-3′; VHJ558 antisense GLT forward, 5′-ATGGGATGGAGCTGGATCTT-3′; VHJ558 antisense GLT reverse, 5′-GACACACTCAGGATGTGTTTGTAG-3′; VH7183 GLT forward, 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′; VH7183 GLT reverse, 5′-GTCTCTCCGCGCCCCCTGCTGGTCC-3′; PAIR4 forward, 5′-TCCATGTTAGTGGTGGCAGA-3′; PAIR4 reverse, 5′-GTGACGACGGCTCATGACTA-3′; PAIR6 forward, 5′-TCCATGTTAGTGGTGGCAGA-3′; PAIR6 reverse, 5′-TCTGCAGTGTGTGACGACAG-3′; Iμ forward, 5′-TTCCAATACCCGAAGCATTTAC-3′; Iμ reverse, 5′-GTCCATGAGCAGCCAGGTG-3′; μ0 forward, 5′-AACATCTGAGTTTCTGAGGCTTGG-3′; μ0 reverse, 5′-TCATCTGAACCTTCAAGGATGCTC-3′; p53 forward, 5′-CCTCCCGGTCCCTTCTGCTGCC-3′; p53 reverse, 5′-ATGCAGACAGGCTTTGCAGAATG-3′; Jκ-Cκ forward, 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′; Jκ-Cκ reverse, 5′-CTCATTCCTGTTGAAGCTCTTGACAATGGG-3′; κo forward, 5′-CAGTGAGGAGGGTTTTTGTACAGCCAGACAG-3′; and κo reverse, 5′-TTTCCAGCTTGGTCCCCCCTCCGAA-3′.
The following primers were used for real-time qPCR analysis: p53 mature mRNA forward, 5′-TATCAGCCTCGAGCTCCCTCTG-3′; p53 mature mRNA reverse, 5′-CAGCAACAGATCGTCCATGCAG-3′; p53 pre-mRNA forward, 5′-CATGCCCAACGCTCTTTGGTTC-3′; p53 pre-mRNA reverse, 5′-CTGCAGCCAAAGGTCCAGTTAC-3′; Puma forward, 5′-ATGGCGGACGACCTCAAC-3′; Puma reverse, 5′-AGTCCCATGAAGAGATTGTACATGAC-3′; Bax forward, 5′-GGAGCAGCTTGGGAGCG-3′; Bax reverse, 5′-AAAAGGCCCCTGTCTTCATGA-3′; p21 forward, 5′-CCGTTGTCTCTTCGGTCCC-3′; p21 reverse, 5′-CATGAGCGCATCGCAATC-3′; Cnot3 forward, 5′-CGCAAGAAGAAAGGCGACAAGG-3′; Cnot3 reverse, 5′-CGAGGTCCAGGTCGTCATAGAGG-3′; Gapdh forward, 5′-ATGGTGAAGGTCGGTGTGAACGGATTTGGC-3′; Gapdh reverse, 5′-AGCTTCCCATTCTCGGCCTGGACTGTTCTG-3′; Hprt forward, 5′-TCCTCCTCAGACCGCTTTT-3′; Hprt reverse, 5′-CCTGGTTCATCATCGCTAATC-3′; VHJ558 GLT forward, 5′-ATGGGATGGAGCTGGATCTT-3′; VHJ558 GLT reverse, 5′-GACACACTCAGGATGTGTTTGTAG-3′; VH7183 GLT forward, 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′; VH7183 GLT reverse, 5′-GTCTCTCCGCGCCCCCTGCTGGTCC-3′; Mcl1 forward, 5′-TCAAAGATGGCGTAACAAACTGG-3′; and Mcl1 reverse, 5′-CCCGTTTCGTCCTTACAAGAAC-3′.
FLAG-tagged RNA-based immunoprecipitation assay.
Preparation of FLAG peptide–tagged RNAs was performed as described previously (Adachi et al., 2014). The PD31 Abelson virus–transformed pro–B cell line (provided by M. Schlissel, University of Michigan, Ann Arbor, MI) was expanded as described previously (Muljo and Schlissel, 2003) and lysed with a buffer containing 20 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, 1% digitonin, protease inhibitor cocktail (Roche), and phosphatase inhibitor cocktail (Nacalai Tesque). FLAG-tagged RNA (10 pmol) was mixed with anti–FLAG M2 agarose (Sigma-Aldrich) and subjected to immunoprecipitation with the cell lysate. The immunoprecipitates eluted with 3× FLAG peptide (Sigma-Aldrich) were analyzed by Western blotting.
RNA-seq and pathway analysis.
Total RNA was extracted from sorted cells by using TRIzol reagent. The DNA library for RNA-seq analysis was constructed with a TruSeq RNA sample prep kit (Illumina) as instructed by the supplier. The size range of the resulting DNA library was estimated on a 2100 Bioanalyzer (Agilent Technologies). After checking the molar concentration by qPCR using a LightCycler 480 (Roche), the DNA library was subjected to sequencing on a HiSeq 1000 sequencer (Illumina) in a 100-bp single-end read mode. The raw data were processed with CASAVA 1.8.2 (Illumina) to generate fastq files. The sequence reads were aligned to the Mus musculus reference genome (Build 37) using TopHat version 2.0.13 (Trapnell et al., 2009). According to the mapped data, Cufflinks version 2.2.1 (Trapnell et al., 2010) was used to calculate the fragments per kilobase of exon per million reads (FPKM) with the M. musculus genome annotation NCBI build 37.2 downloaded from its website. The statistical significance of the differential gene expression between cell populations was evaluated using the Cuffdiff software version 2.2.1 (Trapnell et al., 2010). Genes with false discovery rate (FDR) <0.05 were selected as differentially expressed ones. Four biological replicates were used in each genotype. For pathway analysis of the differentially expressed genes, we used DAVID resources version 6.7. The RNA-seq data are available at Gene Expression Omnibus database under accession no. GSE64455.
V(D)J recombination analysis.
Genomic DNA was isolated from sorted pro–B or pre–B cells by phenol extraction and ethanol precipitation. PCR analyses were performed using published primers as described previously (Schlissel et al., 1991; Fuxa et al., 2004), and PCR conditions were adjusted to be in the linear amplification range by serial dilution of template DNA. Samples from Rag1−/− pro–B cells serve as a negative control for DH to JH, VH to DHJH, and Vκ to Jκ recombination experiments. The PCR products were separated on agarose gels and visualized by ethidium bromide staining.
The following primers were used for V(D)J recombination analysis: VHJ558 forward, 5′-CGAGCTCTCCARCACAGCCTWCATGCARCTCARC-3′; VHGam3.8 forward, 5′-CAAGGGACGGTTTGCCTTCTCTTTGGAA-3′; VH7183 forward, 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′; DH forward, 5′-TTCAAAGCACAATGCCTGGCT-3′; JH3 reverse, 5′-GTCTAGATTCTCACAAGAGTCCGATAGACCCTGG-3′; Vκ forward, 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′; Jκ1 reverse, 5′-GCCACAGACATAGACAACGGAAGAA-3′; Cμ forward, 5′-TGGCCATGGGCTGCCTAGCCCGGGACTT-3′; and Cμ reverse, 5′-GCCTGACTGAGCTCACACAAGGAGGA-3′.
LM-PAT assay.
LM-PAT assay was performed as described previously (Sallés et al., 1999) with some modifications. In brief, the poly(A) tails of total RNAs (500 ng) were first saturated with 5′-phosphorylated oligo(dT)12–18 at 42°C in the presence of T4 DNA ligase. Then, the excess amount of oligo(dT) anchor primer (5′-GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT-3′) was added to the reaction to anneal at the end of poly(A) tails and incubated for 2 h at 12°C to complete ligation. This ligated primer was subjected to prime RT using SuperScript II (Invitrogen). PCR was performed with an anchor primer (5′-GCGAGCTCCGCGGCCGCG-3′) and a p53 3′-UTR-specific sense primer (5′-CTGGCACCTACAATGAAATCTCAC-3′). Specificity of the PCR reaction was confirmed by digestion of the PCR products with a restriction enzyme (ApaI). The PCR products were resolved on a 10–20% gradient polyacrylamide gel, stained with ethidium bromide, and visualized with an ImageQuant LAS 4000 mini imager (GE Healthcare).
Western blot.
Cells were lysed in RIP assay buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 0.05% SDS) containing protease inhibitor cocktail and phosphatase inhibitor cocktail. Equal amounts of each sample were separated by SDS-PAGE and transferred to PVDF membranes (EMD Millipore). Immunoblotting was performed using the following antibodies: anti-CNOT1, CNOT6, CNOT6L, CNOT8, CNOT9, and CNOT10 antibodies described previously (Morita et al., 2007; Chen et al., 2011; Ito et al., 2011); and commercially obtained anti-CNOT2 (Cell Signaling Technology), CNOT3 (clone 4B8; Abnova), CNOT7 (clone 2F6; Abnova), and GAPDH (clone 6C5; EMD Millipore) antibodies.
3D DNA-FISH.
3D DNA-FISH was performed essentially as described previously (Sayegh et al., 2005). In brief, 4 × 104 sorted pro–B cells were attached to poly-l-lysine–coated coverslips. Cells were fixed in paraformaldehyde, permeabilized, and hybridized with fluorescently labeled BAC probes (provided by C. Bossen and C. Murre, University of California, San Diego, La Jolla, CA). BAC RP24-189H12 (distal VHJ558) or RP23-404D8 (proximal VH7183) was labeled with Cy3-dUTP (GE Healthcare), and BAC RP23-109B20 (CH) was labeled with Alexa Fluor 488–dUTP (Invitrogen) using a Nick Translation kit (Roche). The nuclear periphery was stained with anti–Lamin B1 antibody (M-20; Santa Cruz Biotechnology, Inc.) and Alexa Fluor 647–anti–goat antibody (Invitrogen). Image stacks were captured with an inverted microscope IX71 with an oil immersion objective (UPlanSApo 100× NA 1.40; Olympus) and a high-speed spinning disc confocal unit (CSU-X1; Yokogawa Electric Corp.) equipped with a CCD camera (ORCA-AR; Hamamatsu Photonics). Images were collected at 65-nm pixels in X-Y and 200-nm steps in Z with MetaMorph software (Universal Imaging Corp.) as described previously (Isono et al., 2013). Only cells containing signals of both Igh loci were evaluated. The intralocus distances between the probes were calculated using ImageJ software (National Institutes of Health). Statistical analysis was performed with the unpaired Student’s t test using Prism software (GraphPad Software).
Statistical analysis.
Statistical analyses were performed by a two-tailed unpaired Student’s t test using Prism software.
Online supplemental material.
Table S1 shows the list of differentially expressed genes between control and bKO pro–B cells identified by the RNA-seq analysis. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150384/DC1.
Supplementary Material
Acknowledgments
We thank E. Hobeika and M. Reth for Mb1-cre mice, M.C. Nussenzweig for B1-8hi mice, M. Schlissel for the PD31 pro–B cell line, C. Bossen and C. Murre for sharing the 3D DNA-FISH protocol and providing BAC clones, P.D. Burrows for critical reading of the manuscript, and A. Arakawa and S. Ito for technical assistance.
This work was supported by grants to T. Inoue and T. Kurosaki from the Ministry of Education, Culture, Sports, Science and Technology in Japan and a grant to T. Kurosaki from the Japan Science and Technology Agency, Core Research for Evolutional Science and Technology. M. Morita is supported by the Canadian Diabetes Association Postdoctoral Fellowship and a Canadian Institutes of Health Research–funded Chemical Biology Postdoctoral Fellowship.
The authors declare no competing financial interests.
Author contributions: T. Inoue performed most of the experiments with the assistance of K. Isono, T. Ikawa, H. Kawamoto, and H. Koseki; M. Morita, S. Adachi, T. Natsume, and T. Yamamoto contributed to new reagents; A. Hijikata, Y. Fukuda-Yuzawa, T. Fukao, and O. Ohara helped with transcriptome analyses; and T. Inoue and T. Kurosaki designed the research and wrote the paper.
Footnotes
Abbreviations used:
- 3D DNA-FISH
- three-dimensional DNA fluorescence in situ hybridization
- 3′-UTR
- 3′ untranslated region
- ES
- embryonic stem
- EU
- 5-ethynyl uridine
- FPKM
- fragments per kilobase of exon per million reads
- GLT
- germline transcript
- Igh
- Ig heavy chain
- Igl
- Ig light chain
- LM-PAT
- ligase-mediated poly(A) test
- miRNA
- microRNA
- NMD
- nonsense-mediated mRNA decay
- PAIR
- Pax5-associated intergenic repeat
- RBP
- RNA-binding protein
- RIP
- RNA-immunoprecipitation
References
- Adachi S., Homoto M., Tanaka R., Hioki Y., Murakami H., Suga H., Matsumoto M., Nakayama K.I., Hatta T., Iemura S., and Natsume T.. 2014. ZFP36L1 and ZFP36L2 control LDLR mRNA stability via the ERK-RSK pathway. Nucleic Acids Res. 42:10037–10049. 10.1093/nar/gku652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badarinarayana V., Chiang Y.C., and Denis C.L.. 2000. Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics. 155:1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C., Paillard L., and Osborne H.B.. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138–7150. 10.1093/nar/gki1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarski J.J., Nickless A., Bhattacharya D., Amin R.H., Schlissel M.S., and Sleckman B.P.. 2012. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre–B cell survival and limit proliferation. J. Exp. Med. 209:11–17. 10.1084/jem.20112078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolland D.J., Wood A.L., Johnston C.M., Bunting S.F., Morgan G., Chakalova L., Fraser P.J., and Corcoran A.E.. 2004. Antisense intergenic transcription in V(D)J recombination. Nat. Immunol. 5:630–637. 10.1038/ni1068 [DOI] [PubMed] [Google Scholar]
- Braun J.E., Huntzinger E., Fauser M., and Izaurralde E.. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol. Cell. 44:120–133. 10.1016/j.molcel.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Chen C., Ito K., Takahashi A., Wang G., Suzuki T., Nakazawa T., Yamamoto T., and Yokoyama K.. 2011. Distinct expression patterns of the subunits of the CCR4–NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 411:360–364. 10.1016/j.bbrc.2011.06.148 [DOI] [PubMed] [Google Scholar]
- Chicoine J., Benoit P., Gamberi C., Paliouras M., Simonelig M., and Lasko P.. 2007. Bicaudal-C recruits CCR4-NOT deadenylase to target mRNAs and regulates oogenesis, cytoskeletal organization, and its own expression. Dev. Cell. 13:691–704. 10.1016/j.devcel.2007.10.002 [DOI] [PubMed] [Google Scholar]
- Collart M.A., and Panasenko O.O.. 2012. The Ccr4–Not complex. Gene. 492:42–53. 10.1016/j.gene.2011.09.033 [DOI] [PubMed] [Google Scholar]
- Collart M.A., Panasenko O.O., and Nikolaev S.I.. 2013. The Not3/5 subunit of the Ccr4-Not complex: A central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell. Signal. 25:743–751. 10.1016/j.cellsig.2012.12.018 [DOI] [PubMed] [Google Scholar]
- Cooke A., Prigge A., and Wickens M.. 2010. Translational repression by deadenylases. J. Biol. Chem. 285:28506–28513. 10.1074/jbc.M110.150763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., and Parker R.. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632–1643. 10.1101/gad.7.8.1632 [DOI] [PubMed] [Google Scholar]
- Deluen C., James N., Maillet L., Molinete M., Theiler G., Lemaire M., Paquet N., and Collart M.A.. 2002. The Ccr4-Not complex and yTAF1 (yTafII130p/yTafII145p) show physical and functional interactions. Mol. Cell. Biol. 22:6735–6749. 10.1128/MCB.22.19.6735-6749.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A., McManus S., Tagoh H., Medvedovic J., Salvagiotto G., Novatchkova M., Tamir I., Sommer A., Jaritz M., and Busslinger M.. 2011. The distal VH gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro–B cells. Immunity. 34:175–187. 10.1016/j.immuni.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Fortunel N.O., Hatzfeld A., and Hatzfeld J.A.. 2000. Transforming growth factor-β: pleiotropic role in the regulation of hematopoiesis. Blood. 96:2022–2036. [PubMed] [Google Scholar]
- Frischmeyer-Guerrerio P.A., Montgomery R.A., Warren D.S., Cooke S.K., Lutz J., Sonnenday C.J., Guerrerio A.L., and Dietz H.C.. 2011. Perturbation of thymocyte development in nonsense-mediated decay (NMD)-deficient mice. Proc. Natl. Acad. Sci. USA. 108:10638–10643. 10.1073/pnas.1019352108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M., Skok J., Souabni A., Salvagiotto G., Roldan E., and Busslinger M.. 2004. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18:411–422. 10.1101/gad.291504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondo Y., Nakamura K., Nakao K., Sasaoka T., Ito K., Kimura M., and Katsuki M.. 1994. Gene replacement of the p53 gene with the lacZ gene in mouse embryonic stem cells and mice by using two steps of homologous recombination. Biochem. Biophys. Res. Commun. 202:830–837. 10.1006/bbrc.1994.2005 [DOI] [PubMed] [Google Scholar]
- Hesslein D.G., Pflugh D.L., Chowdhury D., Bothwell A.L., Sen R., and Schatz D.G.. 2003. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 17:37–42. 10.1101/gad.1031403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., and Elledge S.J.. 2009. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23:837–848. 10.1101/gad.1769609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Isono K., Endo T.A., Ku M., Yamada D., Suzuki R., Sharif J., Ishikura T., Toyoda T., Bernstein B.E., and Koseki H.. 2013. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev. Cell. 26:565–577. 10.1016/j.devcel.2013.08.016 [DOI] [PubMed] [Google Scholar]
- Ito K., Inoue T., Yokoyama K., Morita M., Suzuki T., and Yamamoto T.. 2011. CNOT2 depletion disrupts and inhibits the CCR4–NOT deadenylase complex and induces apoptotic cell death. Genes Cells. 16:368–379. 10.1111/j.1365-2443.2011.01492.x [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S., van Zelm M.C., Peak M.M., Cutchin S., Riblet R., van Dongen J.J., Grosveld F.G., Knoch T.A., and Murre C.. 2008. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 133:265–279. 10.1016/j.cell.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., and Alt F.W.. 2004. Unraveling V(D)J recombination: Insights into gene regulation. Cell. 116:299–311. 10.1016/S0092-8674(04)00039-X [DOI] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., and Alt F.W.. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570. 10.1146/annurev.immunol.23.021704.115830 [DOI] [PubMed] [Google Scholar]
- Khanna K.K., and Jackson S.P.. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247–254. 10.1038/85798 [DOI] [PubMed] [Google Scholar]
- Kitamura D., Roes J., Kühn R., and Rajewsky K.. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 350:423–426. 10.1038/350423a0 [DOI] [PubMed] [Google Scholar]
- Koralov S.B., Muljo S.A., Galler G.R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B.S., Merkenschlager M., Rajewsky N., and Rajewsky K.. 2008. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 132:860–874. 10.1016/j.cell.2008.02.020 [DOI] [PubMed] [Google Scholar]
- Kosak S.T., Skok J.A., Medina K.L., Riblet R., Le Beau M.M., Fisher A.G., and Singh H.. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. 10.1126/science.1068768 [DOI] [PubMed] [Google Scholar]
- Kruk J.A., Dutta A., Fu J., Gilmour D.S., and Reese J.C.. 2011. The multifunctional Ccr4–Not complex directly promotes transcription elongation. Genes Dev. 25:581–593. 10.1101/gad.2020911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laribee R.N., Shibata Y., Mersman D.P., Collins S.R., Kemmeren P., Roguev A., Weissman J.S., Briggs S.D., Krogan N.J., and Strahl B.D.. 2007. CCR4/NOT complex associates with the proteasome and regulates histone methylation. Proc. Natl. Acad. Sci. USA. 104:5836–5841. 10.1073/pnas.0607996104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M., and Collart M.A.. 2000. The TATA-binding protein-associated factor yTafII19p functionally interacts with components of the global transcriptional regulator Ccr4-Not complex and physically interacts with the Not5 subunit. J. Biol. Chem. 275:26925–26934. [DOI] [PubMed] [Google Scholar]
- Li S., and Wilkinson M.F.. 1998. Nonsense surveillance in lymphocytes? Immunity. 8:135–141. 10.1016/S1074-7613(00)80466-5 [DOI] [PubMed] [Google Scholar]
- Liu H., Schmidt-Supprian M., Shi Y., Hobeika E., Barteneva N., Jumaa H., Pelanda R., Reth M., Skok J., Rajewsky K., and Shi Y.. 2007. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 21:1179–1189. 10.1101/gad.1529307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh B., Jonas S., and Izaurralde E.. 2013. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 27:2125–2138. 10.1101/gad.226951.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Lejtenyi D., and Osmond D.G.. 1999. Regulation of cell survival during B lymphopoiesis: suppressed apoptosis of pro–B cells in P53-deficient mouse bone marrow. Eur. J. Immunol. 29:2484–2490. [DOI] [PubMed] [Google Scholar]
- Lutz J., Heideman M.R., Roth E., van den Berk P., Müller W., Raman C., Wabl M., Jacobs H., and Jäck H.M.. 2011. Pro–B cells sense productive immunoglobulin heavy chain rearrangement irrespective of polypeptide production. Proc. Natl. Acad. Sci. USA. 108:10644–10649. 10.1073/pnas.1019224108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malin S., McManus S., Cobaleda C., Novatchkova M., Delogu A., Bouillet P., Strasser A., and Busslinger M.. 2010. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro–B cell development. Nat. Immunol. 11:171–179. 10.1038/ni.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn B.A., Yancopoulos G.D., Barth J.E., Bona C.A., and Alt F.W.. 1990. Biased expression of JH-proximal VH genes occurs in the newly generated repertoire of neonatal and adult mice. J. Exp. Med. 171:843–859. 10.1084/jem.171.3.843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey T.A., Zhang C.L., and Olson E.N.. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40–47. 10.1016/S0968-0004(01)02031-X [DOI] [PubMed] [Google Scholar]
- Medvedovic J., Ebert A., Tagoh H., Tamir I.M., Schwickert T.A., Novatchkova M., Sun Q., Huis In ’t Veld P.J., Guo C., Yoon H.S., et al. . 2013. Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity. 39:229–244. 10.1016/j.immuni.2013.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meffre E., Casellas R., and Nussenzweig M.C.. 2000. Antibody regulation of B cell development. Nat. Immunol. 1:379–385. 10.1038/80816 [DOI] [PubMed] [Google Scholar]
- Miller J.E., and Reese J.C.. 2012. Ccr4-Not complex: the control freak of eukaryotic cells. Crit. Rev. Biochem. Mol. Biol. 47:315–333. 10.3109/10409238.2012.667214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Suzuki T., Nakamura T., Yokoyama K., Miyasaka T., and Yamamoto T.. 2007. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol. Cell. Biol. 27:4980–4990. 10.1128/MCB.02304-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M., Oike Y., Nagashima T., Kadomatsu T., Tabata M., Suzuki T., Nakamura T., Yoshida N., Okada M., and Yamamoto T.. 2011. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/− mice. EMBO J. 30:4678–4691. 10.1038/emboj.2011.320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder K.W., Brenkman A.B., Inagaki A., van den Broek N.J., and Timmers H.T.. 2007. Regulation of histone H3K4 tri-methylation and PAF complex recruitment by the Ccr4-Not complex. Nucleic Acids Res. 35:2428–2439. 10.1093/nar/gkm175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., and Schlissel M.S.. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus–transformed pre–B cell lines. Nat. Immunol. 4:31–37. 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Yao R., Ogawa T., Suzuki T., Ito C., Tsunekawa N., Inoue K., Ajima R., Miyasaka T., Yoshida Y., et al. . 2004. Oligo-astheno-teratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat. Genet. 36:528–533. 10.1038/ng1344 [DOI] [PubMed] [Google Scholar]
- Neely G.G., Kuba K., Cammarato A., Isobe K., Amann S., Zhang L., Murata M., Elmén L., Gupta V., Arora S., et al. . 2010. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 141:142–153. 10.1016/j.cell.2010.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadaki O., Milatos S., Grammenoudi S., Mukherjee N., Keene J.D., and Kontoyiannis D.L.. 2009. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J. Immunol. 182:6779–6788. 10.4049/jimmunol.0900377 [DOI] [PubMed] [Google Scholar]
- Quong M.W., Martensson A., Langerak A.W., Rivera R.R., Nemazee D., and Murre C.. 2004. Receptor editing and marginal zone B cell development are regulated by the helix-loop-helix protein, E2A. J. Exp. Med. 199:1101–1112. 10.1084/jem.20031180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajewsky K. 1996. Clonal selection and learning in the antibody system. Nature. 381:751–758. 10.1038/381751a0 [DOI] [PubMed] [Google Scholar]
- Reichlin A., Hu Y., Meffre E., Nagaoka H., Gong S., Kraus M., Rajewsky K., and Nussenzweig M.C.. 2001. B cell development is arrested at the immature B cell stage in mice carrying a mutation in the cytoplasmic domain of immunoglobulin β. J. Exp. Med. 193:13–24. 10.1084/jem.193.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D., Demarco I.A., Reddy K.L., Schjerven H., Bertolino E., Chen Z., Smale S.T., Winandy S., and Singh H.. 2008. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat. Immunol. 9:927–936. 10.1038/ni.1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallés F.J., Richards W.G., and Strickland S.. 1999. Assaying the polyadenylation state of mRNAs. Methods. 17:38–45. 10.1006/meth.1998.0705 [DOI] [PubMed] [Google Scholar]
- Sayegh C.E., Jhunjhunwala S., Riblet R., and Murre C.. 2005. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 19:322–327. (published erratum appears in Genes Dev. 2008. 22:1717) 10.1101/gad.1254305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M.S., Corcoran L.M., and Baltimore D.. 1991. Virus-transformed pre–B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J. Exp. Med. 173:711–720. 10.1084/jem.173.3.711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih T.A., Roederer M., and Nussenzweig M.C.. 2002. Role of antigen receptor affinity in T cell–independent antibody responses in vivo. Nat. Immunol. 3:399–406. 10.1038/ni776 [DOI] [PubMed] [Google Scholar]
- Su I.H., Basavaraj A., Krutchinsky A.N., Hobert O., Ullrich A., Chait B.T., and Tarakhovsky A.. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. 10.1038/ni876 [DOI] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., and Salzberg S.L.. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 25:1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., and Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma-Gaur J., Torkamani A., Schaffer L., Head S.R., Schork N.J., and Feeney A.J.. 2012. Noncoding transcription within the Igh distal VH region at PAIR elements affects the 3D structure of the Igh locus in pro–B cells. Proc. Natl. Acad. Sci. USA. 109:17004–17009. 10.1073/pnas.1208398109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., and Winkler G.S.. 2013. RNA decay machines: deadenylation by the Ccr4–Not and Pan2–Pan3 complexes. Biochim. Biophys. Acta. 1829:561–570. 10.1016/j.bbagrm.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Wang Y.H., Stephan R.P., Scheffold A., Kunkel D., Karasuyama H., Radbruch A., and Cooper M.D.. 2002. Differential surrogate light chain expression governs B-cell differentiation. Blood. 99:2459–2467. 10.1182/blood.V99.7.2459 [DOI] [PubMed] [Google Scholar]
- Washio-Oikawa K., Nakamura T., Usui M., Yoneda M., Ezura Y., Ishikawa I., Nakashima K., Noda T., Yamamoto T., and Noda M.. 2007. Cnot7-null mice exhibit high bone mass phenotype and modulation of BMP actions. J. Bone Miner. Res. 22:1217–1223. 10.1359/jbmr.070411 [DOI] [PubMed] [Google Scholar]
- Watanabe C., Morita M., Hayata T., Nakamoto T., Kikuguchi C., Li X., Kobayashi Y., Takahashi N., Notomi T., Moriyama K., et al. . 2014. Stability of mRNA influences osteoporotic bone mass via CNOT3. Proc. Natl. Acad. Sci. USA. 111:2692–2697. 10.1073/pnas.1316932111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G.D., and Alt F.W.. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 40:271–281. 10.1016/0092-8674(85)90141-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.